Summary
Wall-anchored surface proteins are critical for the in vivo survival of Streptococcus pyogenes. Cues in the signal sequence direct the membrane translocation of surface proteins: M protein to the septum, and SfbI to the poles. Both proteins are subsequently anchored to the wall by the membrane bound enzyme sortase A. However, the cellular features of these pathways are not fully understood. Here we show that M protein and SfbI are anchored simultaneously throughout the cell cycle. M protein is rapidly anchored at the septum, and in part of the cell cycle, is anchored simultaneously at the mother and daughter septa. Conversely, SfbI accumulates gradually on peripheral peptidoglycan, resulting in a polar distribution. Sortase is not required for translocation of M protein or SfbI at their respective locations. Methicillin-induced unbalanced peptidoglycan synthesis diminishes surface M protein but not SfbI. Furthermore, overexpression of the division regulator DivIVA also diminishes surface M protein but increases SfbI. These results demonstrate a close connection between the regulation of cell division and protein anchoring. Better understanding of the spatial regulation of surface anchoring may lead to the identification of novel targets for the development of anti-infective agents, given the importance of surface molecules for pathogenesis.
Introduction
Streptococcus pyogenes is an important human pathogen, responsible for 500,000 deaths per year worldwide (Carapetis et al., 2005). Infection ranges from a mild strep throat or skin infection, to severe invasive conditions, including toxic shock syndrome, septicemia, and necrotizing fasciitis or “flesh-eating disease”. Untreated infection may lead to sequela including rheumatic heart disease and glomerulonephritis (Cunningham, 2000). S. pyogenes employs an impressive array of wall-anchored virulence factors that are critical for its in vivo survival (Bisno et al., 2003, Marraffini et al., 2006, Nobbs et al., 2009). Wall-anchored surface proteins possess a conserved C-terminal anchor domain comprised of an LPXTG motif followed by a hydrophobic region and a few positively charged amino acids (Fischetti et al., 1990, Schneewind et al., 1992). The anchoring domain is recognized by sortase, a membranal transpeptidase, which cleaves the LPXTG motif between the threonine and glycine residues (Mazmanian et al., 1999), and connects the freed threonine to the peptidoglycan precursor lipid II (Perry et al., 2002, Marraffini et al., 2006).
While the biochemical aspects of the sorting reaction have been studied in detail, the spatial regulation of this process is not as well understood (Marraffini et al., 2006, Hendrickx et al., 2011, Spirig et al., 2011). Proteins are anchored to the wall of S. pyogenes at two distinct locations, the septum and the poles (Carlsson et al., 2006). Anchoring of M protein to the cell wall takes place exclusively at the septum (Cole & Hahn, 1962, Swanson et al., 1969), where newly anchored M protein localizes within areas of active lipid II export and wall synthesis (Raz & Fischetti, 2008). Since the synthesis of new cell wall is restricted to the septum of S. pyogenes, septal anchoring leads to the coating of the entire cell surface with M protein. In contrast to M protein, SfbI (also known as protein F, or PrtF), which is a major fibronectin binding protein in certain streptococcal strains (Talay et al., 1992, Hanski & Caparon, 1992), displays a polar distribution (Ozeri et al., 2001).
The signal sequence directs surface proteins for translocation at their ultimate cellular location (Carlsson et al., 2006, DeDent et al., 2008). Signal sequences containing a YSIRK-G/S motif are targeted to the septum, while signal sequences lacking this motif are targeted to the poles. Although the YSIRK-G/S motif is required for efficient signal sequence processing (Bae & Schneewind, 2003), mutations in this motif do not affect targeting to the septum (Carlsson et al., 2006, DeDent et al., 2008). Surface proteins possessing a YSIRK-G/S motif are found in S. pyogenes (Carlsson et al., 2006), Staphylococcus aureus (DeDent et al., 2008) and Streptococcus pneumoniae (Tsui et al., 2011), are rare in Enterococcus faecalis (Kline et al., 2009), and are absent from Listeria monocytogenes (Bruck et al., 2011).
S. pyogenes sortase A (SrtA), which is involved in the anchoring of both M protein and SfbI (Barnett & Scott, 2002), localizes to a number of membranal foci (Raz & Fischetti, 2008). These foci are preferentially associated with the division septum, near sites of active M protein anchoring, and are recruited to daughter septa at an early stage of the division cycle. The distribution pattern of S. pyogenes pilus-specific sortases, SrtB and SrtC (Barnett & Scott, 2002, Barnett et al., 2004, Mora et al., 2005), is unknown at present.
In this study we use deconvolution immunofluorescence microscopy to study the two anchoring pathways of S. pyogenes. We show that M protein and SfbI are anchored simultaneously throughout the cell cycle. The anchoring of M protein is restricted to the septum, and occurs simultaneously at the closing mother septum and the forming daughter septa at certain stages of the cell cycle. SfbI on the other hand, is anchored at large peripheral areas, and its gradual accumulation on peptidoglycan results in polar distribution. Sortase is not required for the correct localization of M protein and SfbI translocation. Methicillin-induced unbalanced peptidoglycan synthesis, disrupts the proper assembly of the septum, and results in a marked reduction in surface M protein, but not SfbI. Overexpression of DivIVA also disrupts the septum, and results in a decrease in surface M protein, but an increase in SfbI.
Results
Localization of M protein and SfbI anchoring sites
In an attempt to better understand the two anchoring pathways in S. pyogenes, we used 3D structured illumination microscopy (3D-SIM) to follow the anchoring of M protein and SfbI (Fig. 1A). As expected, M protein (red) covers the entire surface of log phase D471 cells, although at this high resolution the distribution appears somewhat irregular, while SfbI (green) is present at the poles. It is of note however, that while the majority of cells displayed a similar level of M protein fluorescence, the SfbI fluorescence varied greatly between different poles.
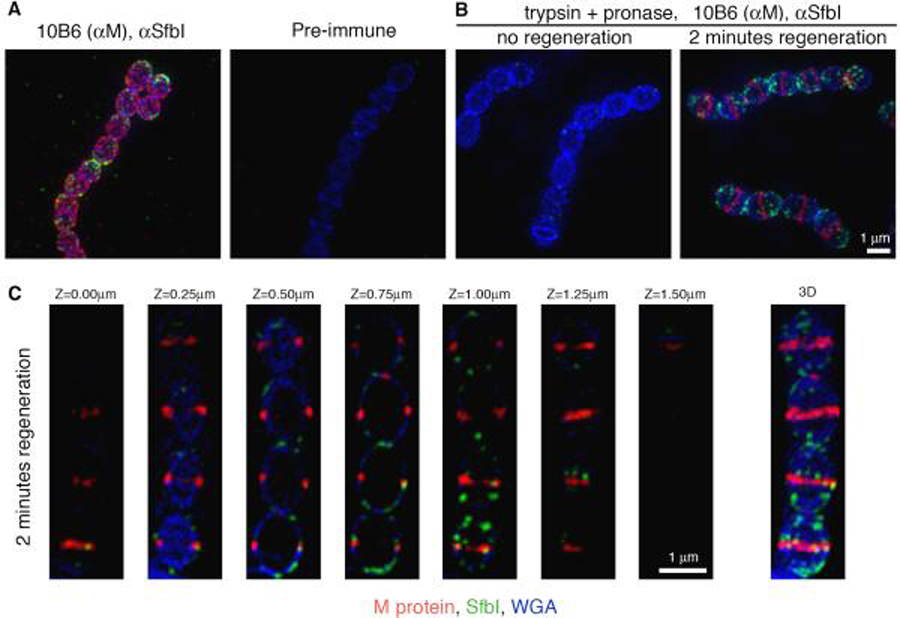
Figure 1. The relative anchoring patterns of M protein and SfbI.
An overnight S. pyogenes D471 culture was diluted 1:100 into TH+Y (A), or TH+Y containing trypsin and pronase (B), and fixed upon reaching OD600 0.5 (A, and B “no regeneration”). Protease treated cells were also harvested at OD600 0.5, washed, and resuspended in TH+Y for 2 minutes at 37°C prior to fixation (B “2 minutes regeneration”, and C). Specific antibodies were used to label M protein (red) and SfbI (green). The cell wall was stained with WGA marina blue (blue). 3D-SIM microscopy images are presented as maximum intensity projections composed of all the Z-sections (A and B), or as sequential Z-sections (C).
The cellular location of active M protein anchoring has previously been studied by digesting existing surface proteins with trypsin, and following the regeneration of new M protein in medium without trypsin (Cole & Hahn, 1962, Swanson et al., 1969). We found that complete removal of SfbI required the use of pronase in addition to trypsin, both of which were included in the growth medium (Fig. 1B, no regeneration). These proteases cannot cross the S. pyogenes cell wall, and their effect is therefore limited to the outer surface of the bacterium. Following a wash and 2 minutes regeneration in medium without proteases, M protein was detected strictly at the septum, while SfbI was detected in patches over a relatively large peripheral area (Fig. 1B, 2 minutes regeneration). A Z-stack view detailing the distribution of M protein and SfbI through the different layers of a representative streptococcal chain is presented in Figure 1C.
M protein and SfbI are anchored simultaneously throughout the cell cycle
To gain further insight into the anchoring of M protein and SfbI, we followed the localization patterns of these proteins as a function of the cell cycle stage. For this purpose D471 cells were treated with proteases as described above, washed, and resuspended in medium without proteases for two minute, allowing the regeneration of surface proteins. Numerous DeltaVision images were processed as average-intensity 3D-projections. A population of 781 cells was obtained by analyzing all the cells whose growth axis was parallel to the slide (and thus the septum was perpendicular to the slide), and that had no signal interference from other cells. Each cell was assigned to one of 6 groups based on cell length. While this division is arbitrary, it provides useful information about cell populations in different stages of the division cycle. Since S. pyogenes grows in chains, a clear-cut distinction of the end of one cell cycle and the beginning of another is not immediately apparent. The first division stage in our analysis was chosen in a manner roughly analogous to that described by Higgins and Shockman (Higgins & Shockman, 1976), in which most cells reveal only preliminary peptidoglycan assembly at the forming septum, and where the mother septum is often not completely closed. A plot profile was generated for each cell, displaying the fluorescent signal intensity relative to the cellular position along the growth axis (see experimental procedures section). Whenever the chain orientation made it possible to determine which pole represented the previous division site (often for stages 1–4, but less often for stages 5–6), that pole was aligned to the left. A representative cell for each division stage, as well as its individual plot profile, are presented (Fig. 2A, left and middle columns). The mean intensity and standard deviation values for each cellular position among all the cells in the group were calculated, and these values are presented as population plots (Fig. 2A, right column). The experiment was repeated two more times, with smaller cell populations, yielding comparable results (not shown). The major cellular regions referred to in this study are presented in Figure 2B.
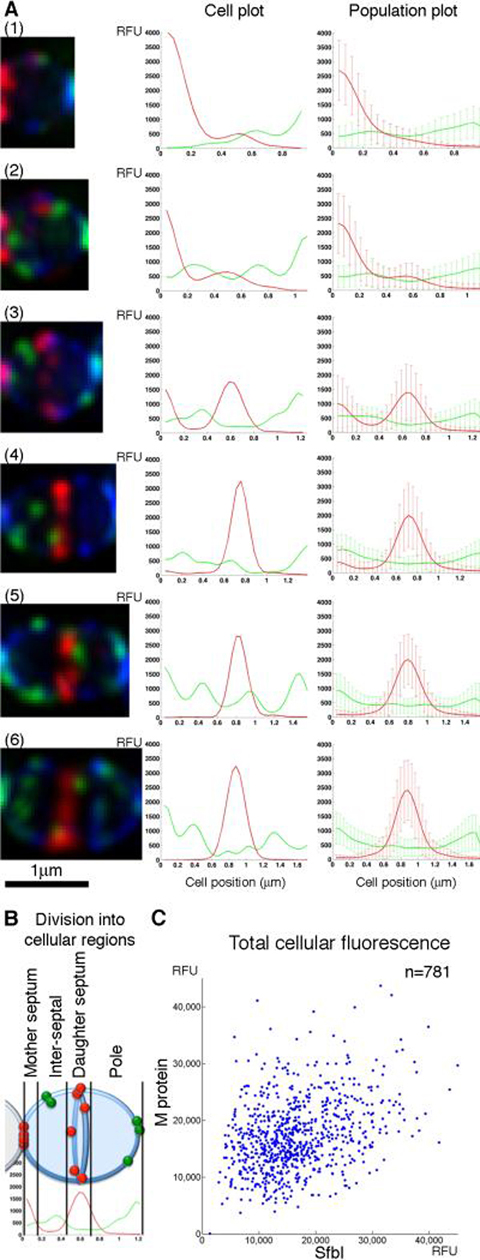
Figure 2. M protein and SfbI are anchored simultaneously throughout the cell cycle.
(A) An overnight S. pyogenes D471 culture was diluted 1:100 into TH+Y containing trypsin and pronase and incubated at 37°C to OD600 0.5, at which point the cells were washed, and resuspended in TH+Y without proteases for 2 minutes at 37°C prior to fixation. The cells were labeled for M protein (red), SfbI (green) and wall (WGA, blue). Average intensity 3D-projections were made for numerous DeltaVision images, resulting in a population of 781 cells. These cells were divided according to cell length to 6 groups, representing different stages of the cell cycle. Each cell was divided into pixel-wide strips, parallel to the division plane, and the total fluorescence intensity was calculated for each strip. These data, when plotted against the relative position in the cell, produced the cellular distribution plots. Representative cells for each division stage are presented on the left, and their respective plots are presented on the middle column. For each division stage, the group's average fluorescent signal distribution and standard deviation values are presented on the right. (B) A representation of the cellular regions of a streptococcal cell, as used in this study. (C) A plot displaying the total M protein fluorescence value for each cell in the population, plotted against its total SfbI fluorescence.
The population plots show that M protein and SfbI are anchored throughout the cell cycle. At the earliest stage (stage 1), M protein is observed primarily at the closing mother septum (here presented on the left). As the cell cycle progresses (stages 2–4), M protein anchoring activity is progressively redistributed to the forming daughter septum, and newly anchored M protein can often be observed simultaneously at both mother and daughter septa. Eventually, M protein is primarily anchored at the daughter septum (stage 5–6). SfbI on the other hand is anchored at a broad peripheral region. In early division (stage 1), SfbI is preferentially anchored at the pole distal to the previous division site. As division progresses (stages 2–4) SfbI is anchored at the old pole as well as the inter-septal region. At the late stages of division (stages 5–6) SfbI is anchored at both poles. Active division septa consistently show the least amount of SfbI anchoring.
To test in a more direct manner whether M protein and SfbI are anchored simultaneously to the surface of individual cells, the total M protein fluorescence of each cell in the population was plotted against the total SfbI fluorescence of the same cell (Fig. 2C). Only relatively few cells were found on the upper-left portion of the plot (representing high M protein fluorescence but no SfbI fluorescence), and the lower right portion (representing high SfbI fluorescence but no M protein fluoresce). Although the cells presented a wide range of fluorescence intensities, most fell in the middle region, indicating that both proteins were anchored. While these data represent the outcome of 2 minutes of protein anchoring activity rather than real-time anchoring, the prevalence of cells displaying both proteins, as well as the anchoring of the two proteins in all stages of the division cycle, indicate that the two proteins are anchored in parallel throughout the cell cycle.
M protein is anchored simultaneously at the closing mother septum, and forming daughter septa
Ovococci are a group of bacteria with slightly elongated coccus morphology, which divide in a single plain, and often form chains of organisms (Zapun et al., 2008). This situation makes the overlap of two successive division cycles possible, with the initiation of daughter septa assembly before the mother septum is completely closed (Higgins & Shockman, 1976, Gibson et al., 1983). Early examinations of the anchoring of M protein to the cell wall of S. pyogenes following trypsinization and 15 minutes of protein regeneration suggested that simultaneous anchoring at the mother and daughter septa is possible (Cole & Hahn, 1962). To study the possibility of simultaneous anchoring at these locations in more detail, we used population-level analysis. While our initial population-level analysis is in agreement with simultaneous anchoring (Fig. 2A), this method averages the fluorescence signal across the cells in the group, and is therefore inadequate for this purpose.
We therefore developed a method for automatically analyzing the distribution of M protein in individual cells. To minimize the possibility that sequential anchoring at the mother and then daughter septa would be interpreted as simultaneous anchoring, protease treated cells were suspended directly in medium without proteases and allowed the regeneration of surface proteins for only 30 seconds before fixation (omitting the one-minute wash step, in which some protein anchoring does occur). Each cell in a population of 1,039 was assigned to one of six groups according to length, and its M protein distribution plot was generated as described above. The resulting plots were analyzed individually using MATLAB (see experimental procedures section), and assigned to one of four categories: M protein anchored at the mother septum alone, the daughter septum alone, both septa, or no septal anchoring (Fig. 3A). To supplement the automated analysis, the entire population was also classified into similar categories by direct observation of the cells (Fig. 3B). While the manual method classified slightly more cells as presenting simultaneous anchoring at both septa, the two methods were generally in good agreement. The majority of cells in the youngest cell population (stage 1) displayed M protein only at the mother septum. With the progression of the cell cycle (stages 2–4) anchoring activity was gradually redistributed to the daughter septum, with roughly 20–30% of stage 2 cells, 30–40% of stage 3 cells, and 20–30% of stage 4 cells displaying M protein at both mother and daughter septa. At the later division stages (stages 5–6) M protein was found almost exclusively at the daughter septum. The analysis of M protein anchoring was repeated with an additional population of 890 cells, resulting from four different cultures, yielding comparable results (Fig. S1A). Additionally, this same analysis was applied to the cell population described in Figure 2, which was allowed two minutes of protein regeneration (Fig. S1B). As expected, this population showed a higher occurrence of M protein detected at both mother and daughter septa. This increase may be attributed in part to the brighter fluorescent signal, yet with longer regeneration time, some sequential anchoring at the mother followed by the daughter septa, cannot be ruled out.
Figure 3. M protein is anchored simultaneously at the mother and daughter septa.
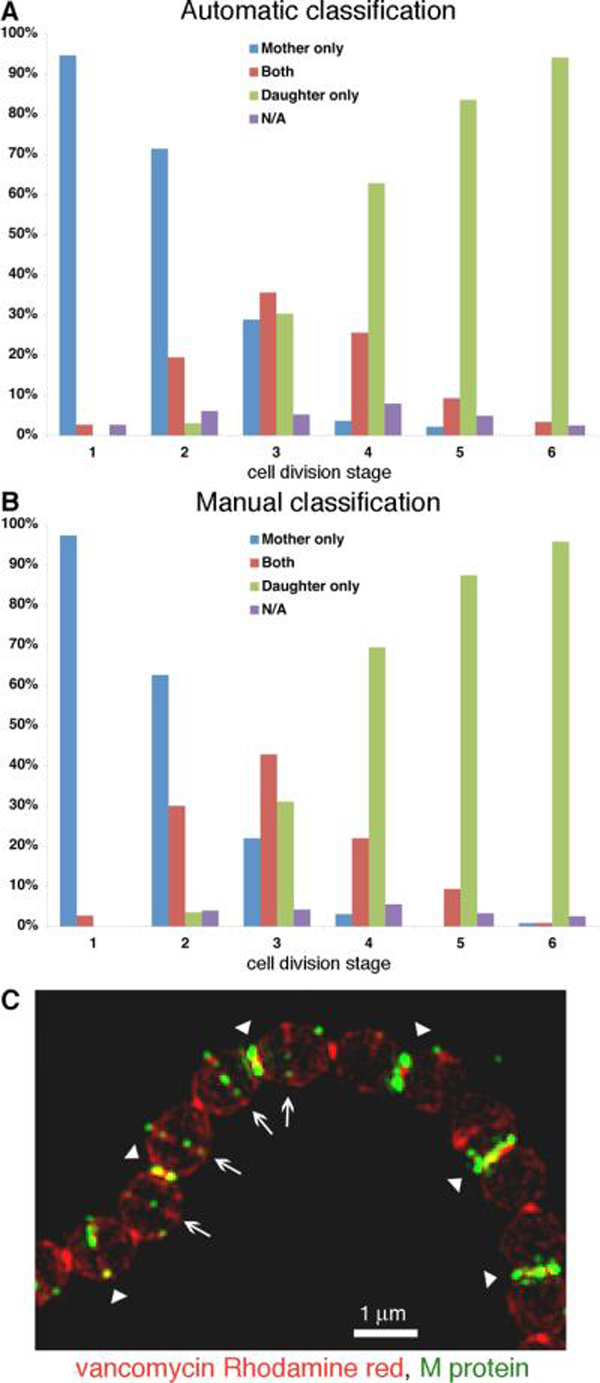
An overnight S. pyogenes D471 culture was diluted 1:100 into TH+Y containing trypsin and pronase and incubated at 37°C to OD600 0.5, at which point the cells were harvested, and resuspended in TH+Y without proteases for 30 seconds at 37°C prior to fixation. M protein was labeled using specific antibodies and average intensity 3D-projections were made for numerous DeltaVision images, resulting in a population of 1,039 cells. The M protein distribution plots of these cells were analyzed using MATLAB (see experimental procedures section) to determine the presence of newly anchored M protein at the mother and daughter septa (A). The location of M protein anchoring on the same cells was determined by direct observation (B). (C) Cells treated in a similar manner were labeled with vancomycin Rhodamine red conjugate (red), and M protein specific antibodies (green), and then visualized by 3D-SIM. Arrowheads represent mother septa and arrows represent forming daughter septa.
M protein anchoring following 30 seconds regeneration was also visualized using 3D Structured Illumination Microscopy (3D-SIM). For this purpose the cells were labeled with M protein specific antibodies (green), and a vancomycin Rhodamine red conjugate (red), which preferentially labels sites of active peptidoglycan synthesis (Daniel & Errington, 2003). Sites of M protein anchoring localized to regions strongly labeled with vancomycin Rhodamine red. In cells displaying both mother and daughter septa, M protein was often observed at both locations (Fig. 3C). These results suggest that in rapidly dividing S. pyogenes cells, simultaneous anchoring of M protein to the mother and daughter septa during the initial stages of daughter septum formation is a common occurrence.
Polar distribution results from gradual accumulation of SfbI on peripheral peptidoglycan
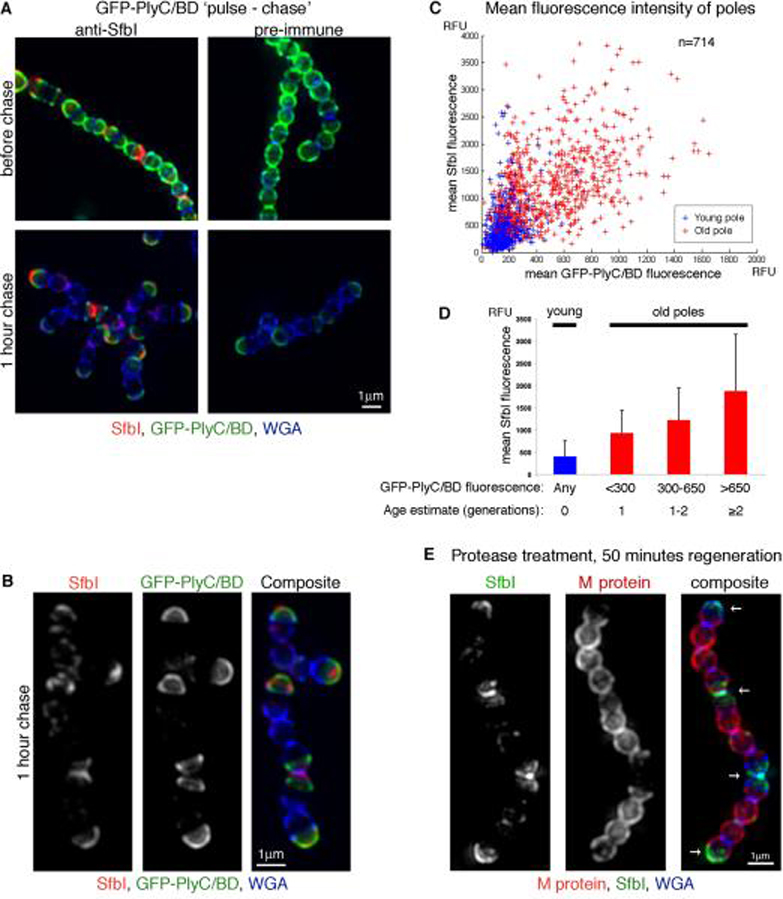
As discussed above, although SfbI was anchored throughout the peripheral region of the cell, the final distribution was distinctly polar, and displayed great variations in fluorescence intensity between different poles. One hypothesis that may explain how diffuse SfbI anchoring could lead to the observed distinctly polar distribution is that SfbI is anchored at a relatively constant pace, and accumulation of more SfbI on older poles leads to the observed difference in fluorescence intensity. To test whether there is a correlation between the amount of SfbI found on the pole and its age we used a fusion protein between Green Fluorescent Protein (GFP) and the binding domain of the phage lysin PlyC (GFP-PlyC/BD) in a “pulse-chase” type experiment. The binding domain of the PlyC lysin binds tightly to the cell wall carbohydrate (Nelson et al., 2006), making it recognizable by its GFP fluorescence. D471 cells were grown in TH+Y media to OD600 0.15, at which stage purified GFP-PlyC/BD was added to the medium for 30 minutes (`pulse'). Cells examined at this time point were brightly fluorescent in the green channel (Fig. 4A, `before chase'). The cells were then washed and incubated for one hour in medium lacking this fusion protein (`chase'), at which time the green fluorescence was limited to poles that were already formed during the `pulse' (Fig. 4A, `1 hour chase'). These poles, which are at least two generations old, showed the most SfbI labeling, while younger poles showed considerably less SfbI labeling. The prevalence of SfbI on old poles labeled with GFP-PlyC/BD is apparent when the two fluorescent signals are examined side by side (Fig. 4B).
Figure 4. Gradual accumulation of SfbI results in polar distribution.
An overnight S. pyogenes D471 culture was diluted 1:100 into TH+Y and grown to OD600 0.15, at which time GFP-PlyC/BD (green) was added to the medium for 30 minutes (“pulse”). The cells were then washed and suspended in TH+Y for one hour at 37°C (“chase”) prior to fixation. SfbI was stained using a specific serum (red), and the cell wall was stained using WGA marina blue (blue). (A) Representative cells before and after the “chase” period. (B) Streptococcal chains following the “chase” period alongside the separate fluorescent channels. (C) Average intensity 3D-projections were made for numerous DeltaVision images, resulting in a population of 714 cells. The signal distribution plots of these cells were analyzed using MATLAB. Each cell was divided into “young pole” (blue) and “old pole” (red) regions. For each pole, the mean GFP-PlyC/BD and SfbI fluorescence values were plotted against each other. (D) The “old pole” group was sub-divided into 3 groups according to GFP-PlyC/BD fluorescence, representing the estimated age of the pole. The mean SfbI fluorescence the standard deviation values are presented for each group, as well as for the “young pole” group in its entirety. (E) An overnight S. pyogenes D471 culture was diluted 1:100 into TH+Y containing trypsin and pronase, and incubated at 37°C to OD600 0.5, at which point the cells were washed, and resuspended in TH+Y without proteases for 50 minutes at 37°C prior to fixation. Specific antibodies were used to label M protein (red) and SfbI (green). The cell wall was stained with WGA marina blue (blue). Arrows denote wall regions that were already assembled at the time of protease treatment. DeltaVision images for (A), (B), and (E), are presented as maximum intensity 3D-projections.
To quantify the relations between the age of the pole and the amount of SfbI fluorescence, the signal distributions of SfbI and GFP-PlyC/BD were analyzed in a population of 714 cells, derived from two separate experiments (each experiment supported the final result, when analyzed individually). Each cell was divided into two regions: a “young pole” consisting of the “mother septum” and “inter-septal” regions (Fig. 2B), and an “old pole” equivalent to the “pole” region (Fig. 2B). The signal at the “daughter septum” region at the middle of the cell (Fig. 2B) was not analyzed, and this region served as buffer between the two poles. For each pole, the average SfbI and GFP-PlyC/BD fluorescence were determined, and the two values were plotted against each other (Fig. 4C). Young poles (blue) were not yet formed at the time of the GFP-PlyC/BD “pulse” and thus did not display substantial GFP-PlyC/BD fluorescence. The majority of these poles displayed only a modest amount of anchored SfbI. Old poles (red) represent a heterogeneous group with ages ranging from one to several generations. The majority of one-generation-old poles was created during the “chase” period, and thus displayed only modest GFP-PlyC/BD fluorescence. Poles that are two or more generations old were already formed during the “pulse” period, and thus displayed a high GFP-PlyC/BD fluorescence. Since the growth of S. pyogenes is not synchronized, a third group of poles are those that were only partly formed during the “pulse”, and thus displayed an intermediate level of GFP-PlyC/BD fluorescence. The average fluorescence values for the poles in each of these four groups are presented in Figure 4D. Both direct examination (Fig. 4C), and comparison of the average SfbI fluorescence associated with each age group (Fig. 4D), reveal a direct correlation between the level of SfbI fluorescence and the age of the pole.
To supplement the “pulse-chase” results, D471 cells were grown in medium containing trypsin and pronase as described above, washed, and incubated in medium without proteases for 50 minutes to regenerate surface proteins. Since M protein is only anchored to newly synthesized peptidoglycan at the septum, areas devoid of M protein represent poles that were already fully formed at the time of protease treatment, and are therefore at least two generations old (Fig. 4E). When the labeling intensity of SfbI in these cells was determined, the “oldest” poles (with the least M protein labeling) displayed the most SfbI labeling, poles that were one generation old displayed weaker labeling, and newly formed poles displayed very weak SfbI labeling (see Fig. 7 for a model representation). The combined results of the two approaches suggest that gradual accumulation of SfbI on peripheral peptidoglycan, leads to the observed polar distribution, with older poles displaying a greater amount of SfbI. These observations also suggest that poles remain active sites of SfbI anchoring for at least two generations following their formation.
Figure 7. A model representation of the anchoring of M protein and SfbI.
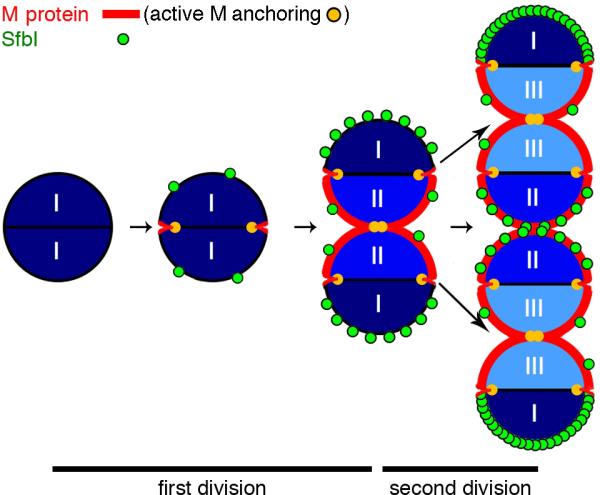
The regeneration of surface proteins during two division cycles is presented. M protein (red) is anchored exclusively to newly synthesized peptidoglycan at the septum (sites of active anchoring labeled yellow). Anchoring of M protein at daughter septa begins before the mother septum is completely closed, resulting in simultaneous anchoring at both locations. Following two generations, M protein is anchored to all newly synthesized, but not pre-existing, peptidoglycan. SfbI (green) is anchored over time in patches to peripheral peptidoglycan, with some preference to the poles. Following two generations, the oldest poles (I) display the most intense SfbI labeling, while one-generation-old poles show less labeling (II), and newly formed poles (III) show little SfbI labeling.
Translocation of M protein and SfbI at distinct locations is maintained in the sortase mutant AR01
Our next aim was to better understand the mechanisms underlying the differences between M protein and SfbI anchoring patterns. We first tested whether deletion of sortase A, which anchors both proteins to the cell wall (Barnett & Scott, 2002), could affect their translocation pattern. The lack of M protein and SfbI anchoring to the cell wall of the sortase mutant strain AR01 was validated by fractionation and Western blot analysis (Fig. S2A). Although no anchoring took place, both proteins were translocated across the plasma membrane, and remained trapped to some extent in the cell wall. M protein and SfbI accumulated at distinct locations on the surface of untreated AR01 cells, and did not substantially colocalize (Fig. S2B). Regeneration of surface proteins following protease treatment revealed that M protein and SfbI first became visible on the surface of AR01 in cellular locations resembling that of wild type D471, namely M protein was translocated at the septum and SfbI at the cell periphery. A Z-stack view of a representative streptococcal chain is presented in Figure S2C. These results indicate that sortase is not required for spatially correct translocation of surface proteins.
Unbalanced peptidoglycan synthesis induced by methicillin results in a marked reduction in the cellular amount of M protein but not SfbI
At the initial stages of protein sorting, a nascent surface protein is cleaved near the C-terminus and covalently attached to lipid II through a sortase-mediated transpeptidation reaction. The ensuing complex serves as substrate for penicillin binding proteins (PBPs), resulting in covalent attachment of the surface protein to the cell wall. PBPs assemble peptidoglycan through two distinct activities, namely the polymerization of glycan chains through transglycosylation, and the cross-linking of these chains through transpeptidation. β-lactam antibiotics, which inhibit the transpeptidation activity but not the transglycosylation activity of PBPs, do not directly affect the sorting reaction (Ton-That & Schneewind, 1999). An interesting feature of β-lactam antibiotics is their ability to bind various PBPs with different affinities, resulting in unbalanced peptidoglycan synthesis (Williamson et al., 1980, Gutmann et al., 1981, Pucci et al., 1986, Lleo et al., 1990). In particular, methicillin was shown to specifically inhibit septal peptidoglycan synthesis in several ovococci, resulting in the formation of rod shaped cells (Lleo et al., 1990, Perez-Nunez et al., 2011). We reasoned that if similar morphological effects could be induced in S. pyogenes, the manner in which M protein and SfbI are anchored during unbalanced peptidoglycan synthesis could shed light on the differences between the two anchoring pathways.
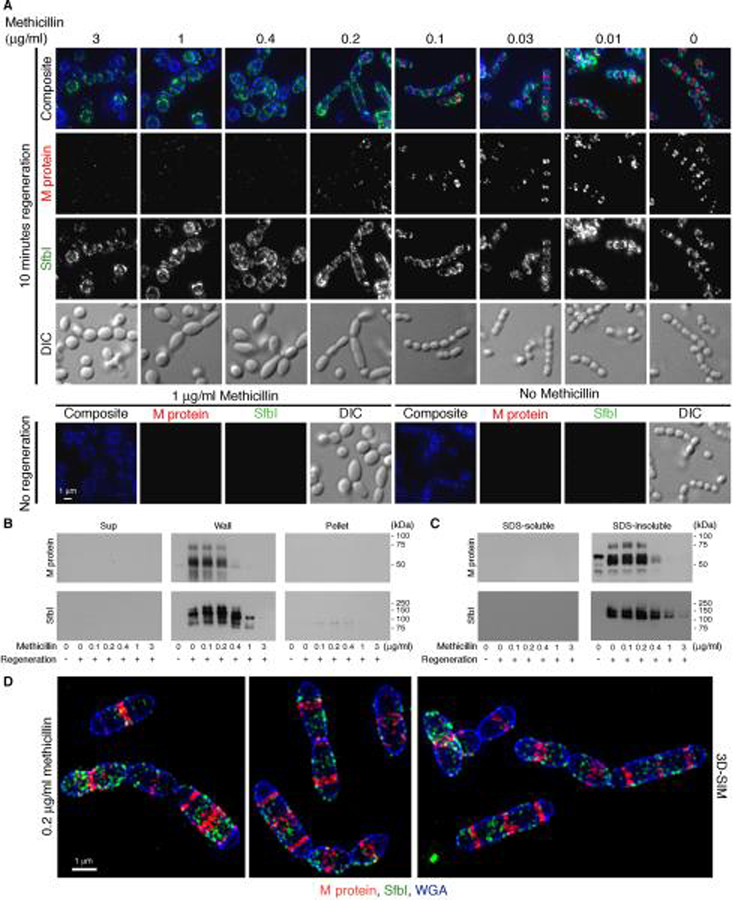
S. pyogenes D471 cells were grown in the presence of trypsin and pronase to digest existing surface proteins until OD600 0.5 was reached, at which time the culture was diluted 1:4 into a similar medium containing ascending concentrations of methicillin. Following one-hour growth, during which time the phenotypic effects of methicillin on peptidoglycan synthesis became apparent, the cells were washed and resuspended in medium containing a similar concentration of methicillin, but lacking proteases. Following 10 minutes of surface protein regeneration, the cells were fixed and the anchoring patterns of M protein and SfbI were determined by immunofluorescence (Fig. 5A). Morphological defects, resulting from unbalanced peptidoglycan synthesis, became apparent at 0.2 μg/ml methicillin, as many cells displayed an elongated or rod-shaped morphology with multiple septa. At higher methicillin concentrations the cells became bulbous with no obvious septa. In rod-shaped cells, M protein was typically anchored at several septa per cell, albeit at a reduced quantity compared to untreated cells, while SfbI was anchored both at the poles and at the inter-septal regions. At higher methicillin concentrations the amount of M protein on the cells was diminished substantially, while significant anchoring of SfbI was still observed. Western blot analysis of fractionated cells revealed a similar pattern, where the amount of M protein was greatly reduced at methicillin concentrations above 0.2 μg/ml, while the amount of SfbI remained substantial (Fig. 5B). At 3 μg/ml methicillin, the level of SfbI anchoring dropped as well, however this reduction may be due to a general decline in cellular functions. To verify that methicillin did not directly interfere with the sorting reaction, cell cultures treated in a similar manner were harvested, and boiled in 2% SDS. The SDS supernatant was collected (Fig. 5C, “SDS-soluble”), and the cells were lysed using the phage lysin PlyC, thereby releasing covalently bound surface proteins (Fig. 5C, “SDS-insoluble”). Treatment with methicillin did not result in the release of substantial quantities of M protein or SfbI following boiling in SDS, indicating that the covalent attachment of surface proteins by sortase was not affected. The anchoring pattern of M protein and SfbI on methicillin-induced rod-shaped cells was further studied by 3D-SIM (Fig. 5D). These images show in greater detail the anchoring of M protein at multiple septa per cell, and the anchoring of SfbI, not only at polar regions, but also at inter-septal regions.
Figure 5. Methicillin-induced unbalanced peptidoglycan synthesis results in a substantial reduction in the cellular amount of M protein compared to SfbI.
An overnight S. pyogenes D471 culture was diluted 1:100 into TH+Y containing trypsin and pronase, and grown at 37°C to OD600 0.5. The cells were then diluted 1:4 into tubes containing TH+Y, trypsin, pronase, and ascending concentrations of methicillin. Following one hour, the cells were washed, and resuspended for 10 minutes in TH+Y containing a similar concentration of methicillin but lacking proteases, and then fixed. (A) M protein (red) and SfbI (green) were labeled using specific antibodies, and the cell wall was stained with WGA marina blue (blue). Deconvolution immunofluorescence images are presented as maximum intensity projections. (B) Similar cultures were fractionated into supernatant, wall, and spheroplast pellet, and processed by Western blot. (C) Additional cultures were harvested, boiled in 2% SDS, and separated into supernatant (“SDS Soluble” fraction), and cell pellet (“SDS insoluble” fraction), which was subsequently treated with the phage lysin PlyC to release wall-anchored proteins, prior to processing by Western blot. (D) Cells treated with 0.2 μg/ml methicillin, were visualized by 3D-SIM and are presented as maximum intensity projections.
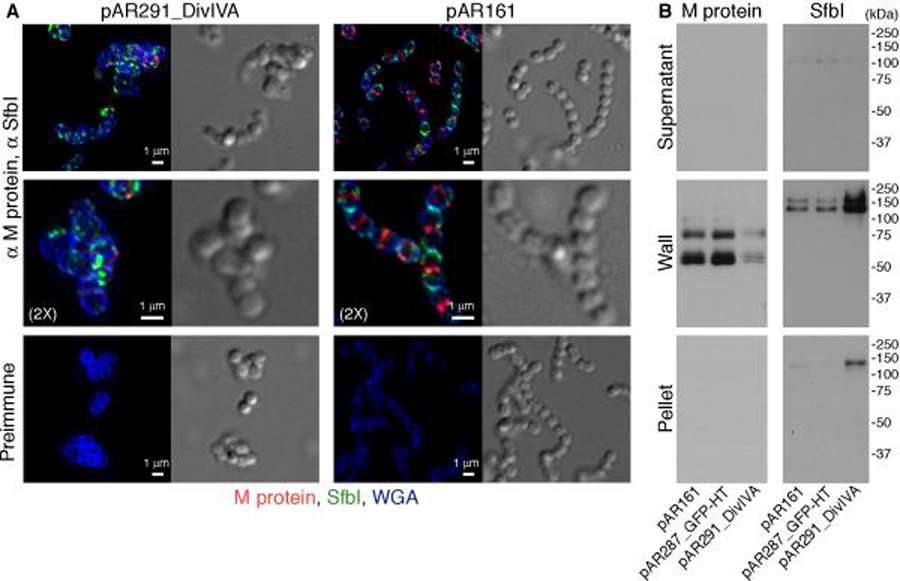
Overexpression of the cell division protein DivIVA results in aberrant cellular morphology, a reduction in the level of M protein, and an increase in SfbI
DivIVA plays a major role in the regulation of septum placement in Gram-positive organisms. Bacillus subtilis DivIVA is targeted to areas of negatively curved membrane at the poles and the septum, recruits MinC and MinD to the poles, and thus prevents the assembly of polar division rings. In the absence of DivIVA, division is severely inhibited and septa are misplaced (Cha & Stewart, 1997, Edwards & Errington, 1997, Marston & Errington, 1999, Lenarcic et al., 2009, Ramamurthi & Losick, 2009). S. pneumoniae and E. faecalis do not possess a MinCD system, nevertheless, DivIVA is important for the correct placement of the septum through a mechanism that is not completely understood (Fadda et al., 2003, Fadda et al., 2007, Ramirez-Arcos et al., 2005). We postulated that overexpression of S. pyogenes DivIVA could similarly interfere with septum placement, and thus provide an additional way to test the dependence of protein anchoring on a functioning septum.
To test the effects of DivIVA overexpression, three plasmids carrying a spectinomycin (spec) selectable marker were constructed: pAR291_DivIVA, which expresses DivIVA under the control of the highly active M protein promoter, pAR287_GFP-HT, which expresses a GFP-HaloTag fusion protein under the control of the same promoter (unrelated protein control), and pAR161, an empty vector. For microscopy studies, overnight cultures of D471 harboring pAR291_DivIVA and pAR161 were diluted 1:50 into TH+Y+spec medium containing trypsin, and pronase. At OD600 0.5, the cells were washed, incubated in medium without proteases for 5 minutes, and the anchoring of M protein and SfbI was examined. Cells harboring pAR291_DivIVA had a reduced growth rate, and displayed aberrant morphology, including enlarged volume and irregular shape (Fig. 6A). The amount of M protein associated with these cells was greatly reduced compared to control samples. When observed on cells, M protein was still associated with the septa despite the aberrant morphology. SfbI was found in wall-associated patches, and displayed no clear cellular localization preference, although the exact distribution pattern was difficult to ascertain due to the irregular shape of these cells. For Western blot analysis, overnight D471 cultures harboring pAR161, pAR287_GFP-HT, or pAR291_DivIVA, were diluted 1:50 into TH+Y+spec medium, and were fractionated upon reaching OD600 0.5. The amount of M protein in cells harboring pAR291_DivIVA was greatly reduced compared to cells containing pAR161, or pAR287_GFP-HT (Fig. 6B). Conversely, the amount of SfbI was increased in cells containing pAR291_DivIVA. The reduction in the level of M protein was not due to the introduction of the M protein promoter on the DivIVA expression plasmid, since cells harboring pAR287_GFP-HT, which possess a similar promoter, did not display altered M protein expression level. When examined by fluorescent microscopy, cells harboring pAR287_GFP-HT were brightly fluorescent in the green channel, confirming the functionality of the M protein promoter used in this study (not shown). It is of note that disruption of proper septum assembly by methicillin or the overexpression of DivIVA, both resulted in a marked reduction in the level of surface M protein.
Figure 6. Overexpression of DivIVA results in a marked decrease in the cellular amount of M protein and an increase in SfbI.
(A) Overnight cultures of D471 cells harboring pAR291_DivIVA, or the empty vector pAR161, were diluted 1:50 into TH+Y+spec containing trypsin and pronase and grown to OD600 0.5. The cultures were then washed, resuspended in medium without proteases for 5 minutes, and then fixed. Specific antibodies were used to label M protein (red) and SfbI (green), and the cell wall was stained with WGA marina blue (blue). DeltaVision images are presented as maximum intensity projections. (B) D471 cells harboring pAR291_DivIVA, pAR287_GFP-HT (non-specific control), or the empty vector pAR161, were grown in TH+Y+spec to OD600 0.5, and fractionated into supernatant, wall, and spheroplast pellet fractions. Samples were examined by Western blot using antibodies specific for M protein and SfbI.
Discussion
Anchoring of surface proteins to the cell wall of streptococci and staphylococci is divided into septal and peripheral anchoring pathways (Carlsson et al., 2006, DeDent et al., 2008). We used S. pyogenes M protein and SfbI as a model system to better understand the relations between the division cycle and protein anchoring through the two pathways, in an attempt to reach a more unified view of these processes. We took two general approaches: the first was largely descriptive and followed the two anchoring pathways throughout the cell cycle using population-level analysis, while the second tested how interference with the proper assembly of the division septum affects protein anchoring. We found that M protein and SfbI are anchored simultaneously throughout the cell cycle. M protein anchoring is strictly limited to the septum and occurs simultaneously at the mother and daughter septa at certain stages of the cell cycle. Conversely, SfbI is anchored in patches throughout the peripheral peptidoglycan, with some preference for the poles, and its accumulation over time results in polar distribution. Figure 7 summarizes these observations in a model form. We also found that sortase A is not required for localized translocation of the two proteins. Methicillin-induced unbalanced peptidoglycan synthesis, disrupts the proper assembly of the septum, and results in a marked reduction in surface M protein, but not SfbI. Overexpression of DivIVA also disrupts the septum, and results in a decrease in surface M protein, but an increase in SfbI.
The relations between the anchoring of surface proteins and the division cycle
The observed overlap in the anchoring of M protein at the mother and daughter septa of S. pyogenes is in agreement with a model proposing the simultaneous synthesis of peptidoglycan at these locations. Electron microscopy studies of cell division in enterococci revealed that initiation of peptidoglycan synthesis at future division sites begins before the mother septum is completely closed, and is independent of the completion of chromosome replication (Higgins & Shockman, 1976, Gibson et al., 1983). The localization pattern of the S. pneumoniae division factors FtsZ (Morlot et al., 2003), FtsA (Lara et al., 2005), and FtsW (Morlot et al., 2004), often shows simultaneous labeling at the mother and daughter septa. The labeling pattern of fluorescent vancomycin, which preferentially labels regions of lipid II export and cell wall synthesis, also shows simultaneous localization at the mother and daughter septa of both S. pneumoniae, and S. pyogenes (Daniel & Errington, 2003, Ng et al., 2004, Raz & Fischetti, 2008). The penicillin binding proteins PBP1b, PBP2a, PBP2b, and PBP2x, all show simultaneous localization at the mother and daughter septa of S. pneumoniae at certain stages of the cell cycle (Morlot et al., 2003, Morlot et al., 2004, Zapun et al., 2008). Examination of the locations where M protein is regenerated following protease digestion, has previously been used to study septal peptidoglycan synthesis (Cole & Hahn, 1962). While this method does not provide real-time results, in this work we have reduced the regeneration time to the bare minimum of 30 seconds, and analyzed the anchoring pattern of this protein in large populations of cells. Our finding that M protein is anchored simultaneously at the mother and daughter septa, strongly supports the model of simultaneous peptidoglycan synthesis at these locations in S. pyogenes.
In contrast to M protein, SfbI is anchored in patches to large peripheral regions of the peptidoglycan, with preference for the poles. The polar distribution of SfbI on S. pyogenes cells is the result of two processes: preference for anchoring at non-septal regions, and gradual accumulation on preassembled peptidoglycan, which results in a correlation between the amount of SfbI anchored and the pole's age. Our results also demonstrate that SfbI is actively anchored at the poles for at least two generations following their formation. Expanding on this basic model, it should be noted that the expression level of non-YSIRK-G/S proteins might also affect their observed distribution. A low expression level may result in substantial labeling only on poles that are far apart, while a high expression level may result in the anchoring of a substantial quantity of proteins even on relatively young poles.
Protein anchoring and the localization of sortase
M protein and SfbI are both anchored to the cell wall by sortase A (Barnett & Scott, 2002). We previously studied the distribution pattern of sortase in S. pyogenes and found that it localizes to a number of membrane-bound foci in each cell. Consistent with the anchoring pattern of M protein described here, sortase foci are preferentially associated with division septa, are recruited to the forming septa at an early stage, and are often present at both mother and daughter septa simultaneously (Raz & Fischetti, 2008). Despite their relative abundance at the division septa, sortase foci are not strictly confined there, and some foci are regularly seen at other cellular locations. One way of explaining the apparent bias in sortase distribution towards septal anchoring is that the streptococcal septum is constantly being split to form peripheral peptidoglycan, which can no longer facilitate the anchoring of YSIRK-G/S proteins, and therefore the time-window available for septal anchoring is very limited. The need for efficient anchoring at the septum is met through the coupling of protein anchoring to septal peptidoglycan synthesis, which ensures a high level of lipid II, PBPs, and possibly other septal factors. Localized translocation of M protein at the septum is also likely to play an important role in ensuring efficient anchoring (Carlsson et al., 2006). The higher prevalence of sortase foci at the septum is therefore likely to play a role in mediating efficient anchoring at this location. SfbI on the other hand, is anchored gradually over a relatively large cellular area, and its anchoring may therefore require a lower concentration of sortase.
The model presented here for the anchoring of surface proteins to the cell wall of S. pyogenes differs substantially from the model proposed for Streptococcus mutans (Hu et al., 2008) and E. faecalis (Kline et al., 2009). These studies showed that sortase A colocalizes with SecA in a single membranal microdomain termed ExPortal. The ExPortal, first described in S. pyogenes, is a membranal microdomain enriched in anionic lipids, in which the secretion related ATPase SecA and the membranal protease/chaperone HtrA localize, and facilitates the secretion and maturation of the streptococcal secreted protease SpeB (Rosch & Caparon, 2004, Rosch & Caparon, 2005, Rosch et al., 2007). A different study however, found that SecA is distributed throughout the streptococcal membrane (Carlsson et al., 2006), and the reason for this difference is not clear. Recently, S. pneumoniae SecA, and SecY were found to be dynamically localized to both the septum and periphery suggesting the absence of an ExPortal in this organism (Tsui et al., 2011). S. pneumoniae sortase A displays a punctate pattern, but does not show preferential distribution to the septum (Tsui et al., 2011). Further study is needed to fully understand the relation between protein translocation and anchoring in S. pyogenes, as well as the molecular mechanisms underlying the distinct anchoring patterns observed in different ovococci.
The effects of septum disruption on protein anchoring
To better understand the mechanisms governing the anchoring of M protein and SfbI to the wall of S. pyogenes, we interfered with two processes: protein sorting, and septum assembly. The deletion of sortase did not alter the cellular locations where M protein and SfbI were translocated, indicating that sortase does not play a role in determining the site of translocation. Conversely, interference with the placement of the septum through the use of methicillin or the overexpression of DivIVA resulted in an increase in cell size, a reduction in the number of septa, and a reduction in the amount of M protein found at the cell surface. The level of SfbI was only little changed following methicillin treatment, and was increased when DivIVA was overexpressed. These data suggest that a functioning septum is critical for the anchoring of YSIRK-G/S-type proteins. When septum formation is prevented, the anchoring of YSIRK-G/S-type proteins does not become delocalized but rather, these proteins are absent from the bacterial surface. The mechanism underlying this phenomenon is currently under investigation. While inhibition of M protein anchoring by the overexpression of DivIVA was linked to severe deformities in cellular morphology, the possibility that DivIVA is involved in the regulation of surface protein anchoring in a more direct manner should not be completely ruled out. Of particular note is that S. pneumoniae DivIVA is localized to both the division septum, and to a certain extent, the poles (Fadda et al., 2007).
One interesting phenomenon was the formation of rod-shaped cells displaying multiple septa, following exposure to an intermediate concentration of methicillin, similar to the effect observed in Lactococcus lactis (Perez-Nunez et al., 2011). In contrast to L. lactis and most other ovococci however, S. pyogenes lacks PBP2b, RodA, MreC and MreD homologues, which are important for peripheral peptidoglycan synthesis in ovococci (Zapun et al., 2008). This distinct peptidoglycan synthesis mechanism adds cell wall material at the splitting septum, and is responsible for the slightly elongated shape of many ovococci. Formation of S. pyogenes rod-shaped cells suggests therefore that the septal mechanism for peptidoglycan synthesis may be sufficient to facilitate the coccus-to-rod transformation. The specific molecular mechanisms involved in this alteration of S. pyogenes shape however, are not fully understood at present.
The formation of rod-shaped S. pyogenes cells provided us with an interesting model, in which to test the relations between surface protein anchoring and cell shape. We found that M protein was regularly anchored simultaneously at multiple septa along the rod, which were analogous to mother and daughter septa in untreated cells. Inhibition of septum closure therefore intensified the propensity to simultaneously anchor M protein at these locations. We also found that SfbI was not anchored solely at the poles of the rod, but was also anchored at the cylindrical inter-septal regions. This suggests that SfbI is not necessarily targeted to the poles as such, but rather, its anchoring pattern may best be described as exclusion from the septum. Interestingly, a distribution pattern of exclusion from the septum was also observed for the S. pneumoniae D,D-carboxypeptidase PBP3 (Morlot et al., 2004). Studying the manner in which such a distribution pattern is achieved may provide clues for the regulation of SfbI translocation. Further insight into the manner by which non-YSIRK-G/S-type proteins are anchored may come from studies dealing with rod-shaped bacteria such as L. monocytogenes (Bierne et al., 2004, Rafelski & Theriot, 2006, Bruck et al., 2011). A direct comparison however, may be complicated by the different manner peptidoglycan synthesis is regulated in these organisms.
Recently, a screen of S. aureus transposon integration library revealed that disruption of proteins containing an abortive infectivity (ABI) domain results in reduced expression level of YSIRK-G/S type surface proteins (analogous to S. pyogenes M protein), but does not affect the expression of proteins that do not contain this motif. Interestingly, these mutants also display thicker septal peptidoglycan, and it is not clear whether the reduction in septal protein expression is a direct effect or the result of defects in septal peptidoglycan synthesis (Frankel et al., 2010).
Conclusions
When considering the two distinct anchoring pathways, each appears to have its own unique characteristics and advantages. Septal anchoring allows efficient coating of the entire surface of the cell from the moment the peptidoglycan is formed. Immediate and extensive coating is likely to be important for the proper function of M protein and possibly other septum-anchored proteins. Consider that anchoring of M protein through the peripheral anchoring pathway may result in large areas of the streptococcal cell wall being devoid of this molecule at any given time. In the absence of M protein and other such molecules, these areas may be subject to opsonization, leading to the elimination of these bacteria through phagocytosis (Perez-Casal et al., 1992). Peripheral anchoring of SfbI on the other hand has the unique characteristic of creating cellular polarity. The extent to which the polar localization of virulence factors is important for the pathogenic process and survival of S. pyogenes is not clear at present. It was shown however, that polar distribution of SfbI resulted in reciprocal clustering of integrins on the surface of host cells (Ozeri et al., 2001), and future studies are likely to uncover additional examples. The ends of a streptococcal chain may have a better opportunity for contact with the host, and thus a higher concentration of certain virulence factors at these locations may be advantageous. Polar localization of proteins is a recurring theme in Gram-negative and Gram-positive bacteria, and has been shown to play a role in the general functions of the bacteria, as well as in pathogenesis (Shapiro et al., 2002, Shapiro et al., 2009, Rudner & Losick, 2010).
The data presented here suggest that factors related to the division ring are likely to play a role in promoting the anchoring of M protein. Over the recent years, the function of many of these division factors has been elucidated (Zapun et al., 2008, Adams & Errington, 2009, Shapiro et al., 2009, Rudner & Losick, 2010), and this knowledge may facilitate direct examination of their importance for surface protein anchoring. Defining the factors critical for the proper regulation of protein sorting could yield novel targets for the development of anti-infective agent, since pathogens lacking surface proteins are greatly impaired in their ability to cause disease (Marraffini et al., 2006, Maresso & Schneewind, 2008).
Experimental procedures
Bacterial strains and culture conditions
Escherichia coli strain DH5α was used for molecular cloning and recombinant protein expression. S. pyogenes strain D471 (an M6 serotype) was from the Rockefeller University collection. AR01 is a srtA knockout strain derived from D471 (Raz & Fischetti, 2008). E. coli was grown in Luria-Bertani (LB) medium, supplemented with 100 μg/ml ampicillin, when needed. S. pyogenes strains were grown in Todd-Hewitt medium (Difco) supplemented with 1% yeast extract (Fisher Scientific) at 37°C. Erythromycin was used at 15 μg/ml for S. pyogenes, and spectinomycin was used at 20 μg/ml for E. coli and 120 μg/ml for S. pyogenes when appropriate.
Reagents and antibodies
The M protein specific 10B6 monoclonal antibody (Jones et al., 1985) was used at a 1:10,000 dilution for Western blot analysis, and 1:1000 for immunofluorescence. SfbI-specific rabbit serum and pre-immune serum (Molinari et al., 1997), were used at a 1:1000 dilution. Goat anti-mouse IgG conjugated to either Rhodamine red (Jackson ImmunoResearch), or FITC (Sigma), were used at 1:1000. Goat anti-rabbit IgG, conjugated to either FITC (Sigma) or Alexa Fluor 647 (Invitrogen) were used at 1:1000. Wheat germ agglutinin (WGA) Marina Blue conjugate (Invitrogen) was used at 5 μg/ml. DAPI (Sigma) was used at 1 μg/ml. Vancomycin was conjugated to NHS Rhodamine red according to manufacturer's instructions (Thermo Scientific), and separated from unbound dye by thin layer chromatography. GFP-PlyC/BD (see below) was used at a final concentration of 50 μg/ml. All other reagents were purchased from Sigma unless otherwise noted.
DNA manipulation
Standard procedures were used for DNA manipulation and for E. coli transformations (Sambrook et al., 1989). Transformation of S. pyogenes was performed according to Perez-Casal (Perez-Casal et al., 1991). Plasmid DNA was isolated using QIAprep spin miniprep kit (Qiagen). PCR amplification procedures were performed using either Vent or Phusion DNA polymerase (New England Biolabs). Oligonucleotides were from Eurofins. Restriction enzymes were from New England Biolabs. T4 DNA ligase was from Invitrogen.
Construction of the plasmids
For the construction of the GFP-PlyC/BD expression vector, pAR159, the binding domain of the PlyC phage lysin was amplified using the following primers : 5 _ PlyC-BD_XbaI (5'-GGCTCTAGAATGAGCAAGATTAATGTAAACGTAGAAAATG-3') 3 _ PlyC-BD_PstI (5'-CGCCTGCAGTTACTTTTTCATAGCCTTTCTGATAGCC-3'), and inserted into the XbaI and PstI sites of pBAD24 (Guzman et al., 1995). Primers 5_H6_GFP_EcoRI (5'-CGCGAATTCATGAGTAAAGGAGAACTTCATCATCATCATCATCATTCCTCCGCCATGAGTAAAG GAGAAGAACTTTTC-3') and 3_GFP_KpnI (5'-GAGGGTACCTTTGTATAGTTCATCCATGCC-3') were used to amplify the GFP_mut2 gene (Cormack et al., 1996). An N-terminal hexahistidine tag is encoded on the upstream primer. This PCR product was inserted into the EcoRI and KpnI sites of the above plasmid, yielding pAR159.
A modified version of the S. pyogenes shuttle vector pLZ12-spec (Husmann et al., 1995) was constructed by replacing the original multiple cloning site (MCS) between the EcoRI and SphI sites, with a DNA fragment formed by aligning primers 5 _ new _ plz MCS (5'-AATTCCCCAAGCTTCCCAGATCTAAACCGCGGAAACAGCTGAAACCATGGAAAGCATG-3') and 3 _ new _ plz MCS (5'-CTTTCCATGGTTTCAGCTGTTTCCGCGGTTTAGATCTGGGAAGCTTGGGG-3'), which contain the new MCS (EcoRI-HinDIII-BglII-SacII-PvuII-NcoI-SphI). The resulting plasmid, termed pAR161, was used for the construction of pAR287_GFP-HT and pAR291_DivIVA.
For the construction of pAR287_GFP-HT, The HaloTag gene was amplified from pFN18A HaloTag T7 Flexi Vector (Promega) using primers 5_HaloL1_SacII (5'-CCCCCGCGGGGTGCATCTGCCGGCATGGCAGAAATCGGTACTGGC-3') and 3_Halo_NcoI (5'-CCCCCATGGCTATCAGCCGGAAATCTCGAGCGTC-3'), and inserted into the SacII and NcoI sites of pAR161. The GFP_mut2 gene (Cormack et al., 1996) was amplified using primers 5 _ GFP _ BglII (5'-GAGAGATCTATGAGTAAAGGAGAAGAACTTTTC-3') and 3 _ GFP _ SacII (5'-GAGCCGCGGTTTGTATAGTTCATCCATGCC-3'), and inserted into the BglII and SacII sites of the resulting plasmid. Finally, the M protein upstream region was amplified from genomic D471 DNA using primers 5 _ Mp _ UTR _ EcoRI (5'-CGCGAATTCACAGCCTAGCCGCAGAAACTC-3') and 3 _ Mp _ UTR _ BglII (5'-CCCAGATCTGCTCCTTATGTTATCATTTTTTAGG-3'), and inserted into the EcoRI and BglII sites, yielding pAR287_GFP-HT.
The construction of pAR291_DivIVA begun with a derivate of pAR161 containing a myc-tag (that is not expressed in the final construct), formed by aligning primers 5_myc_tag (5'-GGGAACAAAAACTTATTTCTGAAGAAGACCTGTAGC-3') and 3_myc_tag (5'-CATGGCTACAGGTCTTCTTCAGAAATAAGTTTTTGTTCCCGC-3'), and inserting the resulting double stranded DNA fragment into the SacII and NcoI sites pAR161. The M protein upstream region was inserted into the EcoRI and BglII sites of this plasmid as described for pAR287_GFP-HT. The DivIVA gene, including a stop codon, was amplified from the genome of D471 using primers 5_DivIVA_BamHI (5'-CCCGGATCCATGGCACTTACAACGCTAGAAATTAAAG-3') and 3_DivIVA_SacII (5'-CCCCCGCGGTTAGATATTTAATTTAAACGTTTGTGTTTCACTGAG-3'), and inserted into the BamHI and SacII sites of the resulting plasmid, yielding pAR291_DivIVA.
Purification of GFP-PlyC/BD
An overnight culture of E. coli DH5α harboring pAR159 was diluted 1:100 into one liter of LB supplemented with ampicillin, grown to OD600 0.5, and induced with 0.2% L-arabinose at room temperature for 5 hours. The construct was purified on a NiNTA column as previously described (Raz & Fischetti, 2008). The eluted fraction was concentrated using an Amicon Ultra centrifugal filter device with a cutoff limit of 5 kDa, and the buffer was changed to PBS by repeated cycles of dilution in PBS and volume reduction. The final protein concentration was 0.85 mg/ml.
Fractionation of S. pyogenes cells and Western blot analysis
Fractionation of the cells, and Western blot analysis were carried out as previously described (Raz & Fischetti, 2008). Samples were normalized to account for slight variations in OD. Fractionation by boiling in SDS was carried out as follows: One milliliter of culture at OD600 0.5 was harvested and washed with 30 mM Tris, pH 6.3. The bacterial pellet was suspended in 50 μl of 2% SDS, boiled for 10 minutes, and centrifuged for 2 minutes at 16000 rcf. The supernatant, containing non-covalently bound proteins, was supplemented with 12 μl of 5× SDS loading buffer. The cell pellet, containing covalently anchored proteins, was washed with 200 μl deionized water, suspended in 50 μl of 30 mM Tris, pH 6.3, containing 300 U/ml PlyC (Nelson et al., 2006) for 15 minutes at room temperature, and then supplemented with 12 μl 5× SDS loading buffer.
Regeneration of surface proteins following protease treatment
For protein regeneration studies, overnight cultures were diluted 1:100 in TH+Y containing 0.35 mg/ml trypsin (Sigma) and 0.04 mg/ml pronase (Sigma). Unless otherwise noted, the cells were harvested upon reaching OD600 0.5 by one-minute centrifugation at 16,000 rcf, resuspended in TH+Y without proteases, and immediately spun again. The cells were immediately resuspended in medium without proteases, and incubated at 37°C for the stated amount of time. Each experiment typically consisted of three to four repeats, and was performend several times.
Fluorescent microscopy
Fluorescent microscopy procedures were carried out as previously described (Raz & Fischetti, 2008); however, the membrane and cell wall permeabilization steps were omitted since the antigens studied are exposed on the bacterial surface. Immunofluorescence experiments involving the use of methicillin or the overexpression of DivIVA were performed using polyclonal SfbI sera pre-adsorbed with the M1 serotype S. pyogenes strain SF370, which lacks a sfbI gene (Ferretti et al., 2001). In those cases the cells were incubated sequentially with the M-protein-specific mouse monoclonal 10B6, anti-mouse Rhodamine red, rabbit anti-SfbI, and anti-rabbit FITC antibodies, for improved signal over background. Structured Illumination microscopy was performed on a DeltaVision OMX Blaze 3D-Structured Illumination Microscopy (3D-SIM) system (Applied Precision) fitted with an Olympus 100×/1.40 NA UPLSAPO objective, Photometrics Evolve EMCCD cameras, and 405, 488 and 568 lasers. 3D-SIM reconstruction and channel alignment were performed using the SoftWoRx algorithms and reconstructed images were exported as maximum projections.
Analysis of protein anchoring using MATLAB
For the purpose of signal analysis, average intensity projections were produced containing the signal data from all the Z-sections, using SoftWoRx (Applied Precision). These projections were converted into tiff format using ImageJ (http://rsb.info.nih.gov/ij/), and the signal distribution data were subsequently analyzed using MetaMorph offline (64-bit) version 7.7.5.0 (Molecular Devices). In each image, all the cells whose growth axis paralleled with the slide (and therefore the septum was perpendicular to the slide), and that presented no signal interference from adjacent cells, were analyzed. Each cell was confined in a rectangle whose long dimension parallels with the cell's growth axis. To analyze the fluorescence intensity of the antigens in different cellular regions, each rectangle was then sub-divided into pixel-wide cross-sections, and the total fluorescence of each section was calculated. These raw fluorescence distribution data were further analyzed using MATLAB version 7.6.0 R2008a (MathWorks). To create cell-specific plots the fluorescence intensity of each section was plotted as a function of its cellular position, represented as the distance from the younger of the two poles, or mother septum. Population plots were obtained by aligning all the cells in each group, and calculating the mean fluorescence signal and standard deviation values for each cellular position.
For the analysis of M protein anchoring at the mother and daughter septa, presented in Figure 3, each cell was divided into 4 regions: (1) mother septum, defined as the 0.13 μm region (3 pixels wide) adjacent to the previous division site (on the left); (2) daughter septum, defined as the 0.21 μm region (5 pixels wide) at the middle of the cell; (3) inter-septal region, located between the mother and daughter septa; and (4) old pole, defined as the remainder of the cell (see Figure 2B). A mother septum region was defined as positive for the anchoring of M protein if it passed a threshold of 200 fluorescence units, and displayed double the minimal signal of the inter-septal region. A daughter septum region was defined as positive for M protein anchoring if, in addition to the above conditions, it also displayed double the minimal signal of the polar region. In the youngest cell population, resolution constraints sometime resulted in protrusion of the mother septum fluorescent signal into the inter-septal region, preventing the achievement of the two-fold fluorescence difference required for the recognition of mother septum anchoring. To address this situation, cells that were negative for M protein anchoring using the above definitions, but passed the fluorescence threshold and displayed 5-fold mother septum fluorescence compared to the old pole, were defined as positive for mother septum anchoring.
For the analysis of SfbI signal distribution following the GFP-PlyC/BD “pulse-chase” experiment, the signal distribution of the different fluorescent channels was acquired as described above. A 5-pixel-wide region in the middle of the cell was left unanalyzed, while the regions on both sides were defined as “young pole” and “old pole” according to the GFP-PlyC/BD distribution along the chain. The average fluorescence intensity of each channel was calculated for each pole. The code used for all the MATLAB operations is available upon request.
Supplementary Material
Acknowledgements
We are grateful to Sung Lee for valuable advice and support, and to Aurélia Delauné, Rolf Lood, and Bryan Utter for critically reading the manuscript. We thank Alison North, Tao Tong, and other members of the Rockefeller University Bio-Imaging resource center for their comments and advice regarding the immunofluorescent imagery. This work was supported by U.S. Public Health Service Grant AI11822 (to V.A.F.).
Abbreviations
- WGA
Wheat Germ Agglutinin.
- PBP
Penicillin Binding Protein.
- RFU
Relative Fluorescence Units.
- 3D-SIM
3D Structured Illumination Microscopy.
Contributor Information
Assaf Raz, Rockefeller University, Bacterial Pathogenesis and Immunology araz@rockefeller.edu.
Susanne Talay, Helmholtz Centre for Infection Research, Department of Medical Microbiology.
Vincent Fischetti, Rockefeller University, Bacterial Pathogenesis and Immunology.
References
- Adams DW, Errington J. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol. 2009;7:642–653. doi: 10.1038/nrmicro2198. [DOI] [PubMed] [Google Scholar]
- Bae T, Schneewind O. The YSIRK-G/S motif of staphylococcal protein A and its role in efficiency of signal peptide processing. J Bacteriol. 2003;185:2910–2919. doi: 10.1128/JB.185.9.2910-2919.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett TC, Patel AR, Scott JR. A Novel Sortase, SrtC2, from Streptococcus pyogenes Anchors a Surface Protein Containing a QVPTGV Motif to the Cell Wall. The Journal of Bacteriology. 2004;186:5865–5875. doi: 10.1128/JB.186.17.5865-5875.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett TC, Scott JR. Differential recognition of surface proteins in Streptococcus pyogenes by two sortase gene homologs. J Bacteriol. 2002;184:2181–2191. doi: 10.1128/JB.184.8.2181-2191.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne H, Garandeau C, Pucciarelli MG, Sabet C, Newton S, Garcia-del Portillo F, Cossart P, Charbit A. Sortase B, a new class of sortase in Listeria monocytogenes. J Bacteriol. 2004;186:1972–1982. doi: 10.1128/JB.186.7.1972-1982.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisno AL, Brito MO, Collins CM. Molecular basis of group A streptococcal virulence. Lancet Infect Dis. 2003;3:191–200. doi: 10.1016/s1473-3099(03)00576-0. [DOI] [PubMed] [Google Scholar]
- Bruck S, Personnic N, Prevost MC, Cossart P, Bierne H. Regulated shift from helical to polar localization of Listeria monocytogenes cell wall-anchored proteins. J Bacteriol. 2011;193:4425–4437. doi: 10.1128/JB.01154-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group A streptococcal diseases. The Lancet Infectious Diseases. 2005;5:685–694. doi: 10.1016/S1473-3099(05)70267-X. [DOI] [PubMed] [Google Scholar]
- Carlsson F, Stålhammar-Carlemalm M, Flärdh K, Sandin C, Carlemalm E, Lindahl G. Signal sequence directs localized secretion of bacterial surface proteins. Nature. 2006;442:943–946. doi: 10.1038/nature05021. [DOI] [PubMed] [Google Scholar]
- Cha JH, Stewart GC. The divIVA minicell locus of Bacillus subtilis. J Bacteriol. 1997;179:1671–1683. doi: 10.1128/jb.179.5.1671-1683.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole RM, Hahn JJ. Cell wall replication in Streptococcus pyogenes. Science. 1962;135:722–724. doi: 10.1126/science.135.3505.722. [DOI] [PubMed] [Google Scholar]
- Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- Cunningham MW. Pathogenesis of group A streptococcal infections. Clin Microbiol Rev. 2000;13:470–511. doi: 10.1128/cmr.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel RA, Errington J. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell. 2003;113:767–776. doi: 10.1016/s0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- DeDent A, Bae T, Missiakas DM, Schneewind O. Signal peptides direct surface proteins to two distinct envelope locations of Staphylococcus aureus. EMBO J. 2008;27:2656–2668. doi: 10.1038/emboj.2008.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards DH, Errington J. The Bacillus subtilis DivIVA protein targets to the division septum and controls the site specificity of cell division. Mol Microbiol. 1997;24:905–915. doi: 10.1046/j.1365-2958.1997.3811764.x. [DOI] [PubMed] [Google Scholar]
- Fadda D, Pischedda C, Caldara F, Whalen MB, Anderluzzi D, Domenici E, Massidda O. Characterization of divIVA and other genes located in the chromosomal region downstream of the dcw cluster in Streptococcus pneumoniae. J Bacteriol. 2003;185:6209–6214. doi: 10.1128/JB.185.20.6209-6214.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadda D, Santona A, D'Ulisse V, Ghelardini P, Ennas MG, Whalen MB, Massidda O. Streptococcus pneumoniae DivIVA: localization and interactions in a MinCD-free context. J Bacteriol. 2007;189:1288–1298. doi: 10.1128/JB.01168-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti JJ, McShan WM, Ajdic D, Savic DJ, Savic G, Lyon K, Primeaux C, Sezate S, Suvorov AN, Kenton S, Lai HS, Lin SP, Qian Y, Jia HG, Najar FZ, Ren Q, Zhu H, Song L, White J, Yuan X, Clifton SW, Roe BA, McLaughlin R. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc Natl Acad Sci U S A. 2001;98:4658–4663. doi: 10.1073/pnas.071559398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischetti VA, Pancholi V, Schneewind O. Conservation of a hexapeptide sequence in the anchor region of surface proteins from gram-positive cocci. Mol Microbiol. 1990;4:1603–1605. doi: 10.1111/j.1365-2958.1990.tb02072.x. [DOI] [PubMed] [Google Scholar]
- Frankel MB, Wojcik BM, DeDent AC, Missiakas DM, Schneewind O. ABI domain-containing proteins contribute to surface protein display and cell division in Staphylococcus aureus. Mol Microbiol. 2010;78:238–252. doi: 10.1111/j.1365-2958.2010.07334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson CW, Daneo-Moore L, Higgins ML. Cell wall assembly during inhibition of DNA synthesis in Streptococcus faecium. J Bacteriol. 1983;155:351–356. doi: 10.1128/jb.155.1.351-356.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutmann L, Williamson R, Tomasz A. Physiological properties of penicillin-binding proteins in group A streptococci. Antimicrob Agents Chemother. 1981;19:872–880. doi: 10.1128/aac.19.5.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanski E, Caparon M. Protein F, a fibronectin-binding protein, is an adhesin of the group A streptococcus Streptococcus pyogenes. Proc Natl Acad Sci U S A. 1992;89:6172–6176. doi: 10.1073/pnas.89.13.6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickx AP, Budzik JM, Oh SY, Schneewind O. Architects at the bacterial surface - sortases and the assembly of pili with isopeptide bonds. Nat Rev Microbiol. 2011;9:166–176. doi: 10.1038/nrmicro2520. [DOI] [PubMed] [Google Scholar]
- Higgins ML, Shockman GD. Study of cycle of cell wall assembly in Streptococcus faecalis by three-dimensional reconstructions of thin sections of cells. J Bacteriol. 1976;127:1346–1358. doi: 10.1128/jb.127.3.1346-1358.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P, Bian Z, Fan M, Huang M, Zhang P. Sec translocase and sortase A are colocalised in a locus in the cytoplasmic membrane of Streptococcus mutans. Arch Oral Biol. 2008;53:150–154. doi: 10.1016/j.archoralbio.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Husmann LK, Scott JR, Lindahl G, Stenberg L. Expression of the Arp protein, a member of the M protein family, is not sufficient to inhibit phagocytosis of Streptococcus pyogenes. Infect Immun. 1995;63:345–348. doi: 10.1128/iai.63.1.345-348.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KF, Manjula BN, Johnston KH, Hollingshead SK, Scott JR, Fischetti VA. Location of variable and conserved epitopes among the multiple serotypes of streptococcal M protein. J Exp Med. 1985;161:623–628. doi: 10.1084/jem.161.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline KA, Kau AL, Chen SL, Lim A, Pinkner JS, Rosch J, Nallapareddy SR, Murray BE, Henriques-Normark B, Beatty W, Caparon MG, Hultgren SJ. Mechanism for sortase localization and the role of sortase localization in efficient pilus assembly in Enterococcus faecalis. J Bacteriol. 2009;191:3237–3247. doi: 10.1128/JB.01837-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara B, Rico AI, Petruzzelli S, Santona A, Dumas J, Biton J, Vicente M, Mingorance J, Massidda O. Cell division in cocci: localization and properties of the Streptococcus pneumoniae FtsA protein. Mol Microbiol. 2005;55:699–711. doi: 10.1111/j.1365-2958.2004.04432.x. [DOI] [PubMed] [Google Scholar]
- Lenarcic R, Halbedel S, Visser L, Shaw M, Wu LJ, Errington J, Marenduzzo D, Hamoen LW. Localisation of DivIVA by targeting to negatively curved membranes. EMBO J. 2009;28:2272–2282. doi: 10.1038/emboj.2009.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lleo MM, Canepari P, Satta G. Bacterial cell shape regulation: testing of additional predictions unique to the two-competing-sites model for peptidoglycan assembly and isolation of conditional rod-shaped mutants from some wild-type cocci. J Bacteriol. 1990;172:3758–3771. doi: 10.1128/jb.172.7.3758-3771.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresso AW, Schneewind O. Sortase as a target of anti-infective therapy. Pharmacol Rev. 2008;60:128–141. doi: 10.1124/pr.107.07110. [DOI] [PubMed] [Google Scholar]
- Marraffini LA, Dedent AC, Schneewind O. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol Mol Biol Rev. 2006;70:192–221. doi: 10.1128/MMBR.70.1.192-221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston AL, Errington J. Selection of the midcell division site in Bacillus subtilis through MinD-dependent polar localization and activation of MinC. Mol Microbiol. 1999;33:84–96. doi: 10.1046/j.1365-2958.1999.01450.x. [DOI] [PubMed] [Google Scholar]
- Mazmanian SK, Liu G, Ton-That H, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- Molinari G, Talay SR, Valentin-Weigand P, Rohde M, Chhatwal GS. The fibronectin-binding protein of Streptococcus pyogenes, SfbI, is involved in the internalization of group A streptococci by epithelial cells. Infect Immun. 1997;65:1357–1363. doi: 10.1128/iai.65.4.1357-1363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora M, Bensi G, Capo S, Falugi F, Zingaretti C, Manetti AG, Maggi T, Taddei AR, Grandi G, Telford JL. Group A Streptococcus produce pilus-like structures containing protective antigens and Lancefield T antigens. Proc Natl Acad Sci U S A. 2005;102:15641–15646. doi: 10.1073/pnas.0507808102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morlot C, Noirclerc-Savoye M, Zapun A, Dideberg O, Vernet T. The D,D-carboxypeptidase PBP3 organizes the division process of Streptococcus pneumoniae. Mol Microbiol. 2004;51:1641–1648. doi: 10.1046/j.1365-2958.2003.03953.x. [DOI] [PubMed] [Google Scholar]
- Morlot C, Zapun A, Dideberg O, Vernet T. Growth and division of Streptococcus pneumoniae: localization of the high molecular weight penicillin-binding proteins during the cell cycle. Mol Microbiol. 2003;50:845–855. doi: 10.1046/j.1365-2958.2003.03767.x. [DOI] [PubMed] [Google Scholar]
- Nelson D, Schuch R, Chahales P, Zhu S, Fischetti VA. PlyC: a multimeric bacteriophage lysin. Proc Natl Acad Sci U S A. 2006;103:10765–10770. doi: 10.1073/pnas.0604521103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng WL, Kazmierczak KM, Winkler ME. Defective cell wall synthesis in Streptococcus pneumoniae R6 depleted for the essential PcsB putative murein hydrolase or the VicR (YycF) response regulator. Mol Microbiol. 2004;53:1161–1175. doi: 10.1111/j.1365-2958.2004.04196.x. [DOI] [PubMed] [Google Scholar]
- Nobbs AH, Lamont RJ, Jenkinson HF. Streptococcus Adherence and Colonization. Microbiol. Mol. Biol. Rev. 2009;73:407–450. doi: 10.1128/MMBR.00014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozeri V, Rosenshine I, Ben-Ze'Ev A, Bokoch GM, Jou TS, Hanski E. De novo formation of focal complex-like structures in host cells by invading Streptococci. Mol Microbiol. 2001;41:561–573. doi: 10.1046/j.1365-2958.2001.02535.x. [DOI] [PubMed] [Google Scholar]
- Perez-Casal J, Caparon MG, Scott JR. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J Bacteriol. 1991;173:2617–2624. doi: 10.1128/jb.173.8.2617-2624.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Casal J, Caparon MG, Scott JR. Introduction of the emm6 gene into an emm-deleted strain of Streptococcus pyogenes restores its ability to resist phagocytosis. Res Microbiol. 1992;143:549–558. doi: 10.1016/0923-2508(92)90112-2. [DOI] [PubMed] [Google Scholar]
- Perez-Nunez D, Briandet R, David B, Gautier C, Renault P, Hallet B, Hols P, Carballido-Lopez R, Guedon E. A new morphogenesis pathway in bacteria: unbalanced activity of cell wall synthesis machineries leads to coccus-to-rod transition and filamentation in ovococci. Mol Microbiol. 2011;79:759–771. doi: 10.1111/j.1365-2958.2010.07483.x. [DOI] [PubMed] [Google Scholar]
- Perry AM, Ton-That H, Mazmanian SK, Schneewind O. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. III. Lipid II is an in vivo peptidoglycan substrate for sortase-catalyzed surface protein anchoring. J Biol Chem. 2002;277:16241–16248. doi: 10.1074/jbc.M109194200. [DOI] [PubMed] [Google Scholar]
- Pucci MJ, Hinks ET, Dicker DT, Higgins ML, Daneo-Moore L. Inhibition of beta-lactam antibiotics at two different times in the cell cycle of Streptococcus faecium ATCC 9790. J Bacteriol. 1986;165:682–688. doi: 10.1128/jb.165.3.682-688.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafelski SM, Theriot JA. Mechanism of polarization of Listeria monocytogenes surface protein ActA. Mol Microbiol. 2006;59:1262–1279. doi: 10.1111/j.1365-2958.2006.05025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthi KS, Losick R. Negative membrane curvature as a cue for subcellular localization of a bacterial protein. Proc Natl Acad Sci U S A. 2009;106:13541–13545. doi: 10.1073/pnas.0906851106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Arcos S, Liao M, Marthaler S, Rigden M, Dillon JA. Enterococcus faecalis divIVA: an essential gene involved in cell division, cell growth and chromosome segregation. Microbiology. 2005;151:1381–1393. doi: 10.1099/mic.0.27718-0. [DOI] [PubMed] [Google Scholar]
- Raz A, Fischetti VA. Sortase A localizes to distinct foci on the Streptococcus pyogenes membrane. Proc Natl Acad Sci U S A. 2008;105:18549–18554. doi: 10.1073/pnas.0808301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosch J, Caparon M. A microdomain for protein secretion in Gram-positive bacteria. Science. 2004;304:1513–1515. doi: 10.1126/science.1097404. [DOI] [PubMed] [Google Scholar]
- Rosch JW, Caparon MG. The ExPortal: an organelle dedicated to the biogenesis of secreted proteins in Streptococcus pyogenes. Mol Microbiol. 2005;58:959–968. doi: 10.1111/j.1365-2958.2005.04887.x. [DOI] [PubMed] [Google Scholar]
- Rosch JW, Hsu FF, Caparon MG. Anionic lipids enriched at the ExPortal of Streptococcus pyogenes. J Bacteriol. 2007;189:801–806. doi: 10.1128/JB.01549-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudner DZ, Losick R. Protein subcellular localization in bacteria. Cold Spring Harb Perspect Biol. 2010;2:a000307. doi: 10.1101/cshperspect.a000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 1989. [Google Scholar]
- Schneewind O, Model P, Fischetti VA. Sorting of protein A to the staphylococcal cell wall. Cell. 1992;70:267–281. doi: 10.1016/0092-8674(92)90101-h. [DOI] [PubMed] [Google Scholar]
- Shapiro L, McAdams HH, Losick R. Generating and exploiting polarity in bacteria. Science. 2002;298:1942–1946. doi: 10.1126/science.1072163. [DOI] [PubMed] [Google Scholar]
- Shapiro L, McAdams HH, Losick R. Why and how bacteria localize proteins. Science. 2009;326:1225–1228. doi: 10.1126/science.1175685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirig T, Weiner EM, Clubb RT. Sortase enzymes in Gram-positive bacteria. Mol Microbiol. 2011;82:1044–1059. doi: 10.1111/j.1365-2958.2011.07887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson J, Hsu KC, Gotschlich EC. Electron microscopic studies on streptococci. I. M antigen. J Exp Med. 1969;130:1063–1091. doi: 10.1084/jem.130.5.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talay SR, Valentin-Weigand P, Jerlstrom PG, Timmis KN, Chhatwal GS. Fibronectin-binding protein of Streptococcus pyogenes: sequence of the binding domain involved in adherence of streptococci to epithelial cells. Infect Immun. 1992;60:3837–3844. doi: 10.1128/iai.60.9.3837-3844.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton-That H, Schneewind O. Anchor structure of staphylococcal surface proteins. IV. Inhibitors of the cell wall sorting reaction. J Biol Chem. 1999;274:24316–24320. doi: 10.1074/jbc.274.34.24316. [DOI] [PubMed] [Google Scholar]
- Tsui HC, Keen SK, Sham LT, Wayne KJ, Winkler ME. Dynamic distribution of the SecA and SecY translocase subunits and septal localization of the HtrA surface chaperone/protease during Streptococcus pneumoniae D39 cell division. mBio. 2011;2(5):e00202–11. doi: 10.1128/mBio.00202-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson R, Hakenbeck R, Tomasz A. In vivo interaction of beta-lactam antibiotics with the penicillin-binding proteins of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1980;18:629–637. doi: 10.1128/aac.18.4.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapun A, Vernet T, Pinho MG. The different shapes of cocci. FEMS Microbiol Rev. 2008;32:345–360. doi: 10.1111/j.1574-6976.2007.00098.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.