Abstract
Somatic mutations in mitochondrial DNA (mtDNA) are hypothesized to play a role in Parkinson disease (PD), but large increases in mtDNA mutations have not previously been found in PD, potentially because neurons with high mutation levels degenerate and thus are absent in late-stage tissue. To address this issue, we studied early stage PD cases and cases of incidental Lewy body disease (ILBD), which is thought to represent presymptomatic PD. We show for the first time that mtDNA mutation levels in substantia nigra (SN) neurons are significantly elevated in this group of early PD and ILBD cases.
Introduction
Mitochondrial dysfunction and oxidative stress have been implicated in the pathogenesis of PD.1 Complex I inhibitors cause parkinsonism,2 complex I deficiency is observed in PD,3-5 and this complex I defect can be transferred to cell lines by expressing mtDNA from PD patients,6 implicating a role for mtDNA mutations. However, inherited mtDNA mutations associated with parkinsonism7, 8 are rare, and levels of somatic (acquired) mtDNA point mutations9-11 or deletions12 in SN differ by less than 10% between PD and age-matched controls.
A major limitation of prior studies is that post-mortem tissue from known PD cases typically represents late stage disease. The few SN neurons still present late in disease may be those that tend to accumulate fewer somatic mtDNA mutations, while neurons that accumulate high levels of mutations may be the ones that degenerate earlier in the disease course. We therefore examined somatic mtDNA point mutations in SN neurons from early stage PD cases and cases with incidental Lewy body disease (ILBD), which may represent an early presymptomatic stage of PD,13, 14 compared to late stage PD cases or controls.
Subjects and Methods
Human samples
Frozen unfixed human postmortem brain tissue was obtained from the Harvard Brain Tissue Resource Center (McLean Hospital, Boston, MA, USA), the Neuropathology Core of the Massachusetts Alzheimer Disease Research Center and the Neuropathology Service of the Massachusetts General Hospital (Boston, MA, USA), the German Brain-Net Tissue Bank (Center for Neuropathology and Prion Research, Ludwig-Maximillians University, Munich, Germany), and the Queen Square Brain Bank for Neurological Disorders (University College London, Institute of Neurology, Queen Square, London, UK). Data derived from different brain banks were pooled after determining that controls from all sources showed similar mutation levels. A subset of the D-loop data from controls reported here have been reported previously.15 ILBD was defined by the absence of clinical parkinsonism or dementia but Lewy bodies present in the SN (Braak stage 3).16 Controls had no clinical history or pathologic evidence of neurodegeneration.
Laser capture microdissection
The Arcturus PixCell II LCM system (Mountain View, CA) was used to isolate single SN neurons from 8-12 μm unfixed frozen methylene blue stained cryostat sections. SN pars compacta neurons were identified based on larger size, morphology, presence of a nucleolus, and neuromelanin pigment. Glia were identified by smaller size, morphology, and lack of a nucleolus. Single cells or groups of cells were isolated on a high efficiency thermoplastic ethylene vinyl acetate (EVA) transfer cap, and DNA was extracted as previously described15 by exposure overnight to an extraction solution containing proteinase K. The proteinase K was then inactivated by incubating the sample at 95 C for 10 minutes. For single cell studies, this 10 μL solution was used directly as template in PCR reactions, thereby minimizing DNA loss or contamination risk that might occur with further purification steps.
Mutation analyses
To detect mtDNA point mutations, mtDNA was amplified using a high-fidelity PCR protocol followed by TA cloning of the PCR product, and selection of individual clones for sequencing. The method and primers have been reported previously,15 with extensive controls for artifacts including PCR error, nuclear pseudogenes, and contamination.10, 17 A minimum of 15,000 base pairs were sequenced for each sample, and the mutation level was expressed as mutations per million base pairs. Only heteroplasmic variants were counted. Several measures were taken to minimize the risk of contaminant DNA influencing the results. First, if fortuitous homoplasmic marker polymorphisms were found in all clones from a large block of cells, we required that >95% of clones from an LCM-isolated cell sample have the marker polymorphisms. Second, if a known polymorphism reported at http://www.mitomap.org was present in >20% of clones from a single cell, but not from a large group of cells from the same subject, that cell was completely excluded from analysis. Although new mutations could arise at the site of known polymorphisms, we considered contamination more likely, and preferred to err on the side of caution. Based on these criteria, 10 neuronal samples were excluded from analysis (7 from controls and 3 from early PD+ILBD), and one homoplasmic variant in one cell was not counted. Although the number of control samples excluded is higher than for early PD+ILBD, this study included 23 controls and only 9 early PD+ILBD cases, and so the percentage of samples that met our pre-specified criteria for exclusion was similar in both groups. Among glial samples, 2 were excluded (1 control and 1 early PD+ILBD).
Statistical analyses
Overall mutation levels and oxidative mutation levels were fit using linear mixed effects models, with random effects for subjects and fixed effects for age, post-mortem interval, disease state (control, early, late), cell type (neuron, glia, mixed), and gene analyzed (ND5, D-loop) (SAS, proc mixed, Version 9.1, SAS Institute Inc., Cary, NC). All pairwise interactions among disease state, cell type, and gene analyzed were considered initially; only the disease state x cell type interaction was significant (p<0.05) and was retained in the final analysis. Histograms of residuals from the models confirmed their approximate normality. Differences in mean mutation levels by disease state and cell type were tested using Wald tests based on these models. To adjust for three pairwise comparisons (early PD vs control, early PD vs late PD, control vs late PD), significance was set at p=0.05/3 (Bonferroni correction).
Results
Frozen unfixed midbrain SN tissue was obtained from 23 controls, 8 PD cases at advanced stages pathologically (“late PD”), 2 PD cases at early stages pathologically (Braak stage 3)16 and 7 ILBD cases (“early PD+ILBD”). We were able to obtain appropriate unfixed SN tissue from only 2 early stage PD cases. Therefore, to provide a sufficient sample size, and based on data suggesting that ILBD represents presymptomatic PD,13, 14 we obtained ILBD cases and analyzed them together with the early PD cases. Sample characteristics are summarized in Table 1. Further details are available in Supplementary Table 1. Although the difference in post-mortem interval (PMI) among disease states was statistically significant, neither age nor PMI within the ranges for these samples contributed significantly to mutation levels. In the best fit linear mixed effects models, age and PMI contributed at most -2.5 mutation/106 bp/year and - 0.15 mutations/106 bp/hour to mutation levels, respectively (Supplementary Figure 1). These effects are very small compared to mutation levels of hundreds/106 bp, and do not account for the observed differences in mutation levels between groups.
Table 1.
Sample Characteristics
| Disease State | Controls | Early PD+ILBD | Late PD | ||
|---|---|---|---|---|---|
| Number of cases | 23 | 9 | 8 | ||
| Neuronal samples analyzed | 54 | 26 | 22 | ||
| Glial samples analyzed | 20 | 14 | |||
| Age (yr) | mean±SD (range) | 75.9±12.7 (57–95) | 86.2±9.4 (73–99) | 74.9±9.7 (67–94) | p=0.519 (ANOVA) |
| PMI (hr) | mean±SD (range) | 39.7±28.0 (11.0–99.0) | 42.3±19.2 (18.0–74.5) | 12.6±3.4 (8.3–17.2) | p=0.043 (ANOVA) |
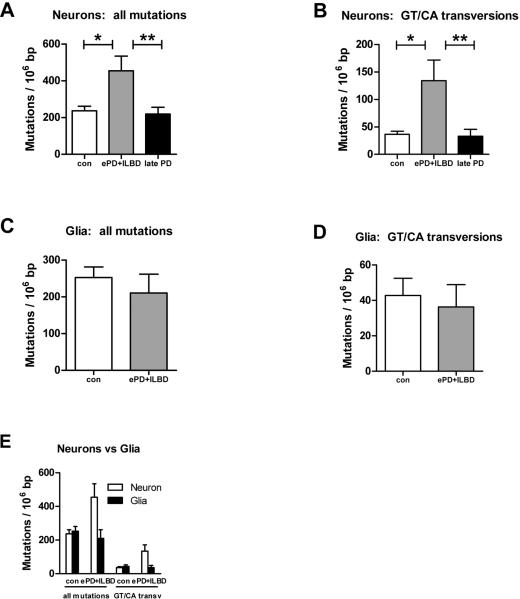
Somatic mtDNA mutation levels were measured using a validated PCR-cloning-sequencing strategy previously described10, 15, 17. Since pigmented (dopaminergic) neurons are the most vulnerable cells in the SN in PD, we analyzed mutation levels specifically in pigmented SN neurons and glia isolated by laser capture microdissection. The distributions of mutations were diffuse throughout the sequenced regions of the ND5 gene and D-loop (Supplementary Figures 2 and 3). In neurons, mean somatic mtDNA point mutation levels were more than 250 mutations/106 bp higher in early PD+ILBD compared to controls (p=0.0001) or late PD (p=0.0003), representing a 2-fold increase, but were similar in controls and late PD (difference of only ~42 mutations/106 bp, p=0.53) (Fig. 1A). We also examined levels of specific mutation subtypes, G→T or C→A transversions. These mutation subtypes are predicted to arise from mispairing induced by 8-hydroxy-2’deoxyguanosine (OH8dG), one of the most common products of oxidative damage to DNA,18 although they also may arise through other mechanisms. Levels of such potential oxidative stress mutations were over 100 mutations/106 bp higher in neurons from early PD+ILBD compared to controls (p<0.0001) or late PD (p=0.0006), representing a 3-fold increase, but again were similar in controls and late PD (difference of only ~4 mutations/106 bp, p=0.89) (Figure 1B). Thus, in SN neurons, overall somatic mtDNA point mutation levels and levels of mutation subtypes that can arise from oxidative stress are both dramatically increased, but only in early PD+ILBD.
Figure 1. Somatic mitochondrial DNA point mutation levels in controls, early PD+ILBD, and late PD.
DNA was extracted from groups of up to 50 neurons or glia isolated by laser capture microdissection; mtDNA regions of interest (D-loop or ND5) were PCR amplified; and point mutations were identified by cloning and sequencing as previously described.10, 15, 17 Bars and error bars indicate means and standard errors. To account for the 3 pairwise comparisons among disease states (early PD+ILBD vs control, early PD+ILBD vs late PD, control vs late PD), significance was set at p=0.05/3.
A. Overall somatic point mutation levels in SN neurons. *p=0.0001, **p=0.0003.
B. Levels of G→T or C→A transversions in SN neurons. *p<0.0001, **p=0.0006.
C. Overall somatic point mutation levels in SN glia.
D. Levels of G→T or C→A transversions in SN glia.
E. mtDNA mutation levels in SN neurons vs glia.
In contrast, overall somatic mtDNA point mutation levels were similar in SN glia from early PD+ILBD and controls (p=0.73) (Figure 1C). Levels of mutation subtypes predicted to arise from oxidative stress were also the same in glia from early PD+ILBD and controls (p=0.95) (Figure 1D). When plotted together (Figure 1E), it is seen that overall somatic mtDNA point mutation levels and levels of mutation subtypes that can arise from oxidative stress are increased preferentially in neurons and only in early PD+ILBD. These results would have been missed without deliberate efforts to procure samples from early stage disease and to analyze specific cell types. However, these results in glia must be interpreted cautiously as, for the early PD+ILBD group, we were able to obtain data on glia only from the ILBD cases.
Discussion
The findings that total and oxidative somatic mtDNA point mutation levels in the SN are dramatically increased preferentially in neurons and only early in PD+ILBD support a role for mtDNA mutations and oxidative stress in the neurodegeneration of PD early in course of the disease. Previously, we reported that levels of somatic mtDNA mutations in the brain correlate with mitochondrial electron transport chain dysfunction.17 Furthermore, in early PD+ILBD SN neurons, average somatic mtDNA point mutation levels are very high (~450 mutations/106 bp). This is similar to mtDNA mutation levels in mice expressing a proof-reading deficient mtDNA polymerase leading to a premature aging phenotype.19, 20 Although comparisons across species must be made cautiously, these mouse data demonstrate that such mutation levels can be functionally significant.
A limitation of this study is that appropriate early stage PD SN tissue was very difficult to obtain, so ILBD cases were used to increase sample size. ILBD shows dopaminergic pathology intermediate between controls and PD,13, 14 and has thus been considered an early, presymptomatic stage of PD, though this is difficult to prove definitively. Of note, average mutation levels (~580 mutations/106 bp) in the 2 early PD cases were among the highest observed, suggesting that the high mutation levels observed in the combined PD+ILBD group was not an artifact of including ILBD cases. An additional limitation is that our methodology assesses only point mutations. MtDNA deletions are modestly elevated in SN neurons in late stage PD compared to controls12, but whether or not deletions also are increased in early PD remains to be determined.
Our data support the hypothesis that mtDNA mutations contribute to neurodegeneration in PD and suggest that strategies to prevent or eliminate mtDNA mutations should be investigated for neuroprotective effects.
Supplementary Material
Supplementary Figure 1. Distributions of mean mutation levels in neurons according to age at death (top) or postmortem interval (bottom).
Supplementary Figure 2. Distribution of somatic mtDNA mutations across the sequenced regions in neurons. Shown are combined data from all clones from neurons from the early PD+ILBD group. Data for the ND5 gene are shown in A and B. Data for the D-loop are shown in B and C. The total numbers of clones in which each particular mutation was identified are shown in A and C. The total numbers of neuronal samples in which each particular mutation occurred at least once are shown in B and D. Nucleotide positions are numbered according to the revised Cambridge reference sequence (Genbank accession number J01415.2 or NC_012920). These data demonstrate a wide distribution of mutations across the sequenced regions.
Supplementary Figure 3. Distribution of somatic mtDNA mutations across the sequenced regions in glia. Shown are combined data from all clones from glia from the early PD+ILBD group. Each somatic mtDNA mutation is indicated by a circle Additional details are as outlined in the legend for Figure 2.
Acknowledgments
Brain tissue was provided by the Harvard Brain Tissue Resource Center (McLean Hospital, Boston, MA, USA, partly supported by PHS grant R24MH068855); the Neuropathology Core of the Massachusetts Alzheimer Disease Research Center (supported by PHS grant P50AG005134) and the Neuropathology Service of the Massachusetts General Hospital (Boston, MA, USA); Brain-Net Germany, Neurobiobank Munich (Center for Neuropathology and Prion Research), Ludwig-Maximilians University Munich, Germany; and the Queen Square Brain Bank for Neurological Disorders (University College London, Institute of Neurology, Queen Square, London, UK). This work was conducted with biostatistical support from Harvard Catalyst | The Harvard Clinical and Translational Science Center (NIH Award UL1 RR025758 and financial contributions from Harvard University and its affiliated academic health care centers). The content is solely the responsibility of the authors and does not necessarily represent the official views of Harvard Catalyst, Harvard University and its affiliated academic health care centers, the National Center for Research Resources, or the National Institutes of Health.
This work was supported by grants from the NINDS (K02NS043311; R03NS053840; R01NS058988; DKS), the William F. Milton Fund of Harvard University (DKS), the NIA (R01AG20729; MTL), the USAMRMC (W81XWH-04-1-0802; MFB), the Paul Beeson Physician Faculty Scholarship (MTL), the MGH/MIT Morris Udall Center of Excellence in PD Research (NIH NS38372), the APDA Advanced Center for Parkinson Research at MGH (IC-C), and the Harvard Neurodiscovery Center and Harvard Catalyst (RAB).
References
- 1.Schapira AH. Mitochondria in the aetiology and pathogenesis of Parkinson's disease. Lancet Neurol. 2008;7:97–109. doi: 10.1016/S1474-4422(07)70327-7. [DOI] [PubMed] [Google Scholar]
- 2.Betarbet R, Sherer TB, MacKenzie G, et al. Chronic systemic pesticide exposure reproduces features of Parkinson's disease. Nat Neurosci. 2000;3:1301–1306. doi: 10.1038/81834. [DOI] [PubMed] [Google Scholar]
- 3.Schapira AH, Cooper JM, Dexter D, et al. Mitochondrial complex I deficiency in Parkinson's disease. Lancet. 1989;1:1269. doi: 10.1016/s0140-6736(89)92366-0. [DOI] [PubMed] [Google Scholar]
- 4.Parker WD, Jr., Boyson SJ, Parks JK. Abnormalities of the electron transport chain in idiopathic Parkinson's disease. Ann Neurol. 1989;26:719–723. doi: 10.1002/ana.410260606. [DOI] [PubMed] [Google Scholar]
- 5.Mizuno Y, Ohta S, Tanaka M, et al. Deficiencies in complex I subunits of the respiratory chain in Parkinson's disease. Biochem Biophys Res Commun. 1989;163:1450–1455. doi: 10.1016/0006-291x(89)91141-8. [DOI] [PubMed] [Google Scholar]
- 6.Swerdlow RH, Parks JK, Miller SW, et al. Origin and functional consequences of the complex I defect in Parkinson's disease. Ann Neurol. 1996;40:663–671. doi: 10.1002/ana.410400417. [DOI] [PubMed] [Google Scholar]
- 7.Thyagarajan D, Bressman S, Bruno C, et al. A novel mitochondrial 12SrRNA point mutation in parkinsonism, deafness, and neuropathy. Ann Neurol. 2000;48:730–736. [PubMed] [Google Scholar]
- 8.Simon DK, Pulst SM, Sutton JP, et al. Familial multisystem degeneration with parkinsonism associated with the 11778 mitochondrial DNA mutation. Neurology. 1999;53:1787–1793. doi: 10.1212/wnl.53.8.1787. [DOI] [PubMed] [Google Scholar]
- 9.Parker WD, Jr., Parks JK. Mitochondrial ND5 mutations in idiopathic Parkinson's disease. Biochem Biophys Res Commun. 2005;326:667–669. doi: 10.1016/j.bbrc.2004.11.093. [DOI] [PubMed] [Google Scholar]
- 10.Simon DK, Lin MT, Zheng L, et al. Somatic mitochondrial DNA mutations in cortex and substantia nigra in aging and Parkinson's disease. Neurobiol Aging. 2004;25:71–81. doi: 10.1016/s0197-4580(03)00037-x. [DOI] [PubMed] [Google Scholar]
- 11.Arthur CR, Morton SL, Dunham LD, et al. Parkinson's disease brain mitochondria have impaired respirasome assembly, age-related increases in distribution of oxidative damage to mtDNA and no differences in heteroplasmic mtDNA mutation abundance. Mol Neurodegener. 2009;4:37. doi: 10.1186/1750-1326-4-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bender A, Krishnan KJ, Morris CM, et al. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet. 2006;38:515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- 13.DelleDonne A, Klos KJ, Fujishiro H, et al. Incidental Lewy body disease and preclinical Parkinson disease. Arch Neurol. 2008;65:1074–1080. doi: 10.1001/archneur.65.8.1074. [DOI] [PubMed] [Google Scholar]
- 14.Dickson DW, Fujishiro H, DelleDonne A, et al. Evidence that incidental Lewy body disease is pre-symptomatic Parkinson's disease. Acta Neuropathol. 2008;115:437–444. doi: 10.1007/s00401-008-0345-7. [DOI] [PubMed] [Google Scholar]
- 15.Cantuti-Castelvetri I, Lin MT, Zheng K, et al. Somatic mitochondrial DNA mutations in single neurons and glia. Neurobiol Aging. 2005;26:1343–1355. doi: 10.1016/j.neurobiolaging.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 16.Braak H, Ghebremedhin E, Rub U, et al. Stages in the development of Parkinson's disease-related pathology. Cell Tissue Res. 2004;318:121–134. doi: 10.1007/s00441-004-0956-9. [DOI] [PubMed] [Google Scholar]
- 17.Lin MT, Simon DK, Ahn CH, et al. High aggregate burden of somatic mtDNA point mutations in aging and Alzheimer's disease brain. Hum Mol Genet. 2002;11:133–145. doi: 10.1093/hmg/11.2.133. [DOI] [PubMed] [Google Scholar]
- 18.Cheng KC, Cahill DS, Kasai H, et al. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G----T and A----C substitutions. J Biol Chem. 1992;267:166–172. [PubMed] [Google Scholar]
- 19.Trifunovic A, Wredenberg A, Falkenberg M, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 20.Kujoth GC, Hiona A, Pugh TD, et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Distributions of mean mutation levels in neurons according to age at death (top) or postmortem interval (bottom).
Supplementary Figure 2. Distribution of somatic mtDNA mutations across the sequenced regions in neurons. Shown are combined data from all clones from neurons from the early PD+ILBD group. Data for the ND5 gene are shown in A and B. Data for the D-loop are shown in B and C. The total numbers of clones in which each particular mutation was identified are shown in A and C. The total numbers of neuronal samples in which each particular mutation occurred at least once are shown in B and D. Nucleotide positions are numbered according to the revised Cambridge reference sequence (Genbank accession number J01415.2 or NC_012920). These data demonstrate a wide distribution of mutations across the sequenced regions.
Supplementary Figure 3. Distribution of somatic mtDNA mutations across the sequenced regions in glia. Shown are combined data from all clones from glia from the early PD+ILBD group. Each somatic mtDNA mutation is indicated by a circle Additional details are as outlined in the legend for Figure 2.