Abstract
Background
The neuropeptide calcitonin gene–related peptide (CGRP) plays a key role in migraine. CGRP gene expression involves an enhancer that is active in neurons, yet inactive in glia. In this report, we analyze epigenetic modifications that allow enhancer activation in glia.
Methods
DNA methylation and histone acetylation states were measured in rat and human-model cell lines and primary cultures of rat trigeminal ganglia glia. The functional consequence of altering the chromatin state was determined by quantitative measurements of both calcitonin (CT) and CGRP mRNAs.
Results
A hypermethylated CpG island flanking the enhancer was identified in glia and non-expressing cell lines. In addition, the chromatin was hypoacetylated. Treatment with the DNA methylation inhibitor 5-aza-2′-deoxycytidine induced CT mRNA ~30-fold in glial cultures. Treatment with a histone deacetylase inhibitor alone had little effect; however, the combination of inhibitors yielded a synergistic ~80-fold increase in CT and ~threefold increase in CGRP mRNA. Treated glia contained CT precursor (pro-CT) immunoreactivity.
Conclusions
Epigenetic modulation is sufficient to induce the CGRP gene in glia. Because the CGRP gene is systemically activated by inflammatory conditions, this suggests that glial pro-CT may be an unexplored biomarker during migraine.
Keywords: CGRP, trigeminal ganglia, glia, epigenetics, pro-calcitonin, gene expression
Introduction
Elevated levels of the neuropeptide calcitonin gene– related peptide (CGRP) have been linked to the onset and pathology of migraine (1–4). The significance of CGRP in migraine has been demonstrated by induction of migraine-like headaches following CGRP injection (5) and effective migraine relief by CGRP receptor antagonists (6–8). CGRP is encoded by the CALCA gene, which yields both CGRP and the hormone calcitonin (CT) as alternative splice products (9). CALCA expression is normally primarily restricted to endocrine and neuronal cells and is not normally expressed by glia. Given the increasing evidence that glia play important roles in pain conditions (10,11), including possibly migraine (12–14), we have investigated the mechanisms that normally prevent expression of the CGRP gene in glia.
Neuronal expression of the CALCA gene has been attributed to an 18-bp enhancer located about 1 kb upstream of the transcription start site. This enhancer is sufficient for cell-specific CALCA gene expression in neuronal-like cell lines and trigeminal ganglia neurons (15–17). The enhancer is activated by the bHLH-Zip upstream stimulatory factor (USF) (17–19). USF can also act with the forkhead protein FoxA2 to synergistically activate the enhancer, although FoxA2 is not present in trigeminal ganglia (19). We previously proposed that the combination of a relatively high USF level in neurons and a less-than-optimal USF-binding site in the 18-bp enhancer could contribute to its neural-specificity (17). However, a similar level of USF1 and USF2 was detected in the CALCA-expressing CA77 cell line as in the non-expressing Rat2 cell line (KY Park and AF Russo, unpublished observation). This indicates that additional mechanisms must restrict CALCA gene expression.
Epigenetic modifications involving DNA methylation and histone deacetylation are well-established mechanisms for repressing gene expression (20,21). DNA methylation at cytosines within a cluster of CpG dinucleotides called a CpG island results in chromatin compaction and gene silencing. Previous studies reported a CpG island extending from about −1.8 kb into exon 1 of the human CALCA gene (22,23). However, correlation of the methylation status with CALCA gene expression was not clear; some “negative” tissues and cell lines (liver and small-cell lung carcinomas) actually express CALCA, and the restriction enzymes used only recognized a subset of CpG sites. In addition to CpG methylation, deacetylation of histones, especially H3 and H4, by histone deacetylases (HDACs) is associated with transcriptional repression (24). To date, histone modifications on the CALCA gene have not been reported.
In this report, we provide evidence that reversal of epigenetic silencing is sufficient for CALCA gene expression. Using rat and human cell lines as model systems, we found that a CpG island near the 18-bp enhancer was hypomethylated in expressing cells, while hypermethylated in non-expressing cells. In addition, histone acetylation was much higher in the CALCA geneexpressing cells than in non-expressing cells. A DNA methylation inhibitor 5-aza-2′-deoxycytidine (Aza-dC) (25) induced the CALCA gene in both non-expressing cell lines and primary glial cultures, demonstrating that DNA methylation represses CALCA expression. Addition of the histone deacetylase inhibitor trichostatin A (TSA) to Aza-dC-treated glial cultures further induced the CALCA gene. These findings are the first evidence that the CALCA gene can be induced in glia.
Methods
Cell lines and trigeminal ganglia glial cultures
Culture conditions for CA77, TT and NCI-H460 cells have been described (18,19). Rat2 cell line (ATCC, Manassas, VA, USA) was maintained in Dulbecco’s Modified Eagle’s Medium (high glucose), 10% fetal bovine serum, penicillin (100 units/ml) and streptomycin (100 μg/ml) at 37°C in 5% CO2. Rat trigeminal ganglia cells were isolated and dispersed as described (17). Rat trigeminal glial cultures were prepared as described (14), with minor modifications. Ganglia were isolated from two-to three-day-old Sprague-Dawley rat pups, treated with 10 mg/ml dispase II (Invitrogen, Carlsbad, CA, USA) at 37°C for 30 minutes, then subjected to vigorous trituration with rapid pipetting, followed by centrifugation at 250 × g for three minutes. The supernatant was transferred to a new tube and centrifuged at 500 × g for five minutes to pellet glia. The pellet was resuspended in L-15 complete medium: L-15 with L-glutamine, 10% fetal bovine serum, penicillin (100 U/ml), streptomycin (100 μg/ml), amphotericin B (2.5 μg/ml), 50mM dextrose, 250 μM ascorbic acid, 10 ng/ml mouse 2.5S nerve growth factor (Alomone Labs, Jerusalem, Israel) and 8 μM glutathione. Cells were plated on 4 cm2 coverslips in a six-well dish. Fresh medium was applied the following day and experiments started two days after plating.
Rat2 and NCI-H460 cells were treated with freshly prepared Aza-dC (Sigma-Aldrich, St. Louis, MO, USA) in dimethylsulfoxide (DMSO) every other day for six days. As a vehicle control, the same volume of DMSO was added (usually 2 μl in 2-ml media). Glial cultures were treated with Aza-dC every day in fresh medium for four to five days. For all cultures, TSA was added 1 day prior to RNA extraction.
Bisulfite sequencing assay
Genomic DNA was isolated from CA77 and Rat2 cells using DNeasy kit (Qiagen, Valencia, CA, USA). Cytosine-to-thymine conversion by sodium bisulfite and DNA purification were performed using EZ DNA Methylation™ Kit (Zymo Research, Orange, CA, USA). The genomic region around the CpG island was amplified with two rounds of polymerase chain reaction (PCR) testing. The first round was: 95°C, 10 minutes; 40 cycles of 94°C, 1 minute, 55°C, 30 seconds, and 72°C, 30 seconds; then 72°C, 30 seconcs. PCR primers were: sense 5′-TTGTTAAGTTTGTTTGTTTGTTTT-3′, antisense 5′-TAAAAAAAAACCAAATACCTCTAA-3′. Ten percent of the reaction was subjected to a second round for 25 cycles at the same settings, but with a new sense primer, 5′-GAGATTTAAGGGTGGGGGTAGGAG-3′, and the same antisense primer. The PCR product was purified and cloned into pSC-A-amp/kan using StrataClone™ PCR Cloning Kit (StrataGen, Kirkland, WA, USA). Plasmids from individual clones were sequenced. Cytosine-to-thymine conversion efficiency was confirmed by monitoring non-CpG cytosine bases. In each cell line, there was 99.6% conversion (622 of 624 cytosine converted to thymine).
For human cell lines, TT and NCI-H460, the same procedure was performed except the PCR was: 95°C, 10 minutes; 40 cycles of 94°C, 1 minute, 59.2°C, 1 minute, 72°C, 1 minute; then 72°C, 10 minutes. For the CpG island 1, primers were: sense 5′-GATTAATTAAGGGTATTTTAGAAGTTAGG-3′, antisense 5′-CCTACCCTACCATCCATCACCTACCTAAAC-3′. For CpG island 2, primers were: sense 5′-AGAGTTGGAAGAGTTTTTATAATTTTGGATT-3′, antisense 5′-ACTTACAATTTAAAACTACTTATCCCTCAAT-3′.
Chromatin immunoprecipitation
The Upstate EZ ChIP™ (chromatin immunoprecipitation) instruction manual (Millipore, Billerica, MA, USA) was followed, with some modifications. Approximately 7 × 106 cells or dispersed cells from 24 neonatal trigeminal ganglia were fixed with 1% formaldehyde for 10 minutes, then quenched with 125mM glycine for 5 minutes. Cross-linked chromatin was sonicated five times (CA77 and ganglia cells) or twice (Rat2 cells) with 20-second pulses (output setting 3) at 1-minute intervals on ice (Ultrasonics, model W-375) to mostly in the 100 bp to ~2 kb size range. Protein G Plus beads (sc-2002, Santa Cruz Biotech, Santa Cruz, CA, USA) were coated with 0.03% bovine serum albumin (BSA) and 0.24 ug/ul sheared salmon sperm DNA (Invitrogen) for 30 minutes at 4°C. For each ChIP sample, 50 μl coated beads was added to cross-linked chromatin extracted from 1.4 × 106 cultured cells or 24 ganglia to preclear the chromatin for 2 hours at 4°C. Antibodies (10 μg, unless otherwise indicated) against histone H3 acetylated at Lys9, 14 (06-599, Millipore) or rabbit IgG (sc-2027, Santa Cruz Biotech) were added to the precleared chromatin and incubated overnight at 4°C. The chromatin-antibody complex was precipitated by incubation with freshly coated beads for 2 hours at 4°C. After extensive washing, chromatin was eluted and crosslinks reversed by incubating in 190mM NaCl at 65°C for 6 hours. The eluate was incubated at 37°C, 30 minutes with 10 μg RNase A (Fermentas, Glen Burnie, MD, USA). Proteins were removed with Proteinase K (Qiagen) at 45°C for 1 hour. DNA was purified using DNeasy® Blood & Tissue kit (Qiagen).
To amplify the 18-bp enhancer region, PCR reactions were: 94°C for 5 minutes; 31 cycles of 94°C, 1 minute, 61°C, 30 seconds, and 72°C, 30 seconds; 72°C, 10 minutes. Primers were: rat 18-bp (GenBank M34090.1) sense 5′-TAAGGGTGGGGGTAGGAG-3′, antisense 5′-CCGCCTGCCTAAGGATTT-3′, rat glyceraldehyde 3-phosphate dehydrogenase (Gapdh) promoter (GenBank AB047300.1) sense 5′-TTCAGATGCATGCTGGGAGCAAAC-3′, antisense 5′-GGACACAAACAACACCCGCTGAAA-3′. PCR products were confirmed by sequencing. To quantify the signals, three independent ChIP assays were performed for CA77 and Rat2 cells with measurement of band intensity using Image J software. The no antibody control was subtracted as background. The signal was normalized to the input band obtained by amplification of 1/100 of the sheared DNA used for cross-linking.
Reverse transcription–PCR and real-time quantitative PCR
RNA was isolated using a QIAshredder column and RNeasy Mini Kit with DNase I treatment (Qiagen). For Rat2 cells: 500 ng RNA was reverse transcribed using a random primer (Applied Biosystems, Carlsbad, CA, USA). Two rounds of PCR were necessary to reliably detect CALCA RNAs. Thirty to 50% of the RT reaction was used for the first PCR, followed by 30% for a second PCR. PCR protocol was: 94°C, 5 minutes; 30 cycles of 94°C, 1 minute, 60°C, 30 seconds, and 72°C, 30 seconds; 72°C, 5 minutes. Primers were: CT/CGRP (GenBank M11597.1) exon 2 sense 5′-CCCTTTCCTGGTTGTCAGCATCTT-3′, exon 3 antisense 5′-CATGCCTGGGCTGC TTTCTAAGGTT-3′, CT (GenBank V01228.1) exon 4 antisense 5′-AGCATGCAGGTACTCAGATTCCCA-3′. CGRP (GenBank M11597) exon 3 sense and exon 5 antisense have been described (17). For NCI-H460 cells: 730 ng of RNA was reversed transcribed. Fifteen percent of the reverse transcription (RT) reaction was used in a single round of PCR to detect CALCA RNAs. PCR protocol was: 95°C, 5 minutes; 40 cycles of 95°C, 1 minutes, 58°C, 30 seconds, and 72°C, 30 seconds; 72°C, 10 minutes. Primers were: CT (126 bp) (RefSeq NM_001741.2) sense 5′-CTGGAGCAGGAGCAAGAGAG-3′, antisense 5′-GGGGAACGTGTGAAACTTGT-3′; CGRP (80 bp) (RefSeq NM_001033953.2) sense 5′-CTGGAG CAGGAGCAAGAGAG-3′, antisense 5′-TGAGTCA CACAGGTGGCAGT-3′; β-actin (142 bp) (RefSeq NM_001101.3) sense 5′-AGAAAATCTGGCACCACACC-3′, antisense 5′-GGGGTGTTGAAGGTCTCAAA-3′. PCR products were verified by sequencing.
For real-time quantitative PCR (Q-PCR), 500 ng DNase I-treated RNA was used for each RT reaction as described above. The Q-PCR reactions contained 10–30% of the RT reaction, 50nM CT or CGRP primers (described above or previously reported (17)) with SYBR Green, as recommended (Applied Biosystems). For normalization, 0.03% of the RT reaction was used for Q-PCR with 18S rRNA (GenBank V0127) primers (17). The Q-PCR protocol was 50°C, 2 minutes, 95°C, 3 minutes, then 40 cycles at 95°C, 15 seconds, 60.7°C, 30 seconds, 72°C, 1 minute. Each sample was measured in triplicate with delta-delta Ct normalization to 18S rRNA Ct values as described (17). Controls included samples with no RT enzyme added to the RT reaction and samples with H2O added in place of cDNA. Both controls yielded Ct values greater than 35, while experimental samples generally yielded Ct values of about 31 for CT and 22 for CGRP from vehicle-treated cultures.
Immunocytochemistry
Immunostaining of rat trigeminal glial cells was done as described (14), unless noted. Two days after culturing, cells were rinsed with phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde for 30 minutes, then permeabilized in 0.2% triton X-100 in PBS for 15 minutes at room temperature. Cells were blocked with 10% BSA in PBS for 30 minutes before overnight incubation at 4°C with mouse monoclonal anti-glial fibrillary acidic protein (GFAP) antibody (G3893, Sigma) (1 : 50 dilution) or β-tubulin III rabbit immunoglobulin G (IgG) (ab15568, Abcam, Cambridge, MA, USA) (1 : 100 dilution). For double immunostaining, anti-pro-CT rabbit IgG (ab53897, Abcam) (1 : 100 dilution) or anti-CT rabbit IgG (ab45007, Abcam) (1 : 100 dilution) was included with the GFAP antibody. All antibodies were diluted in 1.5% BSA. Negative controls without primary antibodies were done. Cells were blocked with 10% BSA for 30 minutes before 1-hour incubation with secondary antibodies fluorescein isothiocyanate (FITC)-donkey-anti-mouse (Jackson Laboratory, Bar Harbor, ME, USA) (1 : 200 dilution in 10% BSA) and RITC-donkey-anti-rabbit (Jackson Laboratory) (1 : 200 dilution in 10% BSA). Images were obtained with a confocal microscope (Zeiss) with the same detection gain settings for FITC and RITC channels.
Statistics
Student’s t-test with two-tailed distribution was used for most analyses. When more than two means were compared, one-way analysis of variance (ANOVA) with Tukey’s post-hoc test was used. Pearson’s chi-square was used to analyze methylation profiles. Clones were assigned to one of three bins based on the number of CpG sites methylated (0–2, 3–11 or 12–14). Expected values were obtained by multiplying the proportion of clones in a bin from the control group to the sum of observed clones of the treatment group. Degrees of freedom were calculated as (r − 1)(c − 1), where r and c are the number of rows and columns, respectively. A p value <.05 was considered significant.
Results
Methylation of CpG islands flanking the 18-bp enhancer region
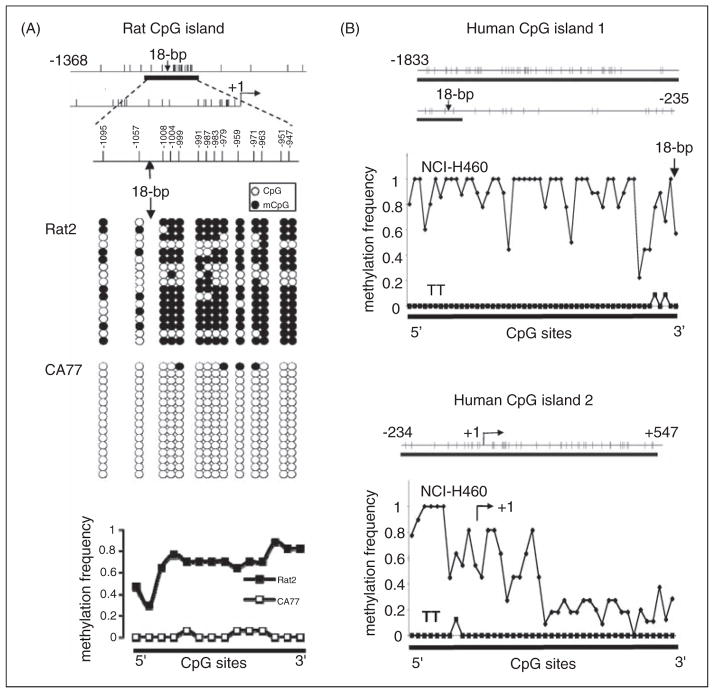
DNA methylation was examined in both rat and human cell lines as models for CALCA expressing and non-expressing cells. The neuronal-like rat CA77 and human TT thyroid C cell lines express the CALCA gene, while the rat embryo-derived Rat2 fibroblast cell line and human non-small-cell lung cancer NCI-H460 cell line do not (19). In the rat CALCA promoter region, we found one CpG island around the 18-bp enhancer at −1129 to −880 bp upstream of transcription start site using CpG island searcher (http://www.cpgislands.com) (Figure 1A). The CpG island meets the conventional criteria (26). It is 250 bp with a 60% G+C content and an observed/expected CpG ratio of 0.6 (calculated as # of nucleotides × # of CpG divided by # of C × G). The methylation status of the CpG island was assessed using the bisulfite cytosine to thymine conversion method followed by sequencing. In Rat2 cells, CpG sites were mostly methylated, with some variability among clones (Figure 1A). In contrast, CA77 cells had nearly all CpG sites unmethylated (Figure 1A). The methylation frequency throughout the CpG island was approximately 70% in Rat2 cells, while below 10% in CA77 cells. Thus, hypermethylation at the 18-bp CpG island is correlated with CALCA gene silencing.
Figure 1.
Methylation status of CpG islands flanking the 18-bp CALCA enhancer. (A) CpG island in the rat CALCA promoter region. Each vertical line represents a CpG dinucleotide within the region shown relative to the transcription start site (at +1 bp). The CpG island is shown as a thick horizontal bar and location of the 18-bp enhancer is indicated. Unmethylated CpG (open circles) and methylated CpG (mCpG, filled circles) sites within the island are shown from 17 Rat2 clones and 16 CA77 clones. Each row represents one clone. Methylation frequency at each CpG in Rat2 and CA77 cell lines is shown from left to right (not to scale). The methylation frequency is the number of clones with methylated CpG divided by the total number of clones. (B) CpG islands in the human CALCA promoter region. CpG island 1 (upper panel) in the distal promoter region and CpG island 2 (lower panel) in the proximal promoter region of CALCA are shown as thick horizontal bars. Each vertical line indicates one CpG site within the indicated region (base pairs relative to transcription start site). The methylation frequencies of CpG sites in NCI-H460 clones (9 and 11 for islands 1 and 2, respectively) and TT clones (11 and 8 for islands 1 and 2, respectively) were calculated as in panel A. If the sequence was not clear, the CpG from that clone was not included in the calculation (>5 clones used for all sites).
We then analyzed the human CALCA gene. Two CpG islands were identified: one at the distal promoter region (from −1833 to −891 bp upstream of transcription start site), which includes the 18-bp enhancer and the other encompassing the proximal promoter and exon 1 (from −234 to +519 bp) (Figure 1B). These sites coincide with the two CpG rich regions within a previously identified CpG island (23). Both CpG islands were highly methylated in the CALCA non-expressing non-small-cell lung cancer NCI-H460 cell line (Figure 1B). Hypermethylation was more pronounced at the 5′ flanking region than in the exon. In contrast, the islands were unmethylated in the CALCA expressing human medullary thyroid carcinoma TT cell line (Figure 1B). The cell-specific pattern of CpG methylation suggests that DNA methylation could account for both rat CALCA and human CALCA gene silencing.
Histone H3 acetylation at the 18-bp enhancer
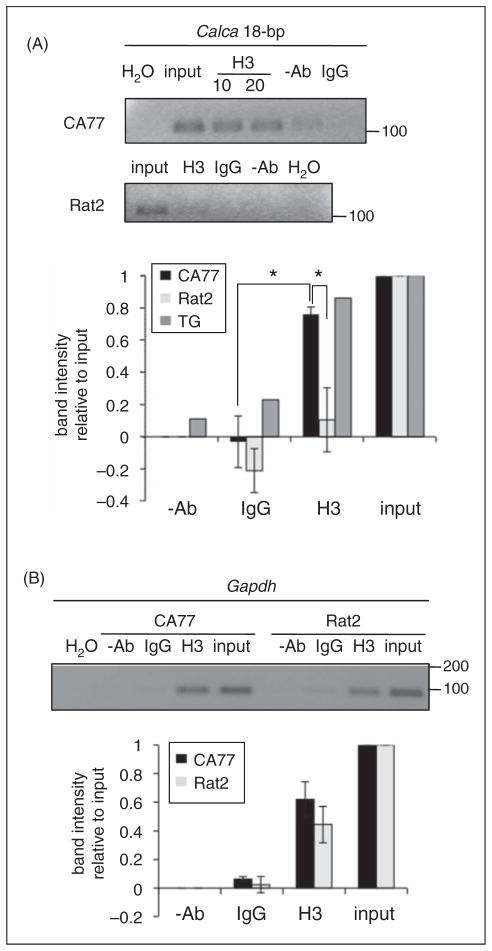
To examine histone acetylation, we performed ChIP assays with the CA77 and Rat2 cell lines. PCR amplification of the 18-bp region following immunoprecipitation with antibodies against histone H3 acetylated at Lys9, 14 (AcH3) yielded a robust signal in CA77 cells, but not in Rat2 cells (Figure 2A). Control immunoprecipitations with IgG and without any antibody yielded only background signals. A similar enrichment was seen with dissociated (not cultured) rat trigeminal ganglia. Enrichment of the 18-bp enhancer with AcH3 antibodies in CA77 cells was greater than in Rat2 cells by almost eightfold (p<.05) (Figure 2A). As a control, the ChIP assay was performed with the same DNA using primers specific for rat GAPDH promoter. Histone H3 and H4 have been reported to be hyperacetylated at the chicken GAPDH promoter (27). A similar signal was observed after ChIP with AcH3 antibodies in both cell lines (Figure 2B). These data indicate that histone H3 acetylation could also be involved in cell-specific CALCA gene expression.
Figure 2.
Acetylation of histone H3 at the 18-bp enhancer. (A) CA77 cells, Rat2 cells or neonatal rat trigeminal ganglia (TG) tissues were harvested for ChIP with antibodies against AcH3K9, K14 (H3). For CA77 cells, two amounts of antibody were used (10 and 20 μg). Chromatin immunoprecipitation (ChIP) with rabbit immunoglobulin G (IgG), without antibodies (-Ab), and with H2O in place of DNA were negative controls. For normalization of the polymerase chain reaction (PCR) signals, 1% of the input DNA was included. Size markers (bp) are indicated. Band intensity was measured with histogram analysis using Image J software. Mean and standard error (SE) from three independent assays with CA77 and Rat2 cells (*, p<.05, Student’s t-test). For trigeminal ganglia, the band intensities are from a single experiment. (B) ChIP assay with primers spanning the GAPDH promoter region. Band intensities were measured as described in panel A, with mean and SE from three independent assays.
Activation of Calca gene expression in Rat2 and NCI-H460 cells by inhibition of DNA methylation
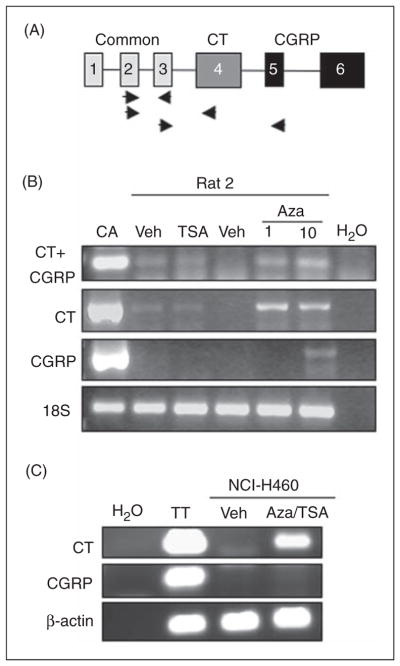
We then examined the functional consequences of CpG methylation and histone acetylation states on CALCA gene silencing in rat and human cell lines. Because the CALCA gene can produce both CT and CGRP through alternative splicing, we used primer sets to detect the common region of CT and CGRP (exons 2 and 3), CT only (exons 2 and 4), and CGRP only (exons 3 and 5) (Figure 3A). There was a detectable induction of CT mRNA when Rat2 cells were treated with 1 μg/ml Aza-dC to block DNA methylation (Figure 3B). As expected, a signal using the common region primers was also detected. When the Aza-dC concentration was increased to 10 μg/ml, CGRP mRNA could also be detected (Figure 3B). By contrast to demethylation, the HDAC inhibitor TSA had very little effect on CALCA expression in Rat2 cells (Figure 3B). Treatment with another HDAC inhibitor, sodium butyrate, also had little or no effect (not shown). Thus, DNA methylation, but not histone acetylation, appears to be a major factor controlling cell-specific CALCA gene expression in Rat2 cells.
Figure 3.
Induction of the CALCA gene by Aza-dC in Rat2 cells. (A) Schematic of the CALCA gene showing the three common exons 1–3 shared by calcitonin (CT) and calcitonin gene–related peptide (CGRP), CT-specific exon 4, and CGRP-specific exons 5, 6. Locations of primers used to measure CT+CGRP (exons 2 and 3), CT (exons 2 and 4), and CGRP (exons 3 and 5) mRNAs are shown. Note that exon 4 is removed from CGRP mRNA. (B) Rat2 cells were treated with 1 or 10 μg/ml Aza-dC (Aza) for six days, or 10 nM trichostatin A (TSA) for one day.
Dimethylsulfoxide (DMSO) was added as a vehicle control (Veh) for the appropriate number of days. RNA from CA77 cells (CA) was used as a positive control. As a polymerase chain reaction (PCR) control, H2O was added in place of cDNA. Amplification of 18S rRNA served as a control for total RNA levels. (C) NCIH460 cells were treated with 10 μg/ml Aza-dC for five days, then with 10 nM TSA for 1 day (Aza/TSA) prior to harvest. DMSO was added as a vehicle control (Veh). RNA from TT cells was a positive control. As PCR controls, H2O was added in place of cDNA and β-actin RNA was amplified from all samples.
We then tested the effect of Aza-dC (10 μg/ml) on NCI-H460 cells. Treatments were done with and without TSA (10nM). There was induction of CT mRNA by both Aza-dC alone (not shown) and the combination of Aza-dC and TSA (Figure 3C). There was no detectable CGRP mRNA. Thus, in both rat and human heterologous cell lines, the gene can be induced by epigenetic modifications and the primary splice product is CT mRNA.
Effects of Aza-dC and TSA on CALCA expression in glia
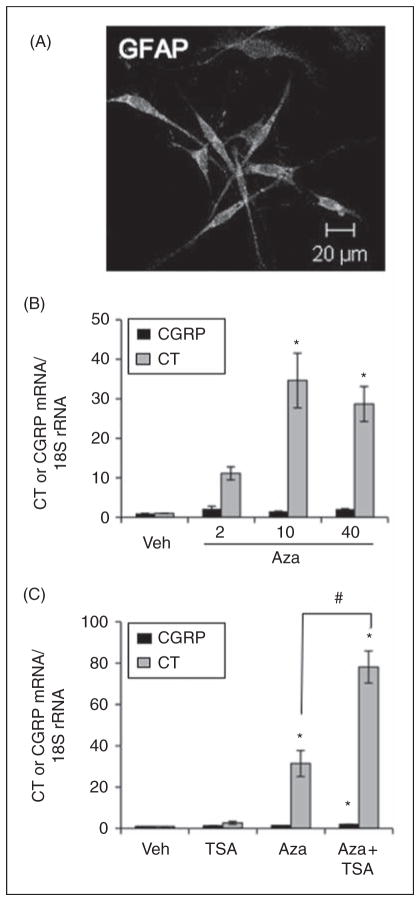
To expand the findings from the Rat2 cell line to a more physiological system, glial-enriched primary cultures from neonatal rat trigeminal ganglia were used. Under our culture conditions, we have previously shown that nearly all trigeminal ganglia neurons express CGRP immunoreactivity and display CALCA promoter activity in culture, but the co-cultured glia do not (16,17). In addition, early studies demonstrated that trigeminal ganglia neurons express CGRP, but not CT, immunoreactivity and that glia express neither peptide (9). A very small amount of CT RNA (0.1% of CGRP) was detected from intact ganglia, although whether this was from neurons or glia was not determined. Because an inherent complication of primary cultures is cellular heterogeneity, we first estimated the purity of the glial cultures. About 70–80% of the cultured cells had a satellite glia morphology and immunoreactivity for the glial marker GFAP (Figure 4A). The number of neurons defined by immunoreactivity for the neuronal marker β-tubulin III varied among cultures, with a range of 5–20% of the cells.
Figure 4.
Induction of the CALCA gene in glial cultures by AzadC. (A) Cultured satellite glia were identified by glial fibrillary acidic protein (GFAP) immunoreactivity. Magnification bar is indicated. (B) Glial cultures were treated with dimethylsulfoxide (DMSO) (Veh) or 2, 10 or 40 μg/ml of Aza-dC for four days followed by reverse transcription and real-time Q-PCR (quantitative polymerase chain reaction) to measure CGRP and calcitonin (CT) mRNA. The Q-PCR data were normalized to 18S rRNA. Mean and standard error (SE) are shown from at least three sets of independent experiments with one-way analysis of variance (ANOVA) followed by Tukey’s post-hoc tests for comparisons to Veh (*, p<.01). (C) Glial cultures were treated with dimethylsulfoxide (DMSO) (Veh) or 2 μg/ml Aza-dC for four days. For the last 1 day, 10 nM trichostatin A (TSA) was added to the vehicle or Aza-dC treated (Aza+TSA) cultures. Mean and SE are shown from three different cultures with one-way ANOVA followed by Tukey’s post-hoc tests with comparisons to Veh (*, p<.01) or between Aza and Aza+TSA treatments, as indicated (#, p<.01).
Bisulfite conversion and sequence analysis of the 14 CpG sites in clones from freshly dissociated glia revealed a bimodal distribution pattern. Of the 12 clones, eight had relatively high methylation (12–14 methylated sites) and three had low methylation (0–2 methylated sites). Only one clone was in the intermediate range (3–11 methylated sites). Similar results were obtained from 16 clones analyzed from vehicle-treated glia after five days in culture (11 high, 2 intermediate, 3 low). The low methylation clones are presumed to represent neurons, which were about 20% of the cells in these experiments. Treatment with 10 μg/ml Aza-dC for 5 days and 10nM TSA for the last 1 day increased the relative number of clones with intermediate methylation (10 high, 8 intermediate, 1 low). The distribution was significantly different than expected based on a chi-square comparison with vehicle treated cultures (p<.001).
The effect of Aza-dC treatment on CALCA gene expression was then measured. Glial cultures were treated with vehicle or Aza-dC for four to five days, followed by RT and Q-PCR. Both CT and CGRP mRNAs were detected in the vehicle treated cultures above background controls (see “Methods”). CGRP expression is presumed to be from residual neurons based on immunostaining (16). CT expression was much lower, ~0.2% of CGRP RNA levels, as reported with intact ganglia (9). Following incubation with Aza-dC, there was a dose-dependent ~30-fold increase in CT mRNA compared to vehicle (Figure 4B). There was also increased CGRP mRNA; however, in contrast to the robust increase of CT mRNA, the CGRP increase was<threefold, independent of dose, and not statistically significant.
Because histone acetylation and DNA methylation can work in a cooperative manner (28), we asked whether the combination of TSA and Aza-dC treatment would further increase CALCA expression in glia. In agreement with data from Rat2 cells, TSA alone did not induce the CALCA gene (Figure 4C). However, the CT mRNA level was greatly increased 78-fold by the combined treatment of Aza-dC and TSA (Figure 4C). The CGRP mRNA level was significantly increased threefold by the combination. This synergistic effect suggests that histone acetyltransferases might readily act on the CALCA gene upon DNA demethylation to enhance gene expression.
Induction of pro-CT expression in glia
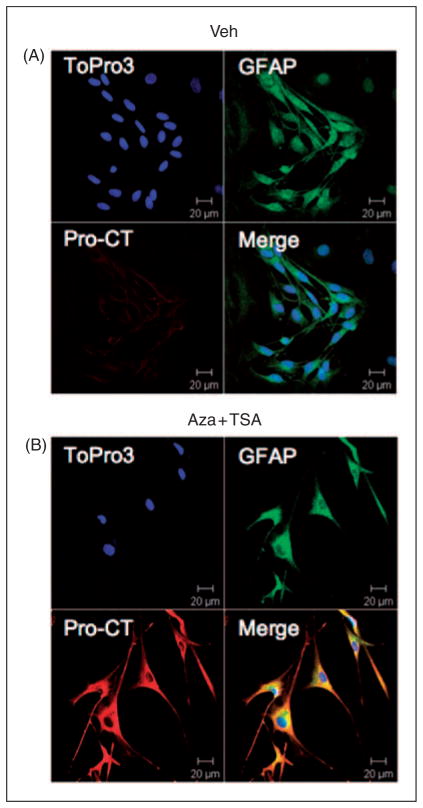
Given the large increase in CT mRNA, we asked if there was a corresponding increase in peptide expression. We could not detect mature CT immunoreactivty in the Aza-dC- and TSA-treated cultures (data not shown). We reasoned that glia may not have sufficient machinery for post-translational processing of the CT precursor protein (pro-CT), so we analyzed the cultures for pro-CT by immunocytochemistry. Immunoreactivity for pro-CT was detected in both control and Aza-dC-treated cultures. While there was some variability, glia treated with the combination of Aza-dC and TSA generally had greater pro-CT immunoreactivity compared to vehicle-treated cultures (Figure 5). From a random sampling of images (each with more than 10 glia), Aza-dC-and TSA-treated cultures had 11 pro-CT positive and three negative images, while DMSO-treated cultures had only three images with a pro-CT signal and nine negative images. Similarly, cultures treated with Aza-dC alone had greater pro-CT immunoreactivity (data not shown). These data indicate that Aza-dC-treated glial cells produce pro-CT.
Figure 5.
Pro-CT (calcitonin precursor) immunoreactivity in glia. Cultures were treated with (A) dimethylsulfoxide (DMSO) (Veh) or (B) combination of 10 μg/ml Aza-dC and 10 nM TSA (Aza+TSA) as in Figure 4C. Cultures were stained with anti–glial fibrillary acidic protein (GFAP) antibodies (glial marker, green), ToPro3 (nucleus marker, blue) and anti-pro-CT antibodies (red). Merged images and magnification bars are shown.
Discussion
Epigenetic regulation is important in development, tissue-specific gene expression and disease (28–30). In this study, we investigated whether epigenetic mechanisms underlie cell-specific CALCA gene expression. We found that CpG island methylation and histone H3 acetylation at the 18-bp cell-specific enhancer correlates with CALCA gene expression. The role of these epigenetic phenomena was tested using the DNA methylation inhibitor Aza-dC and the HDAC inhibitor TSA. While TSA failed to induce the CALCA gene, Aza-dC induced the gene in both human- and rat-cell lines and cultured glia. Interestingly, the combination of TSA and Aza-dC showed a synergistic effect on CALCA gene induction in glia. This indicates that DNA demethylation is required for the effect of histone acetylation to be manifested. This result indicates that CpG methylation around the 18-bp enhancer is a key determinant of cell-specific gene expression.
The observed splicing preference for CT over CGRP once the CALCA gene was activated in trigeminal glia is consistent with previous studies on the CT/CGRP splice choice. Transgenic mice engineered to express a metallothionein-CALCA fusion gene in all tissues had predominantly CT mRNA in non-neuronal cells, including Bergmann glia (31). Furthermore, the Fox-1/Fox-2 proteins required for neuronal production of CGRP mRNA are not present in mouse brain glia (32,33). The relative abundance of pro-CT over mature CT suggests there is insufficient peptide processing machinery in glia.
CpG methylation could conceivably reduce CALCA expression by interfering with USF activation of the 18-bp enhancer. Unfortunately, a USF complex could not be reliably detected by ChIP assays (data not shown), possibly because of the weaker affinity of the non-consensus USF site in the 18-bp enhancer (18). Nonetheless, a direct interference of USF binding, as seen with a hibernation-specific gene (34), seems unlikely since the USF site within the 18-bp enhancer does not have a consensus CpG dinucleotide. Presumably, methyl-binding proteins are recruited near the enhancer and recruit co-repressors to the CALCA gene. ChIP assays testing one candidate, MeCP2, did not detect significant binding of MeCP2 in Rat2 cells (data not shown). Future studies will be needed to examine other methyl-binding proteins and the growing list of other epigenetic silencing factors (35).
The mechanisms controlling dynamic regulation of CpG methylation are only recently becoming understood (36–39). In some cases, extracellular signals have been identified that can induce demethylation. Perhaps best characterized is demethylation of an alternative BDNF promoter in neurons by activity-dependent phosphorylation of MeCP2 (40,41). In addition, DNA methylation can be regulated by glucocorticoids (42), neonatal stress (43), vitamin D (44) and MAP kinases (45). While the regulator of CALCA demethylation has not yet been identified, there are some clues that inflammatory signals might play a role. In sepsis and other conditions of severe inflammation and systemic infection, the CALCA gene is widely induced in tissues that do not normally express it and the predominant product is pro-CT (46–49). Pro-CT can have deleterious biological activity based on animal sepsis studies (50,51). Interestingly, unlike mature CT, pro-CT has recently been shown to bind the CGRP receptor as a partial agonist (52).
Neurogenic inflammation and components of the immune system have long been implicated in migraine (53–57). We speculate that neurogenic inflammation with accompanying mast cell degranulation could lead to epigenetic induction of CALCA. Recently, Edvinsson described an adult rat trigeminal ganglia organ culture that undergoes an in situ inflammatory response (58). Under these conditions, there is induction of CGRP and pro-CT production in satellite glia (L Edvinsson, personal communication). Indeed, we also observed some glial pro-CT immunoreactivity in untreated cultures. While we did not detect induction of CT or CGRP mRNA in glia after treatment with an inflammatory cocktail (10 ng/ml lipopolysaccharide, 10 ng/ml TNFa, 10 ng/ml IL-1b for five days (data not shown)), it is possible that additional cytokines identified in the organ cultures, such as IL-6 (58), are needed, or that differences in the culture paradigms and/or tissue age may be important. To our knowledge, pro-CT has not been measured in migraineurs. Likewise, whether CGRP receptor antagonists in migraine clinical trials will prevent pro-CT actions is not known. Our findings suggest a potential mechanism by which the CALCA gene could be induced to produce pro-CT in trigeminal glia. We propose that pro-CT could be a biomarker of inflammatory activation of the trigeminal system in migraine.
Acknowledgments
We thank Adisa Kuburas for assistance, Charles Harata and Anne Kwitek for tissues, Lars Edvinsson for kindly sharing unpublished data and Rick Domann and Lori Wallrath for helpful discussions. Supported by National Institutes of Health Grants R01DE016511 and T32GM073610.
References
- 1.Arulmani U, Maassenvandenbrink A, Villalon CM, Saxena PR. Calcitonin gene-related peptide and its role in migraine pathophysiology. Eur J Pharmacol. 2004;500:315–330. doi: 10.1016/j.ejphar.2004.07.035. [DOI] [PubMed] [Google Scholar]
- 2.Durham PL. Inhibition of calcitonin gene-related peptide function: a promising strategy for treating migraine. Headache. 2008;48:1269–1275. doi: 10.1111/j.1526-4610.2008.01215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edvinsson L. Novel migraine therapy with calcitonin gene-regulated peptide receptor antagonists. Expert Opin Ther Targets. 2007;11:1179–1188. doi: 10.1517/14728222.11.9.1179. [DOI] [PubMed] [Google Scholar]
- 4.Doods H, Arndt K, Rudolf K, Just S. CGRP antagonists: unravelling the role of CGRP in migraine. Trends Pharmacol Sci. 2007;28:580–587. doi: 10.1016/j.tips.2007.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Lassen LH, Haderslev PA, Jacobsen VB, Iversen HK, Sperling B, Olesen J. CGRP may play a causative role in migraine. Cephalalgia. 2002;22:54–61. doi: 10.1046/j.1468-2982.2002.00310.x. [DOI] [PubMed] [Google Scholar]
- 6.Olesen J, Diener HC, Husstedt IW, et al. Calcitonin gene-related peptide receptor antagonist BIBN 4096 BS for the acute treatment of migraine. N Engl J Med. 2004;350:1104–1110. doi: 10.1056/NEJMoa030505. [DOI] [PubMed] [Google Scholar]
- 7.Ho TW, Ferrari MD, Dodick DW, et al. Efficacy and tolerability of MK-0974 (telcagepant), a new oral antagonist of calcitonin gene-related peptide receptor, compared with zolmitriptan for acute migraine: a randomised, placebo-controlled, parallel-treatment trial. Lancet. 2008;372:2115–2123. doi: 10.1016/S0140-6736(08)61626-8. [DOI] [PubMed] [Google Scholar]
- 8.Connor KM, Shapiro RE, Diener HC, et al. Randomized, controlled trial of telcagepant for the acute treatment of migraine. Neurology. 2009;73:970–977. doi: 10.1212/WNL.0b013e3181b87942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenfeld MG, Amara SG, Evans RM. Alternative RNA processing: determining neuronal phenotype. Science. 1984;225:1315–1320. doi: 10.1126/science.6089345. [DOI] [PubMed] [Google Scholar]
- 10.Watkins LR, Maier SF. Beyond neurons: evidence that immune and glial cells contribute to pathological pain states. Physiol Rev. 2002;82:981–1011. doi: 10.1152/physrev.00011.2002. [DOI] [PubMed] [Google Scholar]
- 11.Ohara PT, Vit JP, Bhargava A, et al. Gliopathic pain: when satellite glial cells go bad. Neuroscientist. 2009;15:450–463. doi: 10.1177/1073858409336094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Capuano A, De Corato A, Lisi L, Tringali G, Navarra P, Dello Russo C. Proinflammatory-activated trigeminal satellite cells promote neuronal sensitization: relevance for migraine pathology. Mol Pain. 2009;5:43. doi: 10.1186/1744-8069-5-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Villa G, Fumagalli M, Verderio C, Abbracchio MP, Ceruti S. Expression and contribution of satellite glial cells purinoceptors to pain transmission in sensory ganglia: an update. Neuron Glia Biol. 2010;6:31–42. doi: 10.1017/S1740925X10000086. [DOI] [PubMed] [Google Scholar]
- 14.Li J, Vause CV, Durham PL. Calcitonin gene-related peptide stimulation of nitric oxide synthesis and release from trigeminal ganglion glial cells. Brain Res. 2008;1196:22–32. doi: 10.1016/j.brainres.2007.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tverberg LA, Russo AF. Regulation of the calcitonin/calcitonin gene-related peptide gene by cell-specific synergy between helix-loop-helix and octamer-binding transcription factors. J Biol Chem. 1993;268:15965–15973. [PubMed] [Google Scholar]
- 16.Durham PL, Dong PX, Belasco KT, et al. Neuronal expression and regulation of CGRP promoter activity following viral gene transfer into cultured trigeminal ganglia neurons. Brain Res. 2004;997:103–110. doi: 10.1016/j.brainres.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 17.Park KY, Russo AF. Control of the calcitonin gene-related peptide enhancer by upstream stimulatory factor in trigeminal ganglion neurons. J Biol Chem. 2008;283:5441–5451. doi: 10.1074/jbc.M708662200. [DOI] [PubMed] [Google Scholar]
- 18.Lanigan TM, Russo AF. Binding of upstream stimulatory factor and a cell-specific activator to the calcitonin/calcitonin gene-related peptide enhancer. J Biol Chem. 1997;272:18316–18324. doi: 10.1074/jbc.272.29.18316. [DOI] [PubMed] [Google Scholar]
- 19.Viney TJ, Schmidt TW, Gierasch W, et al. Regulation of the cell-specific calcitonin/calcitonin gene-related peptide enhancer by USF and the Foxa2 forkhead protein. J Biol Chem. 2004;279:49948–49955. doi: 10.1074/jbc.M406659200. [DOI] [PubMed] [Google Scholar]
- 20.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 21.Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 22.Baylin SB, Hoppener JW, de Bustros A, Steenbergh PH, Lips CJ, Nelkin BD. DNA methylation patterns of the calcitonin gene in human lung cancers and lymphomas. Cancer Res. 1986;46:2917–2922. [PubMed] [Google Scholar]
- 23.Broad PM, Symes AJ, Thakker RV, Craig RK. Structure and methylation of the human calcitonin/alpha-CGRP gene. Nucleic Acids Res. 1989;17:6999–7011. doi: 10.1093/nar/17.17.6999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 25.Robert MF, Morin S, Beaulieu N, et al. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat Genet. 2003;33:61–65. doi: 10.1038/ng1068. [DOI] [PubMed] [Google Scholar]
- 26.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 27.Myers FA, Chong W, Evans DR, Thorne AW, Crane-Robinson C. Acetylation of histone H2B mirrors that of H4 and H3 at the chicken beta-globin locus but not at housekeeping genes. J Biol Chem. 2003;278:36315–36322. doi: 10.1074/jbc.M305822200. [DOI] [PubMed] [Google Scholar]
- 28.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 29.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 30.Dulac C. Brain function and chromatin plasticity. Nature. 2010;465:728–735. doi: 10.1038/nature09231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crenshaw EB, 3rd, Russo AF, Swanson LW, Rosenfeld MG. Neuron-specific alternative RNA processing in transgenic mice expressing a metallothionein-calcitonin fusion gene. Cell. 1987;49:389–398. doi: 10.1016/0092-8674(87)90291-1. [DOI] [PubMed] [Google Scholar]
- 32.Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol. 2005;25:10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou HL, Baraniak AP, Lou H. Role for Fox-1/Fox-2 in mediating the neuronal pathway of calcitonin/calcitonin gene-related peptide alternative RNA processing. Mol Cell Biol. 2007;27:830–841. doi: 10.1128/MCB.01015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fujii G, Nakamura Y, Tsukamoto D, Ito M, Shiba T, Takamatsu N. CpG methylation at the USF-binding site is important for the liver-specific transcription of the chipmunk HP-27 gene. Biochem J. 2006;395:203–209. doi: 10.1042/BJ20051802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poleshko A, Einarson MB, Shalginskikh N, et al. Identification of a functional network of human epigenetic silencing factors. J Biol Chem. 2010;285:422–433. doi: 10.1074/jbc.M109.064667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Metivier R, Gallais R, Tiffoche C, et al. Cyclical DNA methylation of a transcriptionally active promoter. Nature. 2008;452:45–50. doi: 10.1038/nature06544. [DOI] [PubMed] [Google Scholar]
- 37.Kangaspeska S, Stride B, Metivier R, et al. Transient cyclical methylation of promoter DNA. Nature. 2008;452:112–115. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- 38.Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma DK, Jang MH, Guo JU, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen WG, Chang Q, Lin Y, et al. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- 41.Martinowich K, Hattori D, Wu H, Fouse S, He F, Hu Y, et al. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- 42.Kress C, Thomassin H, Grange T. Active cytosine demethylation triggered by a nuclear receptor involves DNA strand breaks. Proc Natl Acad Sci USA. 2006;103:11112–11117. doi: 10.1073/pnas.0601793103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murgatroyd C, Patchev AV, Wu Y, et al. Dynamic DNA methylation programs persistent adverse effects of early-life stress. Nat Neurosci. 2009;12:1559–1566. doi: 10.1038/nn.2436. [DOI] [PubMed] [Google Scholar]
- 44.Kim MS, Kondo T, Takada I, et al. DNA demethylation in hormone-induced transcriptional derepression. Nature. 2009;461:1007–1012. doi: 10.1038/nature08456. [DOI] [PubMed] [Google Scholar]
- 45.Maddodi N, Bhat KM, Devi S, Zhang SC, Setaluri V. Oncogenic BRAFV600E induces expression of neuronal differentiation marker MAP2 in melanoma cells by promoter demethylation and down-regulation of transcription repressor HES1. J Biol Chem. 2010;285:242–254. doi: 10.1074/jbc.M109.068668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Snider RH, Jr, Nylen ES, Becker KL. Procalcitonin and its component peptides in systemic inflammation: immunochemical characterization. J Investig Med. 1997;45:552–560. [PubMed] [Google Scholar]
- 47.Muller B, White JC, Nylen ES, Snider RH, Becker KL, Habener JF. Ubiquitous expression of the calcitonini gene in multiple tissues in response to sepsis. J Clin Endocrinol and Metab. 2001;86:396–404. doi: 10.1210/jcem.86.1.7089. [DOI] [PubMed] [Google Scholar]
- 48.Becker KL, Snider R, Nylen ES. Procalcitonin assay in systemic inflammation, infection, and sepsis: clinical utility and limitations. Crit Care Med. 2008;36:941–952. doi: 10.1097/CCM.0B013E318165BABB. [DOI] [PubMed] [Google Scholar]
- 49.Becker KL, Nylen ES, White JC, Muller B, Snider RH., Jr Clinical review 167: Procalcitonin and the calcitonin gene family of peptides in inflammation, infection, and sepsis: a journey from calcitonin back to its precursors. J Clin Endocrinol Metab. 2004;89:1512–1525. doi: 10.1210/jc.2002-021444. [DOI] [PubMed] [Google Scholar]
- 50.Nylen ES, Whang KT, Snider RH, Jr, Steinwald PM, White JC, Becker KL. Mortality is increased by procalcitonin and decreased by an antiserum reactive to procalcitonin in experimental sepsis. Crit Care Med. 1998;26:1001–1006. doi: 10.1097/00003246-199806000-00015. [DOI] [PubMed] [Google Scholar]
- 51.Martinez JM, Wagner KE, Snider RH, et al. Late immunoneutralization of procalcitonin arrests the progression of lethal porcine sepsis. Surg Infect (Larchmt) 2001;2:193–202. doi: 10.1089/109629601317202678. [DOI] [PubMed] [Google Scholar]
- 52.Sexton PM, Christopoulos G, Christopoulos A, Nylen ES, Snider RH, Jr, Becker KL. Procalcitonin has bioactivity at calcitonin receptor family complexes: potential mediator implications in sepsis. Crit Care Med. 2008;36:1637–1640. doi: 10.1097/CCM.0b013e318170a554. [DOI] [PubMed] [Google Scholar]
- 53.Pietrobon D. Migraine: new molecular mechanisms. Neuroscientist. 2005;11:373–386. doi: 10.1177/1073858405275554. [DOI] [PubMed] [Google Scholar]
- 54.Goadsby PJ. Recent advances in understanding migraine mechanisms, molecules and therapeutics. Trends Mol Med. 2007;13:39–44. doi: 10.1016/j.molmed.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 55.Theoharides TC, Donelan J, Kandere-Grzybowska K, Konstantinidou A. The role of mast cells in migraine pathophysiology. Brain Res Brain Res Rev. 2005;49:65–76. doi: 10.1016/j.brainresrev.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 56.Levy D, Burstein R, Kainz V, Jakubowski M, Strassman AM. Mast cell degranulation activates a pain pathway underlying migraine headache. Pain. 2007;130:166–176. doi: 10.1016/j.pain.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Edvinsson L, Ho TW. CGRP receptor antagonism and migraine. Neurotherapeutics. 2010;7:164–175. doi: 10.1016/j.nurt.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kristiansen KA, Edvinsson L. Regulatory effect of inflammation on cytokines in rat trigeminal ganglia. Cephalalgia. 2009;29(Suppl 1):84. [Google Scholar]