Abstract
Purpose of review
We summarize recent progress on GPIHBP1, a molecule that transports lipoprotein lipase (LPL) to the capillary lumen, and discuss several newly studied molecules that appear important for the regulation of LPL activity.
Recent findings
LPL, the enzyme responsible for the lipolytic processing of triglyceride-rich lipoproteins, interacts with multiple proteins and is regulated at multiple levels. Several regulators of LPL activity have been known for years and have been investigated thoroughly, but several have been identified only recently, including an endothelial cell protein that transports LPL to the capillary lumen, a microRNA that reduces LPL transcript levels, a sorting protein that targets LPL for uptake and degradation, and a transcription factor that increases the expression of apolipoproteins that regulate LPL activity.
Summary
Proper regulation of LPL is important for controlling the delivery of lipid nutrients to tissues. Recent studies have identified GPIHBP1 as the molecule that transports LPL to the capillary lumen, and have also identified other molecules that are potentially important for regulating LPL activity. These new discoveries open new doors for understanding basic mechanisms of lipolysis and hyperlipidemia.
Keywords: diabetes mellitus, gene regulation, lipoproteins, triglyceride metabolism
INTRODUCTION
The processing of triglyceride-rich lipoproteins by lipoprotein lipase (LPL) is the central event in plasma lipid metabolism. The hydrolysis of lipoprotein triglycerides occurs mainly in the heart, skeletal muscle, and adipose tissue [1–3], generating fatty acids that are either used for fuel in striated muscle or stored in the form of triglycerides in adipose tissue [4,5]. Aside from releasing lipid nutrients for uptake by tissues, lipolytic processing also produces atherogenic remnant lipoproteins (e.g., LDLs) and provides lipids for the biogenesis of HDLs. Defective lipolysis causes some cases of severe hyper-triglyceridemia. For example, deficiencies in LPL or its protein cofactor, apolipoprotein (apo) CII, cause familial chylomicronemia, a condition in which the plasma triglyceride levels often exceed 2000 mg/dl [1]. Patients with chylomicronemia exhibit eruptive xanthomas, lipemia retinalis, and hepatosplenomegaly, and carry a high risk for developing fatal pancreatitis [1,6].
LPL is expressed most highly in tissues that either use fatty acids for fuel or store large amounts of triglycerides (i.e., heart, skeletal muscle, and adipose tissue), but LPL transcripts and protein can be found in many other tissues, including certain parts of the brain [7–10]. LPL is synthesized by parenchymal cells (e.g., myocytes and adipocytes), assembled into head-to-tail homodimers, and then secreted into the surrounding interstitial spaces. To be relevant to the processing of triglyceride-rich lipoproteins, however, LPL must be transported from the interstitial spaces to the luminal face of capillary endothelial cells (wherein it is accessible to triglyceride-rich lipoproteins). For years, the mechanism by which LPL reached the capillary lumen was unclear, but this mystery was recently solved. LPL is transported to the capillary lumen by glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1 (GPIHBP1), a GPI-anchored protein of capillary endothelial cells.
Because LPL-mediated processing of lipoproteins is so important for plasma lipid metabolism and fuel delivery to tissues, one would expect that its activity would be regulated. Indeed, this is the case; LPL regulation occurs at the transcriptional, post-transcriptional, translational, and post-translational levels [4,5], and also by the intrinsic properties of the enzyme. LPL enzymatic activity is downregulated by high concentrations of fatty acids, a form of regulation that might serve to prevent fatty acids from exceeding the capacity of tissues to take them up [11]. A variety of proteins that regulate LPL have been identified, including apo-CII [12,13], apo-CIII [14], apo-AV [15–17], angiopoietin-like protein 3 (ANGPTL3) [18,19], and ANGPTL4 [18]. ANGPTL4 has been shown to convert catalytically active LPL to inactive monomers [20], and a recent study showed that ANGPTL3 renders LPL more susceptible to proteolytic inactivation by proprotein convertases [21▪]. These topics have been discussed in recent reviews by Wang and Eckel [5] and Olivecrona and Olivecrona [4]. In this update, we focus on GPIHBP1, the endothelial cell LPL transporter, an area of interest within our laboratory. We also highlight several recent papers describing new molecules that appear relevant to the regulation of LPL activity.
LIPOPROTEIN LIPASE TRANSPORT ACROSS CAPILLARY ENDOTHELIAL CELLS
The identification of GPIHBP1 as the endothelial cell transporter of LPL represents a significant advancement in our understanding of LPL biology. As noted earlier, the mechanism by which LPL entered blood vessels remained elusive for years, but recently was shown to involve GPIHBP1, a GPI-anchored protein of endothelial cells. The association of GPIHBP1 with lipolysis began with the discovery of severe chylomicronemia in GPIHBP1 knockout mice (Gpihbp1−/−) [22]. On a chow diet, the plasma triglyceride levels in Gpihbp1−/− mice were 3000–5000 mg/dl, and they increased to greater than 20 000 mg/dl on a Western diet. For such a striking phenotype to occur with a knockout of a GPI-anchored protein, we immediately suspected that GPIHBP1 must be involved in LPL-mediated processing of lipoproteins and GPIHBP1 would have to be located on endothelial cells. Indeed, this was the case. By immunohistochemistry, GPIHBP1 was located exclusively in capillary endothelial cells, and biochemical studies revealed that GPIHBP1-expressing cells bind LPL avidly. These observations prompted speculation that GPIHBP1 serves as a binding platform for lipolysis on the vascular lumen [22,23].
GPIHBP1 is a member of the lymphocyte antigen 6 family of proteins. It is a small protein, with the mouse protein containing only 228 amino acids. The mature protein has several noteworthy features. The first is a striking accumulation of negatively charged amino acids at the protein’s amino terminus. The second is a ‘Ly6 domain’ spanning approximately 80 amino acids and containing 10 cysteines arranged in a characteristic spacing pattern. Disulfide bonds between the 10 cysteines create a three-fingered structure. The Ly6 domain of mouse GPIHBP1 contains an N-linked carbohydrate chain, which is important for efficient trafficking to the cell surface [24]. A series of mutagenesis studies revealed that both the acidic domain and the Ly6 domain are crucial for GPIHBP1’s ability to bind LPL [25▪,26,27]. The third noteworthy structure is the GPI anchor at the carboxyl terminus. Why nature chose to anchor this protein to the surface of cells with a GPI anchor rather than a transmembrane helix is not clear.
Soon after GPIHBP1 was uncovered, Weinstein et al. [28] reported that Gpihbp1−/− mice have normal tissue stores of LPL, but when compared with wild-type mice, LPL in Gpihbp1−/− mice enters the plasma more slowly after an intravenous dose of heparin. That finding prompted speculation that the metabolic defect in Gpihbp1−/− mice might relate to mislocalization of LPL [28]. More recently, Davies et al. [29▪▪] examined the possibility that GPIHBP1 might be involved in moving LPL to the capillary lumen. First, they tested whether GPIHBP1 was capable of transporting a GPIHBP1-specific monoclonal antibody across cells. Indeed, GPIHBP1-expressing endothelial cells efficiently transported the GPIHBP1-specific antibody (but not an irrelevant control antibody) from the basolateral medium to the apical face of the cells. Transport was abolished when GPIHBP1 at the basolateral surface of the cell was released with phosphatidyl-inositol-specific phospholipase C (PIPLC). Davies et al. [29▪▪] went on to show that GPIHBP1 was effective in transporting LPL from the basolateral to the apical surface of endothelial cells [29▪▪], and this transport could be abolished when LPL binding to GPIHBP1 at the basolateral surface of cells was blocked with heparin or PIPLC pretreatment. As expected, endothelial cell monolayers expressing a mutant version of GPIHBP1 — one that lacked the ability to bind LPL — were unable to transport LPL across cells [29▪▪].
Davies et al. [29▪▪] demonstrated that GPIHBP1 is located on both the luminal and abluminal surfaces of capillary endothelial cells in mouse tissues, as would be expected for a transporter protein. They also showed that GPIHBP1 functioned as a transporter in vivo. When they injected a rat monoclonal antibody against GPIHBP1 into the quadriceps of a wild-type mouse, the antibody quickly diffused into the interstitial space, surrounding each myocyte within the muscle bundle. Within minutes, however, the antibody was bound by GPIHBP1-expressing endothelial cells and transported to the capillary lumen. The presence of the GPIHBP1-specific antibody in the lumen of capillaries was detected by immunohistochemistry after an intravenous injection of a fluorescently labeled anti-rat IgG. No transport of the GPIHBP1-specific antibody was observed in tissues of Gpihbp1−/− mice [29▪▪].
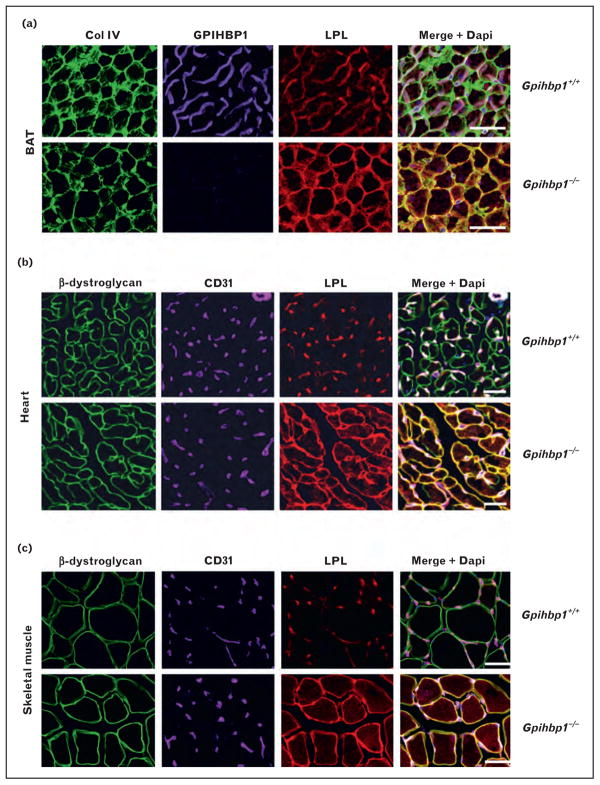
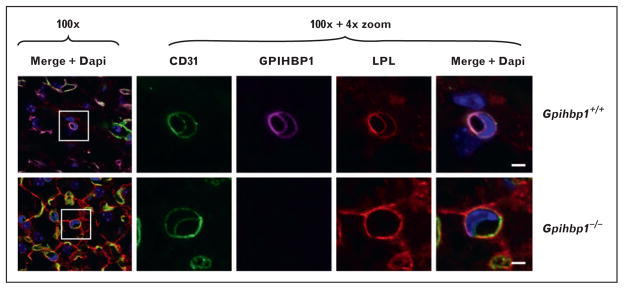
Davies et al. [29▪▪] used confocal microscopy to determine whether the absence of GPIHBP1 caused mislocalization of LPL. In wild-type mice, they found that most of the LPL in tissues was associated with capillaries, colocalizing with GPIHBP1 and CD31 (an endothelial cell marker) (Fig. 1). In contrast, virtually all of the LPL in tissues of Gpihbp1−/− mice was located around parenchymal cells, and little was found associated with capillaries (Fig. 1). When cross-sections of capillaries were examined by confocal microscopy, LPL was easily detectable along the luminal face of endothelial cells in wild-type mice, colocalizing with GPIHBP1 and CD31. In contrast, no LPL was present in the capillary lumen of Gpihbp1−/− mice (Fig. 2). The mislocalization of LPL in Gpihbp1−/− mice, combined with the ability of GPIHBP1 to move antibodies and LPL across endothelial cells, strongly supported the idea that GPIHBP1 is responsible for delivering LPL to the capillary lumen.
FIGURE 1.
Mislocalization of LPL in tissues of Gpihbp1−/− mice. (a) Confocal microscopy showing the binding of GPIHBP1-specific antibodies, LPL-specific antibodies, and collagen IV-specific antibodies to brown adipose tissue (BAT) from wild-type and Gpihbp1−/− mice. Scale bars, 100 μm. (b, c) Confocal microscopy showing the binding of CD31-specific antibodies, LPL-specific antibodies, and β-dystroglycan-specific antibodies to heart (b) and skeletal muscle (c) of wild-type and Gpihbp1−/− mice. The scale bars show a distance of 100 μm (skeletal muscle) or 50 μm (heart). Reproduced with permission from [29■■].
FIGURE 2.
Absence of lipoprotein lipase from the capillary lumen of Gpihbp1−/− mice. Confocal microscopy showing the binding of CD31-specific antibodies, GPIHBP1-specific antibodies, and LPL-specific antibodies to brown adipose tissue (BAT) of a wild-type mouse and a Gpihbp1−/− mouse. Images were taken with a 100× objective without optical zoom or with 4× optical zoom. To visualize both the apical and basolateral surfaces of capillaries, cross-sections of capillaries containing an endothelial cell nucleus (blue) were identified (boxed areas) and viewed at high magnification. GPIHBP1 (purple), LPL (red), and CD31 (green) were detected at the apical and basolateral surfaces of endothelial cells in wild-type mice, but no LPL was present on the apical (luminal) surface of capillaries in Gpihbp1−/− mice. Scale bar, 2.5 μm. Reproduced with permission from [29■■].
GPIHBP1’s crucial role in triglyceride metabolism is not a peculiarity of the mouse. In the past few years, a series of studies has shown that mutations in human GPIHBP1 cause chylomicronemia [30,31▪,32–35]. But these mutations are fairly rare, even in screens of highly selected patients with unexplained hypertriglyceridemia. Most of the clinically significant GPIHBP1 mutations simply abolish GPIHBP1’s ability to bind LPL. However, a recently described mutation was located in GPIHBP1’s carboxyl-terminal hydrophobic domain and presumably interfered with the addition of the GPI anchor [31▪]. Also, one patient had a deletion of the entire GPIHBP1 locus [31▪]. All patients described thus far have had two defective GPIHBP1 alleles; heterozygotes within the families appear to be normolipidemic. Even though GPIHBP1 deficiency is a classic recessive disorder, recent data suggest that a common polymorphism in GPIHBP1 might affect plasma triglyceride metabolism. Charriere et al. [31▪] noted that a common coding polymorphism in GPIHBP1’s signal peptide (C14F) is more common in patients with chylomicronemia than in normolipidemic controls. Another recent study by Voss et al. [36▪] characterized LPL mutations that abolished LPL’s ability to bind to GPIHBP1 and prevented trans-endothelial cell transport. These mutations, which were initially identified in patients with chylomicronemia [37,38], had no effect on the catalytic activity of LPL or its ability to bind to heparin [36▪].
In the mouse, Gpihbp1 transcript levels are modulated by diet, feeding state, and peroxisome proliferator-activated receptor-γ [39], but whether similar regulation occurs in humans is unknown. Also, it is unknown whether the degree of regulation observed in the mouse, invariably less than twofold, has significant effects on LPL transport rates or plasma triglyceride metabolism, particularly, as heterozygous GPIHBP1 deficiency has no impact on plasma triglyceride levels [22,28].
microRNA REGULATION OF LIPOPROTEIN LIPASE
With the recent explosion of interest in microRNAs (miRs), it seemed only a matter of time until a miR affecting LPL was described. In a recent study, Chen et al. [40▪] sought to identify the role of miR-29a (a miR expressed in mature dendritic cells [41]) in inflammation and atherosclerosis. They found that increasing miR-29a expression lowered LPL mRNA and protein levels, an effect mediated through LPL’s 3′ UTR [40▪]. Inhibition of miR-29a led to increased LPL mRNA and protein levels. Treatment of dendritic cells with oxidized LDL increased miR-29a expression, which in turn increased scavenger receptor expression and reduced proinflammatory cytokine release. The authors concluded those effects were mediated by LPL because an siRNA knockdown of LPL mimicked miR-29a’s effects and also attenuated the effects of an miR-29a inhibitor [40▪]. It will be interesting to see whether modulation of LPL transcripts by mIRs is widespread, orconfined to dendritic cells. Because LPL’s regulation is often tissue-specific, it is tempting to speculate that some of this regulation could be mediated by tissue-specific miRs.
SorLA, LIPOPROTEIN LIPASE, AND THE BRAIN
Sortilin-related receptor with A-type repeats (SorLA) is a multifunctional receptor that mediates endocytic and intracellular trafficking [42,43]. Klinger et al. [44▪] found, using surface plasmon resonance analysis, that SorLA binds LPL with high affinity. In cells expressing SorLA, SorLA and LPL colocalized and targeting of LPL to endosomes increased [44▪]. SorLA is expressed highly throughout the brain, and LPL is found in neurons and glia of the brain, particularly in the hippocampus [45]. When Klinger et al. [44▪] examined LPL localization in primary hippocampal neurons and cortical glia, they observed vesicle-like staining, whereas LPL staining in neurons and glia cells from SorLA knockout mice was diffuse, suggesting that SorLA was responsible for the vesicular localization of LPL. The authors speculated that SorLA may be linked to LPL function in the brain. The precise function of LPL in the brain is poorly understood, but a recent study [46▪] found that neuron-specific LPL-deficient mice were hyperphagic and obese. The authors concluded that the brain must use an LPL-dependent mechanism to sense triglyceride-rich lipoproteins and regulate energy balance [46▪]. The discovery of LPL–SorLA interactions could provide a clue to the physiologic function of LPL in the brain.
REGULATION OF LIPOPROTEIN LIPASE BY APOLIPOPROTEINS
Several apolipoproteins affect the efficiency of lipolysis. As noted earlier, apo-CII is a crucial cofactor for LPL and LPL activity is minimal in its absence. apo-AV is also important; high apo-AV expression levels lower plasma triglyceride levels, whereas low levels increase triglyceride levels [15–17]. apo-CIII also has a profound impact on lipolysis; heterozygosity for an APOC3 nonsense mutation lowers plasma triglyceride levels, and a complete deficiency of apo-CIII greatly accelerates lipolysis [14,47]. In a recent study, Lee et al. [48▪▪] identified cyclic AMP-responsive element-binding protein H (CREB-H) as a regulator of several proteins affecting lipolysis, including apo-AV, apo-CII, and apo-CIII. CREB-H, a product of the Creb3L3 gene, is an ER-bound transcription factor that is expressed highly in the liver and small intestine [49]. Creb3L3−/− mice have elevated VLDL levels in the plasma [48▪▪]. VLDL production rates in Creb3L3−/− mice were unchanged, but VLDL clearance was reduced. Although LPL expression levels in adipose tissue and skeletal muscle were normal in Creb3L3−/− mice, the postheparin LPL activity levels were lower. This was most likely due to decreased expression of apo-CII and apo-AV and increased expression of apo-CIII. When the authors generated transgenic mice overexpressing CREB-H in the liver, they observed decreased plasma triglyceride levels and increased expression of Apoc2 and Apoa5 [48▪▪]. To determine whether defects in CREB-H might contribute to hypertriglyceridemia in humans, the authors sequenced CREB3L3 in a cohort of patients with hypertriglyceridemia. In 449 unrelated individuals with hypertriglyceridemia, but no disease-causing mutations in LPL, APOC2, or APOA5, the authors identified 12 nonsynonymous or insertional mutations in CREB3L3. No such mutations were found in normolipidemic controls. A subset of the CREB-H mutants were defective in their ability to activate target genes in a reporter assay [48▪▪].
Perdomo et al. [50▪] recently reported that apoD, an atypical apolipoprotein expressed by many mammalian tissues [51], influences plasma triglyceride levels. apo-D is found predominantly on HDL, but it can also be found on VLDL [52]. Perdomo et al. [50▪] found that adenoviral-mediated overexpression of apo-D lowers plasma triglyceride levels in chow-fed mice and mice with diet-induced obesity. VLDL clearance was increased but VLDL production rates were unchanged. LPL mRNA and protein levels were not altered by apo-D expression, but postheparin plasma LPL activity levels were increased, suggesting that apo-D was somehow capable of increasing LPL activity levels. In fact, when LPL and VLDL were mixed with apo-D (from HepG2 cells that had been infected with an apo-D adenovirus), apo-D increased LPL activity in a dose-dependent fashion. The authors speculated that the ability of apo-D to increase LPL activity might explain the association of APOD missense mutations with elevated plasma triglyceride levels [53].
CONCLUSION
The discovery of new molecules affecting LPL biology has invigorated the triglyceride metabolism field and raised many new questions for investigation. With the discovery of GPIHBP1, the field can formulate fresh hypotheses regarding mechanisms of lipolysis on the surface of endothelial cells. It will be interesting to determine, for example, whether efficient lipolysis requires the clustering of GPIHBP1–LPL complexes on the surface of endothelial cells. It will also be important to determine the cellular mechanisms by which GPIHBP1 transports LPL, and whether transport is perturbed in the setting of acquired hypertriglyceridemia. Also, more work is required to understand how nutrients, hormones, environmental conditions, and metabolic diseases influence triglyceride metabolism. For example, LPL activity is elevated in the hearts of mouse and rat models of diabetes, and several mechanisms have been proposed to explain this observation [54–58,59▪], but our understanding of LPL regulation by insulin and glucose remains incomplete. Also, in a recent study, Bartelt et al. [60▪] found that exposure to cold dramatically increased LPL-mediated plasma triglyceride clearance in mice, but the molecular mechanisms underlying these findings remain unclear. Although the list of physiologic regulators of LPL has grown, the mechanism by which many of the regulators influence LPL activity and lipolysis is incompletely understood. For example, even though the role of apo-CII in activating LPL’s catalytic activity has been known for decades, the precise molecular mechanisms underlying the activation remain unclear. Thus, there are many topics within the field of lipolysis to investigate, and the work is still important. Understanding lipolysis is crucial for understanding hyperlipidemia, obesity, and the delivery to of nutrients to vital tissues such as the heart.
KEY POINTS.
GPIHBP1 transports LPL across endothelial cells.
In mature dendritic cells, LPL is regulated by miRNA-29a.
The sorting protein sortilin-related receptor with A-type repeats (SorLA) binds LPL, and may be involved in regulating LPL in neurons and glia of the brain.
Several activators of LPL, including apo-CII and apo-AV, are induced by CREB-H.
Apo-D increases LPL activity
Acknowledgments
This work was supported by a Postdoctoral Fellowship (to B.S.J.D.) and a Scientist Development Award (to A.P.B.) from the American Heart Association, R01 HL094732 (to APB), P01 HL090553 (to SGY), and R01 HL087228 (to SGY). This work was funded in part by the National Institutes of Health.
Footnotes
Conflicts of interest
There are no conflicts of interest.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
▪ of special interest
▪▪ of outstanding interest
Additional references related to this topic can also be found in the Current World Literature section in this issue (pp. 69–70).
- 1.Brunzell JD, Deeb SS. Familial lipoprotein lipase deficiency, apo C-II deficiency, and hepatic lipase deficiency. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The metabolic and molecular bases of inherited disease. 8. New York: McGraw-Hill; 2001. pp. 2789–2816. [Google Scholar]
- 2.Havel RJ, Kane JP. Introduction: structure and metabolism of plasma lipoproteins. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The metabolic and molecular bases of inherited disease. 8. New York: McGraw-Hill; 2001. pp. 2705–2716. [Google Scholar]
- 3.Tan MH, Sata T, Havel RJ. The significance of lipoprotein lipase in rat skeletal muscles. J Lipid Res. 1977;18:363–370. [PubMed] [Google Scholar]
- 4.Olivecrona T, Olivecrona G. The ins and outs of adipose tissue. In: Ehnholm C, editor. Cellular lipid metabolism. Berlin Heidelberg: Springer; 2009. pp. 315–369. [Google Scholar]
- 5.Wang H, Eckel RH. Lipoprotein lipase: from gene to obesity. Am J Physiol Endocrinol Metab. 2009;297:E271–E288. doi: 10.1152/ajpendo.90920.2008. [DOI] [PubMed] [Google Scholar]
- 6.Reina M, Brunzell JD, Deeb SS. Molecular basis of familial chylomicronemia: Mutations in the lipoprotein lipase and apolipoprotein C-II genes. J Lipid Res. 1992;33:1823–1832. [PubMed] [Google Scholar]
- 7.Ben-Zeev O, Doolittle MH, Singh N, et al. Synthesis and regulation of lipoprotein lipase in the hippocampus. J Lipid Res. 1990;31:1307–1313. [PubMed] [Google Scholar]
- 8.Bessesen DH, Richards CL, Etienne J, et al. Spinal cord of the rat contains more lipoprotein lipase than other brain regions. J Lipid Res. 1993;34:229–238. [PubMed] [Google Scholar]
- 9.Goldberg IJ, Soprano DR, Wyatt ML, et al. Localization of lipoprotein lipase mRNA in selected rat tissues. J Lipid Res. 1989;30:1569–1577. [PubMed] [Google Scholar]
- 10.Vilaro S, Camps L, Reina M, et al. Localization of lipoprotein lipase to discrete areas of the guinea pig brain. Brain Res. 1990;506:249–253. doi: 10.1016/0006-8993(90)91258-i. [DOI] [PubMed] [Google Scholar]
- 11.Bengtsson G, Olivecrona T. Lipoprotein lipase: mechanism of product inhibition. Eur J Biochem. 1980;106:557–562. doi: 10.1111/j.1432-1033.1980.tb04603.x. [DOI] [PubMed] [Google Scholar]
- 12.Catapano AL. Apolipoprotein C-II and lipoprotein lipase activity. Ric Clin Lab. 1982;12:35–40. doi: 10.1007/BF02909307. [DOI] [PubMed] [Google Scholar]
- 13.Kinnunen PK, Jackson RL, Smith LC, et al. Activation of lipoprotein lipase by native and synthetic fragments of human plasma apolipoprotein C-II. Proc Natl Acad Sci U S A. 1977;74:4848–4851. doi: 10.1073/pnas.74.11.4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ginsberg HN, Le NA, Goldberg IJ, et al. Apolipoprotein B metabolism in subjects with deficiency of apolipoproteins CIII and AI. Evidence that apo-lipoprotein CIII inhibits catabolism of triglyceride-rich lipoproteins by lipoprotein lipase in vivo. J Clin Invest. 1986;78:1287–1295. doi: 10.1172/JCI112713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calandra S, Priore Oliva C, Tarugi P, Bertolini S. APOA5 and triglyceride metabolism, lesson from human APOA5 deficiency. Curr Opin Lipidol. 2006;17:122–127. doi: 10.1097/01.mol.0000217892.00618.54. [DOI] [PubMed] [Google Scholar]
- 16.Grosskopf I, Baroukh N, Lee SJ, et al. Apolipoprotein A-V deficiency results in marked hypertriglyceridemia attributable to decreased lipolysis of triglyceride-rich lipoproteins and removal of their remnants. Arterioscler Thromb Vasc Biol. 2005;25:2573–2579. doi: 10.1161/01.ATV.0000186189.26141.12. [DOI] [PubMed] [Google Scholar]
- 17.Pennacchio LA, Olivier M, Hubacek JA, et al. An apolipoprotein influencing triglycerides in humans and mice revealed by comparative sequencing. Science. 2001;294:169–173. doi: 10.1126/science.1064852. [DOI] [PubMed] [Google Scholar]
- 18.Koster A, Chao YB, Mosior M, et al. Transgenic angiopoietin-like (angptl)4 overexpression and targeted disruption of angptl4 and angptl3: regulation of triglyceride metabolism. Endocrinology. 2005;146:4943–4950. doi: 10.1210/en.2005-0476. [DOI] [PubMed] [Google Scholar]
- 19.Shimizugawa T, Ono M, Shimamura M, et al. ANGPTL3 decreases very low density lipoprotein triglyceride clearance by inhibition of lipoprotein lipase. J Biol Chem. 2002;277:33742–33748. doi: 10.1074/jbc.M203215200. [DOI] [PubMed] [Google Scholar]
- 20.Sukonina V, Lookene A, Olivecrona T, Olivecrona G. Angiopoietin-like protein 4 converts lipoprotein lipase to inactive monomers and modulates lipase activity in adipose tissue. Proc Natl Acad Sci U S A. 2006;103:17450–17455. doi: 10.1073/pnas.0604026103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21▪.Liu J, Afroza H, Rader DJ, Jin W. Angiopoietin-like protein 3 inhibits lipoprotein lipase activity through enhancing its cleavage by proprotein convertases. J Biol Chem. 2010;285:27561–27570. doi: 10.1074/jbc.M110.144279. This study identified the mechanism by which ANGPTL3 inhibits LPL activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beigneux AP, Davies BS, Gin P, et al. Glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1 plays a critical role in the lipolytic processing of chylomicrons. Cell Metab. 2007;5:279–291. doi: 10.1016/j.cmet.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Young SG, Davies BS, Fong LG, et al. GPIHBP1: an endothelial cell molecule important for the lipolytic processing of chylomicrons. Curr Opin Lipidol. 2007;18:389–396. doi: 10.1097/MOL.0b013e3281527914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beigneux AP, Gin P, Davies BS, et al. Glycosylation of Asn-76 in mouse GPIHBP1 is critical for its appearance on the cell surface and the binding of chylomicrons and lipoprotein lipase. J Lipid Res. 2008;49:1312–1321. doi: 10.1194/jlr.M700593-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25▪.Beigneux AP, Davies BS, Tat S, et al. Assessing the role of the glycosylphosphatidylinositol-anchored high density lipoprotein-binding protein 1 (GPIHBP1) three-finger domain in binding lipoprotein lipase. J Biol Chem. 2011;286:19735–19743. doi: 10.1074/jbc.M111.242024. This study identified residues in GPIHBP1 that are critical for LPL binding, highlighting the importance of the Ly6 domain for GPIHBP1 function. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beigneux AP, Gin P, Davies BS, et al. Highly conserved cysteines within the Ly6 domain of GPIHBP1 are crucial for the binding of lipoprotein lipase. J Biol Chem. 2009;284:30240–30247. doi: 10.1074/jbc.M109.046391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gin P, Yin L, Davies BS, et al. The acidic domain of GPIHBP1 is important for the binding of lipoprotein lipase and chylomicrons. J Biol Chem. 2008;283:29554–29562. doi: 10.1074/jbc.M802579200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weinstein MM, Yin L, Beigneux AP, et al. Abnormal patterns of lipoprotein lipase release into the plasma in GPIHBP1-deficient mice. J Biol Chem. 2008;283:34511–34518. doi: 10.1074/jbc.M806067200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29▪▪.Davies BS, Beigneux AP, Barnes RH, 2nd, et al. GPIHBP1 is responsible for the entry of lipoprotein lipase into capillaries. Cell Metab. 2010;12:42–52. doi: 10.1016/j.cmet.2010.04.016. This study used both cell culture and in-vivo experiments to show that GPIHBP1 transports LPL across endothelial cell, thus solving a long-standing mystery in the lipolysis field. This finding highlights the important role of endothelial cells in triglyceride metabolism and suggests another possible avenue whereby lipolysis might be regulated. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beigneux AP, Franssen R, Bensadoun A, et al. Chylomicronemia with a mutant GPIHBP1 (Q115P) that cannot bind lipoprotein lipase. Arterioscler Thromb Vasc Biol. 2009;29:956–962. doi: 10.1161/ATVBAHA.109.186577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31▪.Charriere S, Peretti N, Bernard S, et al. GPIHBP1 C89F neomutation and hydrophobic C-terminal domain G175R mutation in two pedigrees with severe hyperchylomicronemia. J Clin Endocrinol Metab. 2011;96:1675–1679. doi: 10.1210/jc.2011-1444. In addition to identifying two new disease-causing mutations in GPIHBP1, this study found that a common SNP in GPIHBP1, C14F, was more common in patients with hypertriglyceridemia – suggesting for the first time that common polymorphisms in GPIHBP1 might contribute to disease. [DOI] [PubMed] [Google Scholar]
- 32.Coca-Prieto I, Kroupa O, Gonzalez-Santos P, et al. Childhood-onset chylomicronaemia with reduced plasma lipoprotein lipase activity and mass: identification of a novel GPIHBP1 mutation. J Intern Med. 2011;270:224–228. doi: 10.1111/j.1365-2796.2011.02361.x. [DOI] [PubMed] [Google Scholar]
- 33.Franssen R, Young SG, Peelman F, et al. Chylomicronemia with low post-heparin lipoprotein lipase levels in the setting of GPIHBP1 defects. Circ Cardiovasc Genet. 2010;3:169–178. doi: 10.1161/CIRCGENETICS.109.908905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olivecrona G, Ehrenborg E, Semb H, et al. Mutation of conserved cysteines in the Ly6 domain of GPIHBP1 in familial chylomicronemia. J Lipid Res. 2010;51:1535–1545. doi: 10.1194/jlr.M002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J, Hegele RA. Homozygous missense mutation (G56R) in glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1 (GPI-HBP1) in two siblings with fasting chylomicronemia (MIM 144650) Lipids Health Dis. 2007;6:23. doi: 10.1186/1476-511X-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36▪.Voss CV, Davies BS, Tat S, et al. Mutations in lipoprotein lipase that block binding to the endothelial cell transporter GPIHBP1. Proc Natl Acad Sci U S A. 2011;108:7980–7984. doi: 10.1073/pnas.1100992108. The study characterized a new class of disease-causing LPL mutations – mutations that abolish LPL binding to GPIHBP1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Henderson H, Leisegang F, Hassan F, et al. A novel Glu421Lys substitution in the lipoprotein lipase gene in pregnancy-induced hypertriglyceridemic pancreatitis. Clin Chim Acta. 1998;269:1–12. doi: 10.1016/s0009-8981(97)00144-7. [DOI] [PubMed] [Google Scholar]
- 38.Henderson HE, Hassan F, Marais D, Hayden MR. A new mutation destroying disulphide bridging in the C-terminal domain of lipoprotein lipase. Biochem Biophys Res Commun. 1996;227:189–194. doi: 10.1006/bbrc.1996.1487. [DOI] [PubMed] [Google Scholar]
- 39.Davies BS, Waki H, Beigneux AP, et al. The expression of GPIHBP1, an endothelial cell binding site for lipoprotein lipase and chylomicrons, is induced by peroxisome proliferator-activated receptor-gamma. Mol Endocrinol. 2008;22:2496–2504. doi: 10.1210/me.2008-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40▪.Chen T, Li Z, Tu J, et al. MicroRNA-29a regulates pro-inflammatory cytokine secretion and scavenger receptor expression by targeting LPL in oxLDL-stimulated dendritic cells. FEBS Lett. 2011;585:657–663. doi: 10.1016/j.febslet.2011.01.027. This study is the first to identify a microRNA that acts on LPL transcripts. This finding suggests that mIRs might play a role in regulating triglyceride metabolism. [DOI] [PubMed] [Google Scholar]
- 41.Holmstrom K, Pedersen AW, Claesson MH, et al. Identification of a microRNA signature in dendritic cell vaccines for cancer immunotherapy. Hum Immunol. 2010;71:67–73. doi: 10.1016/j.humimm.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 42.Nielsen MS, Gustafsen C, Madsen P, et al. Sorting by the cytoplasmic domain of the amyloid precursor protein binding receptor SorLA. Mol Cell Biol. 2007;27:6842–6851. doi: 10.1128/MCB.00815-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nielsen MS, Madsen P, Christensen EI, et al. The sortilin cytoplasmic tail conveys Golgi-endosome transport and binds the VHS domain of the GGA2 sorting protein. EMBO J. 2001;20:2180–2190. doi: 10.1093/emboj/20.9.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44▪.Klinger SC, Glerup S, Raarup MK, et al. SorLA regulates the activity of lipoprotein lipase by intracellular trafficking. J Cell Sci. 2011;124:1095–1105. doi: 10.1242/jcs.072538. This study found that SorLA binds LPL and might be important for LPL localization and turnover in neurons and glia cells. [DOI] [PubMed] [Google Scholar]
- 45.Xian X, Liu T, Yu J, et al. Presynaptic defects underlying impaired learning and memory function in lipoprotein lipase-deficient mice. J Neurosci. 2009;29:4681–4685. doi: 10.1523/JNEUROSCI.0297-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46▪.Wang H, Astarita G, Taussig MD, et al. Deficiency of lipoprotein lipase in neurons modifies the regulation of energy balance and leads to obesity. Cell Metab. 2011;13:105–113. doi: 10.1016/j.cmet.2010.12.006. The authors observe obesity and hyperphagia in mice lacking neuronal LPL. This study addresses the function of LPL in the brain, suggesting an important role in regulating energy balance. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pollin TI, Damcott CM, Shen H, et al. A null mutation in human APOC3 confers a favorable plasma lipid profile and apparent cardioprotection. Science. 2008;322:1702–1705. doi: 10.1126/science.1161524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48▪.Lee JH, Giannikopoulos P, Duncan SA, et al. The transcription factor cyclic AMP-responsive element-binding protein H regulates triglyceride metabolism. Nat Med. 2011;17:812–815. doi: 10.1038/nm.2347. This study showed that CREB-H induces several genes known to activate LPL. The authors also identified several variants in the gene that codes for CREB-H in patients with hypertriglyceridemia. These findings suggest that CREB-H might be a master regulator of triglyceride metabolism, modulating expression of several genes in response to metabolic stimuli such as fasting. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Omori Y, Imai J, Watanabe M, et al. CREB-H: a novel mammalian transcription factor belonging to the CREB/ATF family and functioning via the box-B element with a liver-specific expression. Nucleic Acids Res. 2001;29:2154–2162. doi: 10.1093/nar/29.10.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50▪.Perdomo G, Kim DH, Zhang T, et al. A role of apolipoprotein D in triglyceride metabolism. J Lipid Res. 2010;51:1298–1311. doi: 10.1194/jlr.M001206. In this study, the ability of ApoD to activate LPL activity was described. This study adds another apolipoprotein to those that influence LPL activity and highlights the importance of LPL–apolipoprotein interactions in triglyceride metabolism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Drayna D, Fielding C, McLean J, et al. Cloning and expression of human apolipoprotein D cDNA. J Biol Chem. 1986;261:16535–16539. [PubMed] [Google Scholar]
- 52.McConathy WJ, Alaupovic P. Studies on the isolation and partial characterization of apolipoprotein D and lipoprotein D of human plasma. Biochemistry. 1976;15:515–520. doi: 10.1021/bi00648a010. [DOI] [PubMed] [Google Scholar]
- 53.Desai PP, Bunker CH, Ukoli FA, Kamboh MI. Genetic variation in the apolipoprotein D gene among African blacks and its significance in lipid metabolism. Atherosclerosis. 2002;163:329–338. doi: 10.1016/s0021-9150(02)00012-6. [DOI] [PubMed] [Google Scholar]
- 54.Kim MS, Kewalramani G, Puthanveetil P, et al. Acute diabetes moderates trafficking of cardiac lipoprotein lipase through p38 mitogen-activated protein kinase-dependent actin cytoskeleton organization. Diabetes. 2008;57:64–76. doi: 10.2337/db07-0832. [DOI] [PubMed] [Google Scholar]
- 55.Kim MS, Wang F, Puthanveetil P, et al. Protein kinase D is a key regulator of cardiomyocyte lipoprotein lipase secretion after diabetes. Circ Res. 2008;103:252–260. doi: 10.1161/CIRCRESAHA.108.178681. [DOI] [PubMed] [Google Scholar]
- 56.Kim MS, Wang F, Puthanveetil P, et al. Cleavage of protein kinase D after acute hypoinsulinemia prevents excessive lipoprotein lipase-mediated cardiac triglyceride accumulation. Diabetes. 2009;58:2464–2475. doi: 10.2337/db09-0681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pulinilkunnil T, Abrahani A, Varghese J, et al. Evidence for rapid ‘metabolic switching’ through lipoprotein lipase occupation of endothelial-binding sites. J Mol Cell Cardiol. 2003;35:1093–1103. doi: 10.1016/s0022-2828(03)00205-0. [DOI] [PubMed] [Google Scholar]
- 58.Sambandam N, Abrahani MA, St Pierre E, et al. Localization of lipoprotein lipase in the diabetic heart: regulation by acute changes in insulin. Arterioscler Thromb Vasc Biol. 1999;19:1526–1534. doi: 10.1161/01.atv.19.6.1526. [DOI] [PubMed] [Google Scholar]
- 59▪.Wang Y, Puthanveetil P, Wang F, et al. Severity of diabetes governs vascular lipoprotein lipase by affecting enzyme dimerization and disassembly. Diabetes. 2011;60:2041–2050. doi: 10.2337/db11-0042. This study shows that LPL dimerization increases in the hearts of mouse models of hyperglycemia, leading to increased LPL activity. This study suggests an additional means whereby LPL activity can be modulated in response to disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60▪.Bartelt A, Bruns OT, Reimer R, et al. Brown adipose tissue activity controls triglyceride clearance. Nat Med. 2011;17:200–205. doi: 10.1038/nm.2297. In this study, dramatic increases in triglyceride clearance and uptake were observed in the brown adipose tissue of mice subjected to the cold. This study highlights the importance of brown adipose tissue as a lipolytic tissue. [DOI] [PubMed] [Google Scholar]