Abstract
In an effort to develop pH-sensitive lipoplexes for efficient gene delivery, we report three novel cationic lipids containing a linear ortho ester linker that conjugates either the headgroup (Type I) or one hydrocarbon chain (Type II) with the rest of the lipid molecule. The cationic lipids carry either an iodide or a chloride counterion. Compared to our previously reported cyclic ortho ester linker, the linear ortho ester linker facilitated the construction of cationic liposomes and lipoplexes with different helper lipids. The chloride counterion not only facilitated the hydration of the lipid films during liposome construction, but also enhanced the hydrolysis of the ortho ester linker in the lipoplexes. After incubation at endosomal pH 5.5, the Type I lipoplexes aggregated and destabilized the endosome-mimicking model liposomes, but not the Type II lipoplexes. The helper lipids (DOPE or cholesterol) of the lipoplexes enhanced the pH-sensitivity of the Type I lipoplexes. In CV-1 cells (monkey kidney fibroblast), the Type I ortho ester-based lipoplexes, especially those with the chloride counterion, significantly improved the gene transfection efficiency, in some cases by more than 100 fold, compared to their pH-insensitive counterparts consisting of DOTAP. The gene transfection efficiency of the ortho ester-based lipoplexes was well correlated with their rate of aggregation and membrane destabilization in response to the endosomal pH 5.5.
Keywords: Ortho ester, lipoplex, pH-sensitive, gene delivery, counterion, helper lipid
1. Introduction
The development of gene delivery systems of high efficiency and low toxicity continues to draw strong interest [1] because of its important applications in gene therapy and basic research involving transgenic cell lines or animal models. The reported gene delivery systems fall into two categories, the viral gene delivery systems and the nonviral gene delivery systems [2]. Whereas the viral gene delivery systems offer relatively high gene transfection efficiency, the nonviral gene delivery systems carry the advantages of lower toxicity and more convenient preparation. Complexes of DNA and cationic lipids (lipoplexes) represent one of the most investigated nonviral gene delivery systems, with more than a dozen clinical trials [3-6] and several cationic lipids commercialized for basic research [7, 8].
During gene transfection, the lipoplexes adsorb onto the target cells through electrostatic interactions with the proteoglycans of the extracellular matrix [9] and are subsequently endocytosed and processed into the endosomes [10], where the pH is decreased. A small portion of the DNA molecules in the lipoplexes then escape from the endosome to reach the cytosol [11] and eventually manage to enter the nucleus [12] for gene expression. Conversely, the majority of the lipoplexes are transferred to the lysosome and degraded.
Various approaches have thus been reported to enhance the escape of the lipoplex cargo DNA from the endosomes. Conically shaped lipids [13-15] such as DOPE and cholesterol have long been established as common lipoplex components to help destabilize the endosome membrane. Titratable macromolecules (e.g. GALA peptide) [16, 17] or surfactants (e.g. gemini lipids) [18] that change into membrane-destabilizing conformations at mildly acidic pH have been incorporated into cationic lipoplexes. Furthermore, various acid-labile cationic lipids have been devised to fragment into biomembrane-destabilizing amphiphiles in the endosome [19]. The labile functional groups in such cleavable lipids include vinyl ethers [20], hydrozones [21], and ortho esters [22].
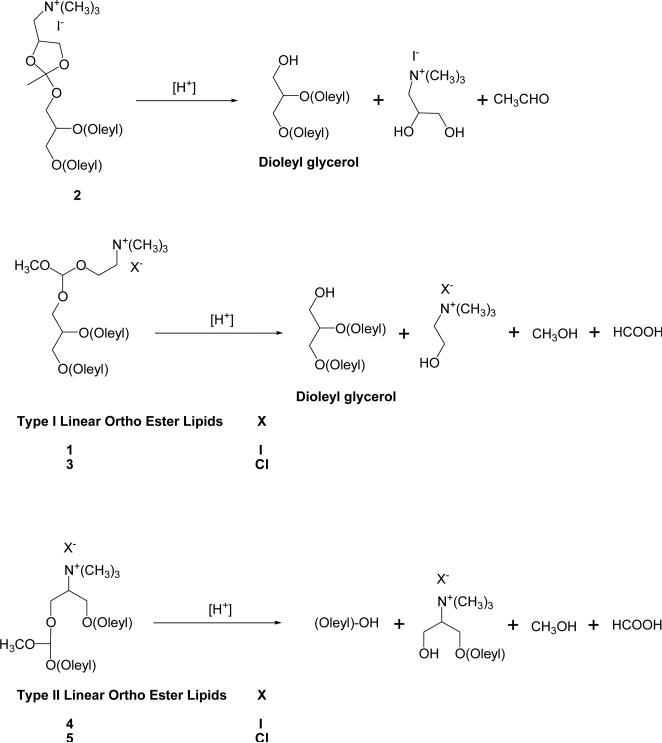
We have been investigating lipids containing ortho ester groups [22-26] because of their higher sensitivity to mildly acidic pHs than most of the chemical groups known in the literature. We reported two cleavable cationic lipids (1 and 2 in Scheme 1) that carry an acid-labile ortho ester linker between the cationic headgroup and the lipid tails [22]. Such lipids are relatively stable at pH 7.4 but are hydrolyzed much more quickly at pH 5.5 to yield the conically shaped lipid dioleylglycerol. After incubation at pH 5.5, lipoplexes consisting of such ortho ester-based cationic lipids underwent Lα → HII phase changes and destabilized endosome-mimicking model liposomes. Among the ortho ester-based lipoplexes under the study, the 2/DOPE/DNA lipoplex mediated the highest gene transfection in cell culture, which was approximately 5-fold as much as the pH-insensitive DOTAP/DOPE/DNA lipoplexes, thus validating the concept that introduction of an acid-labile ortho ester linker to cationic lipids can improve the efficiency of gene delivery by their lipoplexes. However, the improvement of gene transfection by the ortho ester cationic lipids 1 and 2 were modest and the preparation of their liposomes and lipoplexes was often unsuccessful given different helper lipids and buffers.
Scheme 1.
Acid-catalyzed hydrolysis of cationic ortho ester lipids.
In order to further improve the physicochemical properties and the transfection efficiencies of the ortho ester-based lipoplexes, we report three novel cationic lipids carrying a linear ortho ester-linker at different positions of a lipid molecule. We also introduced chloride as well as iodide as the counterion of the cationic lipids. The lipids were designed and synthesized, followed by construction of their lipoplexes together with the helper lipids DOPE or cholesterol. The physicochemical properties of the lipoplexes were characterized including size, ζ-potential and pH-sensitivity. Finally, the ability of such lipoplexes to deliver genes was characterized in cell culture. For comparison, lipoplexes consisting of the previously reported linear ortho ester lipid 1 and those consisting of the pH-insensitive lipid DOTAP were characterized in parallel.
2. Chemistry
2.1. Design of Ortho Ester-based Cationic Lipids
All the three novel ortho ester-based lipids (3, 4, and 5 in Scheme 1) contain a trimethyl ammonium headgroup, two unsaturated oleyl hydrocarbon chains and a linear ortho ester linker. The trimethyl ammonium headgroup carries a formal positive charge regardless of the pH to ensure strong electrostatic interaction with the negatively charged phosphates of plasmid DNA during lipoplex preparation [15]. The two unsaturated lipid chains enhance the fluidity of the lipoplexes and thus facilitate their phase transitions [27]. Based on our prior observations [22], the linear ortho ester linker is expected to be relatively stable at pH 7.4 but hydrolyzes faster at endosomal pH 5.5 to trigger phase changes of the lipoplexes. The linear ortho ester linker is elected over the previously reported cyclic ortho ester linker in compound 2 (Scheme 1) for less steric constraints, which would in turn improve the lipid's ability to arrange into lipid bilayers together with different helper lipids during the construction of liposomes and lipoplexes. In compound 3, the ortho ester linker is introduced between the cationic headgroup and the lipidic dioleylglycerol moiety (hereafter mentioned as Type I ortho ester lipids) so that its hydrolysis would generate the cone-shaped lipid dioleylglycerol (Scheme 1). In the acidic endosomal compartment, this would induce the Lα → HII phase change of the lipoplex to destabilize the endosome membrane. In compounds 4 and 5, the ortho ester linker is introduced between one of the oleyl hydrocarbon chains and the rest of the lipid (hereafter mentioned as Type II ortho ester lipids) so that the hydrolysis would generate an oleyl alcohol and a mushroom-shaped lyso lipid (Scheme 1) that could destabilize the endosome membrane. Structurally similar plasminogen lipids carrying an acid-labile vinyl ether linker between one hydrocarbon chain and the rest of the lipid molecule were shown to cause contents release from their liposomes in response to lowered pH [28]. As to the counterions of these cationic lipids, compound 4 carries the iodide anion as in previously reported ortho ester lipids 1 and 2 whereas lipids 3 and 5 carry the chloride anion that is much more hydratable (ΔGsvo = -319 kJ/mol) than the iodide anion (ΔGsvo = -259 kJ/mol) [29]. We anticipate that the chloride counterion would facilitate the hydration of the lipids and hence the construction of the liposomes and lipoplexes.
2.2. Synthesis
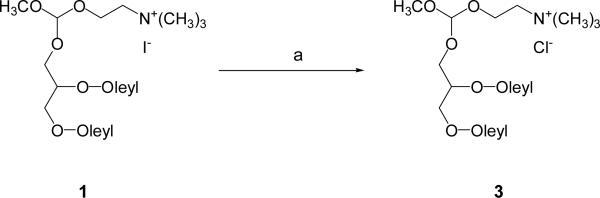
The new Type I linear ortho ester lipid 3 was synthesized (Scheme 2) by exchanging the iodide counterion of the previously reported cationic ortho ester 1 with chloride. A weakly basic resin (Bio-Rex 5, chloride form) was selected for the ion exchange to prevent degradation of the acid-labile ortho ester group during the reaction. The resin was of analytical grade to minimize contamination of the product. Compound 1 was first dissolved with methylene chloride and then loaded onto a column containing the Bio-Rex 5 resin pre-equilibrated with a mixture of CH2Cl2 and MeOH (6/4). The column was then slowly eluted with CH2Cl2/MeOH (6/4) to yield 3 in quantitative yield. We attempted to prepare the chloride analogue of compound 2 using the same method but the product was too unstable for purification or characterization at ambient temperature.
Scheme 2.
Synthesis of Type I ortho ester lipid 3. Reagents and conditions: (a) Bio-Rex 5 resin (chloride form), CH2Cl2/MeOH, rt, 100%.
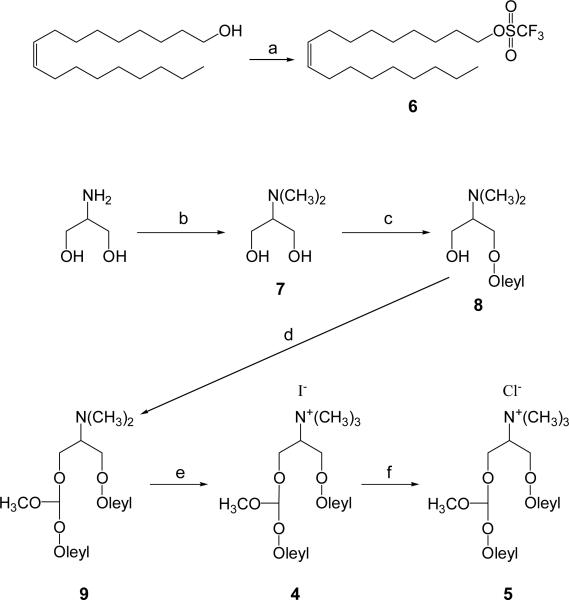
The Type II linear ortho ester lipids 4 and 5 were prepared as shown in Scheme 3. Oleyl alcohol was converted to oleyl triflate (6) as an alkylating agent to build one of the hydrocarbon chains of the lipids. The starting material for the headgroups, 2-amino-1,3-propanediol was methylated following the method of Fernholz and coworkers [30] to give 7. Compound 7 was too hydrophilic to be separated by regular silica gel chromatography but was readily separated by a cationic exchange column (DOWEX® 50WX4-50) in ~60% yield. Compound 7 was deprotonated by two equivalents of the strong base lithium diisopropylamide (LDA), followed by in situ alkylation with one equivalent of oleyl triflate to yield the mono-alkylated amphiphile 8. The bi-functional molecule α,α-dichloromethyl methyl ether [31] was then simultaneously displaced with 8 and oleoyl alcohol to give compound 9 with the ortho ester linkage. Methylation of 9 with iodomethane readily yielded 4 as an iodide salt. A portion of 4 was treated with the ion exchange resin, Bio-Rex 5 (chloride form) to yield the chloride salt 5 in the same manner as the preparation of 3.
Scheme 3.
Synthesis of Type II ortho ester lipids 4 and 5. Reagents and conditions: (a) pyridine/triflic anhydride/CH2Cl2, rt for 1.5 h, 100%; (b) HCOOH/HCHO/H2O, 0 to 65 °C, 10 h, 59%; (c) LDA/THF, -15 to -20 °C for 30 min; 6/THF, -15 to -20 °C for 30 min, rt for 1 h, 54%; (d) oleyl alcohol, NaH, THF, rt for 30 min; α,α-dichloromethyl methyl ether, 45 °C for 10 h, 20%; (e) CH3I, Na2CO3, THF, rt for 8 h, 100%, (f) Bio-Rex 5 resin (chloride form), CH2Cl2/MeOH, rt, 99%.
3. Results
3.1. Construction of Liposomes and Lipoplexes Containing the Ortho Ester-Based Cationic Lipids
Lipidic films containing the ortho ester-based cationic lipids, either by itself or together with a helper lipid (DOPE or cholesterol), were agitated with a pH 7.4 HEPES buffer in order to hydrate the films for liposome formation (Table 1). Whenever a film contained an ortho ester lipid carrying the iodide counterion, the hydration faced significant difficulty. In some cases the film could not be suspended at all whereas in others, the displaced film formed large, wax-like flakes instead of a homogeneous, translucent suspension that is expected for colloidal liposome preparations. Indeed, the ortho ester lipids with the iodide counterion could only be properly hydrated together with equal molar DOPE in the lipidic film and with NaCl in the hydration buffer. The NaCl in the buffer facilitated the hydration presumably by partial replacement of the iodide counterions with the chlorides.
Table 1.
Hydration of lipidic films consisting of ortho ester-based lipids for liposome preparatioin.
| Cationic Lipid | Counterion | Helper Lipid | ||
|---|---|---|---|---|
| None | DOPE | Cholesterol | ||
| 1 | I- | × | √ | × |
| 2 | I- | × | √ | × |
| 3 | Cl- | √ | √ | √ |
| 4 | I- | × | √ | × |
| 5 | Cl- | √ | √ | √ |
| DOTAP | Cl- | √ | √ | √ |
a Lipidic films containing various cationic lipids and helper lipids were agitated with a pH 7.4 buffer (10 mM HEPES, 20 mM NaCl, 4% glucose).
b Mark “√” indicates that the film was hydrated in 15 minutes to form a translucent colloidal suspension.
c Mark “×” indicates that the film could not be suspended or was suspended only into large, wax-like flakes after agitation for more than 1 h in the pH 7.4 buffer.
In contrast, the films containing the lipids with the chloride counterion were hydrated in minutes upon mild agitation regardless of the nature of the helper lipids (DOPE, cholesterol or none). The resultant suspensions where readily extruded through 200 nm polycarbonate membrane to yield liposomes ranging 160 – 200 nm in diameter. The liposomes were then quickly mixed with plasmid DNA (N/P = 5/1, where N/P is the molar ratio of the quaternary ammoniums of cationic lipids and the phosphates of DNA) to form cationic lipoplexes. Lipoplexes consisting of DOTAP, a commercially available analogue of the ortho ester-based cationic lipids, was prepared in the same manner as pH-insensitive controls. All the prepared lipoplexes (Table 2) carried excess positive surface charges, as indicated by their highly positive ζ-potential values. The ζ-potential of the lipoplexes is similar to that of the corresponding liposomes, probably due to the large excess of the cationic lipids (N/P = 5/1) in the lipoplexes. The size of the lipoplexes ranges between 200 nm and 400 nm in diameter. All the lipoplex preparations can be stored at room temperature for several hours with no obvious changes in size or ζ-potential.
Table 2.
Colloidal properties of ortho ester-based liposomes and lipoplexes.
| Liposome Composition | Liposomes | Lipoplexes (N/P = 5/1) | ||||
|---|---|---|---|---|---|---|
| D (nm) | Poly-dispersity index | ζ-Potential (mV) | D (nm) | Poly-dispersity index | ζ-Potential (mV) | |
| 1/DOPE | 188±2.6 | 0.14 | 58.7±1.4 | 248±1.8 | 0.14 | 58.3±0.6 |
| 2/DOPE | 147±1.9 | 0.24 | 61.0±1.1 | 266±2.8 | 0.13 | 52.4±0.4 |
| 3 | 169±2.7 | 0.07 | 55.6±3.5 | 273±3.3 | 0.27 | 59.4±1.6 |
| 3/DOPE | 181±2.8 | 0.12 | 55.6±3.0 | 343±5.0 | 0.26 | 56.8±1.2 |
| 3/Chol | 171±3.0 | 0.25 | 48.7±1.8 | 378±9.3 | 0.29 | 54.4±2.0 |
| 4/DOPE | 187±3.0 | 0.17 | 50.2±2.0 | 285±12 | 0.22 | 54.2±0.9 |
| 5 | 186±2.1 | 0.14 | 57.6±1.0 | 238±1.5 | 0.10 | 62.6±2.2 |
| 5/DOPE | 167±2.7 | 0.07 | 58.8±2.6 | 262±3.3 | 0.20 | 57.0±1.3 |
| 5/Chol | 170±2.7 | 0.07 | 67.4±2.2 | 265±3.0 | 0.02 | 53.9±1.6 |
| DOTAP | 175±2.1 | 0.13 | 59.8±2.0 | 239±2.7 | 0.18 | 63.5±1.9 |
| DOTAP/DOPE | 163±1.8 | 0.17 | 61.9±1.5 | 241±1.5 | 0.16 | 66.3±1.6 |
| DOTAP/Chol | 172±4.3 | 0.08 | 65.2±2.4 | 288±4.1 | 0.21 | 67.7±2.1 |
a Samples were measured in 100 mM HEPES buffer, pH 7.4. The hydrodynamic diameter (D) and the ζ-potential values are reported as mean ± standard deviation of three measurements.
3.2. Hydrolysis of Ortho-Ester-Based Cationic Lipids in Liposomes and Lipoplexes
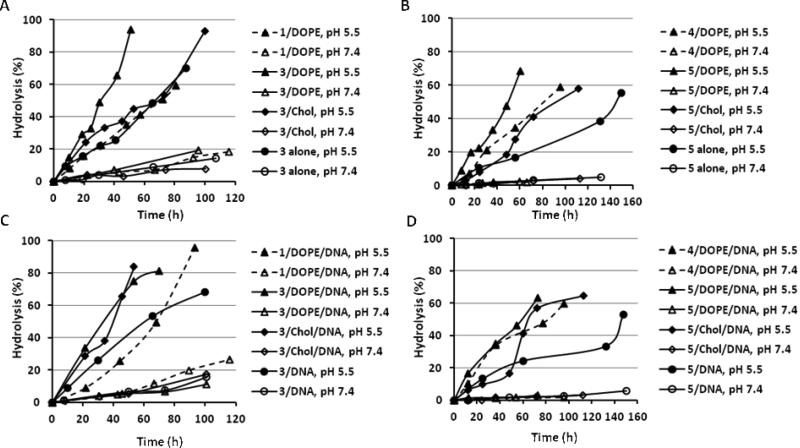
The liposomes and lipoplexes consisting of the ortho ester-based cationic lipids 3, 4, and 5 were incubated at pHs 7.4 and 5.5 and their hydrolysis kinetics were analyzed by HPLC (Fig. 1). The generation of the hydrophobic hydrolysis products (dioleylglycerol for 3; oleyl alcohol for 4 and 5) were monitored by UV to calculate the percentage of hydrolysis because of their much higher resolution in reverse phase chromatography than the corresponding cationic lipids [22]. The hydrolysis kinetics of the previously reported lipid 1, which is the iodide counter part of 3, is also presented for comparison. As expected, the hydrolysis of the ortho ester-based lipids in their liposomes and lipoplexes were much faster at pH 5.5 than at pH 7.4 (Fig. 1, solid symbols versus hollow symbols). For example, approximately 50% of 3 was hydrolyzed after incubation at pH 5.5 for 30 h; at pH 7.4, less than 10% of 3 was hydrolyzed in the same time period.
Fig. 1.
pH-Responsive hydrolysis of ortho ester-based cationic lipids in liposomes and lipoplexes. A) Type I ortho ester lipids 1 and 3 in liposomes, B) Type II ortho ester lipids 4 and 5 in liposomes, C) Type I ortho ester lipids 1 and 3 in lipoplexes, D) Type II ortho ester lipids 4 and 5 in lipoplexes; some of the liposomes and lipoplexes contain 50 mol% DOPE or cholesterol as a helper lipid; N/P = 5/1 in lipoplexes.
All other conditions (counterion, helper lipid, pH) being the same, the ortho ester linker between the headgroup and the lipid chains (as in 1 and 3) was hydrolyzed faster than the ortho ester linker between one lipid chain and the rest of the cationic lipid (as in 4 and 5). For example, it took about 30 h to hydrolyze the first 50% of 3 in the 3/DOPE liposome (Fig. 1A), compared to about 50 h that was needed to hydrolyze the first 50% of 5 in the 5/DOPE liposome (Fig. 1B). For another example, it took about 40 h to hydrolyze the first 50% of 3 in the 3/cholesterol/DNA lipoplex (Fig. 1C), compared to more than 60 h that was needed to hydrolyze the first 50% of 5 in the 5/cholesterol/DNA lipoplex (Fig. 1D).
The chloride counterion enhanced the hydrolysis of the ortho ester-based cationic lipids in their liposomes and lipoplexes compared to the iodide counterion. For example, it took about 30 h to hydrolyze the first 50% of 3 carrying the chloride counterion in the 3/DOPE/DNA lipoplex (Fig. 1C), compared to about 70 h that was needed for the hydrolysis of the first 50% of the iodide counterpart 1 in the 1/DOPE/DNA lipoplex. Another example is the hydrolysis of 5 in the 5/DOPE liposome, which took about 45 h for the first 50% (Fig. 1B), compared to about 80 h that was needed to hydrolyze the iodide counterpart 4 in the 4/DOPE liposome.
The incorporation of the helper lipid DOPE also enhanced the hydrolysis of the ortho ester-based cationic lipids in their liposomes and lipoplexes. For example, the hydrolysis of the first 50% of 3 in the 3/DOPE/DNA lipoplex took about 30 h, compared to about 60 h in the 3/DNA lipoplex (Fig. 1C). Another example is the hydrolysis of 5, which took about 50 h for the first 50% in the 5/DOPE/DNA lipoplex (Fig. 1D), compared to about 150 h in 5/DNA lipoplex.
The effect of the helper lipid cholesterol on the hydrolysis kinetics is more complicated. For lipid 3, which carries an ortho ester linker between the headgroup and the lipidic chains, the incorporation of cholesterol enhanced its hydrolysis in the lipoplex (3/cholesterol/DNA versus 3/DNA in Fig. 1C), but not so in the liposome (3/cholesterol versus 3 only liposome in Fig. 1A). Moreover, both the 5/cholesterol liposome (Fig. 1C) and the 5/cholesterol/DNA lipoplex (Fig. 1D) exhibited a pseudo-sigmoidal hydrolysis curve containing three phases: a relatively slow, linear initial phase from 0 to 50 h, a faster second phase from 50 to 80 h, and a slower third phase after 80 h.
3.3. pH-Triggered Size Changes of Lipolexes Consisting of Ortho Ester-Based Cationic Lipids
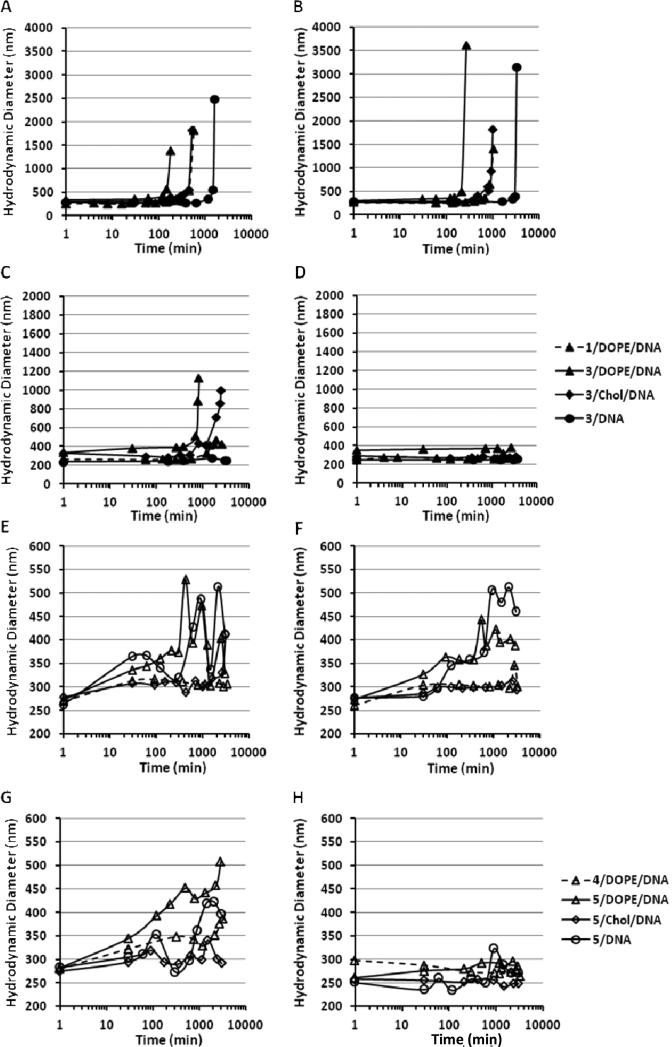
To assess whether the acid-catalyzed hydrolysis of the ortho ester lipids can trigger the changes in the colloidal properties of their lipoplexes, the hydrodynamic diameter of the lipoplexes was monitored after incubation at different pHs (Fig. 2). The lipoplexes under investigation showed little change in size after incubation at pH 7.4 over 48 h, but substantial changes after incubation at pH 6.0 or lower.
Fig. 2.
Acidic pH-triggered aggregation of ortho ester-based lipoplexes (N/P = 5/1). The hydrodynamic diameter of the Type I (A-D) and Type II (E to H) ortho ester-based lipoplexes was measured by dynamic light scattering after incubation at pHs 5.0 (A and E), 5.5 (B and F), 6.0 (C and G) and 7.4 (D and H).
The size change of the lipoplexes consisting of Type I lipids with an ortho ester linker between the headgroup and the lipid chains (1 or 3 in Fig. 2A-D) went through two phases: a lag phase where the size was relatively stable and subsequently an aggregation phase, where the size quickly increased above 1000 nm. In order to compare the length of the lag phase between the lipoplexes, the time needed for the hydrodynamic diameter of a lipoplex formulation to reach 1000 nm is defined as the aggregation time. As shown in Fig. 2A-D, the lag phase as reflected by the aggregation time was shortened when such lipoplexes were exposed to lower pH. For instance, the aggregation time of the 3/DOPE/DNA lipoplex at pHs 6.0, 5.5 and 5.0 was 14 h, 4.5 h and 3 h, respectively. Such observation confirmed the sensitivity of such lipoplexes to acidic pH.
At the same acidic pH, the aggregation time of 3/DOPE/DNA lipoplex was much shorter than that of the 1/DOPE/DNA lipoplex (3 h versus 9.5 h at pH 5.0; 4.5 h versus 17.5 at pH 5.5), indicating that the replacement of the iodide with the chloride counterion accelerated the phase changes of such ortho ester-based lipoplexes in response to acidic pHs. The introduction of the DOPE helper lipid greatly enhanced the phase changes of the lipoplexes as shown by the much shorter aggregation time of 3/DOPE/DNA lipoplex than the 3/DNA lipoplex at the same pH (e.g. 4.5 h versus 56 h at pH 5.5 in Fig. 2B). The introduction of the helper lipid cholesterol also accelerated the pH-sensitive aggregation but the effect was not as strong as DOPE. For example, the aggregation time of the 3/cholesterol/DNA lipoplex at pH 5.0 was 9 h, which was shorter than 27 h for the 3/DNA lipoplex but longer than 3 h for the 3/DOPE/DNA lipoplex at the same pH (Fig. 2A).
After incubation at pH 6.0 or lower, size changes were also observed in lipoplexes consisting of Type II cationic lipids 4 and 5, which carry an ortho ester linker between one lipid chain and the rest of the molecule (Fig. 2E-H). The hydrodynamic diameter of such lipoplexes was little changed during the first 3 to 4 h of incubation at pHs 6.0, 5.5 or 5.0 but increased to 250 – 500 nm in the following 50 h of incubation. However, no precipitation (hydrodynamic diameter larger than 1000 nm) was observed after incubation for 50 h.
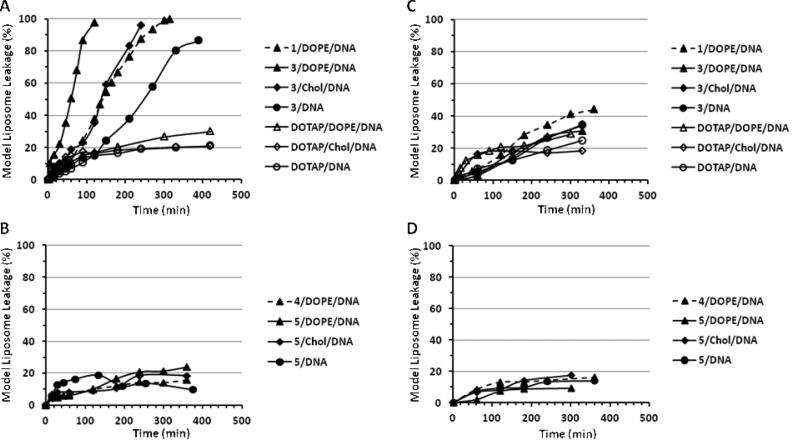
3.4. Destabilization of Model Endosomal Membrane
In order to assess their ability to destabilize endosomal membranes, which is critical for gene delivery, the ortho ester-based lipoplexes were incubated together with model liposomes whose lipid composition mimics that of the endosome membrane [32, 33]. The extent of the model liposome destabilization by the lipoplexes was monitored by the release of the fluorophore ANTS from the model liposomes (Fig. 3). At pH 7.4 (Fig. 3B), the Type I ortho ester-based lipoplexes (consisting of 1 or 3) induced similar level of ANTS leakage from the model liposomes compared to the pH-insensitive lipoplexes consisting of DOTAP. At the endosomal pH 5.5, the Type I ortho ester-based lipoplexes destabilized the model liposomes much more efficiently (> 80% ANTS release in 300 min, Fig. 3A) than the DOTAP-based lipoplexes (< 30% ANTS release in 300 min). The ANTS release induced by 3/DOPE/DNA was faster than both 1/DOPE/DNA and 3/DNA (~60 min, 140 min, and 250 min for 50% ANTS release at pH 5.5, respectively, Fig. 3A), indicating that both the chloride counterion and the DOPE helper lipid enhanced the membrane destabilization. In contrast to the lipoplexes consisting of Type I lipids 1 or 3, the lipoplexes consisting of Type II lipids 4 or 5 that carry an ortho ester linker between one hydrocarbon chain and the rest of the lipid did not destabilize the model membranes more efficiently at pH 5.5 than at pH 7.4 (< 30% ANTS leakage under all conditions, Fig. 3C and 3D). Neither did such lipoplexes destabilize the model membranes more efficiently than the pH-insensitive DOTAP lipoplexes regardless of the pH, the counterion, or the helper lipids.
Fig. 3.
Destabilization of model liposome by Type I (A and B) and Type II (C and D) ortho ester-based lipoplexes at pH 5.5 (A and C) and pH 7.4 (B and D). Lipoplexes consisting of DOTAP serve as pH-insensitive controls.
3.5. Gene Transfection by Lipoplexes Consisting of Ortho Ester-Based Cationic Lipids
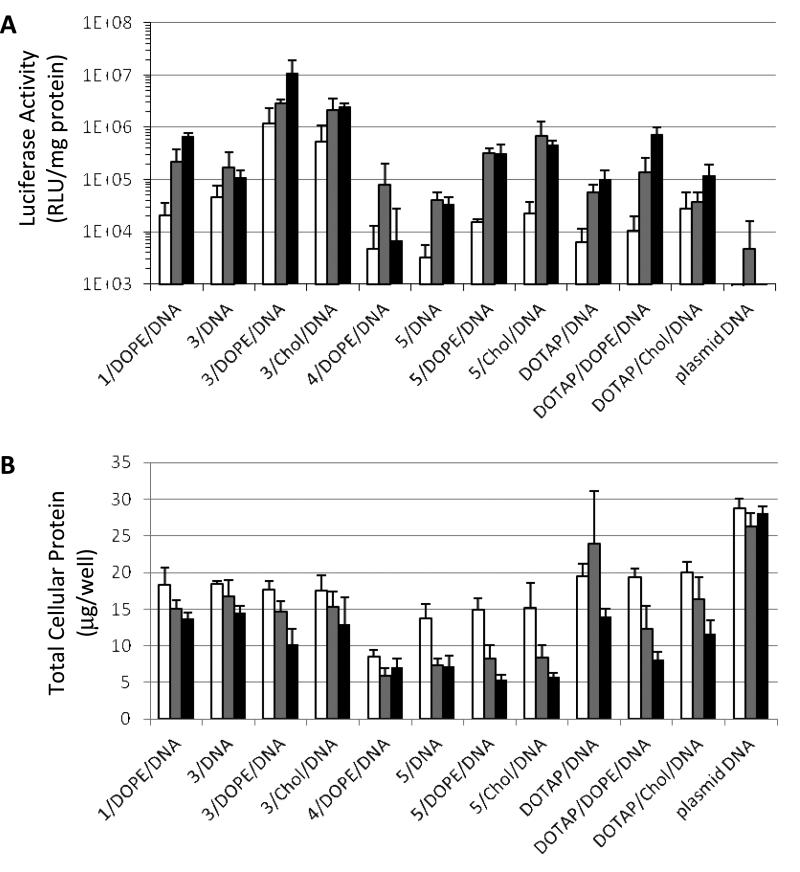
The ability of the ortho ester-based lipoplexes to delivery genes were tested in CV-1 cells (monkey kidney fibroblast), one of the most commonly used cell line for gene transfection studies [34-37]. The commercially available pH-insensitive cationic lipid DOTAP [8] was also tested for comparison. As shown in Fig. 4A, the lipolexes consisting of the Type I lipid 3 carrying a linear ortho ester linker between the headgroup and the lipid tails greatly improved the gene transfection efficiency compared to lipoplexes consisting of the pH-insensitive cationic lipid DOTAP. At 0.3, 1.0 and 3.0 μg/well of plasmid DNA encoding the luciferase reporter gene, 3/DOPE/DNA increased the luciferase expression level by 116-, 21- and 15-fold, respectively, compared to the corresponding pH-insensitive lipoplex DOTAP/DOPE/DNA. At the same three dosage levels, 3/Chol/DNA increased the luciferase gene expression level by 19-, 58- and 21-fold, respectively, compared to the corresponding pH-insensitive lipoplex DOTAP/Chol/DNA.
Fig. 4.
Luciferase gene expression (A) and total protein level (B) of CV-1 cells transfected by ortho-ester based lipoplexes. Lipoplexes consisting of DOTAP are pH-insensitive controls. Empty columns, 0.3 μg DNA/well; grey columns, 1.0 μg DNA/well; solid columns, 3.0 μg DNA/well.
The gene transfection was considerably affected by the counterion of the ortho ester-based cationic lipids (Fig. 4A). The lipoplex 3/DOPE/DNA, in which the cationic lipid 3 contained the chloride counterion, demonstrated superior transfection over the 1/DOPE/DNA lipoplex, in which the cationic lipid 1 contained the iodide counterion. The luciferase gene expression mediated by 3/DOPE/DNA was about 58, 13 and 17 times higher than that by 1/DOPE/DNA at 0.3, 1.0 and 3.0 μg DNA/well dosages, respectively. Among lipoplexes consisting of 3, 3/DOPE/DNA mediated the most efficient reporter gene expression, followed by 3/Chol/DNA, and then by 3/DNA, indicating the enhancing effect of the helper lipids on gene delivery, especially DOPE. It should be noted that the higher gene delivery efficiency of lipoplexes consisting of 3 (3/DOPE/DNA > 3/Chol/DNA > 3/DNA) is well correlated with the faster onset lipoplex aggregation (Fig. 2B) and the more extensive biomembrane destabilization (Fig. 3A) at endosomal pH 5.5. The CV-1 cells treated with the Type I ortho ester lipoplexes exhibited similar or higher total protein levels compared to those treated with the pH-insensitive, DOTAP-based lipoplexes (Fig. 4B), suggesting that the Type I ortho ester lipoplexes impose either comparable or lower toxicities to the CV-1 cells.
Compared to the Type I lipoplexes, the lipoplexes consisting of Type II lipids (4, 5) that carry an ortho ester linker between one of the lipid tails and the rest of the lipid mediated lower reporter gene expression (Fig. 4A). At 0.3, 1.0 and 3.0 μg DNA/well, 4/DOPE/DNA lipoplex mediated only 40%, 60% and 1% of the luciferase gene expression mediated by the pH-insensitive DOTAP/DOPE/DNA lipoplex at the corresponding dosages. Lipoplexes 5/DOPE/DNA and 5/Chol/DNA mediated higher reporter gene expression than the corresponding DOTAP lipoplexes, but only at selected dosages. Among the Type II lipoplexes, the 5/Chol/DNA lipoplex afforded the highest improvement (18 fold) over the corresponding pH-insensitive lipoplex (DOTAP/Chol/DNA) at 1.0 μg DNA/well.
The chloride counterion improved the gene transfection by Type II lipoplexes at higher dosages. For example, at 3.0 μg DNA/well, the 5/DOPE/DNA lipoplex, in which the cationic lipid 5 contained the chloride counterion, mediated more than 40-fold of reporter gene expression than 4/DOPE/DNA, in which the cationic lipid 4 contained the iodide counterion. The introduction of helper lipids also improved the gene transfection of lipoplexes consisting of 5. At 0.3 μg, 1.0 μg and 3.0 μg DNA/well dosages, the 5/DOPE/DNA lipoplex improved the gene transfection by 5-, 8- and 9-fold, respectively, compared to the lipoplex 5/DNA. At the same dosages of the plasmid DNA, 5/Chol/DNA increased the gene transfection by 7-, 17- and 14-fold, respectively, compared to the lipoplex 5/DNA.
The decrease of total cellular protein indicated higher cytotoxicity of the Type II ortho ester-based lipoplexes than the corresponding pH-insensitive DOTAP lipoplexes (Fig. 4B). For example, the total cellular protein after gene transfection by the 5/DOPE/DNA lipoplex at 0.3, 1.0 and 3.0 μg/well DNA dosages was 15.0, 8.3, and 5.4 μg/well, compared to 19.3, 12.4, and 8.2 μg/well after gene transfection by the DOTAP/DOPE/DNA lipoplex, respectively.
4. Discussion
Compared to our prior report on ortho ester-based lipoplexes [22], the current study has a distinctive focus on cationic lipids that carry a linear ortho ester linker. The goal here is to develop ortho ester-based cationic lipids that can be readily formulated into lipoplexes of different lipid compositions in an effort to optimize their physicochemical properties and pH-sensitivity for more efficient gene transfection. The linear ortho ester linkers provide more conformational flexibility to the cationic lipids than the reported cyclic ortho ester linkers [38, 39], and hence would facilitate the assembly of the corresponding liposomes and lipoplexes. In addition, the chloride ion greatly facilitated the hydration of the cationic lipids but in the mean time accelerated the ortho ester hydrolysis both at acidic pH and at pH 7.4, which decreased the stability of the ortho ester lipids. We found that the cationic lipids 3 and 5 (Table 1) with a linear ortho ester linker and a chloride counterion offered a balance between stability and ease of hydration for the preparation of lipoplexes with different helper lipids. The two lipids can be stored as dried solid at -20 °C for up to five months without significant degradation. In contrast, the chloride analogue of lipid 2 carrying a cyclic ortho ester linker was too unstable for purification, let alone lipoplex formulation. Lipid 3 with a Type I linear ortho ester and a chloride counterion also introduced adequate pH-sensitivity to lipoplexes in cooperation with conical helper lipids. Within the time span of endosome maturation (30 min to 60 min [10, 40]), the 3/DOPE/DNA lipoplex was able to release about 50% ANTS from the model liposome, suggesting that, after endocytosis, such optimized Type I lipoplex would significantly facilitate the escape of DNA from the endosomes before their degradation in the lysosomal compartment. Lipoplexes consisting of the Type I lipid 3 eventually enhanced the gene transfection by up to two magnitudes. The ability of 3 to significantly improve transfection by lipoplexes consisting of cholesterol (3/cholesterol/DNA versus DOTAP/cholesterol/DNA in Fig. 4A) also suggests that this lipid has potential to improve gene delivery in vivo. Taken together, our study shows the importance of balancing stability, ease of formulation, and pH-sensitivity in the development of acid-labile lipids for efficient gene delivery.
The major parameter that determines the pH-sensitivity of the ortho ester-based lipoplexes is the rate of hydrolysis of the ortho ester linker of the cationic lipids. Our previous report demonstrated that such rate is affected by the specific configuration of the ortho ester linker in that the cyclic ortho ester linker in 2 was hydrolyzed more quickly than the linear ortho ester linker in 1. Based on studies by Deslongchamps and associates [38, 39], this is most probably due to the stereoelectronic effects from the conformational alignment of the lone pairs of electrons on the oxygens of the ortho ester linkers. The relief of the steric strain of the cyclic ortho ester linker in 2 may also contribute to its faster hydrolysis than the linear linker in 1. In this study, we further demonstrate that the hydrolysis rate of an ortho ester linker in the lipoplex colloidal environment is affected by the position of the ortho ester linker (Type I or Type II lipids), the counterion of the cationic lipid (chloride or iodide), and the helper lipid of the lipoplex (DOPE, cholesterol or none). Based on our observations as well as previous reports on poly ortho ester hydrolysis [41, 42] and those on local pH within lipoplexes [43, 44], we propose that the lipoplex colloidal environment affects the hydrolysis of the ortho ester linker mainly by two physicochemical mechanisms: the hydration effect and the electrostatic field effect. All other conditions being equal, the ortho ester linker of the Type I lipids is closer to the polar headgroup and therefore is more hydrated in their lipoplexes and more accessible to the protons from the aqueous media for faster hydrolysis than the Type II lipids. Similarly, the chloride counterion enhances the hydration of the cationic lipids compared to the iodide counterion, leading to better exposure of the ortho ester linker to protons from the bulk aqueous media and hence the faster hydrolysis of the ortho ester linker. As to the electrostatic field effect, Barenholz and coworkers demonstrated that the local pH at DNA can be 0.7 unit higher after its complexation with cationic liposomes because the protons are repulsed by the excessive positive charges of the cationic lipids [43]. Conversely, incorporation of helper lipids decrease the density of the positive charges in the lipoplexes, thus alleviating the repulsive field effect to yield higher proton concentration in the lipoplexes and consequently faster ortho ester hydrolysis. The conically shaped helper lipids (DOPE or cholesterol) also impose a more negative curvature of the lipid lamellae [15] to further enhance the Lα→HII phase change. Indeed, for lipoplexes containing 50 mol% DOPE or cholesterol, only a small percentage of ortho ester hydrolysis was needed to undergo phase changes (Fig. 1 and 2).
Since the discovery of lipoplexes as a gene delivery system, several mechanisms have been proposed on how the lipoplexes could help transfer the cargo DNA from the endosome to the cytoplasm to enhance the gene transfection. Based on the ability of negatively charged model liposomes to quickly release DNA out of the lipoplexes, Xu and Szoka proposed the “flip-flop” mechanism [45], wherein the negatively charged lipids from the endosome membrane flip to partition into the endocytosed lipoplexes. The negatively charged lipids then pair with the cationic lipids in the lipoplex to displace the DNA out of the lipoplex followed by release of the DNA from the endosome. This mechanism accounts for a variety of observations on cationic lipid/DNA complex-cell interactions but is quite inefficient because the amount of the negatively charged lipids in the endosome membrane is insufficient to displace most of the DNA from the lipoplex [46, 47]. X-ray crystallography studies by Koltover et al [15] later demonstrated that lipoplexes consisting of high mole percentage of conically shaped helps lipids assume an inverted hexagonal phase that destabilizes the biomembrane much better than a lamellar phase, thereby enhancing the endosomal escape. We propose that lipoplexes consisting of the Type I ortho ester lipids enhance the escape of DNA from the endosome by two means. First, the hydrolysis of the Type I ortho ester lipids reduces the excessive positive charges from the lipoplexes, thereby enhancing the release of DNA from the lipoplex, and eventually from the endosome. Second and more importantly, the conically shaped hydrolysis product, dioleylglycerol induces the lipoplex to change from the lamellar phase to the inverted hexagonal phase, which in turn destabilized the endosome membrane to release the DNA into the cytosol. Such proposed mechanisms are supported by the HPLC validation of the hydrolysis products, the dynamic light scattering and P31-NMR measurements [22] of the physicochemical changes of the ortho ester lipoplexes in response to lowered pH, and the spectrofluorometric studies on lipoplex-model liposome interactions.
In contrast, the Type II lipids, whose hydrolysis products are mushroom-shaped amphiphiles with one hydrocarbon chain, did not substantially improve the gene transfection in most of their lipoplex formulations. The model liposome leakage studies suggest that lipoplexes carrying the Type II lipids do not destabilize endosome membranes efficiently. One possible reason for this is that the helper lipids DOPE and cholesterol are conically shaped and therefore would antagonize the surface activity of the mushroom-shaped, detergent like hydrolysis products of the Type II lipids. The detergent-like hydrolysis products may also diffuse out of the endosome to bind and disrupt other cellular membranes to inflict cytotoxicity [48, 49], which would further diminish the transgene expression (Fig. 4B).
Although numerous cationic lipids have been developed to facilitate gene transfection, the relationship between their chemical structure and transfection activity is often elusive. For example, our current study shows that a labile ortho ester linker between the headgroup and the lipid tail (Type I) yields higher transfection than an ortho ester linker between one hydrocarbon chain and the rest of the molecule, whereas Byk and coworkers’ studies on a number of cationic lipids carrying a disulfide linker [50] showed that implementation of the disulfide bond between one carbon chain and the rest of the molecule yielded higher transfection than that between the headgroup and the lipid tail. Furthermore, although our studies show that the chloride counterion facilitates the gene transfection of the Type I ortho ester lipid 3 compared to the iondide counterion of lipid 1, Aberle and coworkers [51] showed that the iodide counterion in DOTAP enhanced gene transfection compared to the chloride counterion. We attribute such apparent discrepancy to the fact that different types of lipoplexes often have differences in the mechanisms of enhancing the transfection so that the same chemical group in different cationic lipids may play different roles during the gene delivery. Based on our physicochemical and cell culture characterizations of ortho ester-based cationic lipids and lipoplexes, we propose that structure-activity relationship studies of cationic lipids need to be carried out in the context of how the structure features of the lipids affect the physicochemical properties of the corresponding lipoplexes and how such physicochemical properties help overcome the biological barriers of gene delivery such as endosome processing coupled with degradation in lysosome.
5. Conclusions
Three novel cationic lipids containing a linear ortho ester linker that conjugates either the headgroup (Type I) or one hydrocarbon chain (Type II) with the rest of the lipid molecule have been developed to enhance endosome escape of lipoplexes during gene delivery. The Type I lipoplexes, especially those with the chloride counterion, significantly improved the gene transfection in CV-1 cells, in some cases by more than 100 fold, compared to the pH-insensitive counterparts consisting of DOTAP. The transfection efficiency of the ortho ester-based lipoplexes was well correlated with their rate of aggregation and membrane destabilization in response to the endosomal pH 5.5. We are exploring the application of lipid 3 in gene delivery in vivo and will report the findings in due course.
6. Experimental protocols
6.1. General Methods and Materials
All chemicals from commercial suppliers were used without further purification unless indicated otherwise. The following lipids, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (sodium salt) (POPS), L-α-phosphatidylinositol (sodium salt) (L-α-PI, soy), and 1,2-dioleoyl-3-trimethylammonium-propane (chloride salt) (DOTAP) were purchased from Avanti Polar Lipids (Alabaster, AL). The fluorophores 8-aminonaphthalene-1,3,6-trisulfonic acid, disodium salt (ANTS) and p-xylene-bis-pyridinium bromide (DPX) were purchased from Invitrogen (Carlsbad, CA, USA). The ortho ester lipids 1 and 2, and 1,2-dioleylglycerol were prepared as previously described [22]. Other reagents and solvents were purchased from Sigma-Aldrich or Fisher. Solvent compositions of chromatography mobile phases are in volume ratios unless indicated otherwise. 1H-NMR (300 MHz), 31P-NMR (75 MHz) and 13C-NMR (75 MHz) were recorded on a Varian Mercury 300 MHz NMR spectrometer and chemical shifts were calibrated against residual solvent signals of CDCl3 (δ 7.26 for 1H-NMR, 77.16 for 13C-NMR) or TMS (δ 0.00 for 1H-NMR) and reported in ppm. ESIMS studies were performed on a Varian 1200L triple-quadruple mass spectrometer, which recorded averaged m/z values. Elemental analyses were performed at the UC Berkeley Micro-Mass Facilities. The pH measurements were made with a Fisher Scientific Accumet® model 15 pH meter. Colloidal size and ζ-potential were measured with a Malvern Zetasizer 3000HSA Dynamic Light Scattering Instrument. Fluorescence spectra were obtained using a QuantaMaster fluorescence spectrofluorometer (Photon Technology International, Lawrenceville, NJ). All the ortho ester-based lipids under investigation were at least 95% in purity as confirmed by elemental analyses and thin layer chromatography.
6.2. 2-[Methoxy-(1,2-dioleylglyceroxy)methoxy]-N,N,N-trimethyl-ethanammonium Chloride (3)
Bio-Rex 5 resin (500 mg, 200-400 mesh, chloride form) [34] was equilibrated in 5 mL CH2Cl2/MeOH (6/4) overnight and then packed in a borosilicate glass Pasteur pipette to form a small plug. A solution of 2-[methoxy-(1, 2-dioleylglyceroxy)methoxy]-N, N, N-trimethyl-ethanammonium; iodide (1) (117.4 mg, 0.15 mmol) in CH2Cl2/MeOH (6/4) was loaded onto the resin plug and slowly eluted with CH2Cl2/MeOH (6/4). Fractions corresponding to the product were pooled and roto-evaporated to yield a clear wax (105 mg, yield 100%). 1H NMR (300 MHz, CDCl3) δ 0.87 (t, 6.5Hz, 6H, (CH2)7CH3), 1.26 (m, 44H, (CH2)6CH3 and OCH2CH2(CH2)5), 1.53 (m, 4H, OCH2CH2(CH2)5), 2.0 (m, 8H, CH2CH=CHCH2), 3.35 (s, 3H, OCH3), 3.38-3.70 (m, 11H, NCH2 and dioleylglyceroxy OCH2/OCH), 3.50 (s, 9H, N(CH3)3), 4.05 (m, 2H, OCH2CH2N), 5.17 (s, 1H, ortho ester O3CH), 5.35 (m, 4H, CH=CH); 13C NMR (75 MHz, CDCl3) δ 14.2, 22.8, 26.3, 26.3, 27.4, 29.5, 29.6, 29.7, 29.8, 29.9, 30.3, 32.0, 32.7, 52.8, 54.8, 57.9, 58.0, 65.3, 65.9, 70.2, 70.8, 72.0, 113.7, 130.8, 130.1. ESIMS: calcd for [C46H92NO5]+, 739.2; m/z found 739.1. Anal. C46H92NO5Cl: calcd C 71.31, H 11.97, N 1.81; found C 71.69, H 12.28, N 1.91.
6.3. 1-O-Trifluoromethanesulfonyl-Z-9-Octadecene (6)
To a stirred solution of pyridine (2.08 mL, 26.2 mmol) in 5.0 mL CH2Cl2 under Argon at room temperature was slowly added triflic anhydride (4.4 mL, 26.1 mmol) in 16 mL CH2Cl2 to form a thick white suspension. Oleyl alcohol (5.9 mL, 18.6 mmol) in 25 mL CH2Cl2 was then injected to the suspension. The reaction mixture was stirred for 1.5 hour at room temperature and washed with 50 mL water. The water layer was extracted with CH2Cl2 (3 × 40 mL) and the extracts were combined with the reaction mixture. The combined organic solution was dried over MgSO4/Na2CO3 (1:1, w/w) and filtered. The filtrate was roto-evaporated to yield thick brown oil (8.0 g, yield 100%). 1H NMR (CDCl3) δ 0.80 (t, 6.6Hz, 3H, (CH2)7CH3), 1.31 (m, 22H, (CH2)6CH3 and OCH2CH2(CH2)5), 1.82 (m, 2H, OCH2CH2(CH2)5), 2.0 (m, 4H, CH2CH=CHCH2), 4.53 (t, 2H, OCH2CH2(CH2)5), 5.36 (m, 2H, CH=CH).
6.4. 2-Dimethylamino-1,3-Propanediol (7) [30]
To an ice cold solution of 2-amino-1,3-propanediol (4.57 g, 50 mmol) in 11 mL formic acid (88% in water, 250 mmol) was added 9.0 mL formaldehyde (37% in water, 120 mmol). The mixture was heated at 80 °C for 10 h. The reaction mixture was cooled to room temperature and loaded onto an acidic cationic exchange column (DOWEX® 50WX4-50) which had been sequentially pre-washed with 5 w% hydrochloric acid in water and then with a mixture of methanol and 35 w% hydrochloric acid in water (6/1). The column was then washed with methanol and eluted with a mixture of methanol and 30 w% NH·3H2O (9/1). Fractions containing the product were pooled and the solvent was evaporated. The residue was dissolved in CH2Cl2, dried over Na2SO4/Na2CO3 (1:1, w/w) and filtered. The filtrate was roto-evaporated to yield a yellowish solid (3.52 g, 59% yield). TLC Rf 0.2 in CH2Cl2/MeOH/NEt3 (10/0.5/0.1). 1H NMR (CDCl3) δ 2.37 (s, 6H, N(CH3)2), 2.60 (m, 1H, CHN(CH3)2), 3.18 (s, 2H, 2OH), 3.66 (d, 6 Hz, 4H, CH(CH2OH)2). ESIMS: calcd for C5H14NO2 [M+H]+, 120.2; m/z found 120.1.
6.5. 2-Dimethylamino-3-oleyloxy-1-propanol (8)
To a solution of 7 (4.37 g, 36.7 mmol) in 100 mL THF at -15 °C to -20 °C was added dropwise a solution of 2 M lithium diisopropylamide in THF (38.2 mmol LDA in 19.1 mL THF). The reaction mixture was stirred at -15 °C to -20 °C for 30 min, followed by the dropwise addition of a THF solution of 6 (1.48 g, 3.69 mmol in 3.5 mL THF). The mixture was stirred at -15 °C to -20 °C for 30 min and then at room temperature for 1 hour. Water (5 mL) was added to stop the reaction. The reaction mixture was roto-evaporated and the red residue was suspended in 100 mL CH2Cl2 and filtered. The filtrate was washed with water (3 × 100 mL), dried over MgSO4, and roto-evaporated. The resultant residue was loaded onto a silica gel column and eluted with CH2Cl2/MeOH/NEt3. Fractions corresponding to the product were pooled and roto-evaporated. The reddish residue was loaded onto a small Amberlite® XAD-7 plug and eluted with hexane/ether (80:20). Fractions containing the product were pooled, and roto-evaporated to give the pure product (734 mg, yield 54%) as yellowish viscous oil. TLC Rf 0.3 in CH2Cl2/MeOH/NH3·H2O (10/1.5/0.2). 1H NMR (CDCl3) δ 0.85 (t, 3H, (CH2)7CH3), 1.28 (m, 22H, (CH2)6CH3 and OCH2CH2(CH2)5), 1.62 (m, 2H, OCH2CH2(CH2)5), 2.0 (m, 4H, CH2CH=CHCH2), 2.45 (s, 6H, N(CH3)2), 2.91 (m, 1H, CHN(CH3)2), 3.23 (bs, 1H, OH), 3.35-3.68 (m, 6H, OCH2), 5.34 (m, 2H, CH=CH).
6.6. 1-Oleyloxy-3-oleyloxy(methoxy)methoxy-N,N-dimethy-2-propanamine (9) [31]
To a solution of 8 (213 mg, 0.58 mmol) and oleyl alcohol (1.09 g, 4.1 mmol) in dry THF (20 mL) under N2 was added sodium hydride (60% dispersion in mineral oil, 204 mg, 5.0 mmol). The mixture was stirred at room temperature for 30 min. α,α-Dichloromethyl methyl ether [31] (0.21 mL, 2.3 mmol) was then injected in one bolus and the mixture was stirred at 45 °C for 10 hours. Triethylamine (0.5 mL) was added to stop the reaction and to stabilize the product. The reaction mixture was roto-evaporated to dryness. The residue was suspended in ethyl acetate (50 mL) and filtered. The filtrate was washed with water (3 × 50 mL), dried over sodium carbonate/magnesium sulfate (1:1, w/w), and roto-evaporated. The resultant residue was loaded onto a silica gel column and eluted with CH2Cl2/Acetone/NEt3 (96:4:0.5). Fractions corresponding to the product were pooled and roto-evaporated to yield clear viscous oil (78.3 mg, yield 20%). TLC Rf 0.4 in CH2Cl2/MeOH/NEt3 (10: 0.4: 0.05) 1H NMR (CDCl3) δ 0.87 (t, 6H, (CH2)7CH3), 1.25 (m, 44H, (CH2)6CH3 and OCH2CH2(CH2)5), 1.57 (m, 4H, OCH2CH2(CH2)5), 2.0 (m, 8H, CH2CH=CHCH2), 2.38 (s, 6H, N(CH3)2), 2.49 (m, 1H, CHN(CH3)2, 3.34 (s, 3H, OCH3), 3.36-3.70 (m, 8H, OCH2), 5.10 (s, 1H, ortho ester O3CH), 5.35 (m, 4H, CH=CH); ESIMS: calcd for C43H85NO4 [M+H]+, 681.1; m/z found 680.7.
6.7. 1-Oleyloxy-3-oleyloxy(methoxy)methoxy-N,N,N-trimethy-2-propanammonium Iodide (4)
To a suspension of 9 (144 mg, 0.21 mmol) in dry THF (3 mL) under N2 was added sodium carbonate (134.6 mg, 1.27 mmol) and iodomethane (0.066 mL, 1.06 mmol). The suspension was stirred under N2 at room temperature for 8 hours. The reaction mixture was filtered and the precipitate washed with CH2Cl2. The filtrate was pooled and roto-evaporated. The residue was dried in high vacuo to give a yellow solid (182 mg, yield 100%). TLC Rf 0.8 in CH2Cl2/MeOH/NH3·H2O (8/2/0.1). 1H NMR (CDCl3) δ 0.87 (t, 6.8Hz, 6H, (CH2)7CH3), 1.26 (m, 44H, (CH2)6CH3 and OCH2CH2(CH2)5), 1.56 (m, 4H, OCH2CH2(CH2)5), 2.0 (m, 8H, CH2CH=CHCH2), 3.35 (s, 3H, OCH3), 3.40-3.60 (m, 4H, oleyloxy O CH2CH2), 3.54 (s, 9H, N(CH3)3), 3.85 (m, 1H, NCH), 3.96-4.41 (m, 4H, NCHCH2), 5.12 (s, 1H, ortho ester O3CH), 5.33 (m, 4H, CH=CH); 13C NMR (75 MHz, CDCl3) δ 14.2, 22.8, 26.2, 26.3, 27.3, 29.4, 29.5, 29.6, 29.7, 29.8, 29.9, 32.0, 32.7, 45.9, 52.7, 54.5, 60.4, 66.1, 67.8, 71.3, 72.4, 113.5, 113.6, 129.9, 130.1. ESIMS: calcd for [C44H88NO4]+, 695.1; m/z found 694.8. Anal. C44H88NO4I: calcd C 64.28, H 10.79, N 1.70; found C 64.42, H 10.99, N 1.73.
6.8. 1-Oleyloxy-3-oleyloxy(methoxy)methoxy-N,N,N-trimethy-2-propanammonium Chloride (5)
Bio- Rex 5 resin (500 mg, 200-400 mesh, chloride form) was pre-equilibrated in 5 mL CH2Cl2/MeOH (6/4) overnight and then packed in a borosilicate glass Pasteur pipette to form a small plug. The plug was loaded with a solution of 4 (85 mg, 0.10 mmol) in CH2Cl2/MeOH (6/4) and then slowly eluted by dropwise addition of CH2Cl2/MeOH (6:4) on the top. Fractions corresponding to the product were pooled and roto-evaporated to yield a clear wax (75 mg, yield 99%). TLC Rf 0.7 in CH2Cl2/MeOH/NH3·H2O (8/ 2/0.1). 1H NMR (CDCl3) δ 0.87 (t, 6.5Hz, 6H, (CH2)7CH3), 1.26 (m, 44H, (CH2)6CH3 and OCH2CH2(CH2)5), 1.54 (m, 4H, OCH2CH2(CH2)5), 2.0 (m, 8H, CH2CH=CHCH2), 3.30 (s, 3H, OCH3), 3.42-3.60 (m, 4H, oleyloxy OCH2CH2), 3.56 (s, 9H, N(CH3)3), 3.85 (m, 1H, NCH), 3.94-4.40 (m, 4H, NCHCH2), 5.11 (s, 1H, ortho ester O3CH), 5.33 (m, 4H, CH=CH); 13C NMR (75 MHz, CDCl3) δ 14.2, 22.8, 25.9, 26.0, 26.3, 27.4, 28.7, 29.3, 29.4, 29.5, 29.6, 29.7, 29.8, 29.9, 32.0, 32.7, 54.5, 59.3, 63.2, 64.2, 67.9, 72.3, 73.8, 113.5, 129.9, 130.1. ESIMS: calcd for [C44H88NO4]+, 695.1; m/z found 694.8. Anal. C44H88NO4Cl: calcd C 72.33, H 12.14, N 1.92; found C 72.38, H 12.41, N 1.96.
6.9. Preparation of Cationic Liposomes and Lipoplexes
A CH2Cl2 solution of a cationic lipid alone or a cationic lipid together with a helper lipid (DOPE or cholesterol at 1/1 molar ratio) in a Pyrex glass test tube was roto-evaporated to form a lipidic film at the bottom of the test tube. The residual solvent of the lipidic film was removed under high vacuum at room temperature for 2 hours. The lipidic film was then hydrated by a pH 7.4 buffer (10 mM HEPES, 20 mM NaCl, 4% glucose) into a 7.5 mM (total lipid) suspension followed by 5 min agitation by a bench top vortex machine. The hydrated lipids were extruded 11 times through a 200 nm membrane to yield a cationic liposome preparation. The liposome preparation was left at room temperature for 30 min, after which an equal volume of a plasmid DNA solution (0.25 mg DNA/mL, pH 7.4, 10 mM HEPES, 20 mM NaCl) was added to the liposome preparation followed by brief agitation to form a lipoplex suspension at N/P = 5/1, where N/P is defined as the molar ratio of the quaternary ammonium of cationic lipids and the phosphates of DNA.
6.10. Hydrolysis of Ortho Ester-based Cationic Lipids in Liposomes and in Lipoplexes
To characterize the hydrolysis of a cationic ortho ester lipid in its liposomes at pH 7.4, a liposome suspension freshly prepared at pH 7.4 was incubated at 37°C. Small aliquots (100 μL) of the liposome suspension were extracted at different time points and diluted with 900 μL of 90% MeOH in water. An aliquot (100 μL) of each diluted sample was immediately analyzed by HPLC.
To characterize the hydrolysis of a cationic ortho ester lipid in its liposomes at pH 5.5, one volume of 100 mM NaOAc/HOAc buffer (initial pH 5.07, with 50 mM NaCl) was mixed with three volumes of a liposome suspension freshly prepared at pH 7.4. The final pH 5.5 of the mixture was confirmed with a pH meter. The mixture was incubated at 37 °C and small aliquots (100 μL) of the liposome suspension were extracted at different time points and diluted sequentially with 100 μL HEPES buffer (100 mM, pH 7.4) and 800 μL MeOH. An aliquot (100 μL) of each diluted sample was immediately analyzed by HPLC.
To characterize the hydrolysis of a cationic ortho ester lipid in its cationic lipoplexes at pH 7.4, a lipoplex suspension freshly prepared at 7.4 was split into 100 μL aliquots, all of which incubated at 37°C. At different time points, one of the aliquots was sequentially mixed with 250 μL methanol, 125 μL methylene chloride, another 125 μL methylene chloride, and 125 μL of 0.9% NaCl. The final mixture was centrifuged at 9300 μg RCF for 30 seconds to give a two-layer solution. The organic layer at the bottom was extracted and evaporated with a gentle nitrogen flow. The residue was dissolved in 500 μL of 100 mM HEPES in 80% MeOH (pH 7.4). An aliquot (100 μL) of the solution was immediately analyzed by HPLC.
To characterize the hydrolysis of a cationic ortho ester lipid in its lipoplexes at pH 5.5, one volume of 100 mM NaOAc/HOAc buffer (50 mM NaCl, initial pH 5.07) was mixed with four volumes of a lipoplex suspension freshly prepared at pH 7.4. The final pH 5.5 of the mixture was confirmed with a pH meter. The mixture was immediately split into 100 μL aliquots, all of which incubated at 37°C. At different time points, one of the aliquots was sequentially mixed with 250 μL methanol, 125 μL methylene chloride, another 125 μL methylene chloride, and 125 μL 0.9% NaCl. The final mixture was centrifuged at 9300 μg RCF for 30 seconds to give a two-layer solution. The organic layer at the bottom was extracted and evaporated with a gentle nitrogen flow. The residue was dissolved in 500 μL 100 mM HEPES in 80% MeOH (pH 7.4). An aliquot (100 μL) of the solution was immediately analyzed by HPLC.
The lipids were separated at room temperature by a reversed-phase C4 Dionex 214TP™ column (4.6 × 150 mm, flow rate = 0.5 mL/min) on a Beckman Coulter 125 HPLC system. The solvent gradient started from 80% MeOH in water to 100% MeOH in 10 min, followed by 100% MeOH for 30 min. The column was then re-equilibrated at 80% MeOH before the analysis of another sample. The lipids were detected by UV absorbance at 210 nm using a deuterium lamp-based System Gold 166 Detector. Under such chromatographic conditions, the ortho ester-based cationic lipids gave only broad streaking peaks, which were too weak for quantifications. Instead, the resolved peaks for the major hydrolysis products [dioleylglycerol (retention time = 15.4 min) for 1 and 3; oleoyl alcohol for 4 and 5 (retention time = 9.0 min)] were monitored for quantifications. Dioleylglycerol and oleyl alcohol were estimated by HPLC to be more than 85% of the hydrolysis products of their corresponding ortho ester lipids. The helper lipids [DOPE (retention time = 13.6 min) or cholesterol (retention time = 12.4 min)] were monitored as internal standards. The helper lipids were confirmed by HPLC to be stable through the aforementioned procedures of hydrolyzing ortho ester lipids.
The extent of hydrolysis is expressed as percentage of hydrolysis (H%), which is defined and calculated by H% = (AHPt/AISt)/(AHP100/AIS100) × 100%, where AHPt is the peak area of the major hydrolysis product (dioleylglycerol or oleyl alcohol) in a sample that has been incubated for a certain period of time (t), AISt is the peak area of the internal standard (DOPE or cholesterol) in the same sample as that of AHPt, AHP100 is the peak area of the major hydrolysis product in the corresponding control liposome preparation of which the ortho ester hydrolysis has been driven to completion (see below for details), AIS100 is the peak area of the internal standard in the same control liposome preparation as that of AHP100.
To prepare a control liposome in which a cationic ortho ester lipid was completely hydrolyzed, 100 μL of a liposome preparation was mixed with 100 μL glacial acetic acid followed by 2 hours of incubation at 37°C. The completion of the ortho ester hydrolysis was confirmed by TLC. The control liposome preparation was then diluted with 800 μL MeOH and an aliquot (100 μL) of the diluted sample was analyzed by HPLC. The data of a control liposome preparation were used to normalize liposome and lipoplex samples of the same original lipid composition.
6.11. Lipoplex Aggregation Assay
Three volumes of a freshly prepared lipoplex suspension in a pH 7.4 buffer (10 mM HEPES, 20 mM NaCl, 4% glucose) was mixed with one volume of an acidic buffer (100 mM NaOAc/HOAc, 50 mM NaCl) of pH 4.69, 5.07 or 5.30 to obtain a mixture at a final pH of 5.0, 5.5 or 6.0, respectively. The final pH of each mixture was confirmed by a pH meter. To characterize the stability of lipoplex at pH 7.4, one volume of a freshly prepared lipoplex suspension was mixed with three volumes of a 100 mM HEPES buffer (pH 7.4, 50 mM NaCl). The mixture was incubated at 37 °C and a small aliquot (45 μL) was extracted at different time points and diluted into 2.5 mL 100 mM HEPES buffer (pH 7.4) followed by an immediate measurement of the particle size on a Malvern light scattering instrument (Zetasizer 3000 HSA) following the manufacturer's recommendations.
6.12. Model Liposome Preparation
The model liposomes mimicking the lipid composition of biomembranes were prepared using the freeze-thaw method [32]. A chloroform solution of POPC (22.5 μmol, 50 mol%), POPE (9 μmol, 20 mol%), POPS (2.25 μmol, 5 mol%), L-α-PI (4.5 μmol, 10 mol%), and cholesterol (6.75 μmol, 15 mol%) was evaporated under reduced pressure to form a lipid film. The residual chloroform in the lipidic film was removed in high vacuo overnight at room temperature. The lipid film was hydrated with a 1.5 ml solution of 50 mM ANTS, 50 mM DPX and 10 mM HEPES at pH 7.4 by 20 min of intermittent agitation to obtain a suspension at 30 mM total lipid concentration. The tube containing the lipid suspension was then filled with N2 and sealed. The lipid suspension in the tube was rapidly frozen in dry ice/acetone for 5 min, followed by melting in water at room temperature for 5 min. The freeze-thawing was repeated for 10 times. The resultant liposome suspension was extruded 11 times through a polycarbonate membrane with pores 0.4 μm in diameter. The size of the extruded liposomes was 284.8 nm in diameter (polydispersity index ~ 0.2) as measured by Malvern Zetasizer 3000 HSA. The liposomes were separated from the unencapsulated fluorescent dyes with a Sephadex® G-75 column eluted with an isotonic buffer (10 mM HEPES, 140 mM NaCl, pH 7.4). The lipid concentration of the eluted liposome was determined by a phosphorous assay [52].
6.13. Model Liposome Leakage Assay
Lipoplex was mixed with the model liposome at a total lipid molar ratio of 1:1. To carry out the leakage assay at pH 5.5, one volume of an acidic buffer (pH 5.15, 100 mM NaOAc/HOAc, 50 mM NaCl) was mixed with three volumes of the lipoplex-model liposome mixture to obtain a final mixture at pH 5.5. To carry out the leakage assay at pH 7.4, a HEPES buffer (pH 7.4, 100 mM HEPES, 50 mM NaCl) was used in place of the acidic NaOAc/HOAc buffer. The resultant mixture was incubated at 37 °C and a small aliquot (50 μL) was transferred into a cuvette containing 2 mL of a pH 7.4 buffer (100 mM HEPES, 50 mM NaCl) at different time points. The fluorescence (λex = 370 nm, λem = 550 nm) of the diluted sample was measured by a QuantaMaster fluorescence spectrofluorometer (Photon Technology International, Lawrenceville, NJ). The data were processed and expressed as percentage of leakage as previously reported [26].
6.14. Gene Transfection in Cultured Cells
The gene transfection procedure is similar to a method described by Felgner and co-workers [53]. The CV-1 cells (a monkey fibroblast cell line, ATCC-CCL70) were cultured in T25 flasks containing MEM (Eagle), which consists of 10% FBS, 2 mM L-glutamine, 1.5 g/L sodium bicarbonate, 0.1 mM nonessential amino acids, 1.0 mM sodium pyruvate, and antibiotics (50 IU/mL penicillin and 50 μg/mL streptomycin) based on ATCC recommendations.
Twelve hours prior to gene transfection, when cells were at approximately 90% confluence, the cells were seeded onto 24-well plates (3 × 104 CV-1 cells/well; cell counting performed with a Coulter Z1 particle counter; 1 mL complete growth medium per well) and cultured overnight at 37 °C in a humidified atmosphere containing 5% CO2 to approximately 60% confluence. The complete growth medium was aspirated. The cells were washed once with FBS and antibiotics free medium (150 μL/well) and supplemented with serum free medium (500 μL/well). Appropriate volumes of freshly prepared lipoplexes (less than 50 μL/well) were added to the cells followed by mild agitation of the 24-well plate. After incubation at 37 °C, 5% CO2 for 5 h, the serum free medium was aspirated, and the cells were washed three times with PBS (150 μL/well). The cells were re-supplemented with complete growth medium (1 mL/well) and cultured for an additional 24 h before the assessment of reporter gene expression.
6.15. Reporter Gene Expression Assay
Reporter gene (firefly luciferase) activity was assessed with the luciferase assay system (Promega, E1500) following a modified protocol based on the manufacturer's recommendations. Briefly, the growth media were aspirated and the cells gently rinsed with PBS buffer (Mg2+- and Ca2+-free). To each plate well was added 150 μL M-PER Reagent (Pierce) instead of the CCLR reagent recommended by the manufacturer. The M-PER Reagent (Pierce) minimized the background interference in the total cellular protein assay. The luciferase activity was measured using a TD-20/20 Luminometer (Turner Design) with a 2-s delay followed by a 10-s data acquisition. Luciferase activity is expressed as relative light units (RLU) per well. The total cellular protein was determined by the BCA protein assay (Pierce) following the manufacturer's recommendations.
Highlights.
▶Novel cationic lipid/lipoplexes with linear ortho ester linkers were developed.
▶Their pH-sensitivity was affected by lipid structure, counterion and helper lipids.
▶Lipid 3 enhanced the gene transfection for up to 100 fold in CV-1 cells.
▶The transfection efficiency correlated strongly with membrane destabilization at endosomal pH.
Acknowledgements
This work was supported by NIH grants GM074614 (HC, HZ, XG), NHLBI-R15HL71520 (RR, DT), NIDCR-R15DE016587 (RR, DT), and the Scholarly/Artistic Activities Grant (HC, HZ, XG) from University of the Pacific.
Abbreviations
- ANTS
8-aminonaphthalene-1,3,6-trisulfonic acid, disodium salt
- DPX
p-xylene-bis-pyridinium bromide
- DOPE
1,2-dioleoyl-sn-glycero-3-phosphoethanolamine
- DOTAP
1,2-dioleoyl-3-trimethylammonium-propane (chloride salt)
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPE
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine
- POPS
1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine (sodium salt)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Petkar KC, Chavhan SS, Agatonovik-Kustrin S, Sawant KK. Nanostructured materials in drug and gene delivery: a review of the state of the art. Crit Rev Ther Drug Carrier Syst. 2011;28:101–164. doi: 10.1615/critrevtherdrugcarriersyst.v28.i2.10. [DOI] [PubMed] [Google Scholar]
- 2.Donkuru M, Badea I, Wettig S, Verrall R, Elsabahy M, Foldvari M. Advancing nonviral gene delivery: lipid- and surfactant-based nanoparticle design strategies. Nanomedicine (Lond) 2010;5:1103–1127. doi: 10.2217/nnm.10.80. [DOI] [PubMed] [Google Scholar]
- 3.Wakabayashi T, Natsume A, Hashizume Y, Fujii M, Mizuno M, Yoshida J. A phase I clinical trial of interferon-beta gene therapy for high-grade glioma: novel findings from gene expression profiling and autopsy. J Gene Med. 2008;10:329–339. doi: 10.1002/jgm.1160. [DOI] [PubMed] [Google Scholar]
- 4.Dow S, Elmslie R, Kurzman I, MacEwen G, Pericle F, Liggitt D. Phase I study of liposome-DNA complexes encoding the interleukin-2 gene in dogs with osteosarcoma lung metastases. Hum Gene Ther. 2005;16:937–946. doi: 10.1089/hum.2005.16.937. [DOI] [PubMed] [Google Scholar]
- 5.Caplen NJ, Alton EW, Middleton PG, Dorin JR, Stevenson BJ, Gao X, Durham SR, Jeffery PK, Hodson ME, Coutelle C, et al. Liposome-mediated CFTR gene transfer to the nasal epithelium of patients with cystic fibrosis. Nat Med. 1995;1:39–46. doi: 10.1038/nm0195-39. [DOI] [PubMed] [Google Scholar]
- 6.Sorscher EJ, Logan JJ, Frizzell RA, Lyrene RK, Bebok Z, Dong JY, Duvall MD, Felgner PL, Matalon S, Walker L, et al. Gene therapy for cystic fibrosis using cationic liposome mediated gene transfer: a phase I trial of safety and efficacy in the nasal airway. Hum Gene Ther. 1994;5:1259–1277. doi: 10.1089/hum.1994.5.10-1259. [DOI] [PubMed] [Google Scholar]
- 7.Zhang S, Xu Y, Wang B, Qiao W, Liu D, Li Z. Cationic compounds used in lipoplexes and polyplexes for gene delivery. J Control Release. 2004;100:165–180. doi: 10.1016/j.jconrel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 8.Felgner PL, Tsai YJ, Sukhu L, Wheeler CJ, Manthorpe M, Marshall J, Cheng SH. Improved cationic lipid formulations for in vivo gene therapy. Ann N Y Acad Sci. 1995;772:126–139. doi: 10.1111/j.1749-6632.1995.tb44738.x. [DOI] [PubMed] [Google Scholar]
- 9.Mounkes LC, Zhong W, Cipres-Palacin G, Heath TD, Debs RJ. Proteoglycans mediate cationic liposome-DNA complex-based gene delivery in vitro and in vivo. J Biol Chem. 1998;273:26164–26170. doi: 10.1074/jbc.273.40.26164. [DOI] [PubMed] [Google Scholar]
- 10.Ziello JE, Huang Y, Jovin IS. Cellular endocytosis and gene delivery. Mol Med. 2010;16:222–229. doi: 10.2119/molmed.2009.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiethoff CM, Middaugh CR. Barriers to nonviral gene delivery. J Pharm Sci. 2003;92:203–217. doi: 10.1002/jps.10286. [DOI] [PubMed] [Google Scholar]
- 12.Dean DA, Strong DD, Zimmer WE. Nuclear entry of nonviral vectors. Gene Ther. 2005;12:881–890. doi: 10.1038/sj.gt.3302534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang L, Farhood H, Serbina N, Teepe AG, Barsoum J. Endosomolytic activity of cationic liposomes enhances the delivery of human immunodeficiency virus-1 trans-activator protein (TAT) to mammalian cells. Biochem Biophys Res Commun. 1995;217:761–768. doi: 10.1006/bbrc.1995.2838. [DOI] [PubMed] [Google Scholar]
- 14.Farhood H, Serbina N, Huang L. The role of dioleoyl phosphatidylethanolamine in cationic liposome mediated gene transfer. Biochim Biophys Acta. 1995;1235:289–295. doi: 10.1016/0005-2736(95)80016-9. [DOI] [PubMed] [Google Scholar]
- 15.Koltover I, Salditt T, Radler JO, Safinya CR. An inverted hexagonal phase of cationic liposome-DNA complexes related to DNA release and delivery. Science. 1998;281:78–81. doi: 10.1126/science.281.5373.78. [DOI] [PubMed] [Google Scholar]
- 16.Duzgunes N, De Ilarduya CT, Simoes S, Zhdanov RI, Konopka K, Pedroso de Lima MC. Cationic liposomes for gene delivery: novel cationic lipids and enhancement by proteins and peptides. Curr Med Chem. 2003;10:1213–1220. doi: 10.2174/0929867033457403. [DOI] [PubMed] [Google Scholar]
- 17.Li W, Nicol F, Szoka FC., Jr. GALA: a designed synthetic pH-responsive amphipathic peptide with applications in drug and gene delivery. Adv Drug Deliv Rev. 2004;56:967–985. doi: 10.1016/j.addr.2003.10.041. [DOI] [PubMed] [Google Scholar]
- 18.Kumar M, Jinturkar K, Yadav MR, Misra A. Gemini amphiphiles: a novel class of nonviral gene delivery vectors. Crit Rev Ther Drug Carrier Syst. 2010;27:237–278. doi: 10.1615/critrevtherdrugcarriersyst.v27.i3.20. [DOI] [PubMed] [Google Scholar]
- 19.Guo X, Szoka FC., Jr. Chemical approaches to triggerable lipid vesicles for drug and gene delivery. Acc Chem Res. 2003;36:335–341. doi: 10.1021/ar9703241. [DOI] [PubMed] [Google Scholar]
- 20.Boomer JA, Thompson DH, Sullivan SM. Formation of plasmid-based transfection complexes with an acid-labile cationic lipid: characterization of in vitro and in vivo gene transfer. Pharm Res. 2002;19:1292–1301. doi: 10.1023/a:1020342523694. [DOI] [PubMed] [Google Scholar]
- 21.Aissaoui A, Martin B, Kan E, Oudrhiri N, Hauchecorne M, Vigneron JP, Lehn JM, Lehn P. Novel cationic lipids incorporating an acid-sensitive acylhydrazone linker: synthesis and transfection properties. J Med Chem. 2004;47:5210–5223. doi: 10.1021/jm0408159. [DOI] [PubMed] [Google Scholar]
- 22.Chen H, Zhang H, McCallum CM, Szoka FC, Guo X. Unsaturated cationic ortho esters for endosome permeation in gene delivery. J Med Chem. 2007;50:4269–4278. doi: 10.1021/jm060128c. [DOI] [PubMed] [Google Scholar]
- 23.Huang Z, Guo X, Li W, MacKay JA, Szoka FC., Jr. Acid-triggered transformation of diortho ester phosphocholine liposome. J Am Chem Soc. 2006;128:60–61. doi: 10.1021/ja057024w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo X, Huang Z, Szoka FC. Improved preparation of PEG-diortho ester-diacyl glycerol conjugates. Methods Enzymol. 2004;387:147–152. doi: 10.1016/S0076-6879(04)87009-5. [DOI] [PubMed] [Google Scholar]
- 25.Guo X, Szoka FC., Jr. Steric stabilization of fusogenic liposomes by a low-pH sensitive PEG-diortho ester-lipid conjugate. Bioconjug Chem. 2001;12:291–300. doi: 10.1021/bc000110v. [DOI] [PubMed] [Google Scholar]
- 26.Guo X, MacKay JA, Szoka FC., Jr. Mechanism of pH-triggered collapse of phosphatidylethanolamine liposomes stabilized by an ortho ester polyethyleneglycol lipid. Biophys J. 2003;84:1784–1795. doi: 10.1016/s0006-3495(03)74986-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szleifer II, Kramer D, Ben-Shaul A, Roux D, Gelbart WM. Curvature elasticity of pure and mixed surfactant films. Phys Rev Lett. 1988;60:1966–1969. doi: 10.1103/PhysRevLett.60.1966. [DOI] [PubMed] [Google Scholar]
- 28.Anderson VC, Thompson DH. Triggered release of hydrophilic agents from plasmalogen liposomes using visible light or acid. Biochim Biophys Acta. 1992;1109:33–42. doi: 10.1016/0005-2736(92)90183-m. [DOI] [PubMed] [Google Scholar]
- 29.Ahrland S. Solvation and complex formation-competing and cooperative processes in solution. Pure Appl.Chein. 1982;54:1451–1468. [Google Scholar]
- 30.Fernholz E, Hinzpeter M, von der Eltz H. Cationic and polycationic amphiphiles, reactives containing the same and their use. 1996 US Patent 6060455.
- 31.Gross H, Rieche A. Derivate der orthoameisensaure aus dichlormethyl-alkyl-athern. Jahrg. 1961;94:538–543. [Google Scholar]
- 32.Bergstrand N, Arfvidsson MC, Kim JM, Thompson DH, Edwards K. Interactions between pH-sensitive liposomes and model membranes. Biophys Chem. 2003;104:361–379. doi: 10.1016/s0301-4622(03)00011-5. [DOI] [PubMed] [Google Scholar]
- 33.Alberts B, Bray D, Lewis J, Raff M, Roberts K. Molecular Biology of the Cell. Garland Publishing Inc.; New York: 1994. Membrane structure; pp. 477–482. e. al. [Google Scholar]
- 34.Wang J, Guo X, Xu Y, Barron L, Szoka FC., Jr. Synthesis and characterization of long chain alkyl acyl carnitine esters-potentially biodegradable cationic lipids for use in gene delivery. J Med Chem. 1998;41:2207–2215. doi: 10.1021/jm950802i. [DOI] [PubMed] [Google Scholar]
- 35.Sperinde JJ, Choi SJ, Szoka FC., Jr. Phage display selection of a peptide DNase II inhibitor that enhances gene delivery. J Gene Med. 2001;3:101–108. doi: 10.1002/jgm.165. [DOI] [PubMed] [Google Scholar]
- 36.Khan Z, Hawtrey AO, Ariatti M. New cationized LDL-DNA complexes: their targeted delivery to fibroblasts in culture. Drug Deliv. 2003;10:213–220. doi: 10.1080/713840398. [DOI] [PubMed] [Google Scholar]
- 37.Jaaskelainen I, Peltola S, Honkakoski P, Monkkonen J, Urtti A. A lipid carrier with a membrane active component and a small complex size are required for efficient cellular delivery of anti-sense phosphorothioate oligonucleotides. Eur J Pharm Sci. 2000;10:187–193. doi: 10.1016/s0928-0987(00)00068-3. [DOI] [PubMed] [Google Scholar]
- 38.Deslongchamps P, Dory YL, Li S. The relative rate of hydrolysis of a series of acyclic and six-membered cyclic acetals, ketals, orthoesters, and orthocarbonates. Tetrahedron. 2000;56:3533–3537. [Google Scholar]
- 39.Li S, Dory YL, Deslongchamps P. Hydrolysis of cyclic orthoesters: experimental observations and theoretical rationalization. Tetrahedron. 1996;52:14841–14854. [Google Scholar]
- 40.Friend DS, Papahadjopoulos D, Debs RJ. Endocytosis and intracellular processing accompanying transfection mediated by cationic liposomes. Biochim Biophys Acta. 1996;1278:41–50. doi: 10.1016/0005-2736(95)00219-7. [DOI] [PubMed] [Google Scholar]
- 41.Heller J, Barr J, Ng SY, Abdellauoi KS, Gurny R. Poly(ortho esters): synthesis, characterization, properties and uses. Adv Drug Deliv Rev. 2002;54:1015–1039. doi: 10.1016/s0169-409x(02)00055-8. [DOI] [PubMed] [Google Scholar]
- 42.Heller J, Ng SY, Fritzinger BK, Roskos KV. Controlled drug release from bioerodible hydrophobic ointments. Biomaterials. 1990;11:235–237. doi: 10.1016/0142-9612(90)90003-9. [DOI] [PubMed] [Google Scholar]
- 43.Meidan VM, Cohen JS, Amariglio N, Hirsch-Lerner D, Barenholz Y. Interaction of oligonucleotides with cationic lipids: the relationship between electrostatics, hydration and state of aggregation. Biochim Biophys Acta. 2000;1464:251–261. doi: 10.1016/s0005-2736(00)00151-6. [DOI] [PubMed] [Google Scholar]
- 44.Zuidam NJ, Barenholz Y. Characterization of DNA-lipid complexes commonly used for gene delivery. Int J Pharm. 1999;183:43–46. doi: 10.1016/s0378-5173(99)00041-1. [DOI] [PubMed] [Google Scholar]
- 45.Xu Y, Szoka FC., Jr. Mechanism of DNA release from cationic liposome/DNA complexes used in cell transfection. Biochemistry. 1996;35:5616–5623. doi: 10.1021/bi9602019. [DOI] [PubMed] [Google Scholar]
- 46.Medina-Kauwe LK, Xie J, Hamm-Alvarez S. Intracellular trafficking of nonviral vectors. Gene Ther. 2005;12:1734–1751. doi: 10.1038/sj.gt.3302592. [DOI] [PubMed] [Google Scholar]
- 47.Kobayashi T, Stang E, Fang KS, de Moerloose P, Parton RG, Gruenberg J. A lipid associated with the antiphospholipid syndrome regulates endosome structure and function. Nature. 1998;392:193–197. doi: 10.1038/32440. [DOI] [PubMed] [Google Scholar]
- 48.Perez MJ, Briz O. Bile-acid-induced cell injury and protection. World J Gastroenterol. 2009;15:1677–1689. doi: 10.3748/wjg.15.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aranzazu Partearroyo M, Ostolaza H, Goni FM, Barbera-Guillem E. Surfactant-induced cell toxicity and cell lysis. A study using B16 melanoma cells. Biochem Pharmacol. 1990;40:1323–1328. doi: 10.1016/0006-2952(90)90399-6. [DOI] [PubMed] [Google Scholar]
- 50.Byk G, Wetzer B, Frederic M, Dubertret C, Pitard B, Jaslin G, Scherman D. Reduction-sensitive lipopolyamines as a novel nonviral gene delivery system for modulated release of DNA with improved transgene expression. J Med Chem. 2000;43:4377–4387. doi: 10.1021/jm000284y. [DOI] [PubMed] [Google Scholar]
- 51.Aberle AM, Bennett MJ, Malone RW, Nantz MH. The counterion influence on cationic lipid-mediated transfection of plasmid DNA. Biochim Biophys Acta. 1996;1299:281–283. doi: 10.1016/0005-2760(95)00230-8. [DOI] [PubMed] [Google Scholar]
- 52.Bartlett GR. Phosphorus assay in column chromatography. J Biol Chem. 1959;234:466–468. [PubMed] [Google Scholar]
- 53.Felgner JH, Kumar R, Sridhar CN, Wheeler CJ, Tsai YJ, Border R, Ramsey P, Martin M, Felgner PL. Enhanced gene delivery and mechanism studies with a novel series of cationic lipid formulations. J Biol Chem. 1994;269:2550–2561. [PubMed] [Google Scholar]