Abstract
The UDP-N-acetylglucosamine (UDP-GlcNAc) acyltransferase, encoded by lpxA, catalyzes the first step of lipid A biosynthesis in Gram-negative bacteria, the R-3-hydroxyacyl-ACP dependent acylation of the 3-OH group of UDP-GlcNAc. Recently, we demonstrated that the Arabidopsis thaliana orthologs of six enzymes of the bacterial lipid A pathway produce lipid A precursors with structures similar to Escherichia coli lipid A precursors (Li, C., et al. (2011) Proc Natl Acad Sci U S A 108, 11387–11392). To build upon this finding, we have cloned, purified, and determined the crystal structure of the A. thaliana LpxA ortholog (AtLpxA) to 2.1 Å resolution. The overall structure of AtLpxA is very similar to that of E. coli LpxA (EcLpxA) with an alpha helical rich C-terminus and characteristic N-terminal left-handed parallel β-helix (LβH). All key catalytic and chain-length determining residues of EcLpxA are conserved in AtLpxA, however the AtLpxA has an additional coil and loop added to the LβH not seen in EcLpxA. Consistent with the similarities between the two structures, the purified AtLpxA catalyzes the same reaction as EcLpxA. In addition, A. thaliana lpxA complements an E. coli mutant lacking the chromosomal lpxA and promotes the synthesis of lipid A in vivo similar to the lipid A produced in the presence of E. coli lpxA. This work shows that AtLpxA is a functional UDP-GlcNAc acyltransferase able to catalyze the same reaction as EcLpxA, and supports the hypothesis that lipid A molecules are biosynthesized in Arabidopsis and other plants.
Introduction
Lipid A is a glucosamine-based phospholipid that anchors lipopolysaccharide (LPS) to the outer monolayer of the outer membrane of Gram-negative bacteria(1). The synthesis of lipid A in most of Gram-negative bacteria is essential for survival, making it a target for antibacterial drugs to combat Gram-negative bacterial infections. The structure of the Kdo2-lipid A portion of LPS is relatively conserved, and most Gram-negative bacteria encode single copy homologs of the nine Escherichia coli enzymes that catalyze the biosynthesis of the Kdo2-lipid A moiety(1).
While Gram-positive bacteria, archaea, fungi, insects, worms and vertebrates do not contain any of the genes for lipid A synthesis, the majority of higher plants, including Arabidopsis thaliana, encode full-length nuclear orthologs of six of the nine key enzymes of the E. coli system responsible for the biosynthesis of Kdo2-lipid A(2). The lpx genes of A. thaliana are functional(2). When RNAi or chromosomal knockout inactivates the lpx genes, the expected lipid A-like precursors are detectable and accumulate as predicted in the total lipid fraction when analyzed by mass spectrometry. Currently, neither the structure nor the function of the lipid A-like molecules generated by plants is known. Considering the low level of lipid A intermediates observed in plants (less than 0.01% of the total lipid), these molecules may play a role in signal transduction or regulation of the immune response(2).
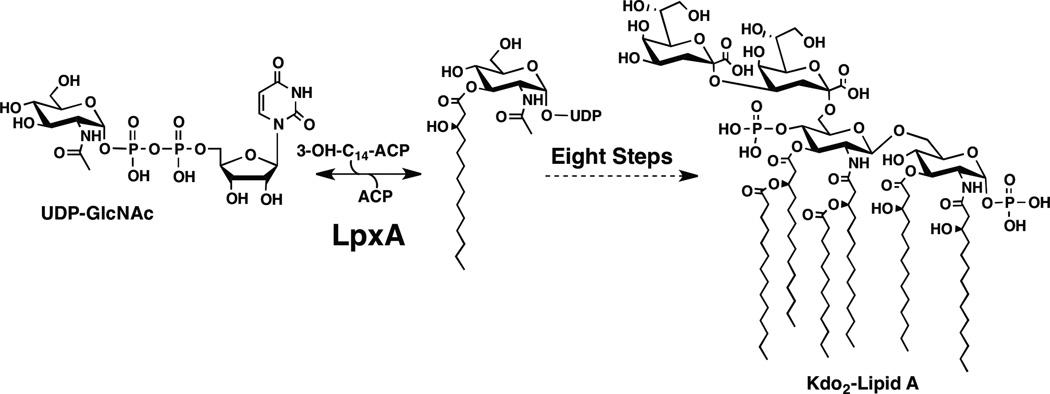
In Gram-negative bacteria, the UDP-N-acetylglucosamine (UDP-GlcNAc) acyltransferase, LpxA, catalyzes the first step in lipid A biosynthesis (Figure 1)(3) and in most systems this gene is required for growth(4). In E. coli, LpxA mediates the reversible transfer of the R-3-hydroxymyristoyl moiety from R-3-hydroxymyristoyl-acyl carrier protein (ACP) to the glucosamine 3-OH group of UDP-GlcNAc (Figure 1). UDP-GlcNAc acyltransferases, such as LpxA, are soluble cytoplasmic proteins, and they usually require R-hydroxyacyl-ACP as donor substrates(5). LpxAs from different bacteria share high sequence homology but can show different acyl chain and sugar selectivity. For example, E. coli LpxA (EcLpxA) prefers R-3-hydroxymyristoyl-ACP, while the Chlamydia trachomatis LpxA prefers myristoyl-ACP(6), and Helicobacter pylori LpxA (HpLpxA) prefers R-3-hydroxypalmitoyl-ACP(7). Leptospira interrogans LpxA uses UDP-2-acetamido-3-amino-2,3-dideoxy-α-D-glucopyranose as a substrate(8).
Figure 1. Reaction catalyzed by LpxA.
Both the EcLpxA and AtLpxA catalyze the acylation of UDP-GlcNAc, the first step in the biosynthesis of Kdo2-lipid A. A. thaliana has orthologs to six of the nine enyzmes required to biosynthesize Kdo2-lipid A in E. coli (2).
The A. thaliana LpxA (AtLpxA) and EcLpxA share significant sequence homology. The initial sequence alignment of the two proteins with ClustalW(9) showed 38% identity and 53% similarity with 6 gaps over 293 amino acid residues (Figure S1). Among the 6 gaps in the alignment, two located in the left-handed parallel β-helix (LβH) region(10), suggesting the existence of two extra inserts in addition to the two loops found in bacterial LpxAs. The first gap was 18 amino acid residues (Asn73 – Gly90), and the second gap was 4 amino acid residues (Glu104 – Gly107) (Figure S1). Based on this sequence alignment, which suggested a structural deviation from the bacterial LpxA structures determined so far(7, 8, 11), we investigated the biochemical properties of the AtLpxA in vitro and in vivo as well as determined its crystal structure to 2.1 Å resolution.
Experimental Procedures
Materials
[α-32P]-UTP was purchased from PerkinElmer (Waltham, MA). Oligonucleotide primers were obtained from Integrated DNA Technologies (Coralville, IA). Unless stated otherwise, all enzymes for cloning were from New England Biolabs (Ipswich, MA), and the E. coli strain for cloning was XL1-Blue (Table 1, Stratagene, Cedar Creek, TX). The crystallography reagents, Crystal screen HT, ammonium sulfate, lithium sulfate, and the StockOptions sodium citrate buffer kit, were from Hampton Research (Aliso Viejo, CA). Reactive Green 19 agarose beads were from Sigma (St. Louis, MO). All other chemicals were obtained from VWR (Radnor, PA) or prepared as described below.
Table 1.
Bacterial strains and plasmids
| Strain/plasmid | Description | Source or reference |
|---|---|---|
| E. coli Strains | ||
| W3110 | Wild type, F−, λ− | E. coli Genetic Stock Center, Yale University |
| DY330 | W3110 ΔlacU169 gal490 λcI857 Δ(cro-bioA) | (20) |
| XL1-Blue | Δmcr ABC recA1 endA1 gyrA96 relA1 supE44 thi-1 lac | Stratagene |
| C41(DE3) | F ompT hsdSB (rB−mB−) gal dcm (DE3) D (srl-recA)306::Tn10 | (16) |
| SJS-1D | DY330 (lpxA::kan)/pHpLpxA | This work |
| SJS-2W | W3110 (lpxA::kan)/pAtLpxA | This work |
| SM101 | thr-1 araC14 lpxA2(ts) tsx-78 Δ(galK-attLAM)99 hisG4(Oc) rfbC1 rpsL136(strR) xylA5 mtl-1 thi-1 | (4) |
| Plasmids | ||
| pET-21b(+) | Expression vector containing a T7 promoter, AmpR | EMD chemicals |
| pET-30a(+) | Expression vector containing a T7 promoter, CamR | EMD chemicals |
| pBAD33 | Medium copy vector, CamR | (13) |
| pCS92 | Expression vector pET-23c(+) containing HpLpxA, AmpR | Dr. C. Sweet |
| SJD-01 | pET-21b(+) containing AtLpxA | This work |
| pHpLpxA | pBAD33 containing HpLpxA | This work |
| pAtLpxA | pBAD33 containing AtLpxA | This work |
Plasmid Construction and Molecular Biology Techniques
The AtlpxA gene, excluding the region encoding the predicted signal peptide (residues 2–32) (12), was amplified by polymerization chain reaction (PCR) from a cDNA library of A. thaliana (var. Columbia) with primers SJP-017F and SJP-017B (Table 2). The PCR product was digested with restriction enzymes NdeI and BamHI, and gel purified from a 1% agarose gel using QIAquick Gel Extraction Kit from Qiagen (Valencia, CA). The digested gel-purified fragment was ligated into similarly digested pET-21b(+) that was treated with calf intestinal phosphatase to construct SJD-01 (Table 1). The AtlpxA gene was subcloned from SJD-01 into pBAD33(13) using the XbaI and HindIII sites to construct pAtLpxA. pHpLpxA (pBAD33 harboring Helicobacter pylori lpxA) was prepared by subcloning from pCS92, the pET-23 vector containing HplpxA (kind gift of Dr. C. Sweet, United States Naval Academy), using the XbaI and HindIII sites. Plasmids were isolated with the QIAprep Miniprep kit from Qiagen, and the bacterial genomic DNAs were purified with the Easy-DNA kit from Invitrogen (Carlsbad, CA). All DNA constructs were confirmed by dideoxy sequencing at the DNA Analysis Facility of Duke University.
Table 2.
Oligonucleotide primers used in this study
| Primers | |
|---|---|
| SJD-01* | |
| SJP-017F | 5’-gagatatacatATGGATTCGAGGGATTCTGAAGTCTTAATACACC-3’ |
| SJP-017B | 5’-cgaattcggatccTAAGTTGTGGAATCGAGCCATTGTCTG-3’ |
| kan cassette† | |
| SJP-030F | 5’-GCGAAGCAACGATGATGTGTGCTCGTAGCCGGGAGGCCTGA tac ATGAGCCATATTCAACGGGAAACGTCTTG-3’ |
| SJP-030B | 5’-CGGCGACCAGGGCAATCGTTAATGGACGCTGTTCAGTCAT ggtatat TTAGAAAAACTCATCGAGCATCAAATGAAAC-3’
TTAGAAAAACTCATCGAGCATCAAATGAAAC-3’ |
| ~ 100 bp above and below lpxA open reading frame region in E. coli chromosome | |
| SJP-028F | 5’-CGCCGCGGCCTGACC-3’ |
| SJP-028B | 5’-CGTGGCCCGGCAACACCAAC-3’ |
| inside kan cassette | |
| SJP-029F | 5’-AATATTGTTGATGCGCTGGC-3’ |
| SJP-029B | 5’-AGCGAGACGAAATACGCGA-3’ |
| sequencing primer | |
| T7P | 5’-TAATACGACTCACTATAGGG-3 |
| T7T | 5’-GCTAGTTATTGCTCAGCGG-3 |
| pBAD-F | 5'-ATGCCATAGCATTTTTATCC-3' |
| pBAD-B | 5'-GATTTAATCTGTATCAGG-3' |
Open reading frames are capitalized, and the restriction sites, NdeI (CATATG) and BamHI (GGATCC), are underlined.
Open reading frames are capitalized, with underlines for lpxA, and double-underline for lpxB. The ribosome binding site (boxed sequence) was inserted during primer design for lpxB(36) expression.
AtLpxA Expression and Purification
The AtLpxA protein was overexpressed and purified by modifying the previously described method for purification of EcLpxA(14, 15). E. coli C41(DE3) cells(16) harboring SJD-01 were grown overnight in LB broth(17) supplemented with 100 µg/mL ampicillin at 37 °C shaking at 220 rpm. The overnight culture was diluted hundredfold to fresh LB broth containing 100 µg/mL ampicillin, and grown to an A600 of ~0.6 at 37 °C. Protein expression was induced for 4 h with 0.2 mM isopropyl-β-d-thiogalactopyranoside at 37 °C (Figure S2A). Cells from a 1 L culture were harvested by centrifugation for 15 min at 4 °C and 4000 ×g, and washed with phosphate buffered saline (PBS, 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4). The cell pellet was resuspended in 20 mL of ice-cold lysis buffer (10 mM potassium phosphate buffer, pH 7.0, and 20% glycerol). The cells were disrupted by one passage through French pressure cell at 18,000 psi. The suspension of lysed cells was centrifuged at 4 °C and 140,000 ×g for 1 h to remove un-lysed cells and cell membranes. The supernatant (~20 mL, ~180 mg total protein) was loaded onto a Reactive Green-19 Agarose column (25 mL bed volume), pre-equilibrated with lysis buffer. After washing the column with 50 mL of lysis buffer, the bound proteins were eluted with 50–100 mL each of lysis buffer containing 100 mM, 200 mM, 400 mM, 1 M of KCl. Each fraction was analyzed with SDS-PAGE to determine the fractions containing AtLpxA (Figure S2B). The fractions containing AtLpxA (4 fractions totaling ~200 mL), were pooled and concentrated to approximately 20 mL (~1 mg/mL) with an Amicon stirred cell concentrator (Millipore, Billerica, MA). A portion (10 mL) was loaded onto a HiLoad 26/60 Superdex 200 prep grade (320 mL bed volume, GE Healthcare), pre-equilibrated with 10 mM potassium phosphate buffer, pH 7.0, containing 200 mM KCl and 20% glycerol. Fractions containing AtLpxA (Figure S2B) were pooled and concentrated to ~18 mg/mL for storage. The concentration of protein was determined by both absorbance at 280 nm and the BCA assay kit (Pierce, Rockford, IL). The purity of the protein was evaluated by SDS-PAGE. The purified protein was flash frozen in liquid nitrogen and stored at −80 °C.
LpxA Activity Assays
R,S-3-hydroxymyristoyl-ACP was obtained as described previously(18, 19) and the LpxA assay conditions were similar to those reported previously(14, 15) with minor modifications. Each reaction (typically 10 µL) contained 40 mM Hepes, pH 7.4, 1 mg/mL bovine serum albumin, 10 µM R,S-3-hydroxymyristoyl-ACP, and 10 µM [α-32P]UDP-GlcNAc at 9 × 105 µCi/mmol. The reaction was started by the addition of enzyme (typically 0.5–20 µg/mL) and incubated at 30 °C for times indicated. The reactions were terminated by spotting 1 µL portions onto a 0.25 mm silica gel 60 thin layer chromatography plate (EMD Chemicals, Gibbstown, NJ). The plates were developed in CHCl3/CH3OH/H2O/CH3COOH (25:15:4:2, v/v/v/v). The plate was exposed to a PhosphorImager screen overnight, and the extent of acylation of [α-32P]UDP-GlcNAc was determined by scanning with a Storm 840 PhosphorImager (GE Healthcare, Piscataway, NJ).
Construction of a conditional lpxA chromosomal knockout strain
To construct a conditional lpxA chromosomal knockout strain, we replaced the E. coli chromosomal copy of lpxA with a kanamycin resistance cassette in the presence of an appropriate covering plasmid, pHpLpxA or pAtLpxA (Table 1). The kanamycin resistance cassette (kan) was amplified from the plasmid pET30a(+) (EMD Chemicals) with primers, SJP-030F and SJP-030B (Table 2), using KOD hot start DNA polymerase following protocol provided by manufacturer (EMD Chemicals). In addition to sequences homologous to the kan gene, primer SJP-O3OF contains 44 base pairs of genomic DNA upstream of the 5′ end of lpxA and primer SJP-030B contains a ribosome binding site followed by the first 34 bp of the downstream lpxB gene on the 3′ end. The resulting PCR product was resolved on a 1% agarose gel and purified with the QIAquick gel kit from Qiagen (Valencia, CA).
The initial conditional mutant was constructed using E. coli DY330 strain (Table 1) that carries the λ RED recombinase(20) and pHpLpxA. Electrocompetent DY330/pHpLpxA cells were prepared by published protocol(21). Briefly, DY330/pHpLpxA were grown at 30 °C in 50 mL LB medium containing 25 µg/mL chloramphenicol and 0.2% L-arabinose until A600 of the cells reached about 0.4. Then the culture was shifted to 42 °C for 15 min to activate the λ RED genes(20). Next, the cells were washed twice with 30 mL of ice-cold water, and then resuspended in 500 µL of ice-cold water. A portion of cells (100 µL) was used for electroporation with the lpxA::kan PCR product generated as described above. Following electroporation, the cells were grown for 2 h at 30 °C in 1 mL of LB broth containing 0.2% L-arabinose. The cells were harvested and re-suspended to a final volume of 100 µL of LB and then plated onto LB-agar plate, containing 0.2% L-arabinose, 25 µg/mL chloramphenicol, 20 µg/mL kanamycin (LB-ara-cam-kan). The plate was incubated at 30 °C overnight. The resulting colonies were re-purified by streaking on LB-ara-cam-kan. To confirm chromosomal lpxA deletion, the lpxA region on the chromosome of DY330 lpxA::kan/pHpLpxA was amplified with primers SJP-028F and SJP-028B (see Table 2 and Figure S3B) and sequenced with primer SJP-028F at the Duke University DNA Analysis Facility. The resulting strain was named as a SJS-1D (Table 1).
A P1 lysate was prepared from SJS-1D to transduce lpxA::kan into the E. coli W3110 cells harboring pAtLpxA (pBAD33 carrying AtLpxA) or pBAD33 as a control. W3110 lpxA::kan pAtLpxA was selected and purified twice from a LB-ara-cam-kan plate containing 100 mM sodium citrate and named as SJS-2W (Table 1). Consistent with lpxA being required for growth(4), transductants were only viable with cells harboring pAtLpxA and not with cells containing the control vector pBAD33. For in vivo complementation tests, SJS-2W and W3110/pBAD33 were streaked on the LB-cam plates containing 0 %, 0.02 %, or 0.2 % L-arabinose.
Isolation and characterization of lipid A species
An overnight culture of E. coli SJS-2W cells and W3110/pBAD33 were grown at 37 °C and diluted hundred-fold to fresh LB media supplemented with 25 µg/mL chloramphenicol and 0.2% L-arabinose. Cells were grown at 37 °C shaking at 220 rpm until an A600 of ~1.0. Cells from 200 mL culture were harvested by centrifugation at 4 °C, 4000 ×g, and washed with 50 mL of PBS. The cell pellet was resuspended in 20 mL PBS, and then 50 mL of CH3OH and 25 mL of CHCl3 were added to generate a single-phase Bligh-Dyer extraction mixture CHCl3/CH3OH/PBS, (1:2:0.8, v/v/v) (22). The single-phase mixture was incubated at room temperature with stirring for 1 hour. The pellet was recovered by centrifugation at room temperature, 2500 ×g for 20 min, and the pellet was washed again with 40 mL of CHCl3/CH3OH/PBS, (1:2:0.8, v/v/v). The pellet was air dried, then resuspended in 25 mL of 50 mM sodium acetate. The pH of the suspension was adjusted to 4.5 by adding 2–3 drops of glacial acetic acid. The suspension was then incubated at 100 °C for 30 min to hydrolyze the Kdo-lipid A linkage(23). The hydrolysis mixture was cooled to room temperature, and then CH3OH and CHCl3 (28 mL each) were added to form 2-phase Bligh-Dyer mixture(22). The lipid A extracted into the lower phase was collected and concentrated with a rotary evaporator. The dried sample was dissolved in a CHCl3/CH3OH mixture (2:1, v/v) and then subjected to mass spectrometry analysis using QSTAR XL electrospray ionization quadrupole time-of-flight mass spectrometer (Applied Biosystems, Foster City, CA) in negative-ion mode as described previously(24, 25). Data analysis was done with the Analyst QS software suite (Applied Biosystems). Exact masses were determined using ChemBioDraw Ultra 12.0.3.
Crystallization and Structure Determination
Initial crystals of AtLpxA were found by screening against the Crystal Screen HT matrix (Hampton Research) using the sitting-drop vapor diffusion method with the Phoenix RE robotics system (Art Robbins Instruments, Sunnyvale, CA). Purified AtLpxA (18 mg/mL in 10 mM potassium phosphate buffer, pH 7.0, 200 mM KCl, and 20% glycerol) was mixed at a 1:1 ratio with a precipitant solution containing 0.5 M ammonium sulfate, 0.1 M sodium citrate tribasic dihydrate (pH 5.6), and 1.0 M lithium sulfate monohydrate, and allowed to equilibrate at 20 °C. Hexagonal-shaped crystals appeared after 1–2 days and grew to a maximal size that measured ~60 µm × 60 µm × 25 µm. Crystals were frozen in liquid N2. X-ray diffraction data were collected on the SER-CAT 22-BM beamline at the Advanced Photon Source (APS) at Argonne National Laboratory. The data were processed and scaled using HKL2000(26) (Table 3). The AtLpxA structure was solved by the molecular replacement method using PHASER(27, 28) and a poly-alanine search model prepared from the coordinates of LpxA from E. coli (PDB id: 1LXA)(10). Regions outside the hexapeptide repeats were removed from the initial MR solution and manually rebuilt using the program COOT(29). Iterative restrained refinement was carried out using REFMAC5(30) within the CCP4 software suite(31) with TLSMD options selected(32). The final model yielded Rwork = 20.7% and Rfree = 24.1% and was validated using MOLPROBITY(33), which reported 96.15% Ramachandran favored region, and 0.35% outlier. The data statistics are summarized in Table 3. Secondary structure elements were analyzed with the help of STRIDE(34), along with a comparison to the known structures of LpxA from several bacteria. Structural alignments and figures were prepared using PyMol(35). Atomic coordinates and structure factors for AtLpxA have been deposited in the Protein Data Bank (3T57).
Table 3.
Data Collection and Refinement Statistics
| AtLpxA | |
|---|---|
| Data Collectiona | |
| Source/Detector | SER-CAT 22-BM /Mar225 CCD |
| Space Group | P6322 |
| a, b, c (Å) | 86.2, 86.2, 200.3 |
| Wavelength (Å) | 1.00 |
| Oscillation Range/Δ,° | 0 ~ 90 /0.75 |
| Resolution (Å) | 50.00 – 2.10 (2.14 – 2.10) |
| No. of Reflections | |
| Measured | 663184 |
| Unique | 26374 |
| Completeness (%) | 100.0 (100.0) |
| Average I/σ(I) | 21.4 (3.1) |
| Redundancy | 10.5 (9.1) |
| Rmerge (%)1 | 11.0 (55.0) |
| Refinement | |
| Resolution (Å) | 24.88 – 2.10 (2.16 – 2.10) |
| No. reflections | 24836 |
| Rwork (%) | 20.7 |
| Rfree (%)b | 24.1 |
| r.m.s. deviations | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.163 |
| No. of protein residues in A.U. | 288 |
| No. of water molecules | 165 |
| Average B-factor (Å2) | |
| Protein | 18.0 |
| Water | 25.9 |
| Ramachandran (%) | |
| Favored regions | 96.15 |
| Allowed regions | 99.65 |
| Disallowed regions | 0.35 |
| All-atom clash score | 6.95 |
| MolProbity score | 1.64 |
Highest resolution shell is shown in parenthesis. Data were collected from a single crystal.
Rfree is the R value obtained for a test set of reflections consisting of randomly selected 5% subset for the data excluded from refinement.
Results
In vitro Activity of the AtLpxA enzyme
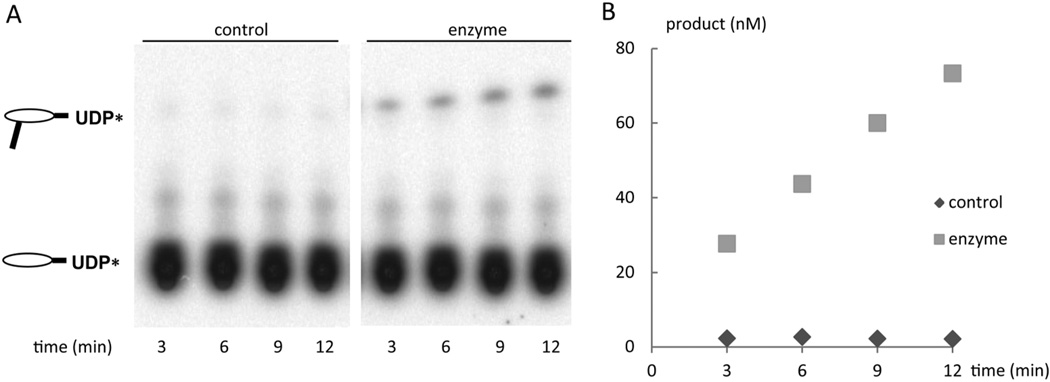
AtLpxA was purified to homogeneity (Figure S2B) and was active under in vitro assay condition for EcLpxA (Figure 2). In vitro assays show AtLpxA transfers a 3-hydroxymyristoyl group from 3-hydroxymyristoyl-ACP to the 3-OH of UDP-GlcNAc with a specific activity of 5.1 nmol min−1 mg−1, about ten times lower than that of the purified EcLpxA when assayed in parallel. While the activity of AtLpxA was lower than that of EcLpxA, it is comparable to the enzymatic activities of other LpxA orthologs from Francisella novicida and Helicobacter pylori purified from E. coli expression system (S. Joo et al., in preparation).
Figure 2. Activity of the purified AtLpxA expressed in E. coli.
Purified AtLpxA (1 µg/mL) was assayed in a reaction mixture containing 40 mM Hepes buffer, radiolabeled UDP-GlcNAc (10 µM) and R,S-3-hydroxymyristoyl-ACP (10 µM) for the time indicated at 30 °C. Panel A shows the reaction products in the presence and absence of AtLpxA after separation of using thin layer chromatography. The formation of [α-32P]-UDP-3-O-(R-3-hydroxymyristoyl)-UDP-GlcNAc was quantified by PhosphorImager as described in Experimental Procedures and plotted versus time in panel B.
Complementation of E. coli lpxA knockout mutant by A. thaliana lpxA
lpxAs from other Gram-negative bacteria are able to rescue the phenotype of a temperature-sensitive mutant of E. coli lpxA, SM101(4, 6). However, attempts to complement SM101 with AtlpxA were unsuccessful. To test whether AtlpxA could functionally replace EclpxA, we constructed a conditional mutant with a deletion of the chromosomal copy of E. coli lpxA. In this mutant, only lpxA provided on the covering plasmid would be expressed preventing the formation of LpxA heterotrimers of EcLpxA and AtLpxA.
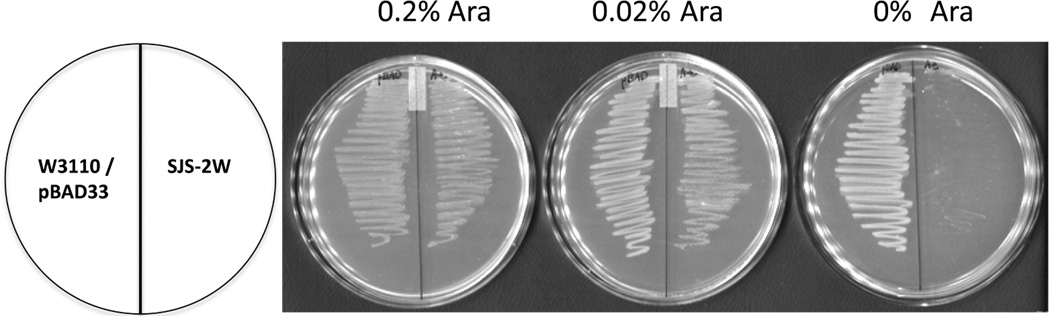
A plasmid containing AtlpxA under control of an arabinose inducible promoter, pAtLpxA, supported the growth of the E. coli lpxA::kan mutant on LB plates but only when AtlpxA expression was induced by the presence of L-arabinose (0.02% or 0.2%) (Figure 3). In the absence of L-arabinose, AtlpxA expression was not induced and the cells did not survive consistent with lpxA being required for growth(4, 6). This result clearly shows that a plant lpxA can functionally replace EclpxA. Combined with the in vitro assay data described above, it is extremely likely that AtLpxA functions analogously to EcLpxA and may function similarly in A. thaliana.
Figure 3. Complementation of the E. coli lpxA::kan with AtlpxA.
Cells, W3110/pBAD33 and SJS-2W were streaked, as indicated by the schematic, onto LB agar plates containing 25 µg/mL chloramphenicol and 0%, 0.02% or 0.2% L-arabinose and incubated overnight at 37 °C.
Characterization of lipid A species
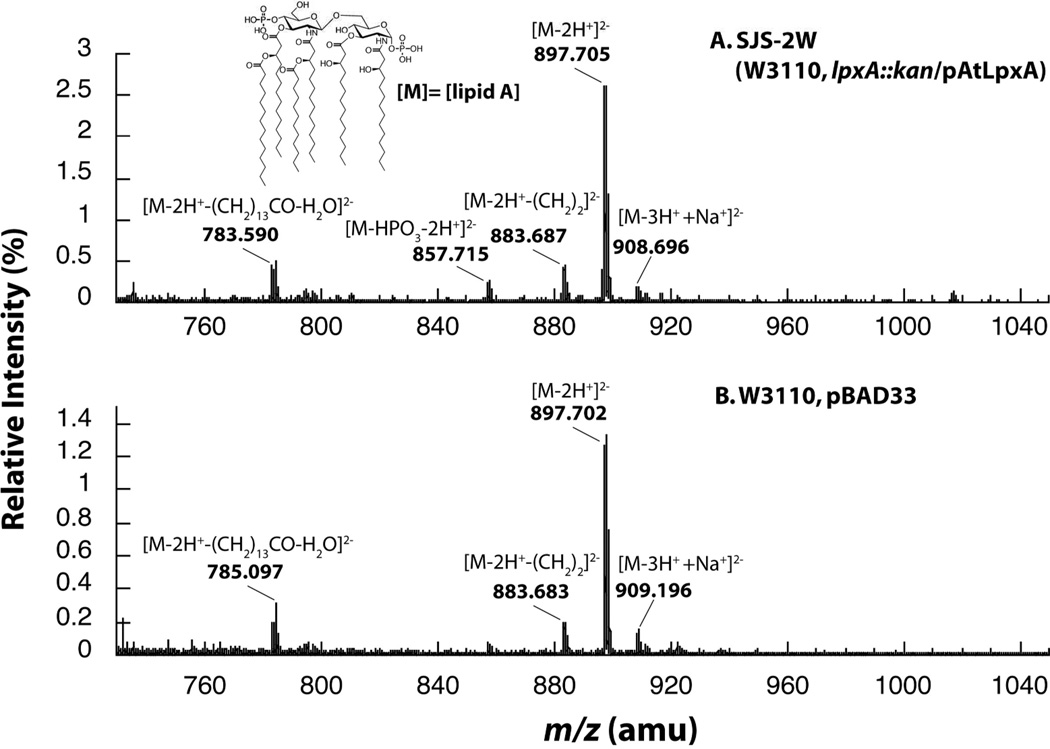
In addition to possessing in vitro enzyme activity analogous to EcLpxA, AtLpxA was able to functionally replace EcLpxA by supporting the biosynthesis of the expected lipid A anchor of LPS. LPS from SJS-2W cells and W3110/pBAD33 was subjected to mild acid hydrolysis to release lipid A for characterization using negative-ion electrospray ionization quadrupole time-of-flight mass spectrometry (ESI-MS). The overall lipid A profiles of SJS-2W, lpxA::kan covered by pAtLpxA, and W3110/pBAD33, are remarkably similar. The major negative-ions observed are doubly charged [M−2H+]2− ions(24). The [M−2H+]2− at m/z 897.70 corresponds to the major hexa-acylated lipid A species (exact mass, m/z 897.6024) (Figure 4, inset) and is the prominent ion in both spectra. The [M−2H+]2− at m/z 883.68 corresponds to the [M−2H+]2− ion of a minor species of hexa-acylated lipid A in which one of the six acyl chains is shorter by two methylenes (exact mass, m/z 883.5867). These shorter chains are likely from the lack of acyl chain specificity of one of the late acyl transferases(24). For example, the secondary fourteen carbon myristate chain added by LpxM to the 3′-hydroxymyristate maybe replaced with a twelve carbon laurate moiety. The [M−2H+]2− ion at m/z 783.59 corresponds to the loss of the myristate group as a free acid (RCOOH) (exact mass, m/z 783.4979), likely occurring during the mass spectrometry analysis(24). The minor [M−2H+]2− ion at m/z 857.72 corresponds to the dephosphorylated lipid A species (exact mass, m/z 857.6195) which is likely produced during mild acid hydrolysis of LPS. Taken together, AtLpxA promotes the formation of the same lipid A species in vivo as EcLpxA. These data also give insight into the acyl-chain specificity of AtLpxA. The predominant lipid A formed in the presence of AtLpxA is exactly the same as the lipid A formed by EcLpxA indicating that AtLpxA has the preference for R-3-hydroxymyristoyl-ACP as EcLpxA(5).
Figure 4. Negative-ion ESI-MS of analysis of lipid A species purified from SJS-2W and W3110/pBAD33.
The negative-ion ESI-MS from m/z 750 to 1040 of the lipid A species isolated from SJS-2W (panel A) and W3110/pBAD33 (panel B). The lipid A species are remarkably similar between the two samples consistent with AtLpxA and EcLpxA having similar activities in vivo and in vitro. The predominant ions are [M-2H]2− ions, however, [M-3H++Na+]2− ions are also detected at m/z 908.59.
Crystal structure of AtLpxA
Based on the high sequence identity (38%), conservation of key active-site residues and the in vitro and in vivo activity reported above, AtLpxA appears to be biochemically similar to that of EcLpxA. To determine how AtLpxA varies structurally from EcLpxA we investigated the structure of AtLpxA. Like EcLpxA, AtLpxA is a homotrimer in aqueous solution, as judged by size-exclusion chromatography of the catalytically active enzyme (Figure S2B). Crystals of AtLpxA were grown and diffracted to 2.1 Å. Similar to EcLpxA(10), AtLpxA crystallized with a single polypeptide chain in the asymmetric unit, and the homotrimer is formed about a crystallographic 3-fold axis of symmetry. Although the majority of AtLpxA could be built, two regions were omitted from the final model due to insufficient, noncontiguous electron density. The first region consists of eight residues (Asp33-Val39) localized to the N-terminus, and the second constitutes nine residues positioned at the C-terminus (Phe328-Thr336).
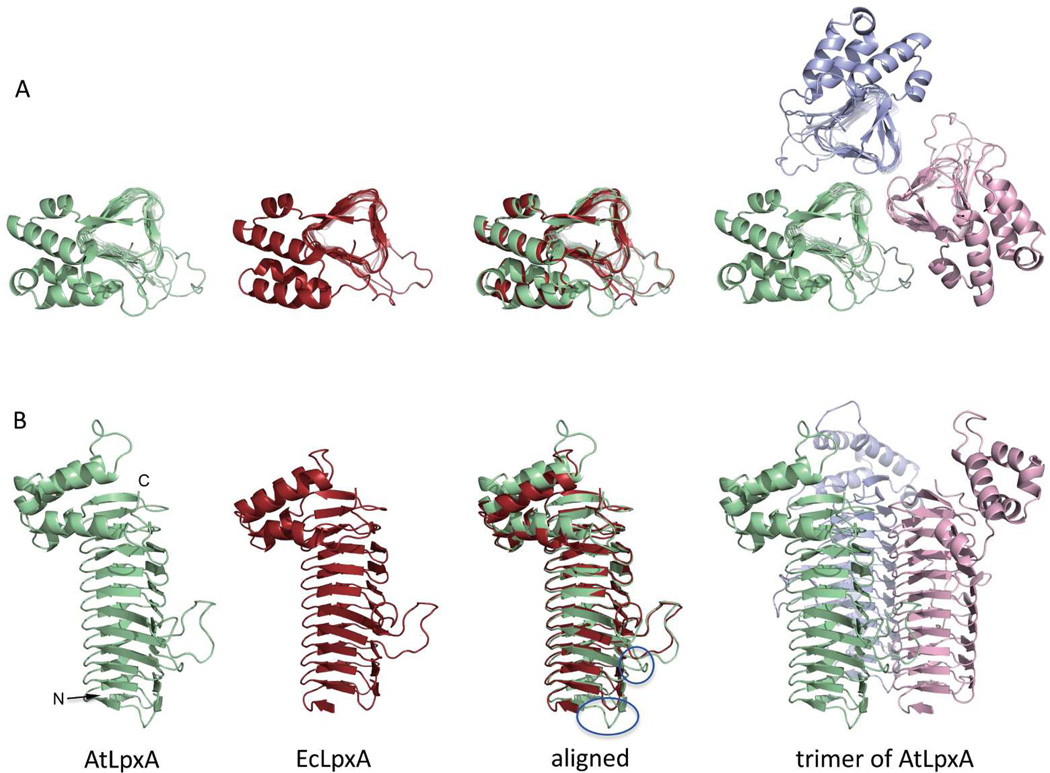
The overall structure of AtLpxA is similar to the structures of the EcLpxA and other LpxAs solved thus far with the characteristic N-terminal LβH domain, in a shape of triangular prism, connected to the C-terminal portion that is comprised mainly of α-helices (Figure 5)(7, 10, 11).
Figure 5. Structure of AtLpxA.
Panel A: Top-down views of the asymmetric units of AtLpxA (green), EcLpxA (red) and the biological trimer of AtLpxA (pale green, light pink and light blue). Panel B: Side views of the molecules as indicated. Circled areas correspond to the coil C0 and loop 0 of AtLpxA.
The N-terminal LβH domain of the AtLpxA protein (residues Leu40 –Met241) consists of 31 β-strands; 29 of these comprise 10 β-helical coils C1 through C10 similar to those found in EcLpxA. Two additional β-strands shape the partial coil C0. As seen in other LpxA proteins, two loops, loop 1, and loop 2, interfere with the LβH domain. Loop 1 is inserted after the fourth coil C4 and loop 2 is inserted after the fifth coil C5 of AtLpxA. Loop 1 and loop 2 are one amino acid shorter and one amino acid longer, respectively, compared to the corresponding loops found in EcLpxA but otherwise have similar conformations to those of EcLpxA. In addition to these two loops, the initial sequence alignment predicted two extra loops (Figure S1). Of the two additional loops, the smaller, 4-residue loop forms loop 0 (residues Glu104 – Gly107) and interrupts the β-helix right after the third helical coil C3. This small loop contains a Glu104 residue, the OE1 atom of which is 3.9 Å apart from the hydroxyl oxygen of Tyr 78 belonging to the adjacent monomer. The triangular rod of LβH domain is bent about 7 degrees in counterclockwise direction at loop 0, when seen from the amino terminal side of 3-fold axis (Figure S4). The larger, 18-residue insert (Asn73 – Gly90) predicted from the initial sequence alignments to interrupt the β-helix actually turned out to form a coil of three hexapeptide repeats. These additional hexapeptide repeats result in the extra coil, C0, discussed above.
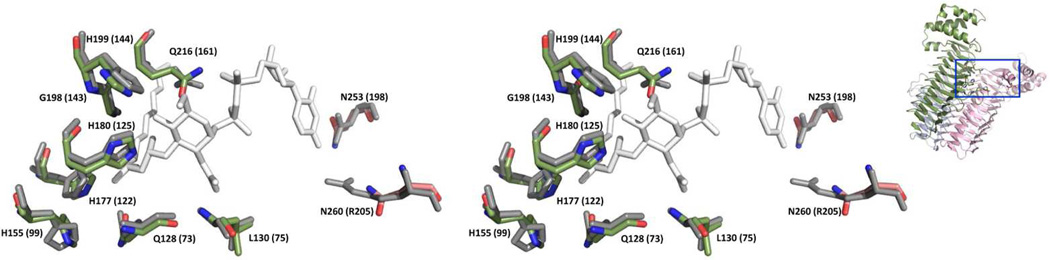
The C-terminal portion of AtLpxA (residues Val242 –Thr336) consists of five α-helices that have slightly different tilts and loops of different length as compared to EcLpxA but is overall very similar. Superposition of EcLpxA and AtLpxA yielded a RMS deviation of 0.9 Å for 180 Cα atom pairs, as determined by analysis in PyMol. In addition, key LpxA active site residues(11, 15) are similarly positioned when the AtLpxA is compared with EcLpxA bound with UDP-3-O-(R-3-hydroxymyristoyl)-GlcNAc(11) (Figure 6).
Figure 6. Overlay of the conserved residues within the active site of AtLpxA on that of EcLpxA (Stereo view).
Residues in the active site of AtLpxA superimposed with the structure of EcLpxA complexed with its product, UDP-3-O-(R-3-hydroxymyristoyl)-GlcNAc (PDB ID: 2QIA)(11). Water molecules are omitted to highlight the conservation of residues between AtLpxA and EcLpxA. AtLpxA residues, EcLpxA residues and product are colored green, grey and white respectively. The residue names and numbers of AtLpxA are shown, with the corresponding residues from EcLpxA in the parentheses. Inset: The box on the AtLpxA trimer shown to the side indicates the active site area enlarged in the figure.
Discussion
Through the cloning, purification and crystallization of AtLpxA we have shown that this plant enzyme shares significant similarities to the bacterial EcLpxA. The purified AtLpxA catalyzes the same in vitro reaction, the transfer of a R-3-hydroxymyristoyl chain from R-3-hydroxylmyristoyl-ACP to the 3′-hydroxyl of the glucosamine of UDP-GlcNAc. In addition, AtLpxA is able to functionally replace EcLpxA, rescuing an E. coli lpxA knockout and promoting the synthesis of E. coli-like lipid A in vivo. Taken together this work clearly supports the hypothesis that plants, such as A. thaliana that possess lpx genes, synthesize lipid A like molecules similar to those found in Gram-negative bacteria.
The AtLpxA possesses the same in vitro enzyme activity as EcLpxA. While its activity is only 1/10 of the EcLpxA, this level is similar to that of other LpxA orthologs, such as from Helicobacter pylori and Francisella novicida (Joo, et al., in preparation), expressed and purified using E. coli expression systems. It is possible that the lower activity is an inherent property of the AtLpxA, however it is possible that the substrates used in the assay were not optimal for the enzyme. The ACP used in the in vitro assays was from E. coli not A. thaliana. The E. coli acyl-ACP may not bind as efficiently to the AtLpxA leading to a lower specific activity. Our data and previous results point to a glucosamine-based nucleotide diphosphate as the substrate for AtLpxA; glucosamine based lipid A-like molecules are detected in A. thaliana lipid extracts(2).
While we have not studied the in vivo substrate specificity of AtLpxA thoroughly, our data support specificity for R-3-hydroxymyristoyl-ACP over other chain lengths. The lipid A species isolated from SJS-2W, lpxA::kan covered by pAtLpxA, was identical to the lipid A profile isolated from wild-type E. coli. If AtLpxA preferred other acyl chain lengths it is likely that it would impact the lipid A species observed in vivo as has been observed previously(6, 37).
Previous work has shown that LpxA orthologs can rescue the phenotype of a well-characterized temperature-sensitive lpxA mutant, SM101(4, 6). For example, lpxA from Helicobacter pylori support the growth of E. coli lpxA temperature sensitive mutant SM101 (C. Sweet, et al, in preparation) at 42 °C. The AtLpxA did not rescue SM101 but did rescue SJS-2W, a chromosomal knockout of lpxA. The complementation of the lpxA::kan mutant clearly shows that AtlpxA can functionally replace EclpxA despite the fact that SM101 was not complemented by AtlpxA. Perhaps the LpxA trimers formed from the mutant EcLpxA and AtLpxA do not possess sufficient catalytic activity to support growth. This chromosomal knockout of lpxA will serve as a valuable tool to study the functions of other LpxA orthologs.
Our 2.1 Å resolution crystal structure of the AtLpxA shows both conservation and variation of the LpxA structure. At first glance, the overall structure of AtLpxA is similar to the structure of EcLpxA. The conserved residues in AtLpxA align strikingly well with the active site residues of EcLpxA (Figure 6), when superimposed to the EcLpxA complexed with the product UDP-3-O-(R-3-hydroxymyristoyl)-GlcNAc (PDB ID: 2QIA) (11). All the residues important for catalysis of EcLpxA(11, 15) are well conserved in AtLpxA. His180 (His125 in EcLpxA) is proposed to be a catalytic base, activating the GlcNAc 3-OH group to help transfer of the acyl chain from R-3-hydroxymyristoyl-ACP and His199 (His144 in EcLpxA) is proposed to hydrogen bond with the GlcNAc 6-OH of the substrate(11, 15).
As described above, the structure of AtLpxA supports the preference for hydroxyacyl chain over acyl chain. Gly173 in EcLpxA is well characterized as a hydrocarbon ruler(11, 14), allowing a longer hydroxymyristoyl chain in the active site as compared to the hydroxydecanoyl chain allowed by Met169-containing Pseudomonas aeruginosa LpxA. Gly228 in AtLpxA is analogous to Gly173 in EcLpxA. The presence of a small residue in this position supports the hydroxymyristoyl acyl chain selectivity suggested by the in vitro and in vivo data presented here.
While there are striking similarities between the EcLpxA and AtLpxA structures, several differences are worth noting. Initial alignment of the EcLpxA and AtLpxA sequences predicted two additional loops (Figure S1, panel A). The longer 18 amino acid insert does not form a loop but instead forms three additional hexapeptide repeats. The initial sequence alignment did not recognize three of the hexapeptide repeats, [STAV]-X-[LIV]-[GAED]- X2(38), due to the presence of aromatic residues (Tyr and Phe) in the position usually occupied by small, or negatively charged residues (Gly, Ala, Glu, Asp). It is not clear whether AtLpxA has a complete coil C0 of 18 amino acids, as the 8 amino terminal residues, DSRDSE, did not show sufficient electron density. While this sequence is recognized as a hexapeptide repeat in sequence alignment using ClustalW, we suspect this region is rather unstructured. This additional coil, whether complete or not, is a significant difference between the AtLpxA and other LpxA bacterial orthologs. Moreover, this additional coil does not appear unique to Arabidopsis enzyme, as a sequence search finds other plant LβH proteins with additional hexapeptide repeats. For example, a putative LβH proteins from Populus trichocarpa (NCBI reference XP_002308293.1) has three additional hexpeptide repeats probably forming a complete coil of C0, resulting in an LβH of 11 coils. The longest LβH domain reported so far exists in E. coli N-acetylglucosamine-1-phosphate uridyltransferase (GlmU) with 11 coils(39). The functional consequence of this additional coil or if this extra coil is essential has yet to be determined.
The AtLpxA also has an additional 4 amino acid loop, loop 0, (Glu104 -inserted Gly107) after coil C3. The OE1 oxygen atom of Glu104 in loop 0 is approximately 4 Å from hydroxyl oxygen of Tyr78 from the adjacent monomer possibly strengthening the inter-chain interaction. While this additional loop 0 does not appear to be present in the bacterial LpxAs, it exists in other plant LpxAs suggesting an important structural role in these enzymes. Interestingly, the alignment of the AtLpxA and EcLpxA structures is better (RMS deviation of 0.8 Å with 170 Cα atom pairs) when the amino terminal region of AtLpxA up to loop 0 is removed. This is likely due to the fact that the LβH domain of AtLpxA is bent around loop 0 region toward the turn 2 between the sheets 2 and 3 about 7 degrees (Figure S4). Among the LpxA protein structures reported so far, this bent-rod shape for the LβH is seen only in AtLpxA and may be related to the presence of loop 0.
In conclusion, we have firmly established that the AtLpxA is a functional LpxA through in vitro and in vivo analyses of its biochemical activity. While structurally similar to the EcLpxA, the differences between these structures may help illuminate the biological role of this protein and lipid A-like molecules in plants.
Supplementary Material
Acknowledgments
We would like to thank Dr. Nathan Nicely and Mr. Chuljin Lee for assistance with X-ray diffraction experiments and Dr. Jinshi Zhao for assistance constructing the bacterial knockout strains. We would like to thank Dr. Charles W. Pemble IV for his critical reading of the manuscript. Diffraction data were collected at the Southeastern Regional Collaborative Access Team (SER-CAT) 22-BM beamline at the Advanced Photon Source, Argonne National Laboratory. Supporting institutions may be found at www.ser-cat.org/members.html. Use of the Advanced Photon Source, an Office of Science User Facility operated for the U.S. Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the U.S. DOE under contract No. W-31-109-Eng-38.
Funding Source Statement: This research was supported by NIH Grant GM-51310 to C. R. H. Raetz, and by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MEST) to S. H. Joo (2010-0003008).
Abbreviations and Textual Footnotes
The abbreviations are:
- ACP
acyl carrier protein
- EcLpxA
Escherichia coli LpxA
- AtLpxA
Arabidopsis thaliana LpxA
- HpLpxA
Helicobacter pylori LpxA
- LβH
left-handed parallel β helix
- LPS
lipopolysaccharide
- ESI-MS
electrospray ionization quadrupole time-of-flight mass spectrometry
- PBS
phosphate-buffered saline
- UDP-GlcNAc
UDP-N-acetylglucosamine
- LB
Luria Bertani
- TLSMD
Translation/Libration/Screw Motion Determination
- RMS
root mean square
- Hepes
4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid
- Kdo
3-deoxy-D-manno-oct-2-ulosonic acid
- PCR
polymerase chain reaction
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- ara
arabinose
- cam
chloramphenicol
- kan
kanamycin
Footnotes
Supporting Information:
Figures showing the sequence alignment of the EcLpxA and AtLpxA (Figure S1), the overexpression and purification of AtLpxA (Figure S2), the scheme for the construction of ΔlpxA E. coli (Figure S3), and the bent rod-shape of the Lβelix domain of AtLpxA (Figure S4) are included in the supporting information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Raetz C, Reynolds C, Trent M, Bishop R. Lipid A modification systems in Gram-negative bacteria. Annu Rev Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li C, Guan Z, Liu D, Raetz CR. Pathway for lipid A biosynthesis in Arabidopsis thaliana resembling that of Escherichia coli. Proc Natl Acad Sci U S A. 2011;108:11387–11392. doi: 10.1073/pnas.1108840108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raetz C, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galloway SM, Raetz CR. A mutant of Escherichia coli defective in the first step of endotoxin biosynthesis. J Biol Chem. 1990;265:6394–6402. [PubMed] [Google Scholar]
- 5.Anderson MS, Raetz CR. Biosynthesis of lipid A precursors in Escherichia coli. A cytoplasmic acyltransferase that converts UDP-N-acetylglucosamine to UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine. J Biol Chem. 1987;262:5159–5169. [PubMed] [Google Scholar]
- 6.Sweet C, Lin S, Cotter R, Raetz C. A Chlamydia trachomatis UDP-N acetylglucosamine acyltransferase selective for myristoyl-acyl carrier protein. Expression in Escherichia coli and formation of hybrid lipid A species. J Biol Chem. 2001;276:19565–19574. doi: 10.1074/jbc.M101868200. [DOI] [PubMed] [Google Scholar]
- 7.Lee BI, Suh SW. Crystal structure of UDP-N acetylglucosamine acyltransferase from Helicobacter pylori. Proteins. 2003;53:772–774. doi: 10.1002/prot.10436. [DOI] [PubMed] [Google Scholar]
- 8.Robins LI, Williams AH, Raetz CR. Structural basis for the sugar nucleotide and acyl-chain selectivity of Leptospira interrogans LpxA. Biochemistry. 2009;48:6191–6201. doi: 10.1021/bi900629e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Research. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raetz C, Roderick S. A left-handed parallel beta helix in the structure of UDP-N-acetylglucosamine acyltransferase. Science. 1995;270:997–1000. doi: 10.1126/science.270.5238.997. [DOI] [PubMed] [Google Scholar]
- 11.Williams A, Raetz C. Structural basis for the acyl chain selectivity and mechanism of UDP-N-acetylglucosamine acyltransferase. Proc Natl Acad Sci U S A. 2007;104:13543–13550. doi: 10.1073/pnas.0705833104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. European Journal of Biochemistry. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 13.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wyckoff T, Lin S, Cotter R, Dotson G, Raetz C. Hydrocarbon rulers in UDP-N-acetylglucosamine acyltransferases. J Biol Chem. 1998;273:32369–32372. doi: 10.1074/jbc.273.49.32369. [DOI] [PubMed] [Google Scholar]
- 15.Wyckoff T, Raetz C. The active site of Escherichia coli UDP-N acetylglucosamine acyltransferase. Chemical modification and site-directed mutagenesis. J Biol Chem. 1999;274:27047–27055. doi: 10.1074/jbc.274.38.27047. [DOI] [PubMed] [Google Scholar]
- 16.Miroux B, Walker JE. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 17.Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bartling CM, Raetz CR. Steady-state kinetics and mechanism of LpxD, the N-acyltransferase of lipid A biosynthesis. Biochemistry. 2008;47:5290–5302. doi: 10.1021/bi800240r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartling CM, Raetz CR. Crystal structure and acyl chain selectivity of Escherichia coli LpxD, the N-acyltransferase of lipid A biosynthesis. Biochemistry. 2009;48:8672–8683. doi: 10.1021/bi901025v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu D, Ellis HM, Lee E-C, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci U S A. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook JG, Russel DW. Molecular Cloning: A Laboratory Manual, 3rd ed. Plainview, NY: 2001. [Google Scholar]
- 22.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 23.Zhou Z, White K, Polissi A, Georgopoulos C, Raetz C. Function of Escherichia coli MsbA, an essential ABC family transporter, in lipid A and phospholipid biosynthesis. J Biol Chem. 1998;273:12466–12475. doi: 10.1074/jbc.273.20.12466. [DOI] [PubMed] [Google Scholar]
- 24.Raetz CRH, Garrett TA, Reynolds CM, Shaw WA, Moore JD, Smith DC, Ribeiro AA, Murphy RC, Ulevitch RJ, Fearns C, Reichart D, Glass CK, Benner C, Subramaniam S, Harkewicz R, Bowers-Gentry RC, Buczynski MW, Cooper JA, Deems RA, Dennis EA. Kdo2-Lipid A of Escherichia coli, a defined endotoxin that activates macrophages via TLR-4. Journal of Lipid Research. 2006;47:1097–1111. doi: 10.1194/jlr.M600027-JLR200. [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Ribeiro A, Guan Z, Abraham S, Raetz C. Attenuated virulence of a Francisella mutant lacking the lipid A 4'-phosphatase. Proc Natl Acad Sci U S A. 2007;104:4136–4141. doi: 10.1073/pnas.0611606104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otwinowski Z, Minor W. [20] Processing of X-ray diffraction data collected in oscillation mode. In: Charles W, Carter Jr, editors. Methods in Enzymology. Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 27.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallographica Section D. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 29.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 30.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallographica Section D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 31.Collaborative Computational Project, N. The CCP4 suite: programs for protein crystallography. Acta Cryst. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 32.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallographica Section D. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 33.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Research. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frishman D, Argos P. Knowledge-based protein secondary structure assignment. Proteins: Structure, Function, and Bioinformatics. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 35.Schrodinger LLC. The PyMOL Molecular Graphics System. 2010 Version 1.3r1. [Google Scholar]
- 36.Crowell DN, Anderson MS, Raetz CR. Molecular cloning of the genes for lipid A disaccharide synthase and UDP-N-acetylglucosamine acyltransferase in Escherichia coli. J Bacteriol. 1986;168:152–159. doi: 10.1128/jb.168.1.152-159.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Odegaard T, Kaltashov I, Cotter R, Steeghs L, van der Ley P, Khan S, Maskell D, Raetz C. Shortened hydroxyacyl chains on lipid A of Escherichia coli cells expressing a foreign UDP-N-acetylglucosamine O-acyltransferase. J Biol Chem. 1997;272:19688–19696. doi: 10.1074/jbc.272.32.19688. [DOI] [PubMed] [Google Scholar]
- 38.Yoder MD, Keen NT, Jurnak F. New domain motif: the structure of pectate lyase C, a secreted plant virulence factor. Science. 1993;260:1503–1507. doi: 10.1126/science.8502994. [DOI] [PubMed] [Google Scholar]
- 39.Olsen LR, Roderick SL. Structure of the Escherichia coli GlmU pyrophosphorylase and acetyltransferase active sites. Biochemistry. 2001;40:1913–1921. doi: 10.1021/bi002503n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.