SUMMARY
Skeletal muscle atrophy induced by aging (sarcopenia), inactivity and prolonged fasting states (starvation) is predominantly restricted to glycolytic type II muscle fibers and typical spares oxidative type I fibers. However, the mechanisms accounting for muscle fiber type specificity of atrophy have remained enigmatic. In the current study, we that although the Fyn tyrosine kinase activated the mTORC1 signaling complex, it also induced marked atrophy of glycolytic fibers with relatively less effect on oxidative muscle fibers. This was due to inhibition of macroautophagy via an mTORC1-independent but STAT3-dependent reduction in Vps34 protein levels and decreased Vps34/p150/Beclin1/Atg14 complexes. Physiologically, in the fed sate endogenous Fyn kinase activity was increased in glycolytic but not oxidative skeletal muscle. In parallel, Y705-STAT3 phosphorylation increased with decreased Vps34 protein levels. Moreover, fed/starved regulation of Y705-STAT3 phosphorylation and Vps34 protein levels was prevented in skeletal muscle of Fyn null mice. These data demonstrate a novel Fyn/STAT3/Vps34 pathway that is responsible for fiber type specific regulation of macroautophagy and skeletal muscle atrophy.
Keywords: Fyn, LKB1, mTORC1, STAT3, Vps34, autophagy, muscle atrophy
INTRODUCTION
Recent studies have demonstrated convergent signals that control nutrient sensing, cellular energy metabolism, growth, macromolecular synthesis and degradation. The AMP-dependent protein kinase (AMPK) represents a central signaling nexus that responds to changes in intracellular energy status that is activated upon reduction of cellular energy state (Lage et al., 2008; Long and Zierath, 2006; Witczak et al., 2008). AMPK is composed of three subunits. AMP binding to the γ subunit that results in enhanced α subunit activation loop T172 phosphorylation and subsequent activation of protein kinase activity (Witczak et al., 2008). In turn, AMPK downstream targets are quite diverse and include the phosphorylation of acetyl-CoA carboxylase to enhance fatty acid oxidation; the phosphorylation of the tuberous sclerosis complex 2 protein (TSC2) and raptor resulting in the inhibition of mTORC1 (mammalian Target of Rapamycin Complex-1)-dependent protein synthesis; and phosphorylation of ULK1 and FOXO3A to increase macroautophagy (Chiacchiera and Simone, 2010; Luo et al., 2005; Shackelford and Shaw, 2009; He and Klionsky, 2009; Mizushima, 2010). Macroautophagy is an important catabolic process that mediates degradation of intracellular components (proteins and organelles) in lysosomes and has also been shown to contribute to maintenance of cellular energetic balance (Singh and Cuervo, 2011). Although counterintuitive, in several mouse models as well as in human patients, inhibition of macroautophagy in skeletal muscle has been linked to muscle atrophy (Grumati et al., 2010; Masiero et al., 2009; Raben et al., 2008).
In peripheral tissues, AMPK T172 α subunit phosphorylation is primarily dependent upon the tumor suppressor protein LKB1 (Liver Kinase B1) and inactivating mutations are responsible for Peutz-Jeghers syndrome, a disorder characterized by gastrointestinal tract hamartomas and pigmented macules on the skin and mouth (Alessi et al., 2006; Jansen et al., 2009). Recently, LKB1 tyrosine phosphorylation through interaction with the proto-oncogene Fyn has been implicated as an important regulatory mechanism for the control of fatty acid oxidation and insulin sensitivity (Yamada et al., 2010). Fyn is a member of the Src family of non-receptor tyrosine kinases and is composed of two major alternatively spliced isoforms, FynB and FynT, that differ by the inclusion of exon 7A or exon 7B, respectively (Cooke and Perlmutter, 1989; Saito et al., 2010). Previous studies have demonstrated signaling specificities of the two Fyn isoforms, in that FynT but not FynB expression can efficiently rescue T cell signaling events in FynT null T cells suggesting cell context-dependent signaling of the two isoforms (Davidson et al., 1994). Since previous studies examining the role of Fyn in insulin action and energy expenditure in vivo relied on Fyn null mice, it was not possible to distinguish the physiologic contributions of the two Fyn isoforms. To address this issue, we generated transgenic mice that overexpress either FynB or FynT selectively in skeletal muscle. Surprisingly, Fyn transgenic mice display a strong muscle atrophy phenotype with characteristics similar to sarcopenia and the initial muscle wasting that occurs during starvation, with selective loss of glycolytic (white) but not oxidative (red) skeletal muscle fibers. Using Fyn transgenic and Fyn knockout mice, we demonstrate that Fyn functions as part of the nutrient sensing system to regulate skeletal muscle fiber-type specific macroautophagy and muscle degeneration.
RESULTS
Phenotypic characterization of Fyn expressing transgenic mice
Previous studies using Fyn null mice and pharmacological inhibition of Fyn activity demonstrated that, in muscle and adipose tissue, Fyn functionally represses fatty acid oxidation and energy expenditure concomitant with a reduction in AMPK activation through the inhibition of LKB1 (Bastie et al., 2007; Yamada et al., 2010). However, since the single fyn gene expresses two isoforms that can not be functionally distinguished (either pharmacologically or by the currently available genetic mouse model), these data were unable to discriminate between FynT and FynB activity. To address this issue, we first performed qPCR to determine the relative tissue specific expression of FynT and FynB mRNA (Fig. 1A). FynT and FynB were equally expressed in mouse brain, although FynT was the predominant transcript expressed in all peripheral tissues examined, with highest levels found in the spleen, heart and subcutaneous adipose tissue. Intermediate levels of FynT mRNA were also found in epididymal adipose tissue and skeletal muscle.
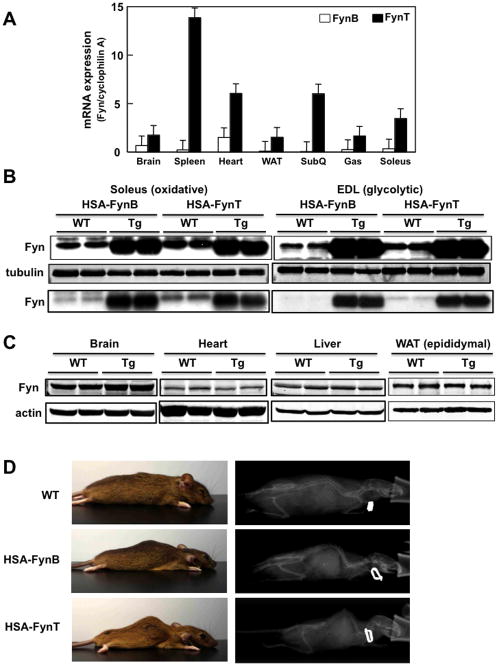
Figure 1. Relative expression level of FynT and FynB in tissues, and generation of skeletal muscle specific FynB and FynT transgenic mice.
A) mRNA expression levels of FynB (open bars) and FynT (solid bars) relative to cyclophilin A in brain, spleen, heart, epididymal (WAT) and subcutaneous (SubQ) adipose tissues, gastrocnemius (Gas) and soleus muscle. B) Fyn protein expression in soleus and extensor digitorum longus (EDL) skeletal muscles of wild type (WT), HSA-FynT and HSA-FynB transgenic mice (upper panels). Tubulin was used as internal loading control. A lighter exposure of the Fyn immunoblot (top panel) is shown below the tubulin immunoblot. C) Fyn protein levels in the brain, heart, liver and epididymal adipose tissue of WT and HSA-FynT transgenic (Tg) mice. Actin was used as internal loading control. D) Physical characteristics (left panels) and microCT pictures (right panels) for wild type (WT), HSA-FynB and FynT transgenic mice.
To specifically express Fyn isoforms in skeletal but not cardiac muscle, we used the human skeletal muscle actin promoter (HSA) to drive FynT or FynB cDNA. As shown in Figure 1B, we selected transgenic mice that expressed approximately equal levels of FynT and FynB in both predominant glycolytic skeletal muscle (extensor digitorum longus (EDL)) and predominantly oxidative skeletal muscle (soleus). Darkened exposure is shown in Figure 1B (top panel) in order to visualize the endogenous Fyn protein and a lightened exposure is shown in the lower panel. Expression specificity was confirmed, as the levels of FynT (Fig. 1C) and FynB (not shown) were similar compared to endogenous levels in the brain, heart, liver and epididymal adipose tissue.
Visual inspection of the transgenic mice clearly indicated two predominant abnormalities (Fig. 1D). The HSA-FynT mice displayed a marked hump behind the neck that was present, although less pronounced in the HSA-FynB mice with a marked reduction in hindquarter mass.
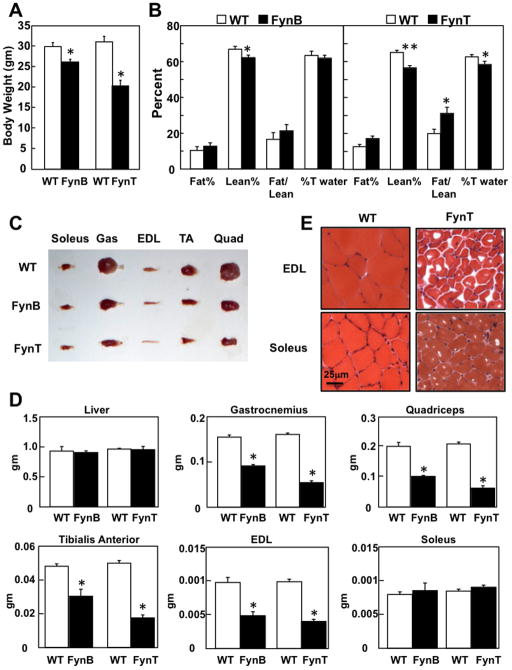
MicroCT scans showed the protrusion on the back was due to spinal cord malformation (kyphosis) and confirmed the marked reduction in hindquarter mass (Fig. 1D). Consistent with these observations, aged-matched HSA-FynB transgenic mice had a relatively smaller (~15%) reduction in body mass whereas the HSA-FynT transgenic mice were nearly 1/3 the weight (10 gm lighter) than control mice (Fig. 2A). Quantitative NMR demonstrated that the relatively reduced weight of the FynT and FynB transgenic mice was due to a reduction in lean body mass (Fig. 2B). Although there was no significant change in fat mass or water content in the HSA-FynB mice, there was a small but significant increase in fat mass with a concomitant decrease in water content in the HSA-FynT mice. The reduced lean mass was accompanied by a decrease in skeletal muscle weight with no change in liver weight (Fig. 2C and D). Interestingly, soleus muscle was not significantly affected. The soleus muscle depot is primarily composed of oxidative (red) muscle fibers whereas the other skeletal muscles are either mostly glycolytic white fibers (i.e., EDL) or a mixture of oxidative and glycolytic muscle fibers (i.e., gastrocnemius; Gas, tibialis anterior; TA, and quadriceps; Quad).
Figure 2. Skeletal muscle atrophic characteristics of HSA-FynT and HSA-FynB transgenic mice.
A) Total body weight of HSA-FynB (FynB) and HSA-FynT (FynT) mice and their respective control littermates (WT). *p< 0.005 vs WT. B) Quantification of fat and lean mass measured by NMR in HSA-FynB (left panel) and HSA-FynT (right panel) mice versus their respective control littermates. *p< 0.05,, **p< 0.01 vs WT, respectively C) Selective muscle sizes from WT, HSA-FynB and HSA-FynT mice. D) Total tissue weight (liver, gastrocnemius, quadriceps, tibialis anterior, EDL and soleus muscles) of WT littermates, HSA-FynB and HSA-FynT transgenic mice. Data are expressed as mean ± SEM. *p<0.005 vs WT. E) Hematoxylin &Eosin (H&E) staining of EDL and soleus muscle from WT and HSA-FynT mice.
Previous studies have reported that FynT, but not FynB, can restore T cell signaling resulting from FynT deficiency and that FynT has higher kinase activity than FynB (Davidson et al., 1994). Recent structural analyses have indicated that linker region of FynB engages the SH3 with higher affinity than the linker region of FynT. This results in a more auto-inhibited and closed conformation state, thereby markedly reducing the ability of FynB to interact with and phosphorylate the Src family kinase substrate Sam68 (Brignatz et al., 2009). To determine the relative interaction of FynT and FynB with LKB1, epitope tagged Flag-LKB1 was co-expressed with epitope tagged FynT-V5 or equal amounts of expressed FynB-V5 in cultured 293 cells (Fig. S1, A). Immunoprecipitation with the Flag antibody (LKB1) resulted in co-immunoprecipitation of FynT-V5 but substantially reduced levels of co-immunoprecipitated FynB-V5. Consistent with reduced substrate kinase activity of FynB versus FynT, muscle extracts from the HSA-FynB mice displayed an approximate 6-fold increase in Fyn peptide substrate kinase activity compared to wild type mice (Fig. S1, B). Despite equal levels of FynT and FynB protein expression (Fig. 1B), muscle extracts from the HSA-FynT mice displayed an approximate 65-fold increase in peptide substrate activity. Thus, the intrinsic substrate recognition and kinase activities of the FynT and FynB isoforms likely account for the degree of phenotypic differences.
Histological analysis of the EDL muscle using hematoxylin & eosin (H&E) staining revealed a remarkable reduction in muscle fiber size (~10 fold) in the HSA-FynT mice with a relatively smaller reduction (~2 to 3-fold) in muscle fiber size in the soleus muscle (Fig. 2E). Both muscle fibers displayed muscle degeneration as seen by nuclear hypertrophy, elongated nuclei and myofibril vacuoles (Fig. S1, C) and was more pronounced in EDL than soleus muscle. The relative absence of central migrating nuclei is also consistent with a muscle wasting phenotype. Cytochrome C oxidase and succinate dehydrogenase staining demonstrated marked increase in mitochondria levels in EDL muscle of HSA-FynT mice with relatively little change in soleus muscle (Fig. S2). Moreover, there was a marked increase in non-specific esterase staining of EDL muscle with only a small increase in acid phosphatase staining. Alterations in the staining patterns for these two enzymes have also been observed in skeletal muscle of other myopathies with compromised autophagic function, such as Danon and X-linked myopathy (Malicdan et al., 2009). Consistent with the relative insensitivity of the soleus muscle to FynT expression, there was a relatively smaller alteration in non-specific esterase in red oxidative soleus muscle fibers and a similarly small increase of acid phosphatase compared to the EDL muscle fibers (Fig. S2).
Ultrastructural disorganization of skeletal muscle fibers in HSA-FynT transgenic mice was also observed by electron microscopy in both mixed (tibialis anterior) fiber muscles (Fig. 3A) and glycolytic EDL muscle (Fig. S3, A) with lesser effects in oxidative soleus muscle fibers (Fig. S3, B). The HSA-FynT mice also displayed intrafiber Z-band breaks with expanded and disorganized mitochondria and sarcoplasmic reticulum structures. Further evidence for glycolytic muscle wasting is based upon the greater extent of fibrosis, trichrome staining for collagen deposition and expression of the pro-inflammatory/fibrosis cytokine, TGFβ1, in EDL versus soleus muscle (Fig. S4).
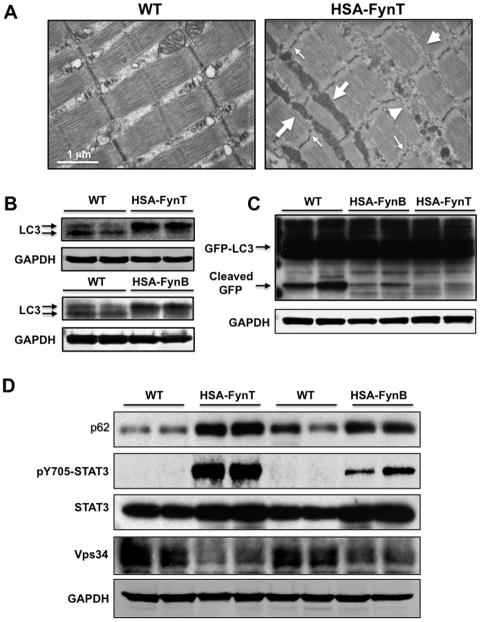
Figure 3. HSA-Fyn mice display defective macroautophagy.
A) Transmission electron microscopy images of tibialis anterior muscle from WT (left) and HSA-FynT (right) mice. Large arrows indicate expanded mitochondria, small arrows indicate Z-line disruption and arrowheads indicate expanded sarcoplasmic reticulum. B) LC3-I and LC3-II protein levels in gastrocnemius muscle of WT and HSA-FynT (lower) compared to WT and HSA-FynB mice (upper) from two independent animals each. C) Tibialis anterior muscle of WT, HSA-FynB and HSA-FynT mice were transfected with GFP-LC3 as described in Experimental Procedures. The free cleaved GFP (lower band) is generated by autophagy from the GFP-LC3 fusion protein (upper bands) was detected by immunoblotting with a GFP mouse monoclonal antibody. Immunoblotting for the GAPDH protein (panels C, D, and E) was used as a loading control. D) Gastrocnemius muscles of WT, HSA-FynT and HSA-FynB mice were immunobotted with p62, phosphoY705-STAT3, total STAT3 and vps34 antibodies from two independent animals. GAPDH was used as internal loading control.
FynT enhances skeletal muscle mTORC1 signaling and inhibits macroautophagy
Recent studies have demonstrated that defective mTORC1 signaling results in muscle atrophy of glycolytic muscle fibers with a lesser effect on oxidative muscle fibers (Bentzinger et al., 2008; Risson et al., 2009). In contrast, AMP-dependent protein kinase (AMPK) deficient mice results in muscle hypertrophy due to enhanced mTORC1 activity (Lantier et al., 2010; Mounier et al., 2009). Since Fyn null mice have increased skeletal muscle AMPK activity, and AMPK is an established inhibitor of mTORC1-dependent signaling (Kimball, 2006; Shackelford and Shaw, 2009), we anticipated that increased Fyn expression would inhibit AMPK and thereby result in mTORC1 activation. In turn, increased Fyn expression would be expected to drive muscle hypertrophy, opposite to the observed muscle atrophy phenotype. Therefore, to determine if increased Fyn expression did in fact result in mTORC1 activation, we examined AMPK and mTORC1 target site phosphorylation (Fig. S5, A). Consistent with previous studies in Fyn null mice that show increased AMPK activation, HSA-FynT transgenic mice displayed reduced skeletal muscle phosphorylation of the T172 AMPK α subunit activation site. In parallel, there was a marked reduction in AMPK specific raptor inhibitory site phosphorylation (S792) and increased mTORC1 specific S6 kinase activation site phosphorylation (S389). In addition, FynT expression resulted in enhanced LKB1 tyrosine phosphorylation, consistent with LKB1 inhibition resulting in reduced AMPK activation (Yamada et al., 2010). Functional activation of AMPK in HSA-FynT mice was further confirmed by down regulation of PGC1α and induction of ACC1α expression (Fig. S5, C and D).
mTORC1 activation would be expected to increase muscle mass, yet we observed an opposite phenotype, suggesting the presence of another overriding mechanism. In this regard, deficiency of skeletal muscle macroautophagy (acid α-glucosidase as seen in Pompe knockout mice) results in marked skeletal muscle atrophy in glycolytic fibers but with less of an effect in oxidative fibers (Raben et al., 2008). In addition, Pompe mice display a similar spinal cord deformation. Further, inhibition of macroautophagy initiation by conditional ATG7 specific knockout in skeletal muscle resulted in similar morphological features of muscle wasting (Masiero et al., 2009). Since our morphological and histological analyses are also consistent with a defect in lysosome function (Fig. S2), we speculated that FynT expression might also inhibit skeletal muscle macroautophagy.
To assess this possibility, we first examined the levels and degradation kinetics of LC3-II, an integral component of the autophagosome membrane that is widely utilized as a marker of this compartment (Kabeya et al., 2000). Levels of LC3-I, a primed form of the protein precursor of LC3-II, were substantially increased and steady-state levels of LC3-II were decreased in the gastrocnemius muscle of both HSA-FynT and HSA-FynB mice (Fig. 3B). Although these findings are supportive of reduced autophagosomes formation, lower autophagosome content may also result from accelerate degradation of these vesicular compartments in lysosomes. To assess autophagic vacuole clearance, we transfected the tibialis anterior muscle in vivo with a GFP-LC3 reporter, which when delivered into lysosomes is proteolytically cleaved to free GFP, with the LC3 insert rapidly degraded in this compartment. Lysosomal cleavage of GFP-LC3 in skeletal muscle was substantially reduced in the HSA-FynT mice with an intermediate amount of cleavage in the HSA-FynB mice (Fig. 3C) as compared to wild type mice. These data suggest that the lower levels of endogenous LC3-II observed in Fyn transgenic mice result from decreased autophagosome formation rather than from accelerated degradation. Similarly, the well-characterized macroautophagy substrate protein SQSTM1/p62 (Masiero et al., 2009; Mizushima et al., 2010) was substantially increased in gastrocnemius (mixed muscle) of HSA-FynT mice with moderate increase in HSA-FynB mice compared to wild type mice (Fig. 3D, top panel). We also confirmed the increased in SQSTM1/p62 protein levels and its presence in aggregate-like structures in the gastrocnemius muscle by immunofluorescence (data not shown). Macroautophagy inhibition was also accompanied by accumulation of ubiquitinated proteins that has also been reported in macroautophagy deficient skeletal muscle (Klionsky et al., 2008) (Fig. S5, B).
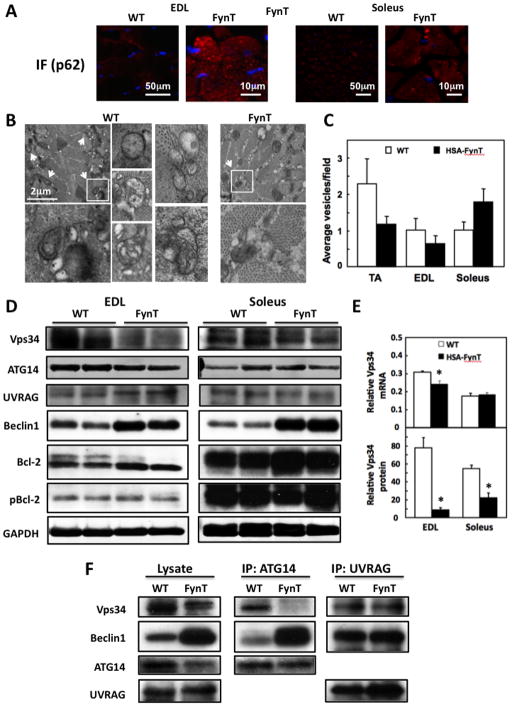
A relative selective perturbation of oxidative muscle macroautophagy was observed based upon SQSTM1/p62 immunofluorescence and the quantification of autophagic vacuoles in the EDL and soleus muscle. SQSTM1/p62 immunofluorescence intensity was increased to a greater extent in the EDL compared to soleus muscles in the HSA-FynT mice (Fig. 4A). Transmission electron microscopy demonstrated the presence of autophagic vacuolar structures as vesicular compartments with double or single limiting membrane containing heterogeneous cytosolic material (Fig. 4B). Morphometric quantification of these autophagic vacuolar structures demonstrated a decrease in tibialis anterior and EDL muscles with an increase in soleus muscle of the HSA-FynT mice compared to wild type mice (Fig. 4C). The lower content of autophagic vacuoles in glycolytic skeletal muscles supports the inhibitory effect of Fyn on autophagosome formation inferred from the cell biological analysis.
Figure 4. Autophagy is differently regulated in fast-twitch glycolytic type II fibers (EDL) and slow-twitch oxidative type I fiber (soleus) of HSA-FynT mice.
A) Representative p62 immunofluorescence and DAPI staining (blue) images from EDL and soleus muscle of WT and HSA-FynT mice. B) Representative transmission electron microscopic images of tibialis anterior muscles from wild type and HSA-FynT (FynT) mice. Examples of autophagic vacuoles (denoted by arrows in the low magnification field) are show at higher magnification in the inserts. C) Quantification of autophagic vacuoles (AV) by transmission electron microscopic in tibialis anterior (TA), EDL and soleus muscles from wild type (open bars) and HSA-FynT (filled bars) mice. The average number of autophagic vacuoles per field was quantified by morphological analyses of at least 15 electron microscopy images from 3 independent mice. D) Beclin1 and Beclin1 complex-related proteins (Vps34, Atg14, UVRAG, Bcl2 and phospho-Bcl-2) expression levels in EDL and soleus muscles of WT and HSA-FynT mice. GAPDH was used as internal loading control. These are representative immunoblots independently performed 3 times E) mRNA expression levels (upper) and protein levels (lower) of Vps34 relative to Hrpt1 or GAPDH in EDL and soleus muscles of WT and HSA-FynT mice. F) Vps34 presence in Beclin1 complex-1 (containing Atg14) and Beclin1 complex-2 (containing UVRAG) in quadriceps muscle of WT and HSA-FynT mice was determined by immunoprecipitation (IP) using either an Atg14 or UVRAG antibody followed by immunoblotting with Vps34, Beclin1, Atg14 and UVRAG antibodies. These are representative immunoblots independently performed 3 times.
Activation of the FOXO family of transcription factors, especially FOXO3a, has been reported to induce muscle mass loss through the induction of atrophy-related muscle-specific E3 ubiquitin ligase genes (MuRF1 and atrogin-1) and by stimulation of autophagy by pro-apoptotic Bcl family members, Bnip3 and Bnip3L (Mammucari et al., 2007; Sandri et al., 2004). Although AMPK has been reported to be an upstream activator of FOXO3a (Chiacchiera and Simone, 2010; Greer et al., 2007), AMPK activity was decreased in the HSA-FynT mice (Fig. S5, A) suggesting a FOXO3a-independent atrophy mechanism. Consistent with this prediction, quantitative PCR demonstrated decreased expression of MuRF1, atrogin-1 as well as the pro-apoptotic BH3 domain proteins Bnip3 and Bnip3L in the EDL muscle of the HSA-FynT mice with no change in soleus muscle) (Fig. S6). Moreover, dystrophin expression was increased in the EDL muscle of HSA-FynT mice and was unchanged in soleus muscle. In contrast, lysosomal proteinase, Cathepsin L, expression is up-regulated in various skeletal muscle atrophy models (Bechet et al., 2005) and was elevated in both the EDL and soleus muscle of the HSA-FynT mice. The relative sensitivity of EDL versus soleus muscle to Fyn expression, likely reflects an intrinsic difference in these muscle fibers, as FynT expression was similar in both depots (Fig. S7, B). In addition, only EDL muscle displayed regulation of STAT3 and Vps34 in both the HSA-FynT (Fig. S7, B; Fig. 4D and 5A) and in wild type mice (Figs. 6A, C and 7A, B and C).
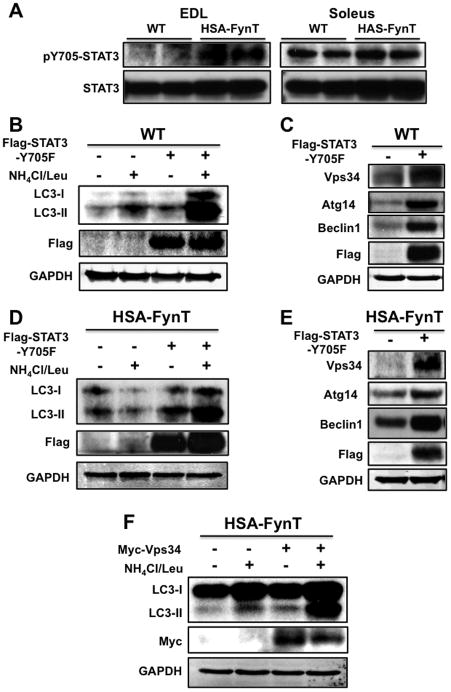
Figure 5. STAT3 is tyrosine phosphorylated in EDL but not soleus muscle of HSA-FynT mice and the dominant negative form of STAT3 (STAT3-Y705F) rescues autophagy deficiency in HSA-FynT mice.
A) EDL and soleus muscle isolated from WT and HSA-FynT mice were immunoblotted with phosphoY705-STAT3 and total STAT3 antibodies from two independent animals. B) Tibialis anterior muscles of WT mice were transfected with either an empty vector (−) or the vector containing the dominant interfering (+) STAT3 mutant (Flag-STAT3-Y705F) and autophagic flow was determined as described in Experimental Methods. C) Beclin1 complex related proteins expression levels in the tibialis anterior muscles of WT mice transfected with empty vector (−) or Flag-STAT3-Y705F (+). D) Tibialis anterior muscles of HSA-FynT mice were transfected with empty vector (−) or the vector containing the dominant interfering (+) STAT3 mutant (Flag-STAT3-Y705F) followed by determination of autophagic flow. E) Beclin1 complex related proteins expression levels in the tibialis anterior muscles of HSA-FynT mice transfected with an empty vector (−) or Flag-STAT3-Y705F (+). These are representative immunoblots independently performed 4 times for panel A and 3 times for panels B–E. F) Tibialis anterior muscles of HSA-FynT mice were transfected with empty vector (−) or the vector containing the Vps34 (Myc-Vps34) followed by determination of autophagic flow.
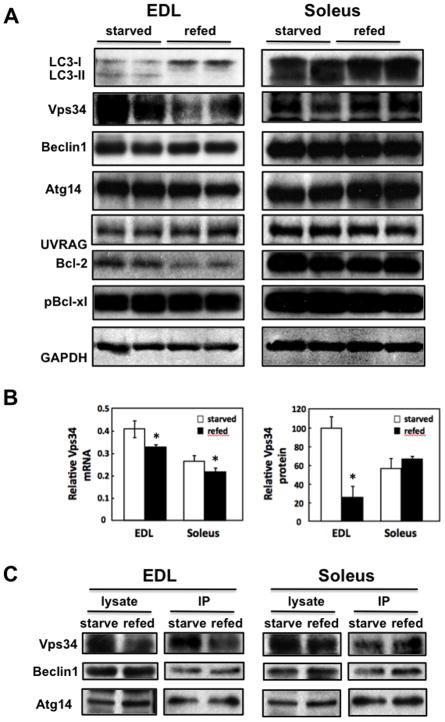
Figure 6. Beclin1 and Beclin1 complex-related proteins are differentially regulated in the EDL and soleus muscles of WT animals in the refed state.
A) WT mice were food restricted for 30 h (starved) and then allowed free access to food for 5 h (refed). The EDL and soleus muscles were isolated and immunoblotted for LC3 in two independent animals. These are representative immunoblots independently performed 3 times. B) mRNA expression levels (left) and protein levels (right) of Vps34 relative to Hrpt1 or GAPDH in EDL and soleus muscles of starved and refed WT mice. C) The EDL and soleus muscle from starved and refed WT mice (lysates) were immunoprecipitated (IP) with an ATG14 antibody and immunoblotted for Vps34, Beclin1 and Atg14. These are representative immunoblots independently performed 3 times.
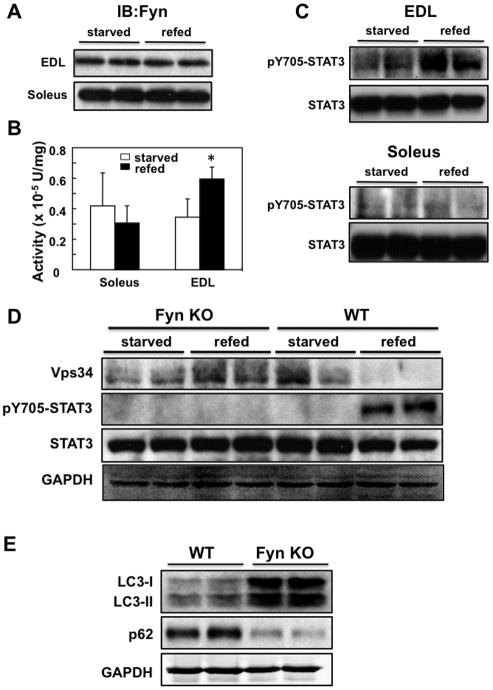
Figure 7. Autophagy is differentially regulated in the EDL and soleus muscles of WT mice in the starved and refed state.
A) EDL and soleus muscle from starved and refed WT mice were immunoblotted for Fyn protein levels in two independent animals. B) Fyn protein kinase activity in the EDL and soleus muscles of starved and refed WT mice was determined as described in Experimental Procedures from 5 independent mice. *p< 0.05 vs. starved. C) EDL (upper) and soleus muscle (lower) from starved and refed WT mice were immunoblotted for pY705-STAT3 and total STAT3 protein levels in two independent animals. D) Quadriceps muscle of FynKO and WT mice were immunoblotted with Vps34, phosphoY705-STAT3 and total STAT3 from two independent animals. GAPDH was used as internal loading control. E) LC3-I, LC3-II and p62 protein levels in quadriceps muscle of FynKO and WT mice from two independent animals each. These are representative immunoblots independently performed 3 times in panels A, C, D and E)
FynT enhances STAT3 tyrosine phosphorylation and alters Vps34 protein levels and the assembly state of the Vps34/p150/Beclin1/Atg14 complex 1
It is well established that mTORC1activation results in inhibition of macroautophagy, and in mammalian cells appears to result from the phosphorylation and inactivation of the ULK1/Atg13/FIP200 complex (He and Klionsky, 2009; Mizushima, 2010). Similarly, recent studies have reported that AMPK directly phosphorylates ULK1 to promote macroautophagy (Egan et al., 2011; Kim et al., 2011). However, activation of mTORC1 through the inhibition of AMPK is known to induce muscle hypertrophy (Lantier et al., 2010; Mounier et al., 2009). Despite mTORC1 activation and reduced AMPK in the HSA-FynT mice, we observed a muscle wasting phenotype. Since the relative contribution of mTORC1 in the regulation of macroautophagy in skeletal muscle remains poorly defined (Sandri, 2010; Zhao et al., 2007), we speculated that the presence of a mTORC1-independent mechanism is responsible for the control of macroautophagy.
To address this issue, we examined protein levels of the class 3 phosphatidylinositol kinase (PIK3C3 gene or Vps34 protein) complex 1 and complex 2 that regulate macroautophagy and endosome trafficking, respectively, independent of mTORC1 function (Funderburk et al., 2010; He and Klionsky, 2009). Vps34 protein levels were decreased in EDL muscle of HSA-FynT mice but not soleus muscle, a finding that is consistent with the inhibition of macroautophagy in glycolytic muscle (Fig. 4D). Although there was no significant change in protein levels of Atg14 and UVRAG, there was an increase in Beclin1 protein with a small increase in Bcl-2 protein, but no change in Bcl-2 phosphorylation. The large decrease in the EDL muscle was predominantly due to down regulation of Vps34 protein with only a small decrease in Vps34 mRNA. Similarly, the smaller reduction in Vps34 protein in soleus muscle occurred without any significant change in Vps34 mRNA (Fig. 4E).
As the relative changes in the individual Beclin1, Vps34, Bcl-2, Atg14 and UVRAG components do not necessarily indicate the status of the required macroautophagy complex-1, Vps34/p150/Beclin1/Atg14 (Funderburk et al., 2010; Itakura and Mizushima, 2009). Co-immunoprecipiation of Atg14 was therefore used to examine the assembly state of complex-1 (Fig. 4F). As previously shown for EDL muscle, mixed fiber quadriceps muscle from HSA-FynT mice had reduced protein levels of Vps34 but increased levels of Beclin1. Immunoprecipitation of Atg14 demonstrated near complete loss of Vps34 from complex 1 in HSA-FynT mice, with a concomitant increase in Beclin1 co-immunoprecipitated with Atg14. As a control, Vps34/Beclin1/UVRAG complex-2 that is involved in Rab5 and 7 positive endosome sorting was essentially unaffected (Backer, 2008; Funderburk et al., 2010). Together, these data demonstrate that FynT expression in skeletal muscle results in the loss of Vps34 from the Vps34/p150/Beclin1/Atg14 complex-1 that is required for macroautophagy that functions independent of mTORC1 regulation (Funderburk et al., 2010; He and Klionsky, 2009).
Blockade of STAT3 signaling reverses FynT inhibition of macroautophagy
To identify potential Fyn substrate targets, we next performed a phosphotyrosine proteome screen comparing skeletal muscle from Fyn null mice with HSA-FynT transgenic mice. Tyrosine phosphorylated STAT3 was significantly increased in the HSA-FynT skeletal muscle compared to Fyn null mice. Increased Y705-STAT3 phosphorylation was confirmed by phosphotyrosine immunoblotting of HSA-FynT and HSA-FynB gastrocnemius muscle (Fig. 3D). Consistent with weaker penetrance of HSA-FynB compared to HSA-FynT mice in terms of kyphosis, muscle mass and macroautophagy markers, Y705-STAT3 phosphorylation and the down regulation of Vps34 proteins was more robust in the HSA-FynT compared to HSA-FynB mice.
As observed in gastrocnemius muscle, EDL muscle from HSA-FynT mice displayed robust Y705F-STAT3 tyrosine phosphorylation but with only a small increase in STAT3 tyrosine phosphorylation in soleus muscle (Fig. 5A). Expression of the dominant-interfering STAT3 mutant (STAT3-Y705F) markedly increased autophagic flow in wild type tibialis anterior muscle (Fig. 5B). In parallel, there was also a marked induction of Vps34 protein expression (Fig. 5C). Surprisingly, there was also increased expression of Beclin1 and Atg14. The fact that Beclin1 was also increased by FynT expression and by expression of the dominant-interfering STAT3 mutant, suggests that the effect on Beclin1 is indirect. Nevertheless, STAT3-Y705F expression in tibialis anterior muscle of HSA-FynT mice reactivated autophagic flow and increased Vps34 protein expression (Fig. 5D and E). In this case, the ability of STAT3-Y705F to increase Beclin1 and Atg14 protein levels was somewhat reduced compared to wild type muscle. Moreover, Vps34 expression also restored autophagic flow to HSA-FynT tibialis anterior muscle (Fig. 5F) Taken together, these data support a model in which STAT3 functions upstream of Vps34.
Skeletal muscle fiber type specific regulation of endogenous STAT3 tyrosine phosphorylation and Vps34 expression by food restriction/refeeding
The data presented above reflect the effect of Fyn overexpression on muscle atrophy and macroautophagy but do not address the role of these events in normal physiological conditions. To address this issue, we examined skeletal muscle fiber-type specific signaling under prolonged food restriction conditions that induce, and following refeeding that suppresses skeletal muscle macroautophagy in wild type mice (Masiero et al., 2009; Ogata et al., 2010). As expected, the upstream mTORC1 activator, Akt displayed enhanced activation site phosphorylation (T308 and S473) in the refed state compared to the starved state. Similarly, mTORC1 activation paralleled the increased activation of Akt as detected by activation site phosphorylation of the mTORC1 downstream target S6 kinase. In contrast, α subunit activation site threonine (T172) of AMPK was increased in starved state and reduced in the refed state. Importantly, the phosphorylation of AMPK, Akt, and the mTORC1 specific substrate S6K1 phosphorylation site were essentially identical between the oxidative soleus and glycolytic EDL muscle (Fig. S7, A).
Although total LC3 protein levels were greater in soleus compared to EDL muscle, in the refed state only LC3-I was detectable in EDL muscle whereas both LC3-I and a smaller amount of LC3-II was presented in the soleus muscle (Fig. 6A). In the starved state, LC3-II levels increased with a concomitant reduction of LC3-I in EDL muscle whereas there was a relatively smaller increase in LC3-II in soleus muscle following starvation. Moreover, 30 h of food restriction increased Vps34 protein levels in EDL muscle with no significant change in Beclin1, Atg14, UVRAG, or Bcl-xl protein levels, although there was also a concomitant increase in Bcl-2 protein levels. Consistent with the soleus muscle being refractory to changes in STAT3 Y705 phosphorylation, prolonged food restriction had no statistically significant effect on Vps34, Beclin1, Atg14 or UVRAG protein levels in soleus muscle, although there was again a small increase in Bcl2 and a non-significant increase in Bcl-xl protein levels. Since macroautophagy is inhibited in the refed state and both Bcl-2 and Bcl-xl can suppress macroautophagy, the decrease in these proteins is not consistent with a block of macroautophagy in the refed state. As observed in HSA-FynT mice, the reduction in Vps34 levels in EDL muscle of fed mice was due to a selective decrease in Vps34 protein levels and was accompanied by only a small decrease in Vps34 mRNA (Fig. 6B).
We next examined the assembly state of the Vps34/p150/Beclin1/Atg14 in EDL and soleus muscle in starved and refed wild type mice (Fig. 6C). Immunoprecipitation of Atg14 from EDL muscle demonstrated reduced co-immunoprecipitation of Vps34 in the refed mice. In contrast, the amounts of Beclin1 and Vps34 co-immunoprecipitated with Atg14 was essentially unchanged in starved and refed soleus muscle. These data demonstrate that in wild type mice the Vps34/Beclin1/Atg14 complex-1 is regulated by starvation/refeeding in a skeletal muscle fiber specific manner analogous to that of transgenic overexpression of Fyn.
Regulation of STAT3 tyrosine phosphorylation and Vps34 protein levels by restriction/refeeding is Fyn-dependent
Although food restriction for 30 h followed by refeeding for 5 h had no significant effect on endogenous Fyn protein expression levels (Fig. 7A), Fyn protein kinase activity was increased approximately 2-fold in starved compared to refed glycolytic EDL muscle (Fig. 7B). In contrast, there was little effect on Fyn kinase activity in the oxidative soleus muscle. In parallel, EDL muscle displayed increased Y705 phosphorylation of STAT3 following refeeding whereas there was no significant change in soleus muscle (Fig. 7C).
To demonstrate that endogenous Fyn functions upstream of endogenous STAT3 and Vps34 genetically, we examined the effect of starvation/refeeding in Fyn null mice. As previously observed, Vps34 protein levels were decreased and Y705-STAT3 phosphorylation was increased in quadriceps muscle in refed wild type mice compared to starved mice (Fig. 7D). In contrast, Vps34 protein levels remained unchanged in starved and refed Fyn null mice. In parallel, there was no detectable increase of Y705-STAT3 phosphorylation in refed Fyn null mice. LC3-II levels were also increased in starved Fyn null mice concomitant with decreased p62 levels compared to wild type mice (Fig. 7E).
DISCUSSION
Muscle mass is maintained over time by two opposing processes, protein synthesis and degradation, that serve to increase and decrease muscle fiber size, respectively. These processes are regulated by a variety of intracellular and extracellular cues, including nutritional status. During states of nutritional deprivation (i.e., prolonged fasting) muscle wasting occurs through degradation of intracellular macromolecular components via the proteasome and lysosome-degradation systems, with the latter also used to generate substrates for energy production. In contrast, during nutritional excess the available substrates are used for macromolecular synthesis to increase muscle mass. The dynamic balance between these opposing processes is critical in maintaining appropriate muscle mass.
A variety of studies have established that the major pathway regulating muscle protein synthesis includes activation of the mTORC1 protein kinase complex, composed of the serine/threonine kinase mTOR plus the regulatory associated proteins Raptor, LST8/GβL, PRAS40 and Deptor (Kazi et al., 2011; Lee et al., 2007). The mTORC1 complex activates the initiation of protein synthesis through phosphorylation and activation of the S6 kinase and phosphorylation and inhibition of 4E-BP1 that suppresses 5′ mRNA capping protein eIF4E (Lee et al., 2007). mTORC1 itself is subject to a complex set of upstream regulators that integrate signaling from hormones, nutrients, amino acids, lipids and redox state (Howell and Manning, 2011). Under normal fed nutritional states, growth factors induce Akt activation that both inhibits TSC2 GAP activity and directly stimulates mTORC1 protein kinase activity. In addition, amino acids, particularly leucine, also activate mTORC1 through an as yet undefined mechanism that involves the Rag family of small GTP binding proteins and lysosomal targeting (Sancak et al., 2010). In contrast, during fasting/starvation or contraction, the decrease in muscle energy (ATP) and increase in AMP results in the activation of the AMP-dependent protein kinase (AMPK) that phosphorylates Raptor and blocks mTORC1 substrate recognition and activates the TSC2 GAP activity thereby suppressing mTORC1 function.
AMPK and mTORC1 also integrate with the macroautophagy system through the ULK1/Atg13/FIP200 complex (He and Klionsky, 2009; Mizushima, 2010). AMPK induces macroautophagy by ULK1 activation site phosphorylation site whereas mTORC1 phosphorylation of ULK1 inhibits its kinase activity and blocks phagophore formation (Egan et al., 2011; He and Klionsky, 2009; Kim et al., 2011). This integrative response between mTORC1 and macroautophagy provides a driving mechanism for the dynamic balance of protein synthesis and degradation in the maintenance of muscle mass in the fed and fasting state. This is consistent with genetic blockade of mTORC1 function resulting in muscle wasting, and in opposition, inhibition of AMPK that activates mTORC1 results in muscle hypertrophy (Bentzinger et al., 2008; Lantier et al., 2010). Surprisingly however, genetic blockade of macroautophagy as in Pompe mice results in muscle wasting in a fiber type specific manner (Raben et al., 2008). Similarly, muscle wasting in aging (sarcopenia) also occurs in a fiber type specific manner, primarily affecting fast-twitch glycolytic type II fibers with sparing of slow-twitch oxidative type I fibers (Bassel-Duby and Olson, 2006).
Interestingly, Fyn transgenic and Pompe mice both display spinal cord curvature (kyphosis) whereas there is no reported indication for this phenotype in ATG7 skeletal muscle deficient mice (Masiero et al., 2009). One possible explanation is that both the Fyn transgenic and Pompe mice have selective inhibition of glycolytic muscle macroautophagy that has little effect in oxidative muscle whereas deletion of ATG7 results in macroautophagy inhibition in both muscle fiber types.
In any case, the data presented in this study provide evidence for an alternative AMPK/mTORC1-independent pathway that may account for the muscle fiber type specificity of macroautophagy regulation and muscle wasting. The non-receptor tyrosine kinase Fyn has previously been shown to function as an upstream regulator of AMPK, fatty acid metabolism and energy expenditure pathways that are integral to muscle mass maintenance and macroautophagy (Hardie, 2011). Since Fyn inhibits AMPK activation and increases mTORC1 activity, we expected and induction of muscle hypertrophy analogous to that seen in AMPK knockout mice (Lantier et al., 2010). Although over expression of Fyn does, in fact, activate mTORC1 concomitant with inhibition of AMPK, these mice display marked muscle wasting. Starvation-induced inhibition and refeeding-induced activation of mTORC1 was essentially identical in both oxidative and glycolytic muscles. Thus, the changes mTORC1 activity per se does not account for the differing sensitivity of the soleus and EDL muscles to macroautophagy regulation. However, Fyn kinase activity was specifically activated in glycolytic muscle in the fed state that directly correlated with STAT3 tyrosine phosphorylation and reduction in Vps34 protein expression. Reduction in Vps34 protein primarily resulted in decreased content of the Vps34/Beclin1 complex-1 that is recruited to the site of autophagosome formation.
The Vps34/Beclin1 complex generates PI3P production necessary for phagophore elongation at the Atg7-dependent conjugation steps (He and Klionsky, 2009). In addition, PI3P plays critical roles in membrane transport, trafficking and membrane fusion events (Backer, 2008; Noda et al., 2010). Consistent with an essential role of Vps34, overexpression of Fyn reduced Vps34 protein levels and the amount of the Vps34/Beclin1 complex-1. The most immediate consequence of the reduced levels of complex-1 in the transgenic mice is a decrease in macroautophagy that appears to occur at multiple steps (including the conversion of LC3-I to LC3-II and reduced autophagic flow with decreased autophagic vacuoles). Importantly, in the refed state in which macroautophagy is inhibited, endogenous Fyn tyrosine kinase activity was unaffected in oxidative muscle but increased in glycolytic muscle. In contrast, in the starved state where macroautophagy is activated, Fyn activity was decreased in glycolytic muscle and again unchanged in soleus muscle. The relative changes in Fyn kinase activity directly correlated with STAT3 tyrosine phosphorylation and Vps34 protein levels. Furthermore, the Fyn-dependent decrease in Vps34 protein levels and inhibition of macroautophagy was reversed by expression of a dominant-interfering STAT3 mutant and by over expression of Vps34.
Importantly, we were able to genetically place Fyn upstream of STAT3 and Vps34 as the Fyn null mice failed to induce STAT3 tyrosine phosphorylation or to decrease Vps34 protein levels in the fed state. Taken together, these data support the presence of a Fyn/STAT3/Vps34 pathway that selectively regulates glycolytic muscle wasting independent of the classical mTORC1 pathway. Future studies are now needed to determine the upstream nutrient/hormone signals that regulate Fyn kinase activity in a muscle fiber type specific manner and the specific signaling mechanisms controlling Vps34 protein synthesis and/or degradation.
EXPERIMENTAL PROCEDURES
Transgenic mice
Mice over expressing murine FynB and FynT cDNA were generated using the human skeletal muscle actin (HSA) promoter with the SV40 polyadenylation and intron sequences (Khan et al., 2001) as detailed in Supplementary Methods. HSA-FynB, HSA-FynT and control littermates were housed in a facility equipped with a 12 h light/dark cycle. Animals were fed ad libitum a standard chow diet (Research Diets, New Brunswick, NJ) containing 67% (Kcal) carbohydrates, 19% protein, and 4% fat. All studies were approved by and performed in compliance with the guidelines of the Albert Einstein College of Medicine Institutional Animal Care and Use Committee (IACUC).
Antibodies and reagents
All antibodies and reagents were obtained from commercial sources as detailed in Supplementary Methods.
Quantitative PCR analysis
mRNA expression levels were determined by SYBR green (Qiagen, Valencia, CA, USA) or by TaqMan (Applied Biosystems, Branchburg, NJ, USA) quantitative RT-PCR as detailed in Supplementary Methods.
Magnetic resonance imaging and micro-Computed Tomography scanning
Fat and lean mass was determined using the EchoMRI-3-in-1™ System from Echo Medical Systems (Houston, TX, USA). Briefly, 10 weeks old animals were placed in a clear plastic holder without anesthesia or sedation and inserted into the EchoMRI-3-in-1™ System. Micro-Computed Tomography (CT) scans were performed on 16 week old mice using the Aloka LaTheta laboratory CT scan machine (Aloka, System Engineering Co., Tokyo, Japan).
Histology and electron microscopy
Muscle samples for histology were stained with H&E, Cytochrome C Oxidase, Succinate Dehydrogenase, Non-specific esterase, Acid phosphatase and Gomori Trichrome as described by Maclidan et al. (Malicdan et al., 2009) as detailed in Supplementary Methods. Muscle samples for electron microscopy were prepared and analyzed as previously described (Koga et al., 2010) as detailed in Supplementary Methods.
Western blotting and immunoprecipitation
Immunoblotting and immunoprecipitation of various skeletal muscle proteins were performed using standard procedures as detailed in Supplementary Methods.
Fyn kinase activity
Fyn tyrosine kinase activity was determined with a Universal Tyrosine kinase activity assay kit (Takara Bio Inc., Shiga, Japan) according to the manufacturer’s instructions as detailed in Supplementary Methods.
Skeletal muscle transfection in vivo
The tibialis anterior muscle was transfected with cDNA vectors by electroporation as detailed in Supplementary Methods.
Autophagic flow
Tibialis anterior muscles of mice fasted for 21 h were cut into pieces and incubated into oxygenated DMEM supplemented with 10%FBS either with or without 40 mM NH4Cl, 100 μM leupeptin for 1 h.
Statistics
Results are expressed as mean ± standard error of the mean (SEM). Differences between animals and/or treatments were tested for statistical significance using Student’s unpaired t test.
Supplementary Material
Acknowledgments
We wish to thank Drs. Jonathan M. Backer, Rani S. Sellers and Rajat Singh for helpful discussions during the course of these studies. We also thank Dr. Xiasong Li for technical advice for microCT analysis, Ms. Risa Yamada and Dr. Haihong Zong for experimental assistance. This work was supported in part by grants DK033823, DK082694 and DK020541 from the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alessi DR, Sakamoto K, Bayascas JR. LKB1-dependent signaling pathways. Annu Rev Biochem. 2006;75:137–163. doi: 10.1146/annurev.biochem.75.103004.142702. [DOI] [PubMed] [Google Scholar]
- Backer JM. The regulation and function of Class III PI3Ks: novel roles for Vps34. Biochem J. 2008;410:1–17. doi: 10.1042/BJ20071427. [DOI] [PubMed] [Google Scholar]
- Bassel-Duby R, Olson EN. Signaling pathways in skeletal muscle remodeling. Annu Rev Biochem. 2006;75:19–37. doi: 10.1146/annurev.biochem.75.103004.142622. [DOI] [PubMed] [Google Scholar]
- Bastie C, Zong H, Xu J, Busa B, Judex S, Kurland IJ, Pessin JE. Role of the Src Kinase Family Member Fyn in the Integrative Metabolic Regulation of Peripheral Tissue Fatty Acid Oxidation. Cell Metab. 2007;5 doi: 10.1016/j.cmet.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Bechet D, Tassa A, Taillandier D, Combaret L, Attaix D. Lysosomal proteolysis in skeletal muscle. Int J Biochem Cell Biol. 2005;37:2098–2114. doi: 10.1016/j.biocel.2005.02.029. [DOI] [PubMed] [Google Scholar]
- Bentzinger CF, Romanino K, Cloetta D, Lin S, Mascarenhas JB, Oliveri F, Xia J, Casanova E, Costa CF, Brink M, et al. Skeletal muscle-specific ablation of raptor, but not of rictor, causes metabolic changes and results in muscle dystrophy. Cell Metab. 2008;8:411–424. doi: 10.1016/j.cmet.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Brignatz C, Paronetto MP, Opi S, Cappellari M, Audebert S, Feuillet V, Bismuth G, Roche S, Arold ST, Sette C, et al. Alternative splicing modulates autoinhibition and SH3 accessibility in the Src kinase Fyn. Mol Cell Biol. 2009;29:6438–6448. doi: 10.1128/MCB.00398-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiacchiera F, Simone C. The AMPK-FoxO3A axis as a target for cancer treatment. Cell Cycle. 2010;9:1091–1096. doi: 10.4161/cc.9.6.11035. [DOI] [PubMed] [Google Scholar]
- Cooke MP, Perlmutter RM. Expression of a novel form of the fyn proto-oncogene in hematopoietic cells. New Biol. 1989;1:66–74. [PubMed] [Google Scholar]
- Davidson D, Viallet J, Veillette A. Unique catalytic properties dictate the enhanced function of p59fynT, the hemopoietic cell-specific isoform of the Fyn tyrosine protein kinase, in T cells. Mol Cell Biol. 1994;14:4554–4564. doi: 10.1128/mcb.14.7.4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funderburk SF, Wang QJ, Yue Z. The Beclin 1-VPS34 complex--at the crossroads of autophagy and beyond. Trends Cell Biol. 2010;20:355–362. doi: 10.1016/j.tcb.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Oskoui PR, Banko MR, Maniar JM, Gygi MP, Gygi SP, Brunet A. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J Biol Chem. 2007;282:30107–30119. doi: 10.1074/jbc.M705325200. [DOI] [PubMed] [Google Scholar]
- Grumati P, Coletto L, Sabatelli P, Cescon M, Angelin A, Bertaggia E, Blaauw B, Urciuolo A, Tiepolo T, Merlini L, et al. Autophagy is defective in collagen VI muscular dystrophies, and its reactivation rescues myofiber degeneration. Nat Med. 2010;16:1313–1320. doi: 10.1038/nm.2247. [DOI] [PubMed] [Google Scholar]
- Hardie DG. AMP-activated protein kinase: a cellular energy sensor with a key role in metabolic disorders and in cancer. Biochem Soc Trans. 2011;39:1–13. doi: 10.1042/BST0390001. [DOI] [PubMed] [Google Scholar]
- He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell JJ, Manning BD. mTOR couples cellular nutrient sensing to organismal metabolic homeostasis. Trends Endocrinol Metab. 2011;22:94–102. doi: 10.1016/j.tem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Mizushima N. Atg14 and UVRAG: mutually exclusive subunits of mammalian Beclin 1-PI3K complexes. Autophagy. 2009;5:534–536. doi: 10.4161/auto.5.4.8062. [DOI] [PubMed] [Google Scholar]
- Jansen M, Ten Klooster JP, Offerhaus GJ, Clevers H. LKB1 and AMPK family signaling: the intimate link between cell polarity and energy metabolism. Physiol Rev. 2009;89:777–798. doi: 10.1152/physrev.00026.2008. [DOI] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. Embo J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazi AA, Hong-Brown L, Lang SM, Lang CH. Deptor knockdown enhances mTOR activity and protein synthesis in myocytes and ameliorates disuse muscle atrophy. Mol Med. 2011 doi: 10.2119/molmed.2011.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AH, Thurmond DC, Yang C, Ceresa BP, Sigmund CD, Pessin JE. Munc18c regulates insulin-stimulated glut4 translocation to the transverse tubules in skeletal muscle. J Biol Chem. 2001;276:4063–4069. doi: 10.1074/jbc.M007419200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball SR. Interaction between the AMP-activated protein kinase and mTOR signaling pathways. Med Sci Sports Exerc. 2006;38:1958–1964. doi: 10.1249/01.mss.0000233796.16411.13. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga H, Kaushik S, Cuervo AM. Altered lipid content inhibits autophagic vesicular fusion. The FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2010;24:3052–3065. doi: 10.1096/fj.09-144519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lage R, Dieguez C, Vidal-Puig A, Lopez M. AMPK: a metabolic gauge regulating whole-body energy homeostasis. Trends Mol Med. 2008;14:539–549. doi: 10.1016/j.molmed.2008.09.007. [DOI] [PubMed] [Google Scholar]
- Lantier L, Mounier R, Leclerc J, Pende M, Foretz M, Viollet B. Coordinated maintenance of muscle cell size control by AMP-activated protein kinase. FASEB J. 2010;24:3555–3561. doi: 10.1096/fj.10-155994. [DOI] [PubMed] [Google Scholar]
- Lee CH, Inoki K, Guan KL. mTOR pathway as a target in tissue hypertrophy. Annu Rev Pharmacol Toxicol. 2007;47:443–467. doi: 10.1146/annurev.pharmtox.47.120505.105359. [DOI] [PubMed] [Google Scholar]
- Long YC, Zierath JR. AMP-activated protein kinase signaling in metabolic regulation. J Clin Invest. 2006;116:1776–1783. doi: 10.1172/JCI29044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z, Saha AK, Xiang X, Ruderman NB. AMPK, the metabolic syndrome and cancer. Trends Pharmacol Sci. 2005;26:69–76. doi: 10.1016/j.tips.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Malicdan MC, Noguchi S, Nishino I. Monitoring autophagy in muscle diseases. Methods Enzymol. 2009;453:379–396. doi: 10.1016/S0076-6879(08)04019-6. [DOI] [PubMed] [Google Scholar]
- Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Masiero E, Agatea L, Mammucari C, Blaauw B, Loro E, Komatsu M, Metzger D, Reggiani C, Schiaffino S, Sandri M. Autophagy is required to maintain muscle mass. Cell Metab. 2009;10:507–515. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22:132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounier R, Lantier L, Leclerc J, Sotiropoulos A, Pende M, Daegelen D, Sakamoto K, Foretz M, Viollet B. Important role for AMPKalpha1 in limiting skeletal muscle cell hypertrophy. FASEB J. 2009;23:2264–2273. doi: 10.1096/fj.08-119057. [DOI] [PubMed] [Google Scholar]
- Noda T, Matsunaga K, Taguchi-Atarashi N, Yoshimori T. Regulation of membrane biogenesis in autophagy via PI3P dynamics. Semin Cell Dev Biol. 2010;21:671–676. doi: 10.1016/j.semcdb.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Ogata T, Oishi Y, Higuchi M, Muraoka I. Fasting-related autophagic response in slow- and fast-twitch skeletal muscle. Biochem Biophys Res Commun. 2010;394:136–140. doi: 10.1016/j.bbrc.2010.02.130. [DOI] [PubMed] [Google Scholar]
- Raben N, Hill V, Shea L, Takikita S, Baum R, Mizushima N, Ralston E, Plotz P. Suppression of autophagy in skeletal muscle uncovers the accumulation of ubiquitinated proteins and their potential role in muscle damage in Pompe disease. Hum Mol Genet. 2008;17:3897–3908. doi: 10.1093/hmg/ddn292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risson V, Mazelin L, Roceri M, Sanchez H, Moncollin V, Corneloup C, Richard-Bulteau H, Vignaud A, Baas D, Defour A, et al. Muscle inactivation of mTOR causes metabolic and dystrophin defects leading to severe myopathy. J Cell Biol. 2009;187:859–874. doi: 10.1083/jcb.200903131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito YD, Jensen AR, Salgia R, Posadas EM. Fyn: a novel molecular target in cancer. Cancer. 2010;116:1629–1637. doi: 10.1002/cncr.24879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri M. Autophagy in skeletal muscle. FEBS Lett. 2010;584:1411–1416. doi: 10.1016/j.febslet.2010.01.056. [DOI] [PubMed] [Google Scholar]
- Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9:563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Cuervo AM. Autophagy in the cellular energetic balance. Cell Metab. 2011;13:495–504. doi: 10.1016/j.cmet.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witczak CA, Sharoff CG, Goodyear LJ. AMP-activated protein kinase in skeletal muscle: from structure and localization to its role as a master regulator of cellular metabolism. Cell Mol Life Sci. 2008;65:3737–3755. doi: 10.1007/s00018-008-8244-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada E, Pessin JE, Kurland IJ, Schwartz GJ, Bastie CC. Fyn-dependent regulation of energy expenditure and body weight is mediated by tyrosine phosphorylation of LKB1. Cell Metab. 2010;11:113–124. doi: 10.1016/j.cmet.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.