Abstract
Cytokines are critical mediators of inflammation and host defenses. Regulation of cytokines can occur at various stages of gene expression, including transcription, mRNA export, and post- transcriptional and translational levels. Among these modes of regulation, post-transcriptional regulation has been shown to play a vital role in controlling the expression of cytokines by modulating mRNA stability. The stability of cytokine mRNAs, including TNFα, IL-6, and IL-8, has been reported to be altered by the presence of AU-rich elements (AREs) located in the 3′-untranslated regions (3′UTRs) of the mRNAs. Numerous RNA-binding proteins and microRNAs bind to these 3′UTRs to regulate the stability and/or translation of the mRNAs. Thus, this paper describes the cooperative function between RNA-binding proteins and miRNAs and how they regulate AU-rich elements containing cytokine mRNA stability/degradation and translation. These mRNA control mechanisms can potentially influence inflammation as it relates to oral biology, including periodontal diseases and oral pharyngeal cancer progression.
Keywords: inflammation, oral cancer, mouth neoplasms, periodontal diseases, RNA stability, microRNAs
Introduction
The regulation of inflammatory cytokines is critical for innate cellular processes such as proliferation and angiogenesis, as well as responses to exogenous stimuli including radiation, stress, and infection (Khabar, 2005). The aberrant expression of cytokines has been correlated with inflammatory diseases, autoimmune disorders, and cancer (Audic and Hartley, 2004). Thus, the expression of cytokines and pro-inflammatory factors, including interleukin-1 (IL-1), IL-4, IL-6, IL-8, IL-10, granulocyte-macrophage colony-stimulating factor (GM-CSF), vascular endothelial growth factor (VEGF), prostaglandin E2 (PGE2), cyclooxygenase-2 (COX-2), matrix metalloproteinases (MMP), and basic fibroblast growth factor (bFGF), is highly regulated at many levels, including gene transcription, messenger ribonucleic acid (mRNA) translation, and mRNA degradation (Stoecklin et al., 2006). All of these regulatory pathways are controlled by multiple biological networks, in particular, post-transcriptional gene regulation, which determines the fate of mRNA in association with RNA-binding proteins (RBPs) and microRNAs (miRNAs). The order of cytokine gene expression and the relative duration of the various inflammatory events are among the hallmarks of the gene activation process. This route is probably the result of interplay among the elements that regulate transcriptional induction, transcriptional repression, and mRNA stability. It has been proposed that the differences in mRNA stability exert a strong influence on the temporal order of gene expression, in some cases overriding that of the transcriptional control elements (Hao and Baltimore, 2009). The Hao and Baltimore study illustrates that the transcripts of cytokines that are expressed early have abundant AU-rich elements in their 3′-untranslated regions (3′UTRs), whereas those expressed later have fewer. The authors conclude that two intrinsic characteristics of genes, mRNA stability and transcriptional control, manage the kinetics of gene expression induced by pro-inflammatory cytokines. The changes in transcriptional regulation that regulate cytokine expression have been reviewed elsewhere (Pries and Wollenberg, 2006; van Kempen et al., 2006). Thus, the review presented here focuses on the relationship between and among RBPs, miRNAs, and cytokine mRNAs in the post-transcriptional regulation of cytokine expression. It also discusses the potential implications of these relationships in oral biology and pathobiology.
AU-Rich Elements Mark Cytokine mRNAs
The regulation of mRNA stability is an important step in the control of overall gene expression. The changes in mRNA stability in most cells are ultimately reflected at the protein level. Thus, mRNA decay of the gene is crucial for homeostasis and normal cell survival. The stability of mRNAs is determined by cis-acting sequences in the 3′UTRs that promote the degradation of mRNA. A well-studied cis-acting element consisting of adenine- and uridine-rich sites dictates the fate of mRNA. The clusters of adenine- and uridine-rich elements (AREs) found in mRNAs encoding cytokines were first identified over 25 years ago (Caput et al., 1986) and were subsequently confirmed by various studies showing that cytokines, chemokines, lymphocytes, proto-oncogenes, and pro-inflammatory genes are subject to ARE-mediated decay (Shaw and Kamen, 1986; Hamilton et al., 2007). AREs provide binding sites for trans-acting factors, such as RBPs, that can subsequently regulate the stability and/or translation of mRNA. The basic motif of which the ARE are comprised are pentamers of AUUUA, nonamers of UUAUUUAUU, and AU-rich clusters composed of linked pentamers and/or nonamers (Wilusz et al., 2001). On the basis of sequences and their decay kinetics, AREs have been grouped into various classes. Typically, Class I AREs contain several AUUUA pentamers scattered throughout the 3′UTR within or near a U-rich region (e.g., c-myc and c-fos). Class II AREs contain overlapping copies of the nonamer UUAUUUAUU within a U-rich region [e.g., cytokine mRNAs such as tumor necrosis factor (TNF) and GM-CSF]. Class III AREs contain mostly U-rich regions but no significant AUUUA pentamers (e.g., c-jun). Bakheet et al. (2006) created a database of human ARE mRNAs (ARED; http://brp.kfshrc.edu.sa/ARED/) based on the patterns of AUUUA motifs. Based on their analysis, the percentage of mRNAs within the human genome that contain AREs is about 8%. Fig. 1 illustrates some of the best examples of AREs located at the distal 3′UTR of the mRNA transcripts of cytokines, such as GM-CSF, TNF, IL-2, IL-3, IL-6, and IL-8, as well as pro-inflammatory factors like COX-2. Thus, AREs are critical elements in controlling the expression of genes at the post-transcriptional level.
Figure 1.
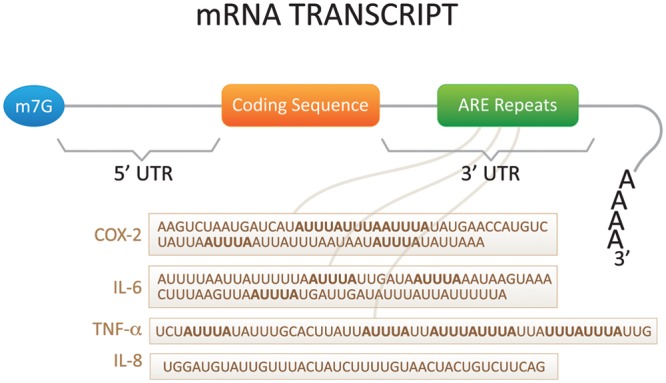
Diagram of cytokine mRNA and sites of post-transcriptional regulation. Inflammatory cytokines are regulated post-transcriptionally through both the 5′- and 3′-untranslated regions (UTRs). The 5′UTR dictates mRNA translation initiation, whereas the 3′UTR dictates mRNA turnover. Cytokines are highly regulated via their AU-rich elements (AREs). TNF-α, IL-6, IL-8, and COX-2 AREs are described in the diagram.
Interaction of Cytokine mRNAs with RNA-Binding Proteins
Cytokine mRNA expression is restricted in resting cells through continuously active mRNA decay mechanisms. Induction of mRNA decay pathways allows for attenuation of the cellular production of cytokines through interactions with RBPs (Anderson, 2009). AREs facilitate the binding of RBPs that degrade or stabilize the mRNA transcripts, often in association with other proteins. Several proteins that can bind to ARE segments have been identified, including tristetraprolin (TTP), human antigen-related protein (HuR), butyrate response factor-1 and butyrate response factor-2 (BRF-1 and BRF-2), ARE/poly(U)-binding/degradation factor (AUF-1), T-cell-restricted intracellular antigen-1 (TIA-1), and TIA-1-related protein (TIAR). However, only subsets of RBPs have been shown to influence the stability or translational efficiency of their target mRNAs. Cytokine mRNAs and their functional interactions with important RBPs are summarized in Table 1. A detailed description of each RBP and their association with cytokine mRNAs are discussed in the Appendix.
Table 1.
RBP Regulation of Specific Cytokine mRNAs
| RBPa | Cytokine/AREb mRNAs | Stability |
|---|---|---|
| TTP | IL-8 | destabilized |
| VEGF | destabilized | |
| IL-3 | destabilized | |
| TNF-α | destabilized | |
| COX-2 | destabilized | |
| IL-10 | destabilized | |
| AUF-1 and 2 | IL-8 | destabilized |
| TNF-α | destabilized | |
| COX-2 | destabilized | |
| GM-CSF | destabilized | |
| HuR | IL-8 | stabilized |
| TNF-α | stabilized | |
| VEGF | stabilized | |
| COX-2 | stabilized | |
| BRF-1 and 2 | COX-2 | destabilized |
| TIAR and TIA-1 | TNF-α | destabilized |
| c-myc | destabilized | |
| CUGBP1 | TNF-α | destabilized |
| CUGBP2 | COX-2 | stabilized |
RBP, RNA-binding protein.
ARE, AU-rich element.
Signaling Pathways Linked To Cytokine mRNA Stability
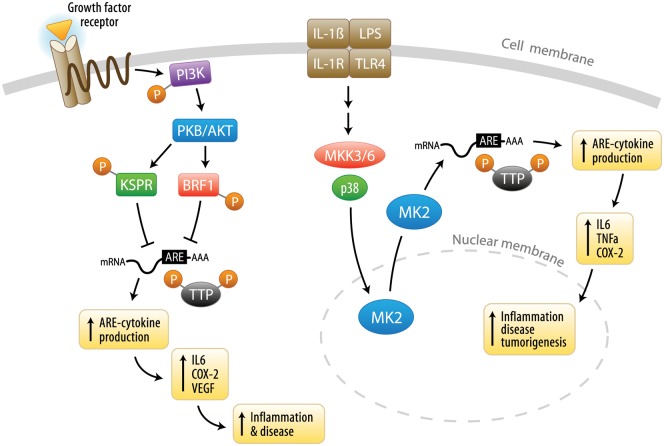
The ability of RBPs to interact with cytokine mRNAs and regulate their expression is also controlled by post-translational modifications. Such modifications are mediated by kinases and phosphatases that may change the binding efficiency of RBPs to the ARE sites within mRNAs and thus alter their gene expression. The p38 MAP kinase pathway is a well-studied system that illustrates how signaling mechanisms direct mRNA stability. As shown in Fig. 2 and highlighted in the Appendix, p38/MK2 signaling is required for TTP phosphorylation, which in turn promotes the sequestration of TTP, an event that is partially dependent upon binding to 14-3-3 proteins. These proteins inhibit the activity of TTP by preventing TTP association with the stress granules in which the mRNA is stored and triaged. Down-regulation of TTP levels then leads to increased cytokine production, because TTP can no longer bind to the AREs and destabilize mRNA. (For additional details, please see the Appendix).
Figure 2.
Signaling pathways that regulate cytokine AREs post-transcriptionally. The p38 pathway is the best-described signaling pathway that regulates AU-rich element (ARE)-mediated cytokine degradation. Activation of IL-1β receptors and the Toll-like receptor (TLR) family stimulates p38 MAPK signaling via signaling intermediates, including MKK3/6. Active p38 dictates mRNA stability through activation of MK2, which subsequently phosphorylates and inactivates TTP. Once inactivated, phosphorylated TTP dissociates from the ARE region of cytokine transcripts to enhance mRNA stability and translation, thereby causing an increase in cytokine secretion that can promote chronic inflammation and inflammatory disease progression if not properly regulated. In addition, the phosphatidylinositol-3 kinase (PI3K) pathway can also post-transcriptionally regulate mRNAs through AKT phosphorylation of RBPs. This phosphorylation alleviates the mRNAs of RBPs that normally destabilize the mRNA and results in increased mRNA stability and thus increased cytokine protein production and secretion, which, if not properly controlled, can induce chronic inflammation.
Cytokines and mRNA Stability
TNF-α
The pro-inflammatory cytokine TNF has been extensively studied for its role in inflammatory diseases. Analysis of TNF reporter gene expression demonstrated that its ARE strictly inhibited translation of the mRNA (Han et al., 1990). Further evidence for the role of the ARE was also observed in a mouse model, where deletion of the ARE from the TNF gene affected mechanisms responsible for TNF mRNA destabilization and translational repression in hematopoietic and stromal cells. In stimulated conditions, TNF ARE was required for both the relief and reinforcement of message destabilization and translational silencing. Notably, deletion of ARE caused chronic inflammatory arthritis and Crohn’s-like inflammatory bowel disease (Kontoyiannis et al., 1999), emphasizing the importance of limiting the expression of critical pro-inflammatory genes through ARE-mediated decay. Recently, we have shown that MAPK phosphatase-1 (MKP-1) is a negative regulator of the host inflammatory response that controls the half-lives of IL-6, IL-10, and TNF-α mRNAs through association with AUF1 (Yu et al., 2011). The results from this study suggested that the half-lives of IL-6, IL-10, and TNF-α mRNAs were significantly increased in bone marrow macrophages derived from MKP-1 knock-out (KO) mice compared with macrophages derived from MKP-1 wild-type (WT) mice after infection with LPS. Thus, our work provides new mechanistic insights into MKP-1 signaling and regulation of cytokine mRNA stability through AUF1 in response to inflammatory stimuli. The interaction between TTP and TNF-α mRNA has been well-studied; specifically, loss of TTP function during the activation of the p38 pathway lengthens the TNF-α mRNA half-life from 37 minutes in unstimulated cells to 90 minutes in stimulated cells (Deleault et al., 2008), dramatically increasing TNF-α cytokine production. Thus, TNF is an excellent target for the prevention of cancer progression. Interestingly, the nuclear factor kappa B (NFκB) has been found to be associated with oral cancer development and plays an essential role in the suppression of TNF-mediated apoptosis (Chen et al., 2002). Hence, targeting signaling kinases associated with TNF provides an additional measure of controlling inflammatory gene expression.
IL-6
Interleukin-6 (IL-6) has many functions in homeostatic regulation, including a role in the immune system, in induction of inflammation, in bone resorption and production, and in various other cellular processes (Keller et al., 1996). Transcriptional and post-translational regulation of IL-6 is critical for maintaining non-pathologic levels of the cytokine and to control both the magnitude and duration of the IL-6 response. Also, loss of IL-6 regulation contributes to the abundance of inflammatory infiltrate in inflammatory diseases and can contribute to the etiology of some cancers (Trikha et al., 2003). In addition, constitutive overexpression of IL-6 is associated with the pathogenesis of rheumatoid arthritis, systemic juvenile arthritis, and Crohn’s disease (Hirth et al., 2002; Souza et al., 2008). TTP and TTP-related proteins, BRF-1 and BRF-2, have a major regulatory role for IL-6. Overexpression studies show that TTP, BRF-1, and BRF-2 induce the degradation of mRNAs containing cytokine AREs (Stoecklin et al., 2003; Sully et al., 2004). IL-6 levels are up-regulated in TTP−/− mice, but not to the same extent as TNF-α, suggesting that redundant pathways exist to regulate IL-6 mRNA stability (Taylor et al., 1996). We have identified that TTP expression in HNSCC was found to be inversely correlated with the secretion of IL-6, and, interestingly, knockdown of TTP increased IL-6 mRNA stability. Conversely, overexpression of TTP in HNSCC cells led to decreased secretion of IL-6 (Van Tubergen et al., 2011). Analysis of these data, together, suggests that TTP plays a critical role in cytokine mRNA stability in HNSCC.
COX-2
Cyclooxygenase-2 (COX-2), expressed at low levels in the stomach, kidney, and intestines (Kujubu et al., 1991), catalyzes the transformation of arachidonic acid to prostaglandin E2 (PGE2), the most important COX-2-produced mediator of inflammation. Because COX-2 is an upstream mediator of several inflammatory cytokines, it is a potential target for inflammatory inhibitors that will treat arthritis, Crohn’s disease, and other inflammatory diseases. COX-2 expression is itself induced by multiple pro-inflammatory mediators such as IL-1, TNF-α, and LPS, which induce COX-2 mRNA transcription and translation. Activation of p38 stabilizes the mRNA transcripts of COX-2 via their effects on TTP, HuR, and AUF-1 (Lasa et al., 2000; Sengupta et al., 2003). The role of HuR in associating with and increasing the stability of COX-2 mRNA has been better described than other RBPs (Subramaniam et al., 2008). Increased cytoplasmic HuR expression has been noted in several cancer types, and in HNSCC it contributed to the increased COX-2 expression observed during tumorigenesis and metastasis (Cha et al., 2011). In particular, the study showed that cytoplasmic HuR expression was significantly associated with COX-2 expression and lymph node metastasis and distant metastasis. Thus, the cytoplasmic expression of HuR appears to be associated with COX-2 expression in OSCCs, and HuR can regulate COX-2 expression in oral cancer. HuR also correlated with COX-2 expression in salivary mucoepidermoid carcinomas (Cho et al., 2007). Thus, HuR plays a major role in controlling COX-2 expression in HNSCC.
IL-8
Interleukin-8 (IL-8) modulates inflammatory response and is a potent angiogenesis stimulator. Tight regulation of IL-8 production is critical for cell function during infection, tissue damage, and cellular homeostasis (Li et al., 2003). Like other cytokines, IL-8 is highly regulated at many stages of expression in the cell, including at the post-transcriptional level (Villarete and Remick, 1996). RBPs target IL-8 mRNA through the AREs in its 3′UTR to increase or decrease cell stability. Multiple proteins have been identified that alter IL-8 mRNA half-life. IL-8 is targeted by HuR, which stabilizes the transcript when activated by inflammatory factors (Choi et al., 2009). Interestingly, HuR and AUF-1 are both capable of stabilizing IL-8 mRNA in human saliva (Palanisamy et al., 2008), and IL-8 mRNA present in human saliva has been shown to be a prognostic marker for oral cancer (St John et al., 2004).
VEGF
Vascular endothelial growth factor (VEGF) facilitates wound healing and tumor progression by promoting angiogenesis. It is regulated post-transcriptionally by AREs in the 3′UTR. VEGF mRNA half-life is stabilized under hypoxic conditions (106 ± 9 min), as compared with normal levels of oxygen (43 ± 6 min) (Levy et al., 1997), through increased expression of HuR (Levy et al., 1998). HuR inhibition in hypoxic cells decreases VEGF expression, and overexpression of HuR stabilizes VEGF only under hypoxic conditions, suggesting that other protein factors and regulatory molecules, such as miRNAs, influence VEGF expression.
Therapeutic Interventions Modulate Cytokine mRNA Stability via ARE-binding Proteins
Several RBPs have been identified since the cloning of the first ARE-binding protein (ARE-BP), AUF1. Studies have begun to elucidate the roles of specific ARE-BPs and their target mRNAs in pathological inflammation. For example, TTP was the first ARE-BP recognized to have an effect on inflammation in intact animals, due to its deleterious effect on TNF mRNA stability (Carballo et al., 1998). More recently, several ARE-BPs have been shown to be involved in the inflammatory response in vivo (Phillips et al., 2004; Katsanou et al., 2005; Sadri and Schneider, 2009). However, because the complete knockout (KO) of these proteins, which are known to be involved in embryonic development and inflammation, is deleterious, there is an urgent need for tissue-specific KO approaches. Conversely, we have shown that overexpression of TTP in an experimental model of inflammatory bone loss results in significant reductions of IL-6, TNF-α, and prostaglandin-E2 (Patil et al., 2008). In fact, our in vivo analyses indicated a significant protective effect from inflammation-induced bone loss and inflammatory infiltrate in animals overexpressing TTP compared with reporter controls. These findings provide experimental evidence that mRNA stability is a valid therapeutic target in inflammatory bone loss. At the same time, small-molecule inhibitors are starting to emerge to inhibit the functions of ARE-BPs. For example, HuR RNA-binding activity is inhibited by small-molecule inhibitors isolated from microbial origins (Meisner et al., 2007). It would thus be fascinating to identify several other inhibitors for different ARE-BPs and test how these proteins interact with ARE containing cytokine messages for the potential prevention of inflammation in head and neck cancers. Based on clinical importance, ARE-mediated mRNA turnover is of high interest for the design and development of novel therapeutics. However, since not many drugs have been identified or developed that directly target ARE-BPs, the modulation of signaling pathways involved in either their synthesis or their intracellular trafficking provides a powerful strategy for chemical interference with ARE-driven mRNA stability. How chemical modulation of ARE-mediated cytokines would affect mRNA stability has already been reviewed in detail elsewhere (Eberhardt et al., 2007; Cheneval et al., 2010). Also, we have previously reviewed the significance of the p38 MAPK pathway in periodontal disease progression and the potential therapeutic consequences of pharmacological antagonism in the treatment of periodontal diseases (Kirkwood and Rossa, 2009). As a result, we will not discuss these points further. However, it is important to note that these studies illustrate that the current most promising targets to modulate ARE-regulated mRNA stability are the various signaling pathways involved in post-transcriptional gene regulation. Thus, interfering with the signaling pathways should alter the pattern of ARE-BPs and regulate the stability of cytokine mRNAs in oral biology and medicine.
MicroRNA-Mediated Regulation of Cytokine Expression Levels
miRNAs are a specific class of evolutionarily conserved small (19-25 nucleotides) endogenous non-coding RNAs that mediate gene expression at the post-transcriptional level (Rana, 2007). By base-pairing to partially or perfectly complementary sites in the 3′UTR of mRNAs, miRNAs can induce either translational repression or mRNA degradation of the target gene (Rana, 2007). However, new evidence suggests that miRNAs act predominantly to decrease target mRNA levels rather than to inhibit translation (Guo et al., 2010). Human miRNAs regulate diverse cellular and molecular processes, including cellular proliferation, differentiation, and apoptosis, and are predicted to regulate > 60% of all protein-encoding genes within the human genome (Rana, 2007; Friedman et al., 2009). Thus, it is not surprising that miRNAs would also be important regulators of the innate immune system and the expression of cytokines during inflammation. Indeed, the dysregulation of miRNAs could potentially contribute to inflammatory diseases and cancer pathogenesis.
miRNAs are believed to regulate cytokine expression through several mechanisms. These include direct miRNA targeting of the cytokine mRNAs, miRNA regulation of the network of cytokine signaling (including receptors and transcription factors), and miRNA-mediated regulation of cytokine mRNAs via their association with RBPs (O’Neill et al., 2011). The evidence for the direct targeting of cytokine mRNAs by miRNAs is limited, with very few targets experimentally verified. The miRNA regulation of cytokine signaling pathways has been extensively examined in several recent review articles (Bak and Mikkelsen, 2010; O’Neill et al., 2011). Therefore, we chose to focus on the areas relating to miRNA-assisted RBP regulation of cytokine mRNAs. More specifically, we examine several mechanisms that demonstrate the cooperative function of miRNAs with RBPs in the repression/activation of shared target cytokine mRNAs (see Table 2). The mechanisms include: (1) mRNA decay and translational inhibition due to miRNA and RBP cooperation; (2) mRNA stabilization due to competition between RBPs and miRNAs; and (3) the environmental effects on mRNA stability, mediated through miRNAs and RBPs.
Table 2.
Mechanisms Demonstrating Cooperative Function between miRNAs and RBPsa in the Repression/Activation of Shared Target Cytokine mRNAs
| Mechanism | Cytokine | miRNA(s) | RBP(s) | Reference(s) |
|---|---|---|---|---|
| 1b | TNFα | miR-16 | TTP | Jing et al., 2005 |
| 1 | TNFα | miR-125bmiR-221miR-579 | TTP and TIAR | El Gazzar and McCall, 2010 |
| 2c | IL10 | miR-4661 | TTP | Ma et al., 2010 |
| 3d,e | TNFα | miR-369-3 | FXR1 and Ago2 | Vasudevan and Steitz, 2007; Vasudevan et al., 2007 |
| 3f | VEGFA | miR-297miR-299 | hnRNP L | Jafarifar et al., 2011 |
RBPs, RNA-binding protein(s).
mRNA decay and translational inhibition due to miRNA and RNA-BP co-dependence.
mRNA stabilization due to competition between RNA-BPs and miRNAs.
Environmental effects on mRNA stability mediated through miRNAs and RNA-BPs.
Dependent on stage of cell cycle.
Dependent on normoxic/hypoxic conditions.
mRNA Decay and Translational Inhibition Due to miRNA and RBP Co-dependence
One of the first examples of miRNA-assisted RBP regulation of a cytokine mRNA was observed for TNF-α. Jing et al. (2005) reported that a miRNA, miR-16, had a partial sequence match with the TNF-α ARE and that the ARE-mediated decay of TNF-α was dependent on both TTP and miR-16. Based on their data, it was hypothesized that miR-16-bound RNA-induced silencing complex (RISC) assisted TTP binding to the ARE, which subsequently induced mRNA degradation by recruiting deadenylation and/or exosomal proteins. Additionally, a more recent study found that three miRNAs, miR-125b, miR-221, and miR-579, were up-regulated during LPS-induced tolerance (a state that causes TNF-α mRNA to be degraded), and that these miRNAs were capable of either associating with TTP to accelerate TNF-α mRNA decay or blocking TNF-α translation, possibly through recruitment of the translational inhibitor TIAR (El Gazzar and McCall, 2010). Alternatively, another study found that HuR binding to the c-Myc 3′UTR repressed c-Myc expression by recruiting let-7/RISC to an adjacent site on the c-Myc 3′UTR (Kim et al., 2011). However, in this case HuR was not found to interact with RISC. Thus, it was proposed that HuR binding to mRNA possibly changes the local conformation of the mRNA, unmasking the let-7 recognition site that triggers a reduction in both mRNA levels and translation.
mRNA Stabilization Due to Competition between RBPs and miRNAs
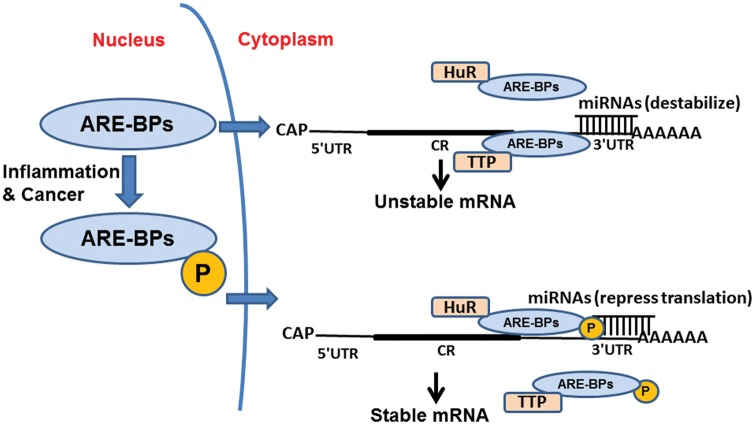
The above examples illustrate how miRNAs and RBPs can function together to promote cytokine mRNA destabilization and/or inhibit translation. Conversely, RBPs and miRNAs can also compete with one another to promote mRNA stability. Bhattacharyya et al. (2006) found that HuR reverses miR-122-mediated repression of CAT-1 translation by binding to a shared site on the ARE-rich CAT-1 3′UTR (Bhattacharyya et al., 2006). Interestingly, a similar mechanism was reported in the regulation of cytokine IL-10 expression levels. miR-4661, which contains a seed region that is complementary to ARE sequences, was demonstrated to up-regulate both IL-10 mRNA and protein levels upon transfection into LPS-stimulated RAW264.7 macrophages by competing with TTP for binding to the ARE sequence in IL-10 mRNA, thus protecting the mRNA from TTP-mediated degradation (Ma et al., 2010). Fig. 3 illustrates the model of competition between ARE-BPs and miRNAs in the regulation of mRNA stability.
Figure 3.
Model of the cooperative function between RNA-binding proteins (RBPs) and miRNAs in the regulation of cytokine mRNAs. Deregulation of miRNAs or RBPs in cancer can be due to the altered expression, localization, activity, or stability of these regulators. The mRNA is presented linearly for simplicity. CR represents a coding region. Under normal conditions, both the AU-rich element-binding protein TTP and miRNAs are involved in the destabilization of cytokine mRNAs, and the non-phosphorylated form of HuR disassociates from mRNAs. However, in response to inflammation and cancer progression, HuR becomes phosphorylated and translocates to the cytoplasm, where it stabilizes mRNAs along with miRNAs that are known to repress translation. This action releases the mRNAs from TTP- and miRNA-mediated destability and repression, respectively. Moreover, inflammation induces TTP phosphorylation and disassociates it from mRNAs along with destabilizing miRNAs.
Environmental Effects on mRNA Stability Mediated through miRNAs and RBPs
In addition to the intricate interplay between miRNAs and RBPs in the regulation of cytokine expression, there is another layer of complexity that needs to be considered—the environment. Vasudevan and colleagues (Vasudevan and Steitz, 2007; Vasudevan et al., 2007) reported that miR-369-3 can bind directly within a region of the TNF-α ARE and activate translation in quiescent cells through the recruitment of two RBPs, FXR1 and AGO2, two factors usually considered negative regulators. In contrast, in proliferating cells, miR-369-3 was found to repress TNF-α expression (Vasudevan et al., 2007). Interestingly, in reporter pull-out experiments, only AGO2 was detected in the repressing ribonucleoprotein (RNP) complex, not FXR1 (Vasudevan et al., 2007; Steitz and Vasudevan, 2009). This implied that FXR1 was associated only with the activating miRNP complex (Steitz and Vasudevan, 2009). In addition to cell cycling, other environmental factors, such as UV exposure and hypoxic stress, can also regulate the crosstalk between miRNAs and RBPs in the repression/activation of shared target mRNAs (Glorian et al., 2011; Jafarifar et al., 2011). In the case of hypoxia, a common feature of neoplastic microenvironments, tumor-associated macrophages induce the expression of the cytokine VEGFA, a critical process for tumor progression and metastasis. Jafarifar et al. (2011) recently reported that two miRNAs, miR-297 and miR-299, normally endogenous negative regulators of VEGFA expression in human monocytic cells, could be negatively modulated by heterogeneous nuclear RNP L (hnRNP L) (Jafarifar et al., 2011). More specifically, it was observed that during normoxia, miR-297 and miR-299 target the CA-rich element (CARE) in the VEGFA 3′UTR and negatively regulate VEGFA expression. However, during hypoxia, miRNA-mediated repression was reversed due to the translocation of hnRNP L from the nucleus to the cytoplasm and its increased binding to the CARE region in the VEGFA 3′UTR.
Conclusions
Overall, mRNA stability is critical for cytokine production. Moreover, it plays an important role in inflammatory disease progression. For example, cytokines, which activate multiple signaling cascades in inflammation, including ERK, JNK, NF-κB, and p38 MAPK, are highly regulated via mRNA stability and translation mechanisms. Moreover, loss of post-transcriptional regulation of cytokine mRNAs can dramatically increase cytokine production, leading to tissue destruction and increased mortality. Given the onset of cytokine production, there are several features of post-transcriptional control that play a critical role in their maintenance (Fig. 2). In summary, the above mechanisms illustrate how the crosstalk between miRNAs and RBPs can regulate the repression/activation of shared target cytokine mRNAs. Moreover, these examples illustrate how specific environmental stressors, such as hypoxia, can promote cancer by modulating the interaction between miRNAs and RBPs with the shared target cytokine mRNA, affecting its expression levels. A better understanding of how miRNAs function together with RBPs, in regulating the expression of shared cytokine mRNAs, could potentially lead to improved therapies for cancer and inflammatory diseases.
Footnotes
This study is supported by grants from the US National Institutes of Health (NIH) [1R01DE018290 (KK), 1R01DE021423 (KK), 2P20 RR017696 (KK), R00DE018165 (VP) R01DE018512 (NJD), R21DE017977 (NJD), K02DE0219513 (NJD), R00DE018191 (AJ), and 5F32DE021305 (EVT)].
The authors declare that there are no conflicts relevant to this manuscript.
A supplemental appendix to this article is published electronically only at http://jdr.sagepub.com/supplemental.
References
- Anderson P. (2009). Intrinsic mRNA stability helps compose the inflammatory symphony. Nat Immunol 10:233-234 [DOI] [PubMed] [Google Scholar]
- Audic Y, Hartley RS. (2004). Post-transcriptional regulation in cancer. Biol Cell 96:479-498 [DOI] [PubMed] [Google Scholar]
- Bak RO, Mikkelsen JG. (2010). Regulation of cytokines by small RNAs during skin inflammation. J Biomed Sci 17:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakheet T, Williams BR, Khabar KS. (2006). ARED 3.0: the large and diverse AU-rich transcriptome. Nucleic Acids Res 34: D111-114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. (2006). Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125:1111-1124 [DOI] [PubMed] [Google Scholar]
- Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. (1986). Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci USA 83:1670-1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo E, Lai WS, Blackshear PJ. (1998). Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science 281:1001-1005 [DOI] [PubMed] [Google Scholar]
- Cha JD, Li S, Cha IH. (2011). Association between expression of embryonic lethal abnormal vision-like protein HuR and cyclooxygenase-2 in oral squamous cell carcinoma. Head Neck 33:627-637 [DOI] [PubMed] [Google Scholar]
- Chen S, Fribley A, Wang CY. (2002). Potentiation of tumor necrosis factor-mediated apoptosis of oral squamous cell carcinoma cells by adenovirus-mediated gene transfer of NF-kappaB inhibitor. J Dent Res 81:98-102 [PubMed] [Google Scholar]
- Cheneval D, Kastelic T, Fuerst P, Parker CN. (2010). A review of methods to monitor the modulation of mRNA stability: a novel approach to drug discovery and therapeutic intervention. J Biomol Screen 15: 609-622 [DOI] [PubMed] [Google Scholar]
- Cho NP, Han HS, Soh Y, Son HJ. (2007). Overexpression of cyclooxygenase-2 correlates with cytoplasmic HuR expression in salivary mucoepidermoid carcinoma but not in pleomorphic adenoma. J Oral Pathol Med 36:297-303 [DOI] [PubMed] [Google Scholar]
- Choi HJ, Yang H, Park SH, Moon Y. (2009). HuR/ELAVL1 RNA binding protein modulates interleukin-8 induction by muco-active ribotoxin deoxynivalenol. Toxicol Appl Pharmacol 240:46-54 [DOI] [PubMed] [Google Scholar]
- Deleault KM, Skinner SJ, Brooks SA. (2008). Tristetraprolin regulates TNF TNF-alpha mRNA stability via a proteasome dependent mechanism involving the combined action of the ERK and p38 pathways. Mol Immunol 45:13-24 [DOI] [PubMed] [Google Scholar]
- Eberhardt W, Doller A, Akool el-S, Pfeilschifter J. (2007). Modulation of mRNA stability as a novel therapeutic approach. Pharmacol Ther 114:56-73 [DOI] [PubMed] [Google Scholar]
- El Gazzar M, McCall CE. (2010). MicroRNAs distinguish translational from transcriptional silencing during endotoxin tolerance. J Biol Chem 285:20940-20951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. (2009). Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19:92-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glorian V, Maillot G, Poles S, Iacovoni JS, Favre G, Vagner S. (2011). HuR-dependent loading of miRNA RISC to the mRNA encoding the Ras-related small GTPase RhoB controls its translation during UV-induced apoptosis. Cell Death Differ 18:1692-1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. (2010). Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466:835-840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton TA, Novotny M, Datta S, Mandal P, Hartupee J, Tebo J, et al. (2007). Chemokine and chemoattractant receptor expression: post-transcriptional regulation. J Leukoc Biol 82:213-219 [DOI] [PubMed] [Google Scholar]
- Han J, Brown T, Beutler B. (1990). Endotoxin-responsive sequences control cachectin/tumor necrosis factor biosynthesis at the translational level. J Exp Med 171:465-475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao S, Baltimore D. (2009). The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nat Immunol 10:281-288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirth A, Skapenko A, Kinne RW, Emmrich F, Schulze-Koops H, Sack U. (2002). Cytokine mRNA and protein expression in primary-culture and repeated-passage synovial fibroblasts from patients with rheumatoid arthritis. Arthritis Res 4:117-125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafarifar F, Yao P, Eswarappa SM, Fox PL. (2011). Repression of VEGFA by CA-rich element-binding microRNAs is modulated by hnRNP L. EMBO J 30:1324-1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, et al. (2005). Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell 120:623-634 [DOI] [PubMed] [Google Scholar]
- Katsanou V, Papadaki O, Milatos S, Blackshear PJ, Anderson P, Kollias G, et al. (2005). HuR as a negative posttranscriptional modulator in inflammation. Mol Cell 19:777-789 [DOI] [PubMed] [Google Scholar]
- Keller ET, Wanagat J, Ershler WB. (1996). Molecular and cellular biology of interleukin-6 and its receptor. Front Biosci 1:d340-d357 [DOI] [PubMed] [Google Scholar]
- Khabar KS. (2005). The AU-rich transcriptome: more than interferons and cytokines, and its role in disease. J Interferon Cytokine Res 25:1-10 [DOI] [PubMed] [Google Scholar]
- Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. (2011). HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev 23:1743-1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkwood KL, Rossa C., Jr (2009). The potential of p38 MAPK inhibitors to modulate periodontal infections. Curr Drug Metab 10:55-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontoyiannis D, Pasparakis M, Pizarro TT, Cominelli F, Kollias G. (1999). Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity 10:387-398 [DOI] [PubMed] [Google Scholar]
- Kujubu DA, Fletcher BS, Varnum BC, Lim RW, Herschman HR. (1991). TIS10, a phorbol ester tumor promoter-inducible mRNA from Swiss 3T3 cells, encodes a novel prostaglandin synthase/cyclooxygenase homologue. J Biol Chem 266:12866-12872 [PubMed] [Google Scholar]
- Lasa M, Mahtani KR, Finch A, Brewer G, Saklatvala J, Clark AR. (2000). Regulation of cyclooxygenase 2 mRNA stability by the mitogen-activated protein kinase p38 signaling cascade. Mol Cell Biol 20:4265-4274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy AP, Levy NS, Iliopoulos O, Jiang C, Kaplin WG, Jr, Goldberg MA. (1997). Regulation of vascular endothelial growth factor by hypoxia and its modulation by the von Hippel-Lindau tumor suppressor gene. Kidney Int 51:575-578 [DOI] [PubMed] [Google Scholar]
- Levy NS, Chung S, Furneaux H, Levy AP. (1998). Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J Biol Chem 273:6417-6423 [DOI] [PubMed] [Google Scholar]
- Li A, Dubey S, Varney ML, Dave BJ, Singh RK. (2003). IL-8 directly enhanced endothelial cell survival, proliferation, and matrix metalloproteinases production and regulated angiogenesis. J Immunol 170:3369-3376 [DOI] [PubMed] [Google Scholar]
- Ma F, Liu X, Li D, Wang P, Li N, Lu L, et al. (2010). MicroRNA-466l upregulates IL-10 expression in TLR-triggered macrophages by antagonizing RNA-binding protein tristetraprolin-mediated IL-10 mRNA degradation. J Immunol 184:6053-6059 [DOI] [PubMed] [Google Scholar]
- Meisner NC, Hintersteiner M, Mueller K, Bauer R, Seifert JM, Naegeli HU, et al. (2007). Identification and mechanistic characterization of low-molecular-weight inhibitors for HuR. Nat Chem Biol 3:508-515 [DOI] [PubMed] [Google Scholar]
- O’Neill LA, Sheedy FJ, McCoy CE. (2011). MicroRNAs: the fine-tuners of Toll-like receptor signalling. Nat Rev Immunol 11:163-175 [DOI] [PubMed] [Google Scholar]
- Palanisamy V, Park NJ, Wang J, Wong DT. (2008). AUF1 and HuR proteins stabilize interleukin-8 mRNA in human saliva. J Dent Res 87:772-776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil CS, Liu M, Zhao W, Coatney DD, Li F, VanTubergen EA, et al. (2008). Targeting mRNA stability arrests inflammatory bone loss. Mol Ther 16:1657-1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips K, Kedersha N, Shen L, Blackshear PJ, Anderson P. (2004). Arthritis suppressor genes TIA-1 and TTP dampen the expression of tumor necrosis factor alpha, cyclooxygenase 2, and inflammatory arthritis. Proc Natl Acad Sci USA 101:2011-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pries R, Wollenberg B. (2006). Cytokines in head and neck cancer. Cytokine Growth Factor Rev 17:141-146 [DOI] [PubMed] [Google Scholar]
- Rana TM. (2007). Illuminating the silence: understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol 8:23-36 [DOI] [PubMed] [Google Scholar]
- Sadri N, Schneider RJ. (2009). Auf1/Hnrnpd-deficient mice develop pruritic inflammatory skin disease. J Invest Dermatol 129:657-670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Jang BC, Wu MT, Paik JH, Furneaux H, Hla T. (2003). The RNA-binding protein HuR regulates the expression of cyclooxygenase-2. J Biol Chem 278:25227-25233 [DOI] [PubMed] [Google Scholar]
- Shaw G, Kamen R. (1986). A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell 46:659-667 [DOI] [PubMed] [Google Scholar]
- Souza LS, Machado SH, Brenol CV, Brenol JC, Xavier RM. (2008). Growth velocity and interleukin 6 concentrations in juvenile idiopathic arthritis. J Rheumatol 35:2265-2271 [DOI] [PubMed] [Google Scholar]
- St John MA, Li Y, Zhou X, Denny P, Ho CM, Montemagno C, et al. (2004). Interleukin 6 and interleukin 8 as potential biomarkers for oral cavity and oropharyngeal squamous cell carcinoma. Arch Otolaryngol Head Neck Surg 130:929-935 [DOI] [PubMed] [Google Scholar]
- Steitz JA, Vasudevan S. (2009). miRNPs: versatile regulators of gene expression in vertebrate cells. Biochem Soc Trans 37(Pt 5):931-935 [DOI] [PubMed] [Google Scholar]
- Stoecklin G, Gross B, Ming XF, Moroni C. (2003). A novel mechanism of tumor suppression by destabilizing AU-rich growth factor mRNA. Oncogene 22:3554-3561 [DOI] [PubMed] [Google Scholar]
- Stoecklin G, Mayo T, Anderson P. (2006). ARE-mRNA degradation requires the 5′-3′ decay pathway. EMBO Rep 7:72-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam D, Ramalingam S, May R, Dieckgraefe BK, Berg DE, Pothoulakis C, et al. (2008). Gastrin-mediated interleukin-8 and cyclooxygenase-2 gene expression: differential transcriptional and posttranscriptional mechanisms. Gastroenterology 134:1070-1082 [DOI] [PubMed] [Google Scholar]
- Sully G, Dean JL, Wait R, Rawlinson L, Santalucia T, Saklatvala J, et al. (2004). Structural and functional dissection of a conserved destabilizing element of cyclo-oxygenase-2 mRNA: evidence against the involvement of AUF-1 [AU-rich element/poly(U)-binding/degradation factor-1], AUF-2, tristetraprolin, HuR (Hu antigen R) or FBP1 (far-upstream-sequence-element-binding protein 1). Biochem J 377(Pt 3):629-639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor GA, Carballo E, Lee DM, Lai WS, Thompson MJ, Patel DD, et al. (1996). A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity 4:445-454 [DOI] [PubMed] [Google Scholar]
- Trikha M, Corringham R, Klein B, Rossi JF. (2003). Targeted anti-interleukin-6 monoclonal antibody therapy for cancer: a review of the rationale and clinical evidence. Clin Cancer Res 9:4653-4665 [PMC free article] [PubMed] [Google Scholar]
- van Kempen LC, de Visser KE, Coussens LM. (2006). Inflammation, proteases and cancer. Eur J Cancer 42:728-734 [DOI] [PubMed] [Google Scholar]
- Van Tubergen E, Vander Broek R, Lee J, Wolf G, Carey T, Bradford C, et al. (2011). Tristetraprolin regulates interleukin-6, which is correlated with tumor progression in patients with head and neck squamous cell carcinoma. Cancer 117:2677-2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Steitz JA. (2007). AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell 128:1105-1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. (2007). Switching from repression to activation: microRNAs can up-regulate translation. Science 318:1931-1934 [DOI] [PubMed] [Google Scholar]
- Villarete LH, Remick DG. (1996). Transcriptional and post-transcriptional regulation of interleukin-8. Am J Pathol 149:1685-1693 [PMC free article] [PubMed] [Google Scholar]
- Wilusz CJ, Wormington M, Peltz SW. (2001). The cap-to-tail guide to mRNA turnover. Nat Rev Mol Cell Biol 2:237-246 [DOI] [PubMed] [Google Scholar]
- Yu H, Sun Y, Haycraft C, Palanisamy V, Kirkwood KL. (2011). MKP-1 regulates cytokine mRNA stability through selectively modulation subcellular translocation of AUF1. Cytokine 56:245-255 [DOI] [PMC free article] [PubMed] [Google Scholar]