Abstract
The peroxiredoxins (Prx) are ubiquitous peroxidases involved in important biological processes; however, details of their enzymatic mechanism remain elusive. To probe potential dynamics-function relationships, molecular dynamics simulations and electrostatic calculations were performed on the atypical 2-cysteine thiol peroxidase (Tpx) from Streptococcus pneumoniae and results compared to a previous study of a typical 2-cysteine Prx from Trypanosoma cruzi. The analyses indicate a commonality between both typical and atypical Prx: dynamic asymmetry. Asymmetry is observed in structure, fluctuations and active site electrostatics. Key residues, including Glu150 and Phe153, play roles in the developing asymmetry; furthermore, in the atypical 2-Cys Tpx, Glu150 exhibits conformation fluctuations suggesting involvement in a proton shuttle. The existence of a pathway of connected residues appears to propagate the asymmetry. The commonality of asymmetry and coupling pathways in both typical and atypical Prxs suggests a driving force towards dimer asymmetry as a common feature that plays a functional role in creating one active site with a lower cysteine pKa.
Keywords: Peroxidatic cysteine, molecular dynamics simulation, half sites reactivity, pKa, electrostatics
INTRODUCTION
Redox signaling and antioxidant defense are critical in maintaining the healthy function of cells. The proteins in the peroxiredoxin (Prx) protein family act as antioxidants and regulate redox signaling in higher organisms. These proteins are ubiquitous peroxidases lacking bound cofactors but exhibiting cysteine-dependent reactivity toward hydrogen peroxide, alkyl hydroperoxides, and peroxynitrite, all important products of oxidative and nitrosative damage and signaling.1 Through their protective role against oxidative damage, and their added roles in eukaryotes as regulators of redox signaling leading to proliferative responses, Prxs are of intense interest for probable roles in human disease.1a As antioxidants, the Prxs from parasitic organisms such as trypanosomatids and Mycobacterium tuberculosis play a critical role in microorganism defense against the reactive oxygen species (ROS) environment imposed by the host immune system.2 Thus, they are also of interest in the development of anti-infective agents.
Although these proteins have been the subjects of a variety of structural and functional studies, many details of their enzymatic mechanism remain elusive. All proteins in the family share a common chemistry that requires the activation of a cysteine, the peroxidatic cysteine (Cp), by deprotonation, followed by reaction with a hydroperoxide leading to the formation of a cysteine-sulfenic acid (Cys-SOH) intermediate.1b, 3 A specific model describing this mechanism has been proposed, which requires that the active site microenvironment somehow lowers the Cp pKa and that an unknown base serves to abstract a proton.1b, 4 The Cp pKa is not known for most Prxs; experimental determinations using peroxidase competition assays yield functional pKa values ranging from 5.2 to 6.3.5 Deprotonation of the cysteine likely involves interaction with an absolutely conserved Arg residue and hydrogen bonding to a highly conserved Thr (or Ser) residue.1b, 6 Crystallographic evidence suggests that the Thr and Arg side chains also play important roles in positioning and activating the incoming peroxide substrates through transition state stabilization within the Prx active site.7 The exact residues involved in, and mechanism of, deprotonation have yet to be identified.4
Following formation of the Cys-SOH, a second cysteine acts to “resolve” the Cys-SOH to a protein disulfide bond in many Prxs and this feature has been used to classify these enzymes into 1-Cys and 2-Cys Prx families.8 1-Cys Prxs are those proteins that do not contain a resolving cysteine. The 2-Cys Prxs contain a resolving cysteine (Cr) within the same protein. This latter group has been further categorized into “typical” and “atypical” classes based on whether the active site disulfide bond of the oxidized family members is formed between adjacent subunits (typical) or within a single subunit (atypical).1b Cellular thiol-containing redox donors such as thioredoxin (Trx) return these enzymes to their activated cysteine-thiol (Cys-SH) state through thiol-disulfide interchange reactions.9
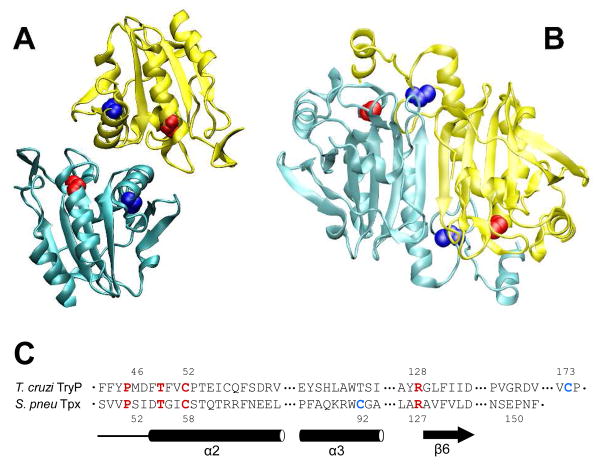
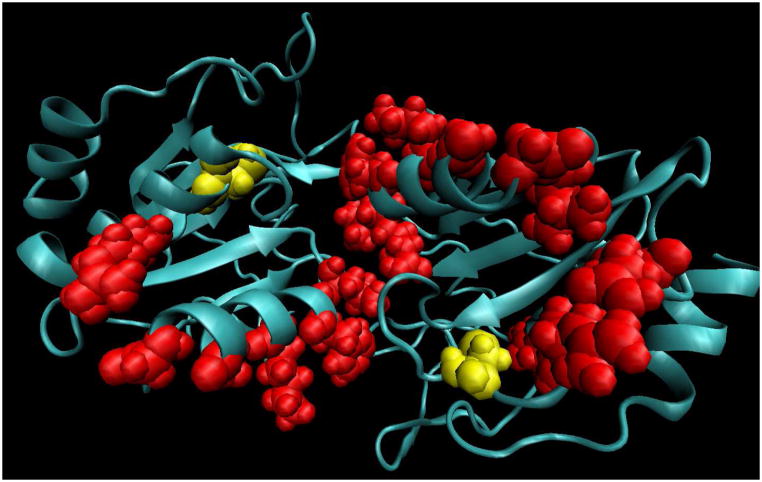
Structurally, all Prxs have a similar topology: a central 5-stranded beta sheet, 5 alpha helices and a 2-stranded beta hairpin.10 This general topology can be seen in the examples of atypical and typical 2-Cys Prxs, such as thiol peroxidase (Tpx) and tryparedoxin peroxidase (Figure 1). Most Prxs fold into interesting quaternary structures, including dimers, octamers, decamers, and dodecamers. Dimers form through one of two interface types: A (“ancestral” or “alternate”) and B (beta sheet).11 The atypical 2-Cys Prxs form homodimers across the A interface in solution (Figure 1A).11a The typical 2-Cys Prxs form dimers through the B interface, as seen for tryparedoxin peroxidase in Figure 1B. These dimers can then associate through the A interface to form larger oligomeric structures,3, 11a, 12 such as the decamer observed for tryparedoxin peroxidase.
Figure 1. Structures and sequence alignment of typical and atypical Prxs.
Cartoon diagrams of the biologically relevant dimers of thiol peroxidase (1psq, A) and tryparedoxin peroxidase (1uul, B). In both panels, monomer A is blue and monomer B is yellow. The peroxidatic cysteines (Cp, residues 58 in 1psq and 52 in 1uul) are shown as red van der Waal spheres; the resolving cysteines (Cr, residues 91 in 1psq and 173 in 1uul) are shown as blue spheres. Tpx, 1psq, is an atypical 2-cysteine Prx, so a complete functional active site, composed of one Cp and one Cr, are within a monomer. Tryparedoxin peroxidase, 1uul, is a typical 2-cysteine Prx, so the biological active sites consist of Cp from one subunit and Cr from the other subunit. Both of these structures have two active sites per dimer. C. Structure-based sequence alignments, with secondary structures (alpha helices and a beta strand; the designation of the secondary structural elements is according to the nomenclature in Hall and coworkers10) shown below the alignment. Four conserved residues across Prxs shown in red (including peroxidatic Cys). The resolving Cys, which differs among Prx subfamilies, is shown in blue. Sequences are for T. cruzi tryparedoxin peroxidase (TryP, 1uul) and S. pneumoniae thiol peroxidase (Tpx, 1psq).
The first step in the Prx reaction mechanism, abstraction of the sulfhydryl proton from Cp, is accomplished by an unknown base.1b, 9 Understanding interactions that lower the Cp pKa should provide insight into enzyme mechanism; however, the Prx active sites assume very different conformations under different redox conditions;6, 13 thus, observation of site-specific dynamics and conformational fluctuations must complement information from static crystal structures. In previous work, we studied the role of dynamics in the electrostatics of the typical 2-Cys Prx tryparedoxin peroxidase active site in both the dimer and decamer form.14 This work demonstrated that asymmetry developed between the active sites in the dimer simulations. In contrast, in individual monomer simulations, the structures evolved similarly.
Here, we extend this work by exploring the details of the asymmetry in Tpx, an atypical 2-Cys Prx, and compare the results to those from the previous study involving a typical 2-Cys Prx. The notion that coupled motions are important to the function of enzymes is not new; there is considerable evidence that protein motions can enhance enzymatic turnover, both through direct vibrational coupling and through varying protein conformations.15 This has been particularly well-explored in DHFR.15–16 The particular type of motions discovered here, ones that propagate asymmetry in the electrostatic environment of the active site, are different that those discovered in other systems. The dynamic asymmetry discovered here also suggests that landscape view of enzyme catalysis should be extended to include the effects of oligomerization.16a, 17 The active sites in typical and atypical 2-cysteine Prxs both contain Cp and Cr in an active site pocket. The two forms differ in that the typical 2-cysteine Prxs are obligate dimers, through B interface interactions, with Cp and Cr each on a different subunit (Figure 1). The atypical Prxs are also dimers, but through the A interface, and Cp and Cr for a single active site are found on a single monomer (Figure 1). Comparing the two active sites should provide insight into common features of the Prx reaction mechanism.
Simulations were performed on the dimers because the complete, functional active site is found within the dimer structures. For this work, monomer simulations were not performed due to our experiences with the typical 2-Cys Prx, in that due to the symmetry the monomers evolved similarly, and due to the additional flexibility that the monomers gained, they accessed conformational states that are not likely to be functional. Electrostatics analysis combined with molecular dynamics (MD) simulations was used to probe the details of the microenvironment around Cp, to observe nanosecond fluctuations that affect the Cp pKa and to suggest mechanisms by which the proton is abstracted from the Cp. The dynamics simulations suggest mechanisms by which the asymmetry is achieved in the typical and atypical 2-Cys Prxs. Specific residues that may be involved in proton abstraction are different in each protein. Asymmetry evolves in both types of Prx, but the pathway residues that potentially connect the active sites are different. We propose that the asymmetry may be important to the biological function of these proteins.
MATERIALS AND METHODS
Starting structures
1psq, a crystal structure of a Tpx, an atypical Prx from Streptococcus pneumoniae,18 was used as the starting point for molecular dynamics simulations. The 1psq structure was solved to a resolution of 2.3 Å and 0.249 Rfree value as part of the New York Structural Genomics Research Consortium. Monomers A and B were selected from the 1psq fully-reduced decamer.
Molecular dynamics (MD) protocol
Simulations require three distinct steps: preparation, equilibration, and production simulations. The standard methods used herein are as reported elsewhere14, 19 and are briefly summarized here. All calculations are performed with the NAMD package20 and the CHARMM param22 parameter set.21 All simulations were performed multiple times. Similar results were seen in each simulation.
The protein was prepared by adding hydrogen atoms and relieving bad steric contacts with short, constrained minimizations, resulting in a 0.09 Å backbone RMSD from the starting crystal structure. The structures were solvated with a solvent-box of TIP3P22 water molecules, then subjected to minimizations with constrained protein atoms and a 225 ps constant-pressure thermal equilibration with the temperature raised from 0 to 300K in 25K cycles. The SHAKE algorithm for all bonds containing hydrogen atoms23 and Particle Mesh Ewald (PME) with a ~1 Å grid24 were both utilized.
Production simulations were run under approximate NVE conditions using the same conditions and parameters as for thermal equilibration, except the temperature was not raised from 300K. The final state of the equilibration simulations was used as initial conditions for the production simulation.
Trajectory analysis
MD trajectories were analyzed by calculating the root mean squared deviation (RMSD) in atomic positions, simulation alpha carbon B-factors, and clustering. All methods were utilized as previously described.19
pKa calculations
Electrostatic calculations were performed using the semi-macroscopic method based on a finite difference approximation to the linear Poisson-Boltzmann equation used in the MEAD program suite25 using the cysteine parameters described previously.26 The combination of using the structures from the protonated simulation to calculate the pKa of the cysteine is approximate in that it does not include explicit conformational coupling between the cysteine deprotonation and the protein; this is likely to result in an underestimate of the actual shift. In principle, it should be possible, with difficulty, to use improved sampling methods such as the orthogonal-space random-walk method27 or pHMD28 to capture the coupling, though such calculations have yet to be done on cysteines such as these. These methods would, in principle, allow for explicit protein-protonation state coupling.
Pathway calculations
The results herein and in our previous publication14 suggest that there is a communication between two active sites at a distance to create electrostatic asymmetry. To determine if the simulation data supports the existence of a pathway that connects the active sites and creates an asymmetry, we developed a specific pathway finding approach to examine fluctuations correlated with the pKa 14 asymmetry. This approach involves: 1) calculating the correlation coefficients between the Cp pKa difference and each residue’s backbone ϕ and ψ angles, and 2) using an average structures to find a pathway between the different active sites that involves these residues whose dihedral angles are correlated with the pKa asymmetry. The specific method developed previously, that we use here, starts at the Cp of subunit A, then each residue with a heavy atoms within a cutoff distance 4.0 Å, and with a correlation above a cutoff correlation coefficient of 0.2 are identified. Then from each of these residues, all the residues with heavy atoms within the same cutoff distance and with the correlation above the same cutoff correlation coefficient are identified. This process is continued until the Cp of subunit B is reached.
RESULTS
For this study, a high quality structural representative of the atypical 2-Cys Prxs was chosen: 1psq, a crystal structure of the Tpx from Streptococcus pneumoniae 18 (Figure 1A). This choice is comparable to that made in our previous study14, in which 1uul, a typical 2-Cys Prx from Trypanosoma cruzi 29 (Figure 1B) was chosen. In 1psq, both the peroxidatic cysteine (Cp), residue 58, and the resolving cysteine (Cr), residue 92, are located at helix termini: Cp at the N-terminus of the helix extending from Gly56 to Glu68 and Cr in the C-terminal turn of a helix that extends from Pro85 to Glu95 (Figure 1A). In 1uul, the Cp and Cr residues 52 and 173, respectively, are also found near the helical termini. After the new molecular dynamics and electrostatics results for the atypical 2-Cys Prx, 1psq, are presented, a detailed comparison between these results and our previous results on the typical 2-Cys Prx, 1uul, will be presented in the Discussion section.
1psq crystal structure monomers are similar, though not identical
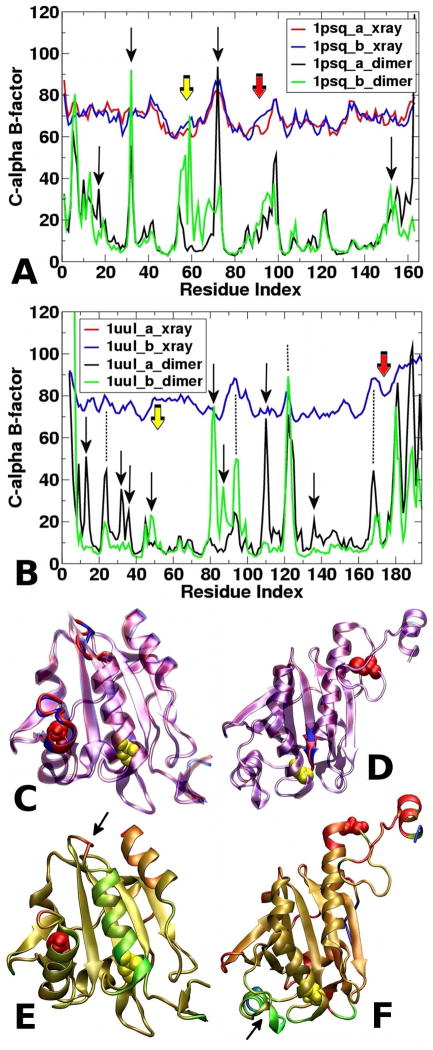
Because this study examines the evolution of asymmetry, we first evaluated the similarity between the two monomers in the crystal structure. 1psq represents an atypical 2-Cys Prx with both peroxidatic and resolving cysteines of a single active site contained within a single monomer (Figure 1A).1b The monomers, A and B, are very similar: the backbone and all-atom RMSDs between the two monomers are 0.37 and 0.84 Å, respectively. Specific backbone structural differences between the two monomers are found in only two locations: at the C-terminus of the helix that contains Cp (though the Cp is located at the helix N-terminus) and at the terminus and turn following the helix that contains Cr (Figure 2C). Comparison of C-alpha B-factors from the 1psq crystal structure show similar, but not identical, positional fluctuations between monomers (Figure 2A, red and blue traces). The largest difference in positional fluctuation (Figure 2A, red arrow, compare red and blue traces) and structural difference (Figure 2C) occurs at residues 92–98; the C-terminus of the helix containing Cr and subsequent short turn is more mobile in the B monomer than the A monomer.
Figure 2. Analysis of the structural fluctuations identified in each monomer during each simulation and comparison to crystal structure B-factors in 1psq (A, C, and E) and 1uul (B, D, and F).
A and B. B-factors for the x-ray structure (red and blue traces) and molecular dynamics simulations (green and black traces) for Tpx (1psq, A) and tryparedoxin peroxidase (1uul, B). Data for monomer A is shown in red and black; data for monomer B is shown in blue and green. Yellow and red solid arrows indicate Cp and Cr, respectively. Dotted lines indicate some regions where x-ray and trajectory B-factors agree; black arrows indicate regions that exhibit asymmetry in the trajectory B-factors or that differ between simulation and x-ray and are discussed in the text. C and D. Structural overlays of monomer A (blue) and monomer B (red) for both Tpx (C) and tryparedoxin peroxidase (D). Side chains of Cp and Cr are yellow and red spheres, respectively. Ribbon is drawn transparently, except in regions that are not superimposable when the crystal structure monomers are overlaid, so the regions that are different in structure between monomers are clearly visible: 1) in Tpx, the C-terminus of the helix containing Cp and around the Cr; and 2) in tryparedoxin peroxidase, at the end of the strand adjacent (in space) to Cp. E and F. Monomer A (following addition of hydrogens and brief minimization) of Tpx (1psq, E) and tryparedoxin peroxidase (1uul, F), colored by the difference in C-alpha B-factor calculated from the MD simulations. (Tryparedoxin peroxidase simulations taken from one MD dimer simulation published previously.14) Red regions exhibit higher Bfactors in monomer A than in B, while green regions exhibit higher B-factors in monomer B than in monomer A; thus, regions of red and green show where the monomers differ in their fluctuations during the dimer simulations. The arrow in (E) indicates the turn containing Leu72; the arrow in (F) indicates residues 81–96, both of which are discussed in the text. Side chains of Cp and Cr are yellow and red spheres, respectively. Comparison of panel C with E (or panel D with F) indicates those regions of the each dimer that show asymmetry in the crystallographic structure of the monomers (C and D) or exhibit asymmetry in fluctuations during the MD simulations (E and F).
The monomers show slight asymmetry in the Cp pKa, as well, calculated to be 6.9 in monomer A and 7.3 in monomer B. No structural difference is observed between monomers in this region (Figure 2C, yellow spheres); thus, this pKa difference appears to be the sum of minor conformational differences in the backbone and non-titratable residues. Overall, the 1psq active sites of the two monomers in the crystal structure exhibit small structural, B-factor, and electrostatic differences.
During MD simulations, structural asymmetry between monomers evolves in the atypical 2-Cys Prx dimer of Tpx
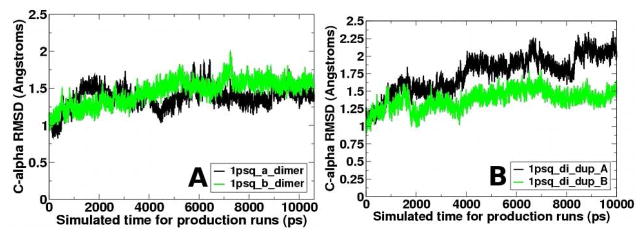
MD simulations (10.6 ns) were performed on the Tpx dimer. Because we expected them to be reduced, Cp and Cr sulfhydryl groups were explicitly protonated in both monomers. The RMSD, calculated between the alpha carbons in the reference structure and each saved structure, indicates that each monomer relaxes to about 1.5 Å from the crystal structure during the first two nanoseconds, then does not change significantly beyond that (Figure 3). This result and others, including radius of gyration and overall change in alpha carbon distances (data not shown), indicate that the simulations are stable over this simulation time.
Figure 3. Comparison of the crystal structure to each structure sampled along the trajectory indicates that the simulations are reasonably stable.
The alpha carbon root mean square deviation (RMSD) between each structure saved during the MD simulation and the reference structure for Tpx (1psq) with data for monomers A and B in black and green, respectively. Data is shown for one simulation. Multiple simulations were consistent. Panels (A) and (B) show two simulations in which either monomer B (panel A) or monomer A (panel B) diverged the most. The data show in panel A are from the same simulation shown in all other results shown in the manuscript.
During the first 2 ns of simulation, the monomer structures diverge. The average monomer structures (with the average structure calculated from 2 ns to the end of the simulation) differ by 1.57 Å backbone RMSD (compared to 0.37 Å in the crystal structure). Modest structural asymmetry has thus developed between the two monomers during the simulation.
The simulation alpha carbon B-factors for each monomer were compared to the crystallographic B-factors (Figure 2A). Many of the B-factor peaks and valleys are consistent between the simulations and the crystal structures; however, a few differences between the crystal structure and simulation B-factors are observed. A small, broad peak which starts at Cr (residue 92) and ends at Asp98 and Asn99 is observed in both trajectories (red arrow, Figure 2A); thus structural fluctuations occur around Cr in both monomers. This is the site of the single difference between monomers in the crystal structure (Figure 2A, red and blue traces). The region around Leu32 also exhibits enhanced flexibility in both monomers during the simulations (Figure 2A, black arrow). The functional relevance, if any, of this flexibility is unclear. This is in a turn between two strands and in principle enhanced flexibility could allow for propagation of motions, but there is no clear evidence for this in the simulation data. More interestingly, asymmetry between the monomers can also be observed in the simulation B-factors, which are mapped to the structure of 1psq in Figure 2E (where red and green regions exhibit higher simulation B-factors in monomers A or B, respectively). In the crystal structure, peak alpha carbon B-factors for both monomers are observed at Leu72. In the MD simulations, this large peak is only observed in the A monomer; it is much smaller in the B monomer (Figure 2A, black arrow). Leu72 is in the loop following the C-terminus of the Cp-containing helix (Figure 2E, arrow). The opposite behavior is observed around the Cp, residue 58, including residues 55–69: the B monomer exhibits larger fluctuations than the A monomer during the simulation (Figure 2A, yellow arrow; Figure 2E, green helix). Overall, these alpha carbon B-factors indicate that these asymmetric backbone fluctuations are located around the active site, suggesting that the fluctuations in the Cp and Cr microenvironments in the monomers differ from each other.
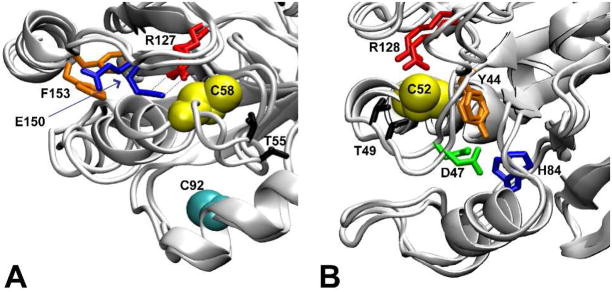
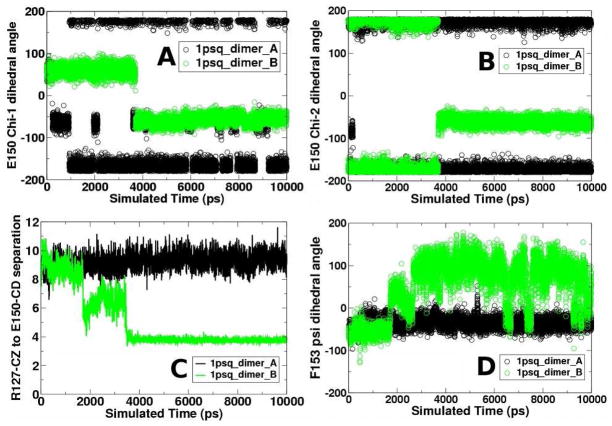
To evaluate the local structures assumed by each monomer, all structures sampled during each simulation were clustered and cluster representatives were compared. In monomer A, only small structural changes are observed around the active site: representative structures of the most diverse clusters superimpose well (not shown). Monomer B, on the other hand, exhibited significant changes at the active site. Comparison of the most distinct cluster representatives are overlaid in Figure 4A. Specifically, Glu150, which is solvent exposed in the monomer A crystal structure and in some monomer B cluster representatives, undergoes side chain dihedral angle rotations at about 3.6 ns, becomes buried, and forms a hydrogen bonded salt bridge with Arg127 (compare blue side chains, Figure 4A). Both Glu150 chi-1 and chi-2 dihedral angles undergo a rotamer flip at 3.6 ns: chi-1 from gauche+ (g+) to gauche− (g−) and chi-2 from trans (t) to g− (Figure 5A and B, green). The salt bridge thus formed in monomer B between Arg127 and Glu150 remains in place the remainder of the simulation (see side chain distances, Figure 5C, green).
Figure 4. A comparison of the most distinct monomer structures from each trajectory reveals the largest structural differences at the protein active sites.
For each trajectory, the two most distinct cluster representatives (as determined by all-atom RMSD between cluster representatives) are overlaid for 1psq monomer B (A) and 1uul monomers B (B). In both panels, Cp is shown as a yellow van der Waal sphere. For Tpx (1psq) the side chains of Thr55 (black), Arg127 (red), Glu150 (blue), and Phe153 (orange) are shown. Cys92, Cr, is shown as a cyan sphere. In 1psq (A), the salt bridge that forms between Glu150 and Arg127 in monomer B and the backbone conformational change that results in a reorientation of Thr55 also in monomer B can clearly be seen. In 1uul (B), the most divergent clusters show the involvement of the Asp47 (green)-Thr49 (black) loop and the motion of His84 (blue), proposed to be involved in a proton shuttle.
Figure 5. Specific angle and distances measured during the Tpx (1psq) simulation provide a structural explanation for the observed electrostatic asymmetry.
The conformation of the Glu150 chi-1 (A) and chi-2 (B) side chain dihedral angles during the Tpx (1psq) simulation; C. distances between Arg127 and Glu150; D. backbone dihedral angles for Phe153. In all cases, data for monomers A and B are shown in black and green, respectively.
Formation of the Arg127-Glu150 salt bridge in monomer B is correlated with a change in the microenvironment of Cp. In both crystal structure monomers, the Arg127 guanidinium group forms a hydrogen bond with the Cp sulfhydryl group, with heavy atom distances between hydrogen bonding residues of 3.2 – 3.7 Å and a hydrogen bond angle of 160°. The salt bridge between Glu150 and Arg127 that forms in the B-monomer at 3.6 ns places the side chain carboxylate and guanidinium moieties of both of these charged residues in close proximity to the Cp sulfur atom (Figure 4A). The salt bridge between Glu150 and Arg127 never forms in monomer A (observe the atomic distances, Figure 5C, black); thus in monomer A only Arg127, and not Glu150, is in proximity to Cp throughout the simulation (not shown).
Observation of the Glu150 side chain in the 1psq monomer simulations provides insights into monomer-specific differences that evolve in the active site microenvironments. As mentioned above, in monomer B the Glu150 side chain, both chi-1 and chi-2, rotates at 3.6 ns and remains there for the remainder of the simulation (Figure 5A). In contrast, Glu150 chi-1 in monomer A, which, like monomer B, starts in the g+ conformation, explores both the g− and trans (t) conformations during the simulation (Figure 5A, black). A rotamer flip of Glu150 to the g− conformation is, thus, insufficient to form a stable salt bridge. Why does the Glu150 chi-1 in monomer A rotate, but never “lock” (with the concomitant rotation of chi-2) into a conformation to form a salt bridge with Arg127?
One possible answer is that the salt bridge in monomer B appears to form in two stages: a first stage at about 1.8 ns, then the second, closer contact at 3.6 ns (Figure 5C). At the second stage, simultaneous rotation of Glu150 chi-1 and chi-2 is observed. We hypothesized that a first stage is a necessary precursor that allows the Glu150 side chain rotation to form the salt bridge with Arg127.
We therefore looked for a conformational change that causes this first stage and found one. A backbone psi dihedral angle change in Phe153 correlates with this first stage (Figure 5D). Initially, this psi angle is −60°, but at 1.9 ns it begins sampling conformations from −50 to +50°. At 2.9 ns, this psi angle transitions a second time to sampling conformations between 50 and 100° (Figure 5D). The initial transition occurs at the same time as the first stage in salt bridge formation (Figure 5C) and the correlation coefficient between the Glu150-Arg127 distance and this backbone dihedral angle is 0.84 in monomer B, suggesting a strong correlation between these two events. During this process, Phe153 effectively transitions from a right-handed alpha helical conformation to a conformation between the beta-gamma regions of the Ramachandran plot, consistent with extended beta strands or inverse gamma turns.30 The intermediate, less stable backbone conformation sampled from 1.9 to 2.9 ns is found in the δR or “bridge” region of the Ramachandran plot31 and is likely stabilized by the developing electrostatic interaction between Glu150 and Arg127 side chains. The complete transition to the beta-conformation then occurs with the additional Phe153 backbone rotation, which allows the full salt bridge to “lock” at 3.6 ns when the Glu side chain next samples the appropriate chi-1 and chi-2 rotamers. This Phe153 backbone conformational change explains the peak in the alpha carbon B-factor plot for monomer B that is not seen in monomer A (Figure 2A, black arrow at residue 151).
The strong salt bridge formed in monomer B has a large structural consequence: the helix containing Cp reorients (Figure 4A) so that Cp shifts relative to these charged residues. This salt bridge is the origin of the difference in simulation B-factor observed in 1psq (Figure 2E, green helix). This helix reorientation has consequences for the interaction of Cp with a number of residues (discussed subsequently).
Overall, during the 10.6 ns simulation, asymmetry clearly evolves between the two monomers of Tpx (1psq). Similar sampling and behavior was observed in replicate simulations, although the exact times of changes were different. Indeed, while we show the example of a simulation where monomer B diverged the most from the starting structure, monomer A was observed to diverge in a different simulation (Figure 3B). Because of the distinct changes around the active site structure during each simulation, the Cp pKa should also change, and change differently in each monomer. Such observations, which we present in the next section, would be significant in terms of understanding the reactivity of the Cp and would allow us to hypothesize mechanisms for the first steps in the atypical 2-Cys Prx reaction mechanism as we shall do.
Electrostatic asymmetry in Tpx: Glu150 replaces Thr55 as a key Cp pKa modulating residue in only one monomer
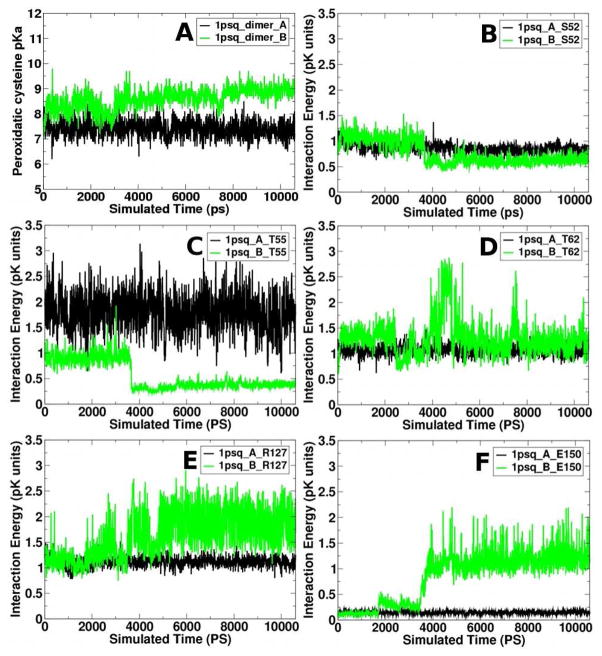
To observe the variation in the Cp pKas, dimer electrostatics were calculated for each structure generated by the MD simulation. Calculated Cp pKas in each monomer continue to diverge during the dimer simulation (Figure 6A). The pKa in monomer A quickly increases 0.5 pK units, from 6.9 to an average of 7.4. It then remains relatively constant throughout the simulation. In monomer B, on the other hand, the pKa increases, from 7.3 to an average of 8.7 (average calculated from 2 to 10.6 ns). If these calculations represent a biologically relevant snapshot (which we argue they do—replicate simulations sample similar conformations, though events do not occur at identical times), the results suggest that at physiological pH (~7.4) the Cp in monomer A is unprotonated 50% of the time whereas monomer B would be approximately 95% protonated. If this asymmetry were lacking and each cysteine had the average pKa of 8.05, then each cysteine would be unprotonated only 18% of the time, and so this asymmetry increases the effective fraction of unprotonated (and therefore active) Cps from 0.18 to 0.28 at physiological pH. This asymmetry is identified even though all simulations were performed with both Cps fully protonated. We suggest that the slight asymmetry captured in the static crystal structure shows only a small bit of asymmetric behavior that would occur in solution; however, ensemble-based solution experimental measurements would be unable to observe such asymmetry.
Figure 6. Evolution of the asymmetric Cp pKa and interaction energies during the Tpx (1psq) simulation.
(A) Peroxidatic cysteine side chain pKa and (B–F) interaction energies during the MD simulation. In all plots, the black and green traces indicate the A and B monomers, respectively. The Cp pKa in the starting crystal structures are 6.9 and 7.3 for monomers A and B, respectively. Interaction energy plots for the interaction of residues Ser52 (B), Thr55 (C), Thr62 (D), Arg127 (E), and Glu150 (F) with Cp, residue 58.
We compare the pKa trace for monomer B (Figure 6A, green trace) to the monomer B dihedral angle results presented earlier (Figure 5, green traces). The pKa rapidly (0.1 ns) increases to about 8.4, then remains relatively constant slightly for about 1.8 ns, then begins a gradual decrease that lasts until about 2.9 ns. It then abruptly increases to its average of 8.7. The timing and shape of the slow decrease from 1.9 to 2.9 ns is consistent with the conformational sampling of 0 to 50° in the Phe153 psi angle (Figure 5D), not with the Glu150 side chain dihedral angles (which do not change during this same time frame; Figure 5A, B). This result indicates that the Phe153 backbone conformational change is important to the change in the Cp pKa in monomer B.
To explore the interactions further, we identified those residues that significantly affect the Tpx Cp pKa by calculating interaction energies between side chains during the simulation (Table 1). Although a few electrostatic interactions are identified across the subunits (Table 1, below dark gray row), most interactions, and all strong ones, are located within the monomer containing Cp itself. Only five residues exhibit large interactions at any time: Thr62 and Arg127 in both monomers; Ser52 and Thr55 in monomer A; and Glu150 in monomer B (Table 1, gray cells). All shift the pKa > 0.8 pK unit for at least some portion of the simulation in at least one monomer and all show asymmetric behavior during the simulation (Figure 6B-F).
Table 1.
Average interaction energies over the simulation for all residues that affect the Cp pKa.
| 1psqA | 1psqB | ||
|---|---|---|---|
| Cp pKa | 7.41 | Cp pKa | 8.63 |
| burial | 1.10 | burial | 1.33 |
| background | −1.33 | background | −0.37 |
| Residue | I. E.1 | Residue | I. E. |
| 48 SER | 0.59 | 48 SER | 0.72 |
| 52 SER | 0.87 | 52 SER | 0.75 |
| 54 ASP | 0.46 | 54 ASP | 0.30 |
| 55 THR | 1.75 | 55 THR | 0.54 |
| 59 SER | 0.44 | 59 SER | 0.45 |
| 60 THR | 0.34 | 60 THR | 0.35 |
| 62 THR | 1.06 | 62 THR | 1.28 |
| 63 ARG | 0.14 | ||
| 67 GLU | 0.13 | ||
| 79 THR | 0.45 | 79 THR | 0.48 |
| 81 SER | 0.70 | 81 SER | 0.63 |
| 83 ASP | 0.58 | 83 ASP | 0.49 |
| 90 ARG | 0.15 | ||
| 92 CYS | 0.17 | 92 CYS | 0.15 |
| 95 GLU | 0.16 | ||
| 104 SER | 0.29 | 104 SER | 0.25 |
| 105 ASP | 0.20 | 105 ASP | 0.17 |
| 127 ARG | 1.11 | 127 ARG | 1.66 |
| 149 SER | 0.14 | 149 SER | 0.19 |
| 150 GLU | 0.89 | ||
| 54 ASP (B) | 0.2 | ||
| 83 ASP (B) | 0.19 | 83 ASP (A) | 0.15 |
| 89 LYS (B) | 0.12 | 89 LYS (A) | 0.10 |
| 90 ARG (B) | 0.22 | 90 ARG (A) | 0.17 |
| 106 TYR (B) | 0.22 | 106 TYR (A) | 0.20 |
I. E.: interaction energy, in pK units, between Cp and residue given. Shaded residues are those with interaction energies above 0.8 pK units. Interactions below the dark gray row are interactions on the opposite subunit. Interaction energy consists of several terms.25b, 35 The burial (Born) term is the contribution to pKa due to the different dielectric constant between protein and water. The solvent accessible area change will usually lead to the change of this term. The background term is the electrostatic interactions between titratable atom and backbone or side chain atoms.
To understand the structural fluctuations that cause the interaction energy asymmetry, the interaction energy plots (Figure 6B-F) can be compared to the Cp pKa trace (Figure 6A) and the structural fluctuations (Figure 5). In monomer A, the interaction energies remain mostly unchanged during the simulation (Figure 6, black traces), reflecting the minimal change observed in Cp pKa. Thr55 exhibits the largest interaction, with an energy of 1.8 pK units, on average, throughout the simulation (Figure 6C). This is likely due to the close interaction between the Thr side chain hydroxyl and the Cys side chain sulfhydryl groups: an average of 3.1 Å for monomer A during the simulation. Detailed analysis of this interaction suggests that a hydrogen bond is formed between Cp and Thr55, with the protonated Cys sulfhydryl group acting as a donor and the Thr side chain oxygen as the acceptor. In monomer A, Arg127 exhibits a smaller and constant interaction energy of, on average, 1.2 pK units (Figures 6E, black trace). These interactions suggest a hydrogen bond between Arg127 and Cp side chains, with Cp acting as the acceptor. Glu150 has little impact on the Cp pKa in monomer A (Figure 6F, black trace).
In monomer B, the interaction energies are not constant, but the fluctuations correlate with the conformational and pKa changes already discussed; ultimately, these fluctuations produce the pKa asymmetry. All of the interaction energies undergo a significant transition at 3.6 ns (Figure 6B-E, green traces), which is consistent with the rotation of the Glu150 side chain chi-1 and 2 dihedral angles (Figure 5A and B, green traces), the Phe153 psi angle rotation (Figure 5D, green trace) and the “lock” on the salt bridge between Arg127 and Glu150.
Thr55 interaction energies are the most simple, starting with a relatively strong interaction with Cp (~0.9 pK units) until 3.6 ns, followed by an abrupt decrease to 0.25 pK units (Figure 6C, green trace). Structural analysis indicates that the initial interaction between Thr55 and Cp in this subunit is not a classical hydrogen bond. The locked salt bridge between Arg127 and Glu150 clearly displaces the Thr55-Cp interaction and the Thr55 side chain rotates away (Figure 4A). The Glu150-Cp interaction energy also abruptly increases at 3.6 ns, data indicating that when it forms a salt bridge with Arg127, its interaction with Cp also significantly increases. The Phe153 backbone conformational change slightly increases the interaction energy between Glu150 and Cp, with a small increase in energy at 1.9 ns and a gradual decrease until about 2.8 ns (Figure 6F, green trace). At 2.8 ns, the interaction energy increases slightly, then gradually decreases until the significant increase at 3.6 ns. In monomer B, the Cp pKa fluctuations and the Cp-Glu150 interaction energy are correlated, with a correlation coefficient of 0.6, indicating the importance of Glu150 in controlling the Cp pKa in this monomer.
The Arg127 to Cp interaction energy is most interesting (Figure 6E). It starts high, above 1 pK unit. At 1.9 ns, correlating with the Phe153 conformational change, the interaction energy begins to increase, gradually increasing up to 2 pK units at 2.9 ns. At 2.9 ns, exactly when Phe153 transitions to the third conformation (Figure 5D), the Arg127-Cp interaction energy decreases abruptly to 1 pK unit. It stays at this low value until 3.6 ns, when it abruptly increases to 2 pK units (Figure 6D). There is a decrease, then increase, in the interaction energy between Arg127 and Cp around 4.2 ns of which we have not yet identified the cause. Thus, in monomer B in this simulation, the backbone conformation of Phe153 has an effect on the Cp pKa by altering its interactions with Arg127 and Glu150. At the same time that Arg127 and Glu150 lock into the salt bridge illustrated in Figure 4, both residues increase their interactions with Cp and largely replace the Cp interaction with Thr55. One set of residues (Arg and Glu) simply replaces another (Arg and Thr).
Thus, both structural and electrostatic characteristics indicate that, starting from a slightly asymmetric structure, Tpx A and B evolve significant asymmetry during the simulation. Specifically, in monomer B, the Phe153 backbone psi angle rotates to generate a conformation (within the bridge region on a Ramachandran plot) that allows a closer interaction between Glu150 and Arg127, then Phe153 transitions to another conformation (in the beta-gamma region of a Ramachandran plot), which causes an increase in the Cp pKa. This increased pKa in monomer B is constant for the rest of the simulation and appears to be “locked in” by rotation of the Glu150 side chain to form a strong hydrogen-bonded salt bridge with Arg127 and, at the same time, replacing Thr55 as the residue important for the maintenance of the Cp pKa in this atypical 2-Cys Prx. These specific observations are consistent with the higher fluctuations observed in monomer B during the simulation. Based on the observations, it is reasonable to hypothesize that Glu150 side chain rotations, facilitated by a Phe153 backbone shift, play a role in the abstraction of a proton from Cp during the Prx reaction.
Exploration of the pathway of residue interactions that cause asymmetry in Tpx, an atypical 2-Cys Prx
The above results indicate that Cp pKas are dynamically asymmetric. This raises the question of how the two active sites communicate with each other, so that in each configuration there is an asymmetry. To answer this question we adopted the same pathway finding approach used in our previous study of 1uul.14 This approach involves calculating the correlation coefficients between the Cp pKa difference and each residue’s backbone ϕ and ψ angles, and then using the structure to find a pathway between the different active sites (see Methods).
Using this approach, we find one pathway that connects the two active sites in Tpx as depicted in Figure 7. This pathway connects the two Cps through the helices containing the resolving cysteines. In contrast to our previous study on a typical 2-Cys Prx,14 we only find one pathway, not two; however, much like the second pathway identified in that work, the pathway we identified here involves the helices containing the resolving cysteines. It also includes more ionizable residues (Figure 7)--four arginines, an aspartic acid, and a glutamic acid--than the pathways found in our previous work.14 The higher prevalence of charged residues in this pathway leads us to speculate, although we have no direct evidence, that this pathway may involve some sort of proton transport. Such a proton shuttle mechanism has been shown experimentally to be important in function and maintenance of asymmetry between two active site cofactors in thiamine-dependent enzymes.32
Figure 7. Proposed pathway coupling the two Cps in Tpx monomers A and B.
The specific residues involved in the pathway that couples the two peroxidatic cysteines (Arg64 (in monomer A), Leu97 (A), Ala94 (A), Arg90 (A), Ala87 (A), Pro85 (A), Leu84 (B), Pro85 (B), Ala87 (B), Arg90 (B), Ala94 (B), Leu97 (B), Asp98 (B), Glu68 (B), Arg64 (B) and Thr62 (B)) are depicted in red bonded representation while the Cps are depicted in yellow. The remainder of the dimer is depicted in a cartoon representation.
A backbone-specific interaction in a typical 2-Cys Prx
We previously published a detailed analysis of molecular dynamics simulations of a typical 2-Cys Prx, tryparedoxin peroxidase (1uul).14 In that work, we showed that, starting from essentially identical monomers, subunit asymmetry develops in this typical 2-Cys Prx, as we show here in Tpx, an atypical 2-Cys Prx. This asymmetry was not observed in the simulations of individual monomers, but was observed in simulations of dimers and decamers. The contributing residues and pathway of development of the asymmetry were identified. (A comparison between the typical and atypical 2-Cys Prx results is elaborated in the Discussion.)
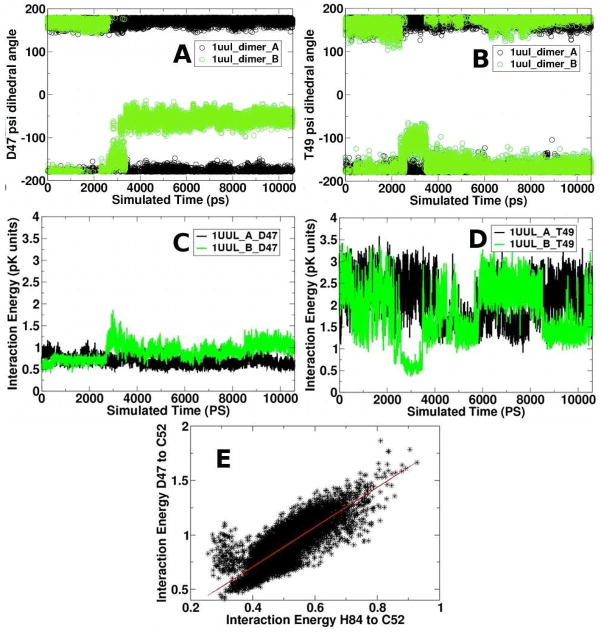
In that paper, we reported that the background electrostatics terms of Met46, Asp47, and Phe48 correlated with increased Cp pKa. A background electrostatics term indicates a change in the impact of non-charged residues on the pKa. Two obvious questions arise as to the structural origin of the correlated background term for these residues: does it contribute to the observed asymmetry, and how does it compare to our current study? To observe specific structural changes causing this background electrostatics term, we re-analyzed our previous results with RMSD-based clustering analysis (Figure 4B). The rearrangement of the environment around the Cp involves substantial changes in the conformation of the loop containing Asp47 and Thr49, caused by a rotation of the Asp47 psi backbone dihedral angle from a t-to a g-rotamer conformation (Figure 8B). This conformational transition occurs from 2.5–3.5 ns in this simulation and is correlated with a change in the backbone psi dihedral angle of Thr49 (Figure 8D). The backbone oxygen atom of Asp47 reorients to a position pointing toward the surface of the protein, while the Thr49 changes position as a result of backbone rearrangement, to closely interact with the Cp sulfhydryl group (Figure 4D). (Asp47 and Thr49 phi angles do not change during these simulations.)
Figure 8. Specific backbone dihedral angles measured during the tryparedoxin peroxidase (1uul) simulation provide a structural explanation for some of the observed electrostatic asymmetry.
The backbone Asp47 (A), Thr49 psi angles (B) and interaction energies between Cp and Asp47 (C) and Thr49 (D) are plotted against the time of simulation. Data for monomers A and B are shown in black and green, respectively. Panel (E) shows the correlation between the Asp47 and His84 interaction energies.
The decrease in the interaction between Thr49 and Cp between 2.5 and 3.5 ns is correlated with the motion of the Asp47 and Thr49 backbone angles (Figure 8). Comparison of the most distinct cluster representatives in the 1uul simulations (Figure 4B) and the correlations in motion (Figures 8A-D) suggests that the motion of Asp47 and Thr49 is also correlated with His84. Indeed, the interaction energies of His84 to Cp and Asp47 to Cp is highly correlated (Figure 8E). The consistency of these observations across simulations suggests the importance of the Asp47-Thr49 loop conformation in maintaining the Cp pKa and the possibility of Asp47 and His84 acting as proton shuttles in the typical 2-Cys Prx, tryparedoxin peroxidase (1uul).
DISCUSSION
Active site asymmetry as a general feature in Prxs
The electrostatic and structural analysis of the simulation involving the S. pneumoniae Tpx, an atypical 2-Cys Prx protein, suggests the existence of active site asymmetry between the two monomer units and provides a putative pathway connecting these two active sites. Furthermore, a comparison between these results on an atypical 2-Cys Prx protein and our previous study14 of a typical 2-Cys Prx allows us to speculate on how these proteins may have evolved differently, yet have generated a similar overall mechanism of creating active site asymmetry to increase the fraction of Cp that are active at a given moment.
These two studies suggest that Cp in both typical and atypical Prxs exhibit dynamic asymmetry that leads to asymmetric electrostatic environments as measured by the different instantaneous (calculated) pKas. Furthermore, in both Prxs, pathways comprised of contacting residues whose fluctuations correlate with the pKa differences between the active sites are found that connect the two active sites within the dimer.
In our study of a typical 2-Cys Prx we found, across 10 dimer simulations, that the average pKa of monomer A was 7.51, while in monomer B it was 7.57, yet the average instantaneous difference between the two active sites was 0.97. We also found that the monomers randomly broke symmetry and, in most simulations, that once the symmetry was broken, it was stable on at least the nanosecond time-scale.
The current study of an atypical 2-Cys Prx supports the same big picture: there is an instantaneous dynamic asymmetry in which one monomer develops a different electrostatic environment that is stable on at least the nanosecond time scale. When taken together, these observations suggest a common behavior among Prx dimers; i.e., the sequence-equivalent homodimers become structural heterodimers with different reactivities, wherein one monomer becomes the highly reactive one, as suggested by the lower pKa, and the other monomer becomes the less reactive one. In other words, these proteins may be described as exhibiting half-site reactivity. The actual enhancement of reactivity, as suggested by the change in protonation fraction, is modest at best 1.5x in this work and ~1.05x in our previous work. Rather, the main physical effect would be to localize the reactivity to a single active site within a dimer on at least a nanosecond timescale. Experimental work has also been described to support asymmetry between subunits in Prxs: according to one report, upon oxidation of one active site within an oligomer, other sites exhibit lower reactivity.33 Our current work extends the experimental results and suggests that, even prior to oxidation, the sites exhibit asymmetry within the homodimer structure. Such behavior would not be dissimilar to other enzymes with multiple active sites which have been shown or suggested to exhibit asymmetry, and thus sometimes cooperativity, between their two active sites.32b
This observation, combined with the discovery of pathways connecting both active sites in each type of dimer, suggests that the two active sites in Prx dimers are in fact a pair of coupled and co-regulated active sites, providing for the possibility of communication upon binding of substrate. While the simulations do not directly demonstrate this, the existence of a pathway, and localization of an enhanced electrostatic microenvironment, suggest that when a substrate binds to one active site, this binding event might be communicated to the other active site, which may allow that active site to respond dynamically to binding and to “prepare” for the next binding event or the next step in catalysis. The specifics of how the asymmetry forms varies between these two types of Prxs and the distinguishing features of each communication pathway suggest that there is a strong driving force towards dimer asymmetry in this family of proteins.
The primary sources for the asymmetry in 1psq appear to be residues Arg127, Glu150, and the backbone of Phe153. This is suggestive of functional roles for these residues in catalysis. Arg127 forms a hydrogen bond with Cp, which would stabilize the deprotonated state, and Glu150 undergoes a significant conformational change (facilitated by the Phe153 backbone conformational change) that both stabilizes Arg127 and increases the electrostatic interaction between Glu150 and Cp, which shifts the peroxidatic cysteine pKa upward. Although we cannot observe proton transfer in our simulations, the Glu150 conformational changes are suggestive, and we hypothesize that this glutamic acid may in fact be the base involved in deprotonation, although the possible role of Glu150 as a base obviously needs further study.
Detailed comparison of asymmetry in typical and atypical 2-Cys Prxs
While overall structural and electrostatic evidence indicate that asymmetry evolves in the both the typical and atypical 2-Cys Prx dimers, the details that cause the asymmetry are different in the two proteins. In the typical 2-Cys Prx, the Cp pKa is largely modulated by changes in the conformation of the residue 46–50, Cp-containing loop, while in the atypical 2-Cys Prx, the changes are largely modulated by the backbone of Phe153 and the side chain of Glu150. In both cases, the backbone changes are in short loops near to Cp. Here we provide detailed comparisons between the typical and atypical 2-Cys Prx, as observed in the illustrations in Figure 2.
As observed in the 1psq Tpx simulation, the 1uul tryparedoxin peroxidase simulation Bfactors exhibit asymmetry between the two monomers (Figure 2B, black arrows) and the differences are localized to specific regions of the structure (Figure 2F). Monomer A simulations exhibit larger fluctuations which differ between monomers at residues 13, 23–24, 31–35 (2 peaks), 110, and 186–187; these are at various turns at the protein surface, the C-terminal helix, and the end of the helix just to the N-terminal side of Cr (Figure 2F, red and green regions). The C-terminal helix contains the YF motif identified as a major difference between the sensitive and robust 2-Cys Prx;34 interestingly, we see larger flexibility in this region only in monomer A. The B monomer exhibits significantly larger fluctuations than the A monomer in just two regions: a small peak at residue 49 and 3 peaks from residues 81 to 96 (green regions, Figure 2F). The 81–96 region (Figure 2F, arrow) is of interest for several reasons: 1) it contains the helix that is homologous to the helix containing Cr in the atypical 2-Cys Prx; 2) it is bounded by the GGLG motif, identified as the second motif that distinguishes the sensitive Prx from the robust Prx;34 and 3) it contains His84, which we suggest (together with Asp47) might act as part of a proton shuttle in this Prx family. This asymmetry in fluctuations reinforces the suggestion that pKa asymmetry may have functional significance as regions of the structure that are either known to be important in reactivity--points 1 and 2 previously--or that may be involved in a proton shuttle--point 3--show asymmetry as well.
In comparing and contrasting the simulations on these typical and atypical Prx proteins, we see general commonalities: 1) dynamic asymmetry between the monomers in the dimers as reflected in fluctuations and the active site predicted pKa; and 2) pathways(s) connecting the two active sites that may propagate the asymmetry between the two subunits. We see differences in the precise identity of the residues involved in the pathways propagating the asymmetry and in the residues whose fluctuations are involved asymmetry as one might expect for different types of Prx proteins. We also see a difference in steric and electrostatic interactions across the subunit interface. In the 1uul, the typical Prx, several interactions were identified (for example, A175 and Y82),14 while no cross subunit interactions were identified the 1psq (atypical Prx) simulations. Furthermore, in the 1psq simulation, there are suggestive fluctuations in a residue (Glu150) that could potentially be involved a proton shuttle, however, in the 1uul simulations, while there is an asymmetry in the fluctuations of a different residue (His84) that could be involved in a proton shuttle, the evidence is weaker there, perhaps reflecting a more complex proton shuttle mechanism. These two residues that may have a role in a proton shuttle are not part of the pathway(s) connecting the active sites, although they are near residues in the pathway(s). This suggests that there may be a distinction between residues responsible for shuttling protons – or at least the initial residues in such a shuttle – and those residues that couple the active sites.
CONCLUSIONS
Both structural and electrostatic characteristics indicate that, starting from a slightly asymmetric structure, Tpx monomers A and B evolve significant asymmetry during MD simulations. Our observations, combined with the discovery of pathways connecting both active sites in each type of dimer, suggests that the two active sites in Tpx dimers are in fact a pair of coupled and co-regulated active sites, providing for the possibility of communication upon binding of substrate. The specifics of how the asymmetry forms varies between these two types of Prxs and the specifics of the communication pathways suggests that there is a strong driving force towards dimer asymmetry in this family of proteins.
Acknowledgments
The authors thank Derek Parsonage for helpful discussions regarding crystallographic refinement, and Stacy (Knutson) Howerton and Brian Kell for assistance with figures. The authors acknowledge partial support from the NSF (MCB 0517343 to JSF) and the NIH (RO1 GM050389 to LBP). The pathway analysis was partially supported by a subaward from P30 CA12197 to FRS. The calculations herein were performed on the WFU DEAC cluster and we thank Wake Forest Provost Office and Information Systems department for their support. Additional disk space was provided via an IBM SUR grant, and we thank IBM for their support.
Abbreviations
- Prx
peroxiredoxin
- Tpx
thiol peroxidase
- Cp
peroxidatic cysteine
- Cr
resolving cysteine
- Trx
thioredoxin
- MD
molecular dynamics
- RMSD
root mean square deviation
References
- 1.(a) Kang SW, Rhee SG, Chang TS, Jeong W, Choi MH. Trends Mol Med. 2005;11:571–578. doi: 10.1016/j.molmed.2005.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Wood ZA, Schroder E, Harris JR, Poole LB. Trends Biochem Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 2.(a) Jacobson FS, Morgan RW, Christman MF, Ames BN. J Biol Chem. 1989;264:1488–1496. [PubMed] [Google Scholar]; (b) Christman MF, Morgan RW, Jacobson FS, Ames BN. Cell. 1985;41:753–762. doi: 10.1016/s0092-8674(85)80056-8. [DOI] [PubMed] [Google Scholar]; (c) Greenberg JT, Demple B. EMBO J. 1988;7:2611–2617. doi: 10.1002/j.1460-2075.1988.tb03111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poole LB, Ellis HR. Methods Enzymol. 2002;348:122–136. doi: 10.1016/s0076-6879(02)48632-6. [DOI] [PubMed] [Google Scholar]
- 4.Poole LB. The catalytic mechanism of peroxiredoxins. In: Flohé L, Harris JR, editors. Peroxiredoxin systems. Springer; New York: 2007. pp. 61–81. [Google Scholar]
- 5.(a) Poole LB, Hall A, Nelson KJ. Curr Protoc Toxicol. 2011;Chapter 7(Unit7):9. doi: 10.1002/0471140856.tx0709s49. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Reeves SA, Parsonage D, Nelson KJ, Poole LB. Biochemistry. 2011;50:8970–8981. doi: 10.1021/bi200935d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wood ZA, Poole LB, Hantgan RR, Karplus PA. Biochemistry. 2002;41:5493–5504. doi: 10.1021/bi012173m. [DOI] [PubMed] [Google Scholar]
- 7.Hall A, Parsonage D, Poole LB, Karplus PA. Journal of molecular biology. 2010;402:194–209. doi: 10.1016/j.jmb.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chae HZ, Robison K, Poole LB, Church G, Storz G, Rhee SG. Proc Natl Acad Sci U S A. 1994;91:7017–7021. doi: 10.1073/pnas.91.15.7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hofmann B, Hecht H-J, Flohé L. Biol Chem. 2002;383:347–364. doi: 10.1515/BC.2002.040. [DOI] [PubMed] [Google Scholar]
- 10.Hall A, Nelson K, Poole LB, Karplus PA. Antioxidants & redox signaling. 2011;15:795–815. doi: 10.1089/ars.2010.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.(a) Sarma GN, Nickel C, Rahlfs S, Fischer M, Becker K, Karplus PA. J Mol Biol. 2005;346:1021–1034. doi: 10.1016/j.jmb.2004.12.022. [DOI] [PubMed] [Google Scholar]; (b) Karplus PA, Hall A. Structural Survey of the Peroxiredoxins. In: Flohé L, Harris JR, editors. Peroxiredoxin Systems. Springer; New York: 2007. pp. 40–61. [Google Scholar]
- 12.Guimaraes BG, Souchon H, Honore N, Saint-Joanis B, Brosch R, Shepard W, Cole ST, Alzari PM. J Biol Chem. 2005;280:25735–25742. doi: 10.1074/jbc.M503076200. [DOI] [PubMed] [Google Scholar]
- 13.Alphey MS, Bond CS, Tetaud E, Fairlamb AH, Hunter WN. J Mol Biol. 2000;300:903–916. doi: 10.1006/jmbi.2000.3881. [DOI] [PubMed] [Google Scholar]
- 14.Yuan Y, Knaggs MH, Poole LB, Fetrow JS, Salsbury FR., Jr Journal of Biomolecular Structure & Dynamics. 2010;28:51–70. doi: 10.1080/07391102.2010.10507343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nashine VC, Hammes-Schiffer S, Benkovic SJ. Current opinion in chemical biology. 2010;14:644–651. doi: 10.1016/j.cbpa.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.(a) Boehr DD, McElheny D, Dyson HJ, Wright PE. Science. 2006;313:1638–1642. doi: 10.1126/science.1130258. [DOI] [PubMed] [Google Scholar]; (b) Agarwal PK, Billeter SR, Rajagopalan PT, Benkovic SJ, Hammes-Schiffer S. P Natl Acad Sci USA. 2002;99:2794–2799. doi: 10.1073/pnas.052005999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benkovic SJ, Hammes GG, Hammes-Schiffer S. Biochemistry. 2008;47:3317–3321. doi: 10.1021/bi800049z. [DOI] [PubMed] [Google Scholar]
- 18.Kniewel R, Buglino J, Solorzano V, Wu J, Lima CD, Burley SK. New York SGX Research Center for Structural Genomics. 2003. [Google Scholar]
- 19.Knaggs MH, Salsbury FR, Jr, Edgell MH, Fetrow JS. Biophysical journal. 2007;92:2062–2079. doi: 10.1529/biophysj.106.081950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalé L, Skeel R, Bhandarkar M, Brunner R, Gursoy A, Krawetz N, Phillips J, Shinozaki A, Varadarajan K, Schulten K. J Comp Physics. 1999;151:283–312. [Google Scholar]
- 21.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. J Comp Chem. 1983;4:187–217. [Google Scholar]
- 22.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 23.van Gunsteren WF, Berendsen HJ. Mol Phys. 1977;34:1311–1327. [Google Scholar]
- 24.Darden T, York T, Pedersen LG. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 25.(a) Bashford D. An object-oriented programming suite for electrostatic effects in biological molecules. In: Ishikawa Y, Odehoeft RR, Reynders JVW, Tholburn M, editors. Lecture Notes in Computer Science. Vol. 1343. Springer; Berlin: 1997. pp. 233–240. [Google Scholar]; (b) Bashford D. Front Biosci. 2004;9:1082–1099. doi: 10.2741/1187. [DOI] [PubMed] [Google Scholar]
- 26.Salsbury FR, Jr, Knutson ST, Poole LB, Fetrow JS. Protein Sci. 2008;17:299–312. doi: 10.1110/ps.073096508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng L, Chen M, Yang W. P Natl Acad Sci USA. 2008;105:20227–20232. doi: 10.1073/pnas.0810631106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee MS, Salsbury FR, Jr, Brooks CL., 3rd Proteins. 2004;56:738–752. doi: 10.1002/prot.20128. [DOI] [PubMed] [Google Scholar]
- 29.Pineyro MD, Pizarro JC, Lema F, Pritsch O, Cayota A, Bentley GA, Robello C. J Struct Biol. 2005;150:11–22. doi: 10.1016/j.jsb.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 30.Milner-White EJ. J Mol Biol. 1990;216:386–397. [PubMed] [Google Scholar]
- 31.Karplus PA. Protein Sci. 1996;5:1406–1420. doi: 10.1002/pro.5560050719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.(a) Frank RA, Titman CM, Pratap JV, Luisi BF, Perham RN. Science. 2004;306:872–876. doi: 10.1126/science.1101030. [DOI] [PubMed] [Google Scholar]; (b) Frank RA, Leeper FJ, Luisi BF. Cellular and molecular life sciences: CMLS. 2007;64:892–905. doi: 10.1007/s00018-007-6423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Budde H, Flohe L, Hecht HJ, Hofmann B, Stehr M, Wissing J, Lunsdorf H. Biological chemistry. 2003;384:619–633. doi: 10.1515/BC.2003.069. [DOI] [PubMed] [Google Scholar]
- 34.Wood ZA, Poole LB, Karplus PA. Science. 2003;300:650–653. doi: 10.1126/science.1080405. [DOI] [PubMed] [Google Scholar]
- 35.Antosiewicz J, McCammon JA, Gilson MK. Biochemistry. 1996;35:7819–7833. doi: 10.1021/bi9601565. [DOI] [PubMed] [Google Scholar]