Abstract
The Thai HIV phase III prime-boost trial (RV144) using ALVAC-HIV® (vCP1521) and AIDSVAX B/E® was, to our knowledge, the first to demonstrate acquisition efficacy. Vaccine-induced, cell-mediated immune responses were assessed. T cell epitope mapping studies using IFN-γ ELISPOT were performed on PBMC from HIV-1 uninfected vaccine (N=61) and placebo (N=10) recipients using HIV-1 Env peptides. Positive responses were measured in 25 (41%) vaccinees and were predominantly CD4+ T cell mediated. Responses were targeted within the HIV Env region, with 15/25 (60%) of vaccinees recognizing peptides derived from the V2 region of HIV-1 Env, which includes the α4β7 integrin binding site. Intracellular cytokine staining confirmed that Env responses predominated (19/30; 63% of vaccine recipients) and were mediated by polyfunctional effector memory CD4+ T cells, with the majority of responders producing both IL-2 and IFN-γ (12/19; 63%). HIV-Env Ab titers were higher in subjects with IL-2 compared to those without IL-2 secreting HIV-Env specific effector memory T cells. Proliferation assays revealed that HIV Ag-specific T cells were CD4+ with the majority (80%) expressing CD107a. HIV-specific T cell lines obtained from vaccine recipients confirmed V2 specificity, polyfunctionality and functional cytolytic capacity. While the RV144 T cell responses were modest in frequency compared to humoral immune responses, the CD4+ T cell response was directed to HIV-1 Env and more particularly the V2 region.
Keywords: Human, Vaccination, Viral, AIDS, HIV-1, T cells
Introduction
The HIV epidemic continues to grow with 33 million persons living with HIV/AIDS and 7,400 new infections daily in 2008 (1). Although recent data suggest a slowing in the rate of new infections, a HIV-1 vaccine in the setting of a comprehensive prevention program remains the most cost-effective and globally applicable public health approach to controlling the epidemic. While no efficacy was observed for the VaxGen (injection drug use and men who have sex with men risk cohorts) and Merck (predominantly men who have sex with men risk cohort in the Step trial) candidate HIV-1 vaccines (2–4), the Thai Phase III trial (RV144) demonstrated 31% reduction in acquisition of infection without effect on post-infection viral load or CD4+ T cell count in infected vaccinees (5). A prior phase I/II trial of the identical vaccine regimen demonstrated robust humoral and cellular immune responses in vaccinees as measured by HIV-specific binding and neutralizing antibody assays and antigen-specific lymphoproliferation assays (6). Cumulative chromium release CD8+ CTL activity was detected in 27% of vaccine recipients (6), and was comparable to that reported with similar vaccine regimens using chromium release (7, 8) or IFN-γ ELISPOT (9).
Despite numerous international canarypox trials with or without an Env protein boost (10, 11), T cell epitope mapping studies following immunization with ALVAC and HIV Env protein(s) have been limited. One study using an identical vaccination schedule to RV144 assessed cellular immune responses in HIV seronegative subjects following ALVAC (vCP205, which expresses HIV-1 subtype B gag, pro and env) alone, in combination with a single boost of gp160 MN/ LAI-2, or gp160 MN/LAI-2 alone, reported stronger and broader responses to vaccine-matched HIV peptides after ALVAC prime/protein boost, compared to volunteers immunized with vCP205 alone (12). CD4+ T cell lines from subjects receiving the combination regimen demonstrated positive responses across the entire gp160, with the notable exception of the V3 region. More extensive T cell epitope mapping studies have been performed with HIV prime-boost vaccine strategies using DNA priming followed by either modified vaccinia virus Ankara (MVA) (13) or recombinant adenovirus serotype 5 vectors (14).
Preliminary studies of RV144 cellular immune responses identified reactivity that was predominantly CD4+ T cell mediated and directed against HIV-1 Env (5). The IFN-γ ELISPOT and intracellular cytokine staining (ICS) for IFN-γ and IL-2 assays following stimulation of PBMC with HIV peptide pools demonstrated a modest cell-mediated immune response. In vaccinees, a greater frequency of IFN-γ ELISPOT responders was observed against Env (21/153; 14%) compared to Gag (9/148; 6%) and ICS responses were CD4+ T cell mediated and focused on Env (47/142; 33%) rather than Gag (2/144; 1%) (5).
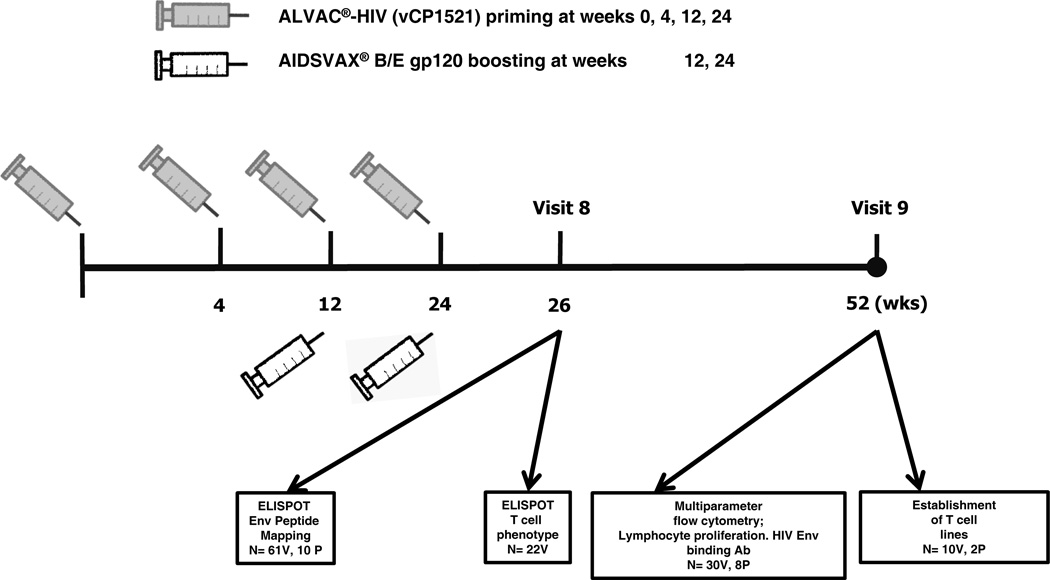
This study was undertaken to assess T cell responses to HIV-1 Env epitopes induced by the RV144 vaccine regimen of ALVAC-HIV® (vCP1521) (Sanofi) and AIDSVAX B/E® (Global Solutions for Infectious Diseases) following completion of immunization. A comprehensive functional analysis of vaccine-induced T cell responses, including quantification of T cell responses by IFN-γ ELISPOT at 2 weeks post final vaccination was performed. Based on the findings at the peak immunogenicity time-point, functional characterization of T cell responses (cytokine, proliferation, CD107a expression and lytic activity) was performed at 6 months following the completion of immunization. Figure 1 shows the time-points following immunization for which cellular immune responses were assessed.
Figure 1.
Immunization regimen and cellular immune assessment time-points for subjects enrolled in RV144. V-Vaccine; P-Placebo
Materials and Methods
Study Design
RV144 was a phase III trial conducted on the Eastern seaboard of Thailand (5). A total of 12,542 volunteers received the full injection series of vaccine or placebo. The vaccine regimen consisted of recombinant canarypox, ALVAC-HIV® (vCP1521), expressing HIV circulating recombinant form (CRF) 01_AE gp120 (92TH023) with subtype B (LAI) transmembrane portion of gp41 and subtype B (LAI) gag and protease, administered at 0, 1, 3 and 6 months. AIDSVAX B/E®, a bivalent HIV gp120 vaccine containing B and CRF01_AE envelopes derived from MN and A244 respectively, was co-administered with the ALVAC at 3 and 6 months. All vaccines were administered into the deltoid muscle. The study was approved by the institutional review boards of the Royal Thai Army, Mahidol University, the Ministry of Public Health, Thailand and the U.S. Army Medical Research and Materiel Command.
PBMC from a subset of 600 HIV-1 uninfected volunteers who completed the RV144 injection regimen were selected for immunogenicity assessment in April 2008. A randomized sample with a 3:1 frequency of vaccine:placebo recipients was drawn for immunogenicity testing after removing both infected individuals and a 4:1 matched random subset of uninfected individuals that were set aside for immune correlate studies by external investigators. Cellular immunogenicity to HIV Env and Gag assessed by IFN-γ ELISPOT (N=200) and IFN-γ/IL-2 ICS (N=200) was previously measured pre- and 24 weeks (V9) following the completion of vaccination in study participants (5). The current study was performed at two laboratories: the United States Military HIV Research Program in Thailand (ELISPOT assays) and the USA (polychromatic flow cytometry, characterization and establishment of T cell lines).
PBMC from subjects with positive IFN-γ ELISPOT responses to Env at 24 weeks post immunization (N=20) were selected for peptide-specific IFN-γ ELISPOT assays at 2 weeks following the completion of immunization. An additional 41 subjects with negative IFN-γ ELISPOT responses at V9 were also tested from the group of 200 subjects. Forty-one subjects were selected as the laboratory was blinded while testing, but knew that the vaccine:placebo ratio was 3:1 and was aiming to test a minimum of 10 placebos for enhanced assay validity. PBMC from an additional 50 RV144 trial participants (40 vaccinees and 10 placebo recipients) collected at 24 weeks following the completion of immunization were tested for polychromatic flow cytometry, lymphoproliferation and establishment of T cell lines. PBMC from the same subjects for the different visits could not be used for the investigation of cell-mediated responses by both ELISPOT and ICS as the PBMC collection in RV144 was extremely limited - 8ml and 16ml of whole blood at V8 and V9, respectively, and the subjects’ V9 PBMC had been previously used in ELISPOT studies (5).
Cell Preparation
PBMC were prepared using 8 ml sodium citrate Vacutainer® CPT™ according to the manufacturer’s instructions (BD) and cryopreserved in RPMI 1640 medium (Invitrogen) containing 20% heat-inactivated fetal-calf serum (Invitrogen) and 10% DMSO (Sigma-Aldrich) in a Cryo 1°C freezing container (Nalgene). Cells were stored at ≤ −130°C. Immunogenicity assessments were performed on cryopreserved specimens; average cell viability was 95% (range: 80%–100%) following thawing. PBMC were thawed and rested overnight prior to all assays with the exception of ICS where they were used immediately after thawing. To evaluate the T cell type responding to the peptides in the IFN-γ ELISPOT assay, overnight rested PBMC were split into three aliquots at 4 ×106 PBMC/ml and treated with immunomagnetic Dynabeads M-450 CD4 (for CD4 depletion), Dynabeads M-450 CD8 (for CD8 depletion) or Dynabeads M-450 anti-mouse immunoglobulin (for sham depletion) (Dynal Biotech ASA). The beads and adhered cells were removed with a magnet and the resulting cell populations were washed twice, resuspended in complete medium composed of RPMI 1640 (Invitrogen) supplemented with L-glutamine 4 mM, penicillin 100 U/ml, streptomycin 100 µg/ml and 10% heat-inactivated normal human serum (Gemini Bioproducts) and tested as for the IFN-γ ELISPOT assay described below. The efficiency of depletion was consistently ≥ 95% (range: 97%–99% for both CD4+ and CD8+ depletion)
Antigens and Peptides
The HIV CRF01_AE derived A244 gp120 was kindly provided by Marc Gurwith (Global Solutions for Infectious Diseases) and was identical to that contained in the AIDSVAX B/E® vaccine. The 165 CRF01_AE 92TH023 Env gp160 peptides of 15-mers overlapping by 11 aa (New England Peptide) were used for stimulation of PBMC in the ELISPOT assays in Thailand. A separate peptide set of 138 peptides of 15–18 aa overlapping by 10–12 aa spanning CRF01_AE isolate CM235 Env protein (JPT Peptide Technologies) was used for PBMC and T cell stimulation in the US. If there were sufficient cells, in addition to PHA, two peptide sets were used as positive controls - a commercial CMV pp65 peptide pool (JPT Peptide Technologies), and the CEF pool representing immunodominant CD8+ T cell epitopes within CMV, EBV and influenza (15). All peptides were used at a final concentration of 1µg/ml.
IFN-γ ELISPOT Assay
Peptides in the HIV-1 Env antigen were mapped using an IFN-γ ELISPOT assay as previously described (5). Briefly, ninety-six-well hydrophobic membrane-bottomed plates (Millipore) were coated overnight at 4°C with anti-human IFN-γ monoclonal antibody (final concentration 5 µg/ml [Mabtech]). Cells (PBMC, CD4+ or CD8+-depleted PBMC) were resuspended in complete medium and plated at a concentration of 2 × 105 /well. In instances where T cell depletions were performed, the pre-depletion PBMC cell count/well was used to avoid CD4+ or CD8+ T cell enrichment. A panel of 26 pools of 12 to 14 (total 165) peptides spanning the 92TH023 gp160 was added to single wells of cells in a matrix format and the plates incubated overnight at 37°C/5%CO2. Wells containing cells and media only were supplemented with the equivalent concentration of DMSO as the peptide pools and served as negative controls. PHA was used as a positive control, and if there were sufficient cells, CMVpp65 and CEF were also included in the assay as additional positive controls. Cells plus PHA were tested in triplicate wells. Negative controls were performed in quadruplicate. After incubation at 37°C in 5% CO2 for 20 to 24 hours, cells were removed by washing with PBS/0.05% Tween-20 (Sigma-Aldrich). Captured IFN-γ was detected by incubation for 2 hours at 37°C with biotinylated anti-human IFN-γ monoclonal antibody (Mabtech) at 2 µg/ml in PBS/0.5% BSA. Following incubation plates were washed with PBS/0.05% Tween-20 (Sigma-Aldrich) and Avidin-Peroxidase-Complex (Vectastain Elite Kit) was added for 1 hour at room temperature. Unbound complex was removed by washing and the peroxidase staining was performed using AEC substrate (Vectastain AEC Kit) according to the manufacturer’s instructions. Spots were counted with a C.T.L. analyzer and software (version 4.0.19, C.T.L. Analyzers). Results are expressed as spot forming cells (SFC)/106 PBMC. A positive IFN-γ response was defined as ≥20 SFC/106 PBMC (uncorrected) and ≥ 4 times the average of the DMSO treated wells. This revised cut-off was based on earlier IFN-γ ELISPOT assays with PBMC from 200 Thai HIV-seronegative samples each tested in quadruplicate. The mean of PBMC treated with media only was 2 SFC/106 PBMC and the standard deviation was 4 (5).
ICS and Immunophenotyping Assay
The method used was based upon a standard ICS format assay for antigen-specific T cells (16). Cryopreserved PBMC were thawed, washed and resuspended in RPMI 1640 containing 10% normal human serum for immediate assay. PBMC (1 × 106) were incubated for 6 h in the presence or absence of a peptide pool (1 µg/peptide/ml) representing the CM235 Env protein or the positive control staphylococcal entertotoxin B (10 ng/ml). Anti-CD28-biotin and anti-CD49 MAbs (BD Pharmingen) were included at set-up, and the protein transport inhibitors Monensin (GolgiStop, BD Pharmingen) and Brefeldin A (Sigma-Aldrich) were added after 2 hours. Cells were washed, stained with Aqua Live/Dead (Invitrogen), washed and resuspended in flow wash buffer (PBS with 0.5% BSA, 0.1% azide), resuspended in 10% mouse IgG (FcR block) for 15 min, centrifuged and surface stained with anti-CD14/CD19-Alexa700 (BD Pharmingen), anti-CD45RA-FITC (BD Pharmingen), anti-CD45RO-eFluor650 (eBioscience),), anti-CD4-QDot605 (Invitrogen) and steptavidin-QDot800 (Invitrogen) to detect the CD28-biotin MAb. Cells were fixed in 2% PFA (Tousimis, Rockville MD) washed and left overnight at 4 °C. The following morning cells were incubated with Perm Wash (Becton Dickinson) for 15 minutes, washed and simultaneously surface/intracellular stained with anti-IFN-γ-eFluor450 (eBioscience), anti-TNF-α-PE-Cy7 (BD Pharmingen), anti-IL-21-PE (eBioscience), anti-CD3-APC-H7 (BD Pharmingen), anti-CD8-ECD (Beckman-Coulter), and anti-IL2-PerCP-Cy5.5 (BioLegend). Cells were acquired on a custom built LSRII flow cytometer (BD Biosciences). At least 250,000 total events were acquired in the lymphocyte light scatter gate and the data analyzed using the following software packages: FlowJo Version 9.1 (Treestar Inc). Polychromatic flow cytometry analysis and presentation of distributions were performed using PESTLE and SPICE version 5.1, downloaded from <http://exon.niaid.nih.gov/spice> (courtesy Mario Roederer, Vaccine Research Center, National Institute of Allergy and Infectious Disease, National Institutes of Health, Bethesda, MD) software. Functional responses were determined for each of the following subsets of CD4+ and CD8+ T cells: Naïve cells (CD45RA+/CD45RO−/CCR7+/CD28+), central memory cells CD45RA−/CD45RO+/CCR7+/CD28+), effector memory cells (CD45RA−/CD45RO+/CCR7−/CD28+/−) and effector cells (CD45RA+/CD45RO−/CCR7−/CD28−). A positive response was defined as ≥ 0.05% gated positive cells for the test Ag and the response exceeding the unstimulated control by ≥3 times.
T-cell Proliferation Assay and Functional Assessment
Measurement of T cell proliferation and their subsequent functional profile was performed using a modified CFSE-based lymphocyte proliferation assay (17). Cryopreserved PBMC were thawed, washed and resuspended in RPMI 1640 containing 1% normal human serum, then labeled with 7.5µM CFSE (Invitrogen) for 8 minutes, washed again and distributed at 1 × 106 cells per stimulation condition in 1 ml of RPMI 1640/10% normal human serum. Cells were incubated for 6 days in the presence or absence of the CM235 Env peptide pool. At the end of the incubation period 2 × 105 autologous EBV-transformed B lymphoblastoid cell lines (B-LCL) that were Env peptide-pulsed overnight and washed, were co-incubated with the PBMC for a further 5 hours in the presence of anti-CD107a-PE (BD Pharmingen) and the protein transport inhibitors Monensin (GolgiStop, BD Pharmingen) and Brefeldin A (Sigma-Aldrich). The cells were washed, stained with Aqua Live/Dead (Invitrogen), washed in flow wash buffer (PBS with 0.5% BSA, 0.1% azide) and resuspended in 10% mouse IgG (FcR block) for 15 min, centrifuged and then surface stained with anti-CD14/CD19-PECy5 (Invitrogen), anti-CD4-Qdot605 (Invitrogen), anti-CD8-ECD (Beckman-Coulter) and anti-CD3-APC-H7 (eBioscience). Cells were fixed in 2% paraformaldehyode (Tousimis, Rockville MD) washed and left overnight at 4 °C, followed by incubation with Perm Wash (Becton Dickinson) for 15 minutes, washed and simultaneously surface/intracellular stained with anti-IFN-γ-eFluor450 (eBioscience), anti-TNF-α-PE-Cy7 (BD Pharmingen), anti-IL-2-APC (eBioscience). Cells were acquired on a custom built LSRII cytometer (BD Biosciences). At least 250,000 total events were acquired in the lymphocyte light scatter gate and the data were generated by gating on the CFSElow population and analyzing the cytokine production in this population. This assay permits the simultaneous measurement of both the proliferative capacity of Ag-specific T cells and the functional profiling of the cells post-proliferation.
Generation and Characterization of CD4+ T cell lines
PBMC from 10 vaccinees and 2 placebo recipients were cultured in complete medium. The CD4+ T cell lines were established as previously described (18). Briefly, PBMC at 1 × 107 cells/ml were pulsed with CRF01_AE strain A244 gp120 (25 µg/ml) for 4 h at 37°C in complete medium. Cells were diluted in complete medium, plated at 2 × 106 cells/ml in 24-well plates (Costar) and incubated at 37°C, 5% CO2. After 4 days the cells were fed with 10 U/ml recombinant IL-2 (rIL-2) (Boehringer-Mannheim). After 15 days, the T cells were harvested, washed, counted and tested in a proliferation assay to assess their specificity to CM235 Env peptides. Epitope mapping was performed using the [3H] lymphoproliferation assay as previously described (12). The proliferative response of the expanded PBMC was measured by incubating 3×104 T cells in a 96-well flat bottom plate (Costar) containing the CM235 Env matrix pooled peptides (24 pools containing 11–13 peptides each) or no peptides and with 2 × 105 irradiated autologous B-LCL. Assays were performed in duplicate. After 2 days of incubation, the cells were pulsed with 1 µCi/well of [3H] for 18 h, harvested using the Tomtec, Mach3M (EG&G Wallac) and counted in a 1450 microbeta trilux (EG& G Wallac). T-scan data are shown as z-scores, i.e. as reactivity in standard deviations (s) from the median reactivity to all the peptides. Standard deviations were determined in each experiment using this equation: s = (median – first quartile)/0.675. Values above z =3.29 s were considered significant in this assay.
Cryopreserved specific T- cell lines were thawed, washed and resuspended in RPMI 1640 with 10% normal human serum at 1×106 cells/ml, co-incubated for 4 h with 5×105 irradiated autologous B-LCL and stimulated with HIV CM235 Env pooled peptides (1 µg/peptide/ml). Anti-CD107a-FITC, anti-CD28/CD49 MAbs (BD Pharmingen), the protein-transport inhibitors Monensin (GolgiStop, BD Pharmingen) and Brefeldin A (Sigma-Aldrich) were included in the assay mix at set-up. Cells were washed, stained with Aqua Live/Dead (Invitrogen), washed and resuspended in FACSwash buffer (0.5% BSA, 0.1% sodium azide), followed by surface staining with anti-CD14/CD19-Alexa700 (BD Pharmingen), and then simultaneous surface/intracellular staining with anti-CD4-ECD (Coulter), anti-IFN-γ-Pacific Blue (eBioscience), anti-TNF-α-PE-Cy7 and anti-MIP-1β-PE (BD Pharmingen), anti-CD3-APC-H7, anti-CD8-PerCPCy5.5, and anti-IL2-APC (BD Biosciences). Cells were acquired on a custom built LSRII cytometer (BD Biosciences). At least 500,000 total events were acquired and the data analyzed using the software packages as for the ICS assay.
CTL activity: Antigen-specific cytolytic activity was measured in a standard [51Cr] release assay. Briefly, CD4+ HIV-Env (CRF01_AE A244 gp120) specific T cell lines, were re-stimulated for 7 days prior to the assay for use as effector cells using purified anti-human-CD3 antibody at 1mg/ml (BD Pharmingen), anti human-CD28 purified antibody (1 mg/ml: BD Pharmingen) and a pool of mismatched irradiated PBMC from healthy donors. Two days after the re-stimulation the cultures were supplemented with 10 IU/ml of rIL-2 (Boehringer-Mannheim). Cultures were maintained at 37°C, 5% CO2 and 95% humidity. On day 7, 2×106 autologous B-LCL were incubated for 18–20 h with either 5µg/ml of CM235 Env peptide pool, CM235 Env peptide #32 (VHALFYKLDIVPIEDNK) or no peptide before being used as targets. The viability of the B-LCL was at least 80% at the time of the overnight incubation. Targets were labeled for 1h at 37°C with 100µCi of [51Cr] (Perkin Elmer). Cells were washed and plated in 96 well U-bottomed plates (Costar). Effectors were added to the target cells at different ratios ranging from 40:1 to 0.5:1 and incubated for 6 h at 37°C. The amount of lysis was calculated by measuring [51Cr] released into the medium. Maximum release was determined by adding 100µl of 10%SDS to the target cells, and spontaneous lysis of the target cells was determined by adding 100µl of 10% normal human serum-RPMI 1640. Supernatants were harvested and transferred to 96 well luma plates (Perkin Elmer) and counted in a Top count NXT gamma counter (Perkin Elmer Microplate Scintillation and Luminescence Counter). Results are expressed as percent specific lysis calculated as follows: [(experimental counts per minute - spontaneous counts per minute)/(maximum counts per minute – spontaneous counts per minute)] X 100.
HIV Envelope Ab ELISA
Frozen plasma samples collected at V9 from subjects on whom ICS and immunophenotyping were performed (30 vaccinees and 8 placebo recipients) were tested for HIV binding antibody titers with the gp120 antigens used in the AIDSVAX B/E® boost (MN and A244) in a standard validated ELISA as previously described (5, 19). Samples were screened for the presence of gp120 B and E antibodies at an initial dilution of 1:50. Non-reactive samples were assigned a reciprocal titer of 25. Reactive samples (optical density >0.200) were titrated beginning at 1:100 at 2-fold dilutions. The reciprocal end-point titer was calculated as the greater OD value of either (2X Mean OD negative control + 2X SD) or 0.100.
Statistics
The magnitude of ELISPOT and ICS responses are expressed after subtraction of background. Data are expressed as mean or median SFC/million PBMC with ranges for the ELISPOT and medians with ranges for the ICS and summary data. Antibody titers were expressed as geometric mean titers. Data analyses were performed using Prism 5 (GraphPad, San Diego, CA). Comparisons of categorical data were made using Fisher’s exact test. Comparisons between continuous variables were performed using parametric (if the data followed a normal distribution) or non-parametric tests. The level of statistical significance was 5% for all analyses. P-values were not adjusted for multiple comparisons.
Results
ELISPOT Matrix Mapping
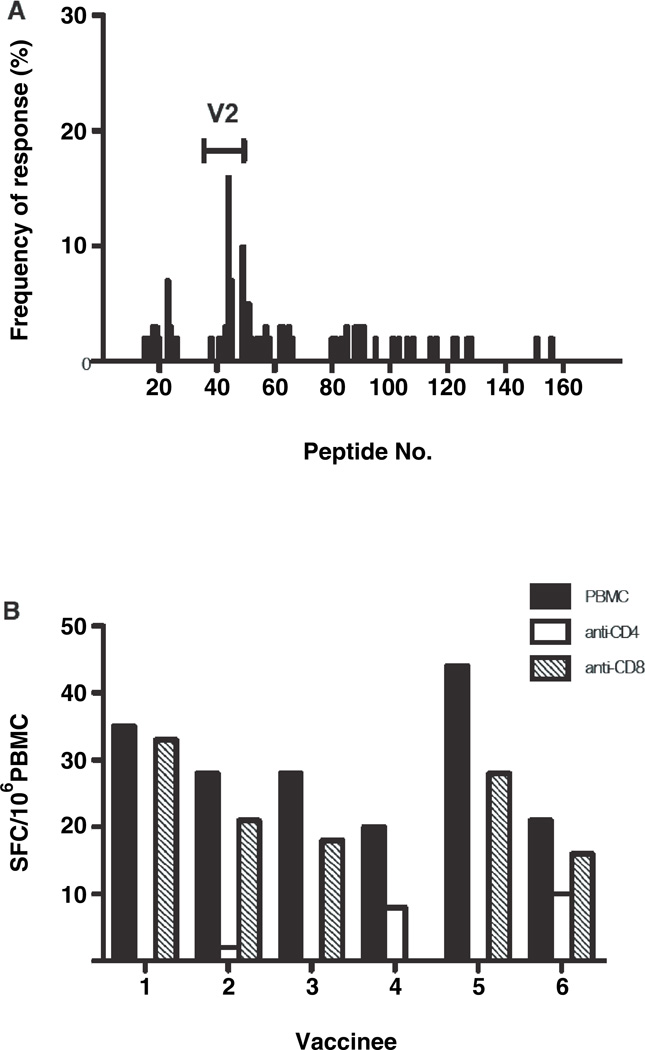
PBMC from 153 HIV-1 uninfected vaccinated subjects were initially tested by IFN-γ ELISPOT to ALVAC-HIV® (vCP1521) Env and Gag matched peptides at 6 months following the completion of immunization (V9), of whom 14% responded to Env (median: 87 SFC/106PBMC; range 47–222) and 6% to Gag (median: 88 SFC/106 PBMC; range 59–99) (5). Selecting 20 with positive ELISPOT responses to 92TH023 Env at V9, 13/20 (65%) subjects were also positive to Env peptides at 2 weeks post vaccination (V8). An additional 41 vaccinees and 10 placebo recipients (Figure 1) were tested with the Env peptide matrix at V8, and overall 25/61 (41%) vaccines and 0/10 placebo recipients had a positive IFN-γ ELISPOT response. Figure 2A shows the frequency of individual peptide responses to the Env gp160 protein for the 61 vaccinees tested at V8. IFN-γ responses were elicited across the entire protein. The predominant response (15/61; 25%) occurred within the Env V2 region – peptides 37–50, corresponding to HXB2 aa numbering 145–208. A substantial proportion (10/25; 40%) of positive responders recognized peptide 44 (VHALFYKLDIVPIED; EnvVD15), corresponding to HXB2 aa numbering 172–186, and a smaller proportion of subjects (6/25; 24%) were reactive to peptide 49 (EYRLINCNTSVIKQA; Env EA15), corresponding to HXB2 aa numbering 190–204. The median number (range) of Env epitopes recognized was 2 (1–24) in the 25 HIV vaccinees.
Figure 2.
HIV Env-specific cellular immune responses in RV144 HIV uninfected vaccine recipients are directed at variable region 2 and predominatly CD4+ T cell mediated. A, Individual HIV Env peptide responses of subjects measured by the IFN-γ ELISPOT assay. The y-axis shows the frequency of positive responders to the individual peptide as a percentage of the total number of vaccinees tested (N=61). The HIV Env V2 region is shown (peptides 37–50). B, Bar graph of IFN-γ ELISPOT responses of whole, CD4+ or CD8+ T-cell depleted PBMC from 6 vaccinees stimulated with TH023 Env pooled peptides. The y-axis shows the spot forming cells (SFC)/106 cells.
Interestingly, the predominant peptide recognized in the vaccinated group – EnvVD15, contains the integrin α4β7 binding motif (LDI/V), which may participate in the initial interaction between HIV and CD4+ target cells, increase HIV viral replication (20–22) and is infrequently recognized in HIV-1 infected Thais (23).
Cell depletion studies were performed to discriminate the T cell type producing IFN-γ. PBMC collected at V8 from 22 HIV-1 uninfected vaccinated subjects (Figure 1) were tested with EnvVD15 and the complete 92TH023 Env peptide pool following sham, CD4+ or CD8+ T cell depletion. Five of 22 subjects were positive in the ELISPOT assay to the whole Env pool (median: 28 SFC/106PBMC; range: 20–44) using the cut-off described for the peptide matrix. Depletion of CD4+ T cells resulted in complete loss of ELISPOT reactivity to the Env pool (median: 0; range 0–8 SFC/106 CD4+ depleted PBMC), while CD8+ cell depletion had minimal impact on the magnitude of the ELISPOT responses, compared to whole PBMC (median: 21; range: 0–33 SFC/106 CD8 depleted PBMC; p=0.063) (Figure 2B). Despite the earlier time-point in the current study, none of the 22 subjects tested to the TH023 Env peptide pool to assess the T cell subset producing IFN-γ in the ELISPOT assay met the criteria for a positive response of ≥ 55 SFC/106 PBMC and ≥ 4 X background used in the initial immunogenicity study performed at 6 months following the completion of immunization (5). However, this is consistent with earlier studies of this vaccine regimen, which reported that novel cellular immune responses arose up to 6 months following the completion of immunization (6).
Multi-functional Intracellular Cytokine Staining (ICS) and immunophenotyping analysis
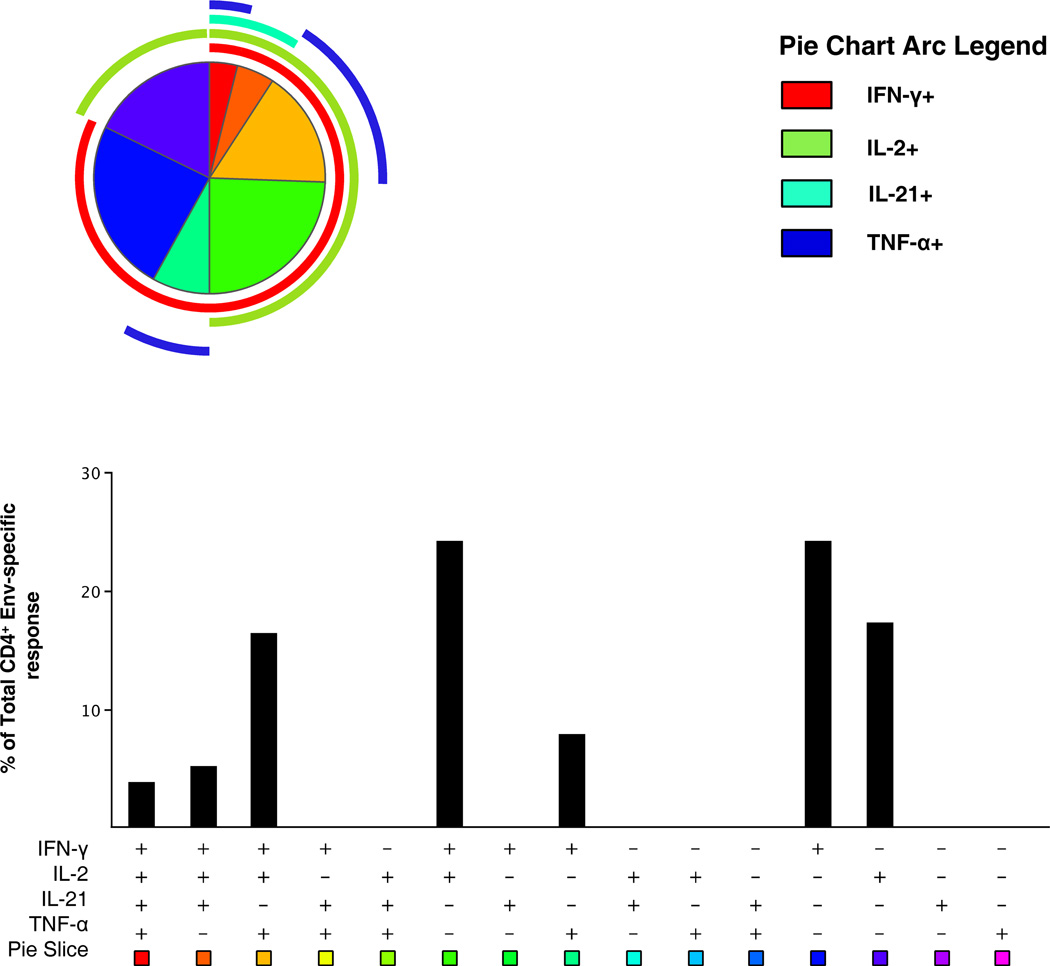
Polychromatic flow cytometry was used to explore both the functional and phenotypic profile of the Env-specific T cells in a direct ICS assay. Supplemental Figure S1 shows the gating strategy used for the multi-parameter flow cytometry assay for functional profiling. PBMC from HIV uninfected subjects receiving the full immunization regimen were assessed for their ability to up-regulate the synthesis of IL-2, IFN-γ, TNF-α and IL-21. IL-21 was included in the panel to identify CD4+ T cells with the capacity to augment B cell maturation and antibody production (24). PBMC were analyzed from 30 HIV-1 uninfected vaccinees and 8 placebo recipients (Figure 1). Among the vaccine group 19 subjects (63%) had detectable positive responses to at least one cytokine (IFN-γ or IL-2) in response to the Env peptide pool. Responding cells were exclusively CD4+ T cells with an effector memory phenotype (CD45RA−/CD45RO+/CCR7−/CD28 +/−). In terms of the total number of responders to each cytokine, IL-2 predominated (57% responders), followed by IFN-γ (43%), with lower numbers of responders producing TNF-α and IL-21 (20% and 13%, respectively). One false positive response for IL-21 was detected in a placebo recipient (Table I). Positive responses to HIV Env for any cytokine were not detected in the naïve, central memory or effector CD4+ T cell populations or any CD8+ T cell population. Multi-functional analysis using boolean gating revealed that the majority of the CD4+ effector memory T cells produced more than 1 cytokine (58%), and, as expected based upon the single cytokine analysis IL-2 and IFN-γ producing cells were predominant (Figure 3). Most of the subjects with positive responses to IL-2 also produced IFN-γ (12/17; 71%). Median response magnitudes within the CD4+ effector memory subset for IL-2 and IFN-γ were 0.12% (range: 0.05%–0.43%) and 0.13% (range: 0.05%–0.46%), respectively. A tri-functional cell subset mainly synthesized IL-2, IFN-γ and TNF-α and represented 22% of the antigen-specific population.
Table I.
Number and frequency (%) of effector memory CD4+ T cell single cytokine responses from vaccine (N=30) and placebo (N=8) recipients following PBMC stimulation with CM235 gp160 Env peptides
| Cytokine | N Vaccinees (%) | N Placebos (%) |
|---|---|---|
| IL-2 | 17 (57) | 0 |
| IFN-γ | 13 (43) | 0 |
| TNF-α | 6 (20) | 0 |
| IL-21 | 4 (13) | 1 (13) |
Figure 3.
Functional profile of vaccine-induced CD4+ T cells. PBMC stimulated with HIV Env (CM235) peptides and the functional composition (IL-2, IFN-γ, IL-21 and TNF-α) of the Env-specific CD4+ T cells 6 months post-vaccination are shown for 19 responders. The majority (>95%) of the antigen-specific cells were of the effector memory phenotype (CD45RA−/CD45RO+/CCR7−/CD28 +/−) and the responses are grouped and color-coded on the basis of the number of functions, corresponding to the Boolean subsets in the graph. Pie arcs show the relative amount of each individual function. All possible combinations of responses are shown as Boolean subsets in the bar graph below on the x-axis, and the median percentages of the functionally distinct cell populations are shown on the y-axis.
HIV-specific Ab responses
Env-specific Ab to both strains of HIV present in the AIDSVAX B/E® was detected in all 30 vaccine recipients and 0/8 placebo recipients, with a reciprocal geometric mean titer (range) of 1131 (100–12800) and 2016 (200–12800) to A244 and MN gp120, respectively. The titers of Env- specific Ab in the vaccine recipients was stratified on HIV-specific positive IL-2 responses in effector memory CD4+ T cells of vaccine recipients. Subjects with IL-2 responses had higher HIV-specific Ab titers compared with the 13 subjects without IL-2 responses to both A244 (geometric mean titer [range]: 1736 [100–12800] versus 646 [100–6400]; p=0.015) and MN (geometric mean titer [range]; 2832 [400–12800] versus 1293 [200–12800]; p=0.045).
Lymphocyte proliferation functional recall assay
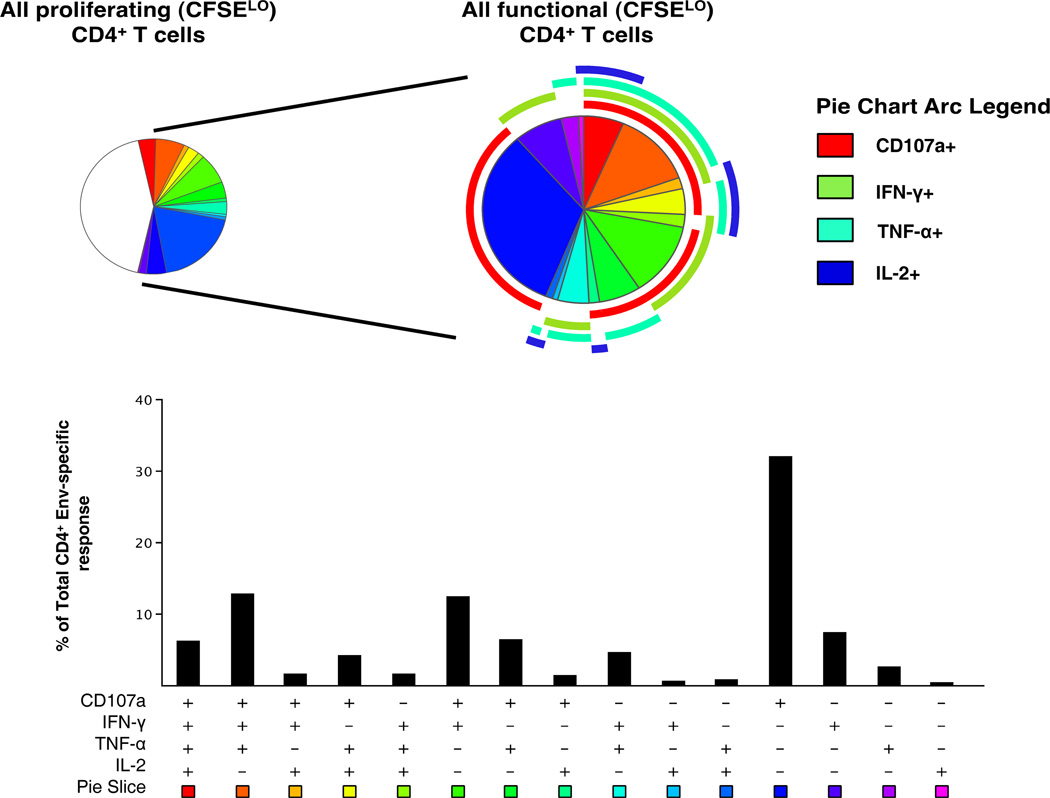
Simultaneous T cell proliferation and functional profiles were measured in PBMC from HIV uninfected vaccine recipients using a CFSE-based flow cytometric assay following re-stimulation with peptide-pulsed autologous B-LCL. This assay again showed that Env-specific CD4+ T cells were the predominant responding cells (17/30; 57% vaccinees). A single placebo recipient (1/8) showed a positive response to Env. Thirteen (43%) vaccinees demonstrated concordance with positive responses in both ICS and CFSE proliferation assays following Env stimulation. Upon restimulation following 6 days in culture, proliferating CD4+ T cells, defined as CFSELO, exhibited an unusual functional phenotype potentially capable of cytotoxicity, as measured by CD107a surface expression, accompanied by IFN-γ and TNF-α production but with minimal IL-2 production (Figure 4). Multi-functional analysis of the proliferating T cells revealed that a large fraction (43%) of these cells did not express any of the four functions assayed. Of the functional cells, CD107a expression was detected in the majority of subsets, as expected from the single function analysis, and was most commonly detected in combination with IFN-α. The majority (80%) of the antigen-specific functional CD4+ T cells expressed CD107a on the cell surface, with IFN-α being the predominant cytokine (50%), followed by TNF-α (42%), with minimal IL-2 production (19%).
Figure 4.
Polyfunctional analysis of proliferating HIV-1 Env specific CD4+ T cells. The pie chart on the left shows the functional profile of all proliferating CD4+ T cells (CFSELO), while that on the right shows only those proliferating CD4+ T cells that are functional after re-stimulation with peptide-pulsed autologous B-LCL. Pie slices correspond to the fraction of cells within the CD4+ T cell population while the pie arcs show the relative amount of each individual function. The bar graph represents Boolean subset analysis of all functionally distinct cells within the total CD4+ population. The x-axis shows all possible combinations of responses, while the y-axis represents the median percentage of each subset within the functionally positive CD4+ population. Results shown are generated from 17 vaccinees with positive proliferative responses by CFSE analysis.
HIV-specific T cell lines
CD4+ T cells were expanded from PBMC of HIV uninfected vaccinees stimulated with gp120 in culture, a procedure that yields oligoclonal, antigen-specific T cell lines (18). These lines were tested in a lymphoproliferation assay with CM235 Env peptides in a matrix format. Expanded CD4+ T cell lines from four subjects yielded responses that were deconvoluted and assigned peptide specificity. The recognized peptides confirmed similar areas of recognition found in the IFN-γ ELISPOT assay (Table II). Epitopes within the C1 and V2 regions were recognized by T cell lines from 3/4 subjects, and 2/4 in the C2 and C5 regions. One subject’s T cell line responded only to the C4 region of CM235 gp120. Despite using a slightly different peptide set (CM235; CRF01_AE. R5-tropic) to that used in the IFN-γ ELISPOT matrix mapping (92TH023; CRF01_AE.R5 tropic), the expanded CD4+ T cells from 2/4 subjects demonstrated peptide reactivity to the analogous region of 92TH023 EnvVD15, which has the identical sequence to peptide 44 and two additional C-terminus residues, asparagine and lysine (Table II).
Table II.
HIV-1 gp120 Env peptides recognized using by PBMC using the IFN-γ ELISPOT assay and T cell lines established from RV144 vaccinees
| 92 TH023 Env gp120 peptides recognized in ELISPOT assay |
CM 235 Env peptides recognized by the expanded T cell lines from RV144 vaccinees |
Location in gp 120 |
|---|---|---|
| NMWKNNMVEQMQEDV | MWKNNMVEQMQEDVISL | C1 |
| NNMVEQMQEDVISLW | C1 | |
| EQMQEDVISLWDQSL | C1 | |
| No Response | TNAKLTNVNNITSVSNT | C1/V1 |
| VHALFYKLDIVPIED | VHALFYKLDIVPIEDNK | V2 |
| QLLLNGSLAEEEIII | HGIKPVVSTQLLLNGS | C2 |
| LNGSLAEEEIIIRSENL | C2 | |
| IIRSENLTNNAKTIIV | C2 | |
| MWQGAGQAMYAPPIS | AMYAPPISGRINCVSNI | C4 |
| MYAPPISGRINCVSN | ISGRINCVSNITGILL | C4 |
| INCVSNITGILLTRDGG | C4 | |
| TFRPGGGNIKDNWRI | TNETFRPGGGNIKDNW | V5/C5 |
Bold font denotes common amino acids between the 2 peptide sets: regular font denotes differences between the analogous peptides
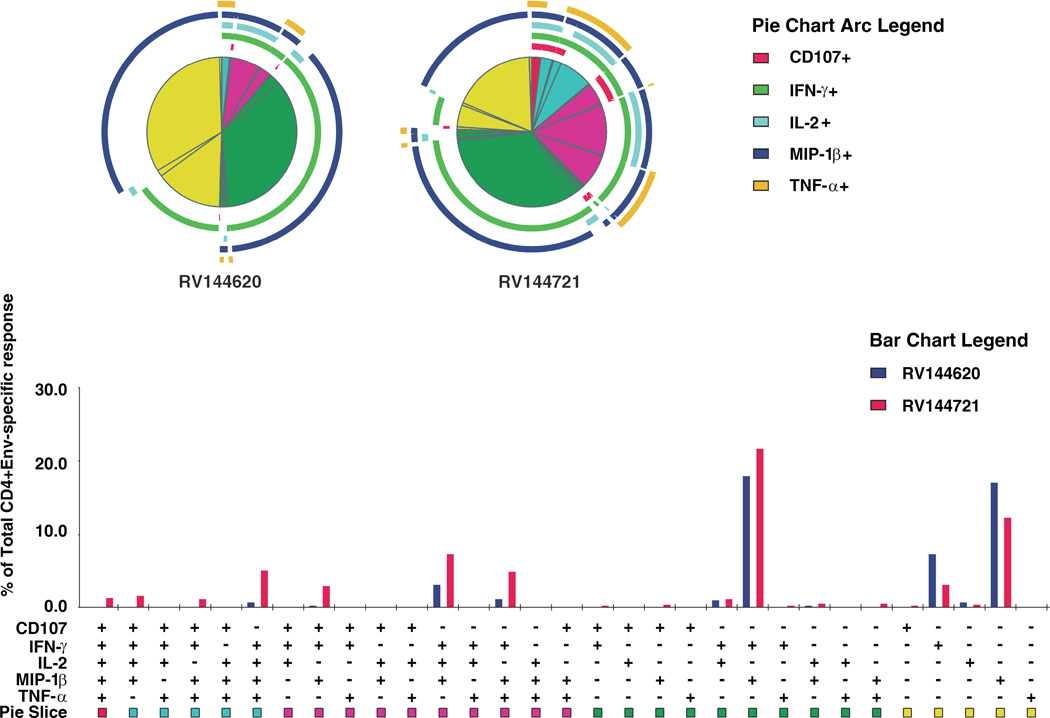
The two expanded T cell lines (144620 and 144721) that recognized the 92TH023 EnvVD15 analogue were tested in a polychromatic ICS assay following stimulation with CM235 Env pooled peptides. The cytokine production profile was predominantly multi-functional and cells showed CD107a surface expression (Figure 5).
Figure 5.
Multifunctional profiles of Env-specific CD4+ T cell lines derived from 2 RV144 vaccine recipients 6 months post-vaccination. Results shown were generated following in vitro stimulation of CD4+ T cell lines expanded using gp120 A244. Each pie chart corresponds to the functional profile for each subject. Responses are grouped and color-coded on the basis of the number of functions. The pie chart summarizes the data, with each slice of the pie corresponding to the fraction of cells within the total CD4+ T cell population. Pie arcs show the relative amount of each individual function. Bars represent the fraction of functionally distinct cells within the total CD4+ T cell population.
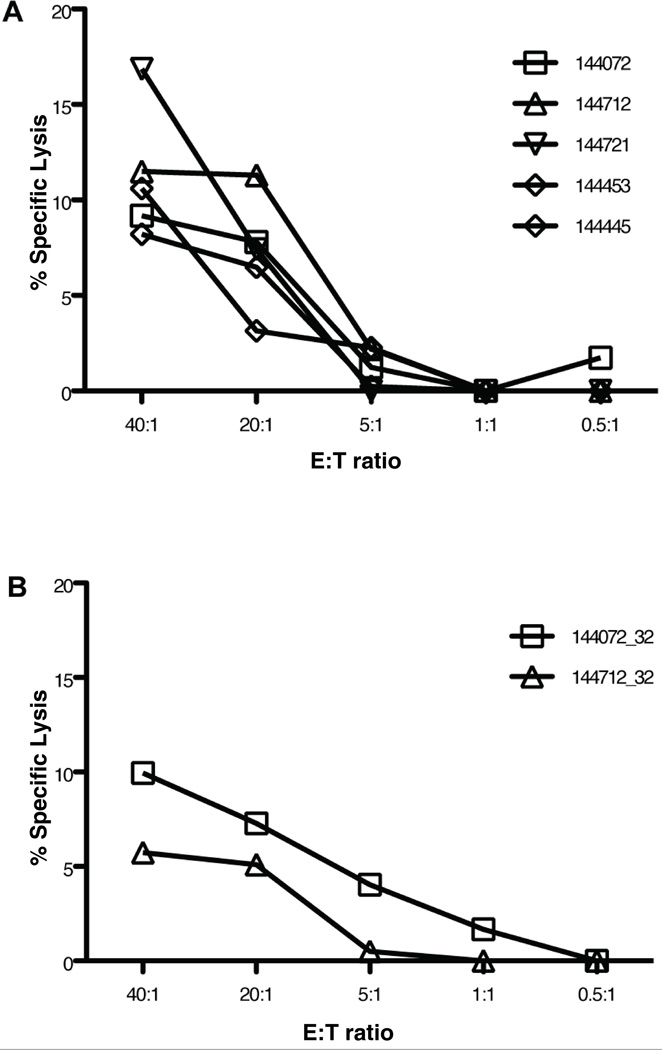
[51Cr] cytotoxicity assays were performed on 144721 and an additional 4 T cell lines. Table III shows the immunophenotype of the 5 CD4+ T cell lines and their Env region specificity. Four of the 5 lines responded to peptides in the V2 region. All lines demonstrated cytolytic activity to the CM235 Env peptide pool (Figure 6A), with 144721 having the greatest lytic activity despite an absence of CD8+ T cells (Table III). T cell lines 144072 and 144712 were further tested for functional cytotoxicity to the TH023 EnvVD15 analogue EnvVK16. Lytic activity to EnvVK16 was observed for both lines, and more strongly for 144072 (Figure 6B). T cell line 144072 also had no measurable CD8+ T cells (Table III).
Table III.
Immunophenotype of T cell lines established from RV144 vaccine recipients and HIV Env recognition pattern. Cells were used as effector cells in cytotoxicity assays to the HIV CM235 Env peptide pool
| T cell phenotype | |||
|---|---|---|---|
| Subject | %CD4+ | %CD8+ | HIV Env region |
| 144072 | 96 | 0 | V2 |
| 144712 | 66 | 11 | V2 |
| 144453 | 71 | 10 | C1 |
| 144721 | 98 | 0 | V1/V2 V2 |
| 144445 | 80 | 6 | C1 V2 |
Figure 6.
Cytotoxic function of CD4+ T cell lines measured by [51Cr] release. A. Lytic activity of T cell lines from 5 vaccinated subjects to autologous B-LCL pulsed with HIV Env (CM235) peptide pool. B. Lytic activity of T cell lines from 2 vaccinated subjects to B-LCL pulsed with the CM 235 Env V2 peptide #32 (VHALFYKLDIPIEDNK). Results are expressed as the percentage of specific lysis (y-axis) of the target cells at various E:T ratios (x-axis).
Discussion
This study assessed T cell responses in RV144 vaccinees by IFN-γ ELISPOT at 2 weeks post immunization using the same HIV Env pool as that used in a previous study of RV144 cellular immune responses at 6 months following the completion of immunization (5).
T cell epitope mapping using peptides matching the ALVAC 92TH023 gp160 demonstrated that PBMC from HIV uninfected vaccinated subjects predominantly recognized epitopes within the V2 region of Env, particularly EnvVD15, which includes the putative integrin α4β7 binding site. One previous study where subjects were immunized with different regimens (a combination of ALVAC and gp120, ALVAC alone or gp160MN/LAI-2 alone) showed that one individual vaccinated with ALVAC (vCP205) only, recognized a peptide in the gp120 V1–V2 region (12). The current study did not observe any gp120 V1 responses in HIV uninfected vaccine recipients. A recent study of DNA priming with a NYVAC boost (EuroVacc 02) using HIV antigens derived from a clade C isolate also reported MHC class II restricted peptide reactivity in the V2 region (VYALFYRLDIVPLTK) encoding the α4β7 binding site, which was the first published report of recognition of this particular Env peptide in either uninfected HIV vaccine recipients or HIV-1 infected subjects (13). One previous study assessing Env and Gag peptide recognition in CRF01_AE HIV infected Thais used the identical Env peptides to those in the current study and reported reactivity to EnvVD15 in 1/20 subjects (23). The EuroVacc clade C peptide EnvVK15 shares 67% homology with the 92TH023 equivalent peptide, EnvVD15, but has the same α4β7 binding motif (LDI) (13). CD8+ T cell depletion studies verified that EnvVD15 was also class II restricted.
The mean number of Env epitopes recognized (3.6) in the RV144 vaccine group with positive responses in the IFN-γ ELISPOT assay is similar to that reported for the EuroVacc 02 phase I study (4.2) (13) and multi-clade HIV-1 DNA prime/recombinant Adenovirus 5 vaccine regimen (3.3) (14), but greater than that reported for the Step trial (1) (25).
Despite measurable CD8+ cytolytic T cells using the chromium release assay (6), direct ex vivo HIV-specific CD8+ T cell responses from RV144 subjects’ PBMC were barely measurable (<10%) in the IFN-γ/IL-2 combination ICS assay and were equivalent to the frequency of responses seen in the placebo recipients (5). However, robust CD4+ T cell responses assessed by both [3H] incorporation and the ICS assays were reported in the vaccine group (5). The finding of direct ex vivo vaccine-induced T cell immune responses being predominantly CD4+ T cell mediated agrees with data reported from DNA prime followed by poxvirus or adenovirus boost studies (10, 11, 13, 14, 26, 27). One DNA prime/adenovirus serotype 5 boost HIV-1 vaccine trial reported a greater frequency (93%) of HIV-specific CD4+ compared to CD8+ T cell (71%) responses four weeks following recombinant adenovirus 5 boosting (14).
Polychromatic flow cytometry of PBMC following stimulation with a heterologous CRF01_AE Env peptide set to that used in the IFN-γ ELISPOT assay again demonstrated that the vaccine elicited direct ex vivo T cell responses that were exclusively CD4+ T cell mediated and also multifunctional. Multifunctional (defined as production of IL-2 in addition to effector function, such as production of IFN-γ) T cells (28) are thought to be important for the control of a number of virus infections (28–31). The combination of IL-2 and IFN-γ production capability is consistent with the phenotypic effector memory classification of these cells (28).
The finding that up to 6 months following the completion of immunization the HIV-specific CD4+ T cells were of the effector memory phenotype, with most cells producing IL-2, would allow both effector function, proliferation and induction of CD8+ T cell antiviral activity (14, 30, 32). Effector memory CD4+ T cells producing IL-2 are thought to be important in the development of IgG-secreting plasma B cells and have been reported to be positively associated with Ab titers to hepatitis B surface Ag up to 9 years following immunization (33), a vaccine which like the RV144 regimen induces robust humoral immune responses with limited direct ex vivo T cell responses. The finding that the vaccine group with IL-2 producing effector memory T cells had higher gp120 Ab titers compared to the group without would support this observation.
To our knowledge, this is the first use of polychromatic flow cytometry in a clinical trial using this vaccine regimen, and demonstrates that the ALVAC + gp120 B/E vaccine regimen induced polyfunctional and potentially cytolytic CD4+ T cell responses up to 6 months following completion of immunization. All CD4+ T cells induced by the vaccine were of the effector memory phenotype, which is similar to that previously reported for a DNA/NYVAC clinical vaccine trial (13).
One limitation of our study was that we did not assess the phenotype of the responding cells at 2 weeks following the completion of immunization, so the profile may differ from that at the 24 week time-point, but the EuroVacc 02 study did not detect a difference in the phenotype of vaccine-induced CD4+ T cells with increased time from the completion of immunization (13). The presence of vaccine-induced CD4+ T cell responses has been reported to correlate with virus control in a vaccine-challenge study using ALVAC gag-pol-env prime/ gp120 boost vaccine regimen and simian-human immunodeficiency virus challenge (34). The lymphocyte proliferation functional recall assay showed that the cytokine profile of functional CD4+ T cells implied a terminal effector function (31, 35).
The establishment of polyfunctional HIV-1 Env specific CD4+ T cell lines that also express CD107a and are capable of cytotoxic activity further verified the findings of the lymphocyte proliferation functional recall assay, although it should be stressed that these findings are not directly ex vivo. However, one pilot study with HIV-infected volunteers compared the delayed type hypersensitivity skin test assay with autologous CD4+ T cell line lymphoproliferation using identical HIV Env V3 peptides and reported excellent concordance, suggesting that the lines may be measuring a relevant in vivo immune function (36), though prolonged culture may change the phenotype associated with a particular T cell receptor genotype.
HIV-1 specific cytotoxic CD4+ T lymphocytes using a variety of assays have been described in HIV-1 infection (37–42). A study of acute HIV infection in the US (presumably HIV subtype B) using CD4+ T cell clones derived from a volunteer infected with HIV for 11 days reported V2 epitope recognition using a 20 aa peptide (RDKMQKEYALLYKLDIVSID) which includes the LDI putative α4β7 binding site; comparison with EnvVD15 shows 64% homology in the area of overlap (42). This clone secreted predominantly IFN-γ and lower levels of IL-4 and IL-10. CD4+ T cell recognition to whole Env protein was lost in this subject at 16 days post-infection just prior to antiretroviral therapy initiation and was not observed again despite repeated testing (up to 683 days post-infection), despite the infecting virus not showing sequence variation within this region (42). Other studies of cellular immune responses in HIV infected cohorts have inferred that this V2 epitope may be recognized, but have not identified it as a highly recognized region (43–45).
Appay et al., speculated that the generation of CD4+ CTL could be to control viruses targeting class II expressing cells and/or that downregulate MHC class I expression (38). In natural infection, these perforin containing CD4+ T cells posessed a terminally differentiated phenotype (CD45RO+/CCR7−/CD28−), with production of TNF-α and IFN-γ, but not IL-2 (38), similar to the cytotoxic CD4+ T cell lines established following HIV-1 Env restimulation in this study. CD4+ CTL have also been described following immunization with HIV gp160 protein subunit although cloned CD4+ T cell lines were used (46). It has been proposed that generation of cytotoxic CD4+ T cells in addition to CD8+ CTL by HIV vaccine regimens would provide the advantage of dual MHC class I and II recognition and the greater diversity of class II epitope recognition would allow greater breadth of virally infected targets (35, 47). Cytolytic CD4+ T cells may also be important in the control of several herpesviruses, measles and dengue viruses (48).
While the direct ex vivo cellular immune response induced by the RV144 vaccine regimen was modest in relation to the humoral immune response as measured by binding antibody, the observation that the RV144 vaccinees recognized T cell epitopes predominantly within the V2 loop of HIV-1 Env is intriguing given the location of a known ligand (LDI) for the α4β7 integrin within this region (20). Substitution of the aspartic acid in the LDI region with hydrophilic basic amino acids has been found to abrogate virus infectivity, whereas substitution with negatively charged amino acids maintained virus infectivity with heightened neutralization sensitivity (49).
The lack of published data from epitope mapping studies on the predominance of EnvVD15 T cell epitope responses in HIV infected subjects to date (43, 44, 50, 51) suggest that this epitope may be masked on the virion surface, not easily produced and/or presented in MHC class II, or that cells with this specificity are preferentially infected with the virus and destroyed during infection (42, 52).
The RV144 vaccine regimen induced modest ex vivo T cell responses with respect to frequency and magnitude as measured by IFN-γ ELISPOT and ICS compared to other HIV vaccine regimens with either poxvirus (13, 53) or adenovirus (54, 55) vector boosting following DNA priming. However, humoral immune responses and antigen-specific lymphoproliferative responses measured by HIV binding antibody to gp120 and [3H], respectively, were robust (5). A similar vaccine regimen combination of ALVAC prime and HIV Env boost has shown some protection in animal models. An infant macaque model of ALVAC-SIV expressing gag-pol-env administered at 0, 1 and 3 weeks followed by multiple low dose oral challenges of simian immunodeficiency virus at 4 weeks also demonstrated partial protection from infection, despite minimal HIV-1 specific IFN-γ ELISPOT responses in immunized animals (56). Another ALVAC-HIV-1 gag-pol-env-nef prime/ HIV-1 Env protein boost study followed by a mucosally administered simian-human immunodeficiency virus as a challenge in macaques reported only modest (33%) HIV specific CTL activity, but there was a reduction in infection in the immunized animals compared to the unimmunized group, in addition to protection from CD4+ T cell loss following virus challenge (34).
Characterizing the immune response in studies such as this can help generate potential correlates for formal testing. Despite the modest T cell responses induced by the RV144 trial regimen, the finding of preferential cellular immune responses to gp120 V2 epitopes led to studies on antibody responses to gp120 V2 induced by the vaccine regimen in HIV-uninfected vaccinees and was found in 92% of subjects at 2 weeks following the completion of immunization (N. Karasavvas, manuscript in preparation). A case-control study using multiple assays for cellular and humoral immune responses in RV144 vaccinees who became HIV-infected compared to those who remained uninfected reported an inverse correlation between antibody levels to a conformational V1V2 epitope and HIV infection, but no other potentially protective immune responses (57).
The comprehensive functional capacity of the Env-specific CD4+ T cells induced by the RV144 vaccine regimen needs to be further assessed. Ongoing work, including additional HIV vaccine trials using poxvirus prime/HIV Env protein boost, is seeking to elucidate correlate(s) of protection induced by the RV144 vaccine regimen by allowing larger specimen collection volumes for more detailed immunogenicity studies (58).
Supplementary Material
Acknowledgements
We would like to thank the RV144 study participants for allowing this study to occur, the Thai Ministry of Public Health staff in Rayong and Chon Buri for their invaluable assistance on this trial and the Vaccine Clinical Trial and Data Management staff at Mahidol University. We would also like to thank Josephine H. Cox, Nicos Karasavva and Peter Gilbert for helpful discussions and review of the manuscript, Kanyasiri Kongnonkok and Boot Keawboon for their work on the ELISPOT assays, Bonnie Slike for assistance with the tables. Gratitude is also expressed to Viseth Ngauy for her continued support of this project.
These studies were supported in part by an Interagency Agreement Y1-AI-2642-12 between U.S. Army Medical Research and Materiel Command (USAMRMC) and the National Institutes of Allergy and Infectious Diseases. In addition this work was supported by a cooperative agreement (W81XWH-07-2-0067) between the Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc., and the U.S. Department of Defense (DOD).
Abbreviations used in this manuscript
- CRF
circulating recombinant form
- ICS
intracellular cytokine staining
- RV144
Thai HIV Phase III vaccine trial
- RV152
HIV break-through infection protocol for participants in the Thai phase III trial
- SFC
spot forming cells
Footnotes
The authors have no conflicting financial interests.
The opinions herein are those of the authors and should not be construed as official or representing the views of the U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, the Department of Defense or Department of the Army.
Presented in Part at: “AIDS Vaccine Meeting 2009”, 19–22 October, 2009, Paris, France (abstract: OA07-04LB); 2010 Keystone Symposium “HIV Vaccines/Viral Immunity”, 21–26 March, 2010, Banff, Canada (abstract X5 159); 4th Vaccine and ISV Annual Global Congress, 3–5 October, 2010, Vienna Austria (abstract O2.1) and Europrise: Rational Design of HIV Vaccines and Microbicides Network Annual Conference, 15–18 November, 2010 Lisbon, Portugal
References
- 1.UNAIDS. AIDS Epidemic Update: December 2009. UNAIDS/0936E/JC1700E 2009 [Google Scholar]
- 2.Buchbinder SP, Mehrotra DV, Duerr A, Fitzgerald DW, Mogg R, Li D, Gilbert PB, Lama JR, Marmor M, Del Rio C, McElrath MJ, Casimiro DR, Gottesdiener KM, Chodakewitz JA, Corey L, Robertson MN. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet. 2008;372:1881–1893. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flynn NM, Forthal DN, Harro CD, Judson FN, Mayer KH, Para MF. Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J Infect Dis. 2005;191:654–665. doi: 10.1086/428404. [DOI] [PubMed] [Google Scholar]
- 4.Pitisuttithum P, Gilbert P, Gurwith M, Heyward W, Martin M, van Griensven F, Hu D, Tappero JW, Choopanya K. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J Infect Dis. 2006;194:1661–1671. doi: 10.1086/508748. [DOI] [PubMed] [Google Scholar]
- 5.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 6.Nitayaphan S, Pitisuttithum P, Karnasuta C, Eamsila C, de Souza M, Morgan P, Polonis V, Benenson M, VanCott T, Ratto-Kim S, Kim J, Thapinta D, Garner R, Bussaratid V, Singharaj P, el-Habib R, Gurunathan S, Heyward W, Birx D, McNeil J, Brown AE. Safety and immunogenicity of an HIV subtype B and E prime-boost vaccine combination in HIV-negative Thai adults. J Infect Dis. 2004;190:702–706. doi: 10.1086/422258. [DOI] [PubMed] [Google Scholar]
- 7.Belshe RB, Stevens C, Gorse GJ, Buchbinder S, Weinhold K, Sheppard H, Stablein D, Self S, McNamara J, Frey S, Flores J, Excler JL, Klein M, Habib RE, Duliege AM, Harro C, Corey L, Keefer M, Mulligan M, Wright P, Celum C, Judson F, Mayer K, McKirnan D, Marmor M, Woody G. Safety and immunogenicity of a canarypox-vectored human immunodeficiency virus Type 1 vaccine with or without gp120: a phase 2 study in higher- and lower-risk volunteers. J Infect Dis. 2001;183:1343–1352. doi: 10.1086/319863. [DOI] [PubMed] [Google Scholar]
- 8.Evans TG, Keefer MC, Weinhold KJ, Wolff M, Montefiori D, Gorse GJ, Graham BS, McElrath MJ, Clements-Mann ML, Mulligan MJ, Fast P, Walker MC, Excler JL, Duliege AM, Tartaglia J. A canarypox vaccine expressing multiple human immunodeficiency virus type 1 genes given alone or with rgp120 elicits broad and durable CD8+ cytotoxic T lymphocyte responses in seronegative volunteers. J Infect Dis. 1999;180:290–298. doi: 10.1086/314895. [DOI] [PubMed] [Google Scholar]
- 9.Russell ND, Graham BS, Keefer MC, McElrath MJ, Self SG, Weinhold KJ, Montefiori DC, Ferrari G, Horton H, Tomaras GD, Gurunathan S, Baglyos L, Frey SE, Mulligan MJ, Harro CD, Buchbinder SP, Baden LR, Blattner WA, Koblin BA, Corey L. Phase 2 study of an HIV-1 canarypox vaccine (vCP1452) alone and in combination with rgp120: negative results fail to trigger a phase 3 correlates trial. J Acquir Immune Defic Syndr. 2007;44:203–212. doi: 10.1097/01.qai.0000248356.48501.ff. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franchini G, Gurunathan S, Baglyos L, Plotkin S, Tartaglia J. Poxvirus-based vaccine candidates for HIV: two decades of experience with special emphasis on canarypox vectors. Expert Rev Vaccines. 2004;3:S75–S88. doi: 10.1586/14760584.3.4.s75. [DOI] [PubMed] [Google Scholar]
- 11.Paris RM, Kim JH, Robb ML, Michael NL. Prime-boost immunization with poxvirus or adenovirus vectors as a strategy to develop a protective vaccine for HIV-1. Expert Rev Vaccines. 2010;9:1055–1069. doi: 10.1586/erv.10.106. [DOI] [PubMed] [Google Scholar]
- 12.Ratto-Kim S, Loomis-Price LD, Aronson N, Grimes J, Hill C, Williams C, El Habib R, Birx DL, Kim JH. Comparison between env-specific T-cell epitopic responses in HIV-1-uninfected adults immunized with combination of ALVAC-HIV(vCP205) plus or minus rgp160MN/LAI-2 and HIV-1-infected adults. J Acquir Immune Defic Syndr. 2003;32:9–17. doi: 10.1097/00126334-200301010-00003. [DOI] [PubMed] [Google Scholar]
- 13.Harari A, Bart PA, Stohr W, Tapia G, Garcia M, Medjitna-Rais E, Burnet S, Cellerai C, Erlwein O, Barber T, Moog C, Liljestrom P, Wagner R, Wolf H, Kraehenbuhl JP, Esteban M, Heeney J, Frachette MJ, Tartaglia J, McCormack S, Babiker A, Weber J, Pantaleo G. An HIV-1 clade C DNA prime, NYVAC boost vaccine regimen induces reliable, polyfunctional, and long-lasting T cell responses. J Exp Med. 2008;205:63–77. doi: 10.1084/jem.20071331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koup RA, Roederer M, Lamoreaux L, Fischer J, Novik L, Nason MC, Larkin BD, Enama ME, Ledgerwood JE, Bailer RT, Mascola JR, Nabel GJ, Graham BS. Priming immunization with DNA augments immunogenicity of recombinant adenoviral vectors for both HIV-1 specific antibody and T-cell responses. PLoS One. 2010;5:e9015. doi: 10.1371/journal.pone.0009015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Currier JR, Kuta EG, Turk E, Earhart LB, Loomis-Price L, Janetzki S, Ferrari G, Birx DL, Cox JH. A panel of MHC class I restricted viral peptides for use as a quality control for vaccine trial ELISPOT assays. Journal of immunological methods. 2002;260:157–172. doi: 10.1016/s0022-1759(01)00535-x. [DOI] [PubMed] [Google Scholar]
- 16.Currier JR, Ngauy V, de Souza MS, Ratto-Kim S, Cox JH, Polonis VR, Earl P, Moss B, Peel S, Slike B, Sriplienchan S, Thongcharoen P, Paris RM, Robb ML, Kim J, Michael NL, Marovich MA. Phase I safety and immunogenicity evaluation of MVA-CMDR, a multigenic, recombinant modified vaccinia Ankara-HIV-1 vaccine candidate. PLoS One. 2010;5:e13983. doi: 10.1371/journal.pone.0013983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abate G, Eslick J, Newman FK, Frey SE, Belshe RB, Monath TP, Hoft DF. Flow-cytometric detection of vaccinia-induced memory effector CD4(+), CD8(+), and gamma delta TCR(+) T cells capable of antigen-specific expansion and effector functions. J Infect Dis. 2005;192:1362–1371. doi: 10.1086/444423. [DOI] [PubMed] [Google Scholar]
- 18.Ratto S, Sitz KV, Scherer AM, Manca F, Loomis LD, Cox JH, Redfield RR, Birx DL. Establishment and characterization of human immunodeficiency virus type 1 (HIV-1) envelope-specific CD4+ T lymphocyte lines from HIV-1-seropositive patients. J Infect Dis. 1995;171:1420–1430. doi: 10.1093/infdis/171.6.1420. [DOI] [PubMed] [Google Scholar]
- 19.Rerks-Ngarm S, Brown AE, Khamboonruang C, Thongcharoen P, Kunasol P. HIV/AIDS preventive vaccine 'prime-boost' phase III trial: foundations and initial lessons learned from Thailand. Aids. 2006;20:1471–1479. doi: 10.1097/01.aids.0000237362.26370.f8. [DOI] [PubMed] [Google Scholar]
- 20.Arthos J, Cicala C, Martinelli E, Macleod K, Van Ryk D, Wei D, Xiao Z, Veenstra TD, Conrad TP, Lempicki RA, McLaughlin S, Pascuccio M, Gopaul R, McNally J, Cruz CC, Censoplano N, Chung E, Reitano KN, Kottilil S, Goode DJ, Fauci AS. HIV-1 envelope protein binds to and signals through integrin alpha4beta7, the gut mucosal homing receptor for peripheral T cells. Nat Immunol. 2008;9:301–309. doi: 10.1038/ni1566. [DOI] [PubMed] [Google Scholar]
- 21.Cicala C, Martinelli E, McNally JP, Goode DJ, Gopaul R, Hiatt J, Jelicic K, Kottilil S, Macleod K, O'Shea A, Patel N, Van Ryk D, Wei D, Pascuccio M, Yi L, McKinnon L, Izulla P, Kimani J, Kaul R, Fauci AS, Arthos J. The integrin alpha4beta7 forms a complex with cell-surface CD4 and defines a T-cell subset that is highly susceptible to infection by HIV-1. Proc Natl Acad Sci U S A. 2009;106:20877–20882. doi: 10.1073/pnas.0911796106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nawaz F, Cicala C, Van Ryk D, Block KE, Jelicic K, McNally JP, Ogundare O, Pascuccio M, Patel N, Wei D, Fauci AS, Arthos J. The genotype of early-transmitting HIV gp120s promotes alphabeta-reactivity, revealing alphabetaCD4+ T cells as key targets in mucosal transmission. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1001301. e1001301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kantakamalakul W, de Souza M, Bejrachandra S, Ampol S, Cox J, Sutthent R. Identification of a novel HIV type 1 CRF01_AE cytotoxic T lymphocyte (CTL) epitope restricted by an HLA-Cw0602 allele and a novel HLA-A0206/peptide restriction. AIDS Res Hum Retroviruses. 2006;22:1271–1282. doi: 10.1089/aid.2006.22.1271. [DOI] [PubMed] [Google Scholar]
- 24.Kuchen S, Robbins R, Sims GP, Sheng C, Phillips TM, Lipsky PE, Ettinger R. Essential role of IL-21 in B cell activation, expansion, and plasma cell generation during CD4+ T cell-B cell collaboration. J Immunol. 2007;179:5886–5896. doi: 10.4049/jimmunol.179.9.5886. [DOI] [PubMed] [Google Scholar]
- 25.McElrath MJ. Immune responses to HIV vaccines and potential impact on control of acute HIV-1 infection. J Infect Dis. 2010;202(Suppl 2):S323–S326. doi: 10.1086/655658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Letvin NL. Progress and obstacles in the development of an AIDS vaccine. Nat Rev Immunol. 2006;6:930–939. doi: 10.1038/nri1959. [DOI] [PubMed] [Google Scholar]
- 27.McMichael AJ. HIV vaccines. Annu Rev Immunol. 2006;24:227–255. doi: 10.1146/annurev.immunol.24.021605.090605. [DOI] [PubMed] [Google Scholar]
- 28.Harari A, Dutoit V, Cellerai C, Bart PA, Du Pasquier RA, Pantaleo G. Functional signatures of protective antiviral T-cell immunity in human virus infections. Immunol Rev. 2006;211:236–254. doi: 10.1111/j.0105-2896.2006.00395.x. [DOI] [PubMed] [Google Scholar]
- 29.Kannanganat S, Kapogiannis BG, Ibegbu C, Chennareddi L, Goepfert P, Robinson HL, Lennox J, Amara RR. Human immunodeficiency virus type 1 controllers but not noncontrollers maintain CD4 T cells coexpressing three cytokines. J Virol. 2007;81:12071–12076. doi: 10.1128/JVI.01261-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pantaleo G, Harari A. Functional signatures in antiviral T-cell immunity for monitoring virus-associated diseases. Nat Rev Immunol. 2006;6:417–423. doi: 10.1038/nri1840. [DOI] [PubMed] [Google Scholar]
- 31.Seder RA, Ahmed R. Similarities and differences in CD4+ and CD8+ effector and memory T cell generation. Nat Immunol. 2003;4:835–842. doi: 10.1038/ni969. [DOI] [PubMed] [Google Scholar]
- 32.Potter SJ, Lacabaratz C, Lambotte O, Perez-Patrigeon S, Vingert B, Sinet M, Colle JH, Urrutia A, Scott-Algara D, Boufassa F, Delfraissy JF, Theze J, Venet A, Chakrabarti LA. Preserved central memory and activated effector memory CD4+ T-cell subsets in human immunodeficiency virus controllers: an ANRS EP36 study. J Virol. 2007;81:13904–13915. doi: 10.1128/JVI.01401-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Litjens NH, Huisman M, Hijdra D, Lambrecht BM, Stittelaar KJ, Betjes MG. IL-2 producing memory CD4+ T lymphocytes are closely associated with the generation of IgG-secreting plasma cells. J Immunol. 2008;181:3665–3673. doi: 10.4049/jimmunol.181.5.3665. [DOI] [PubMed] [Google Scholar]
- 34.Pal R, Venzon D, Letvin NL, Santra S, Montefiori DC, Miller NR, Tryniszewska E, Lewis MG, VanCott TC, Hirsch V, Woodward R, Gibson A, Grace M, Dobratz E, Markham PD, Hel Z, Nacsa J, Klein M, Tartaglia J, Franchini G. ALVAC-SIV-gag-pol-env-based vaccination and macaque major histocompatibility complex class I (A*01) delay simian immunodeficiency virus SIVmac-induced immunodeficiency. J Virol. 2002;76:292–302. doi: 10.1128/JVI.76.1.292-302.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heller KN, Gurer C, Munz C. Virus-specific CD4+ T cells: ready for direct attack. J Exp Med. 2006;203:805–808. doi: 10.1084/jem.20060215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sitz KV, Loomis-Price LD, Ratto-Kim S, Kenner JR, Sau P, Eckels KH, Redfield RR, Birx DL. Delayed-type hypersensitivity skin testing using third variable loop peptides identifies T lymphocyte epitopes in human immunodeficiency virus-infected persons. J Infect Dis. 1997;176:1085–1089. doi: 10.1086/516517. [DOI] [PubMed] [Google Scholar]
- 37.Nemes E, Bertoncelli L, Lugli E, Pinti M, Nasi M, Manzini L, Manzini S, Prati F, Borghi V, Cossarizza A, Mussini C. Cytotoxic granule release dominates gag-specific CD4+ T-cell response in different phases of HIV infection. AIDS. 2010;24:947–957. doi: 10.1097/QAD.0b013e328337b144. [DOI] [PubMed] [Google Scholar]
- 38.Appay V, Zaunders JJ, Papagno L, Sutton J, Jaramillo A, Waters A, Easterbrook P, Grey P, Smith D, McMichael AJ, Cooper DA, Rowland-Jones SL, Kelleher AD. Characterization of CD4(+) CTLs ex vivo. J Immunol. 2002;168:5954–5958. doi: 10.4049/jimmunol.168.11.5954. [DOI] [PubMed] [Google Scholar]
- 39.Norris PJ, Moffett HF, Yang OO, Kaufmann DE, Clark MJ, Addo MM, Rosenberg ES. Beyond help: direct effector functions of human immunodeficiency virus type 1-specific CD4(+) T cells. J Virol. 2004;78:8844–8851. doi: 10.1128/JVI.78.16.8844-8851.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zaunders JJ, Dyer WB, Wang B, Munier ML, Miranda-Saksena M, Newton R, Moore J, Mackay CR, Cooper DA, Saksena NK, Kelleher AD. Identification of circulating antigen-specific CD4+ T lymphocytes with a CCR5+, cytotoxic phenotype in an HIV-1 long-term nonprogressor and in CMV infection. Blood. 2004;103:2238–2247. doi: 10.1182/blood-2003-08-2765. [DOI] [PubMed] [Google Scholar]
- 41.Musey L, Hughes J, Schacker T, Shea T, Corey L, McElrath MJ. Cytotoxic-T-cell responses, viral load, and disease progression in early human immunodeficiency virus type 1 infection. N Engl J Med. 1997;337:1267–1274. doi: 10.1056/NEJM199710303371803. [DOI] [PubMed] [Google Scholar]
- 42.Malhotra U, Holte S, Zhu T, Delpit E, Huntsberry C, Sette A, Shankarappa R, Maenza J, Corey L, McElrath MJ. Early induction and maintenance of Env-specific T-helper cells following human immunodeficiency virus type 1 infection. J Virol. 2003;77:2663–2674. doi: 10.1128/JVI.77.4.2663-2674.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Addo MM, Yu XG, Rathod A, Cohen D, Eldridge RL, Strick D, Johnston MN, Corcoran C, Wurcel AG, Fitzpatrick CA, Feeney ME, Rodriguez WR, Basgoz N, Draenert R, Stone DR, Brander C, Goulder PJ, Rosenberg ES, Altfeld M, Walker BD. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J Virol. 2003;77:2081–2092. doi: 10.1128/JVI.77.3.2081-2092.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frahm N, Korber BT, Adams CM, Szinger JJ, Draenert R, Addo MM, Feeney ME, Yusim K, Sango K, Brown NV, SenGupta D, Piechocka-Trocha A, Simonis T, Marincola FM, Wurcel AG, Stone DR, Russell CJ, Adolf P, Cohen D, Roach T, StJohn A, Khatri A, Davis K, Mullins J, Goulder PJ, Walker BD, Brander C. Consistent cytotoxic-T-lymphocyte targeting of immunodominant regions in human immunodeficiency virus across multiple ethnicities. J Virol. 2004;78:2187–2200. doi: 10.1128/JVI.78.5.2187-2200.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Novitsky V, Flores-Villanueva PO, Chigwedere P, Gaolekwe S, Bussman H, Sebetso G, Marlink R, Yunis EJ, Essex M. Identification of most frequent HLA class I antigen specificities in Botswana: relevance for HIV vaccine design. Hum Immunol. 2001;62:146–156. doi: 10.1016/s0198-8859(00)00236-6. [DOI] [PubMed] [Google Scholar]
- 46.Orentas RJ, Hildreth JE, Obah E, Polydefkis M, Smith GE, Clements ML, Siliciano RF. Induction of CD4+ human cytolytic T cells specific for HIV-infected cells by a gp160 subunit vaccine. Science. 1990;248:1234–1237. doi: 10.1126/science.2190315. [DOI] [PubMed] [Google Scholar]
- 47.Soghoian DZ, Streeck H. Cytolytic CD4(+) T cells in viral immunity. Expert Rev Vaccines. 2010;9:1453–1463. doi: 10.1586/erv.10.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Norris PJ, Rosenberg ES. CD4(+) T helper cells and the role they play in viral control. J Mol Med. 2002;80:397–405. doi: 10.1007/s00109-002-0337-3. [DOI] [PubMed] [Google Scholar]
- 49.O'Rourke SM, Schweighardt B, Phung P, Fonseca DP, Terry K, Wrin T, Sinangil F, Berman PW. Mutation at a single position in the V2 domain of the HIV-1 envelope protein confers neutralization sensitivity to a highly neutralization-resistant virus. J Virol. 2010;84:11200–11209. doi: 10.1128/JVI.00790-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Novitsky V, Cao H, Rybak N, Gilbert P, McLane MF, Gaolekwe S, Peter T, Thior I, Ndung'u T, Marlink R, Lee TH, Essex M. Magnitude and frequency of cytotoxic T-lymphocyte responses: identification of immunodominant regions of human immunodeficiency virus type 1 subtype C. J Virol. 2002;76:10155–10168. doi: 10.1128/JVI.76.20.10155-10168.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Masemola A, Mashishi T, Khoury G, Mohube P, Mokgotho P, Vardas E, Colvin M, Zijenah L, Katzenstein D, Musonda R, Allen S, Kumwenda N, Taha T, Gray G, McIntyre J, Karim SA, Sheppard HW, Gray CM. Hierarchical targeting of subtype C human immunodeficiency virus type 1 proteins by CD8+ T cells: correlation with viral load. J Virol. 2004;78:3233–3243. doi: 10.1128/JVI.78.7.3233-3243.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Douek DC, Brenchley JM, Betts MR, Ambrozak DR, Hill BJ, Okamoto Y, Casazza JP, Kuruppu J, Kunstman K, Wolinsky S, Grossman Z, Dybul M, Oxenius A, Price DA, Connors M, Koup RA. HIV preferentially infects HIV-specific CD4+ T cells. Nature. 2002;417:95–98. doi: 10.1038/417095a. [DOI] [PubMed] [Google Scholar]
- 53.Sandstrom E, Nilsson C, Hejdeman B, Brave A, Bratt G, Robb M, Cox J, Vancott T, Marovich M, Stout R, Aboud S, Bakari M, Pallangyo K, Ljungberg K, Moss B, Earl P, Michael N, Birx D, Mhalu F, Wahren B, Biberfeld G. Broad immunogenicity of a multigene, multiclade HIV-1 DNA vaccine boosted with heterologous HIV-1 recombinant modified vaccinia virus Ankara. J Infect Dis. 2008;198:1482–1490. doi: 10.1086/592507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kibuuka H, Kimutai R, Maboko L, Sawe F, Schunk MS, Kroidl A, Shaffer D, Eller LA, Kibaya R, Eller MA, Schindler KB, Schuetz A, Millard M, Kroll J, Dally L, Hoelscher M, Bailer R, Cox JH, Marovich M, Birx DL, Graham BS, Michael NL, de Souza MS, Robb ML. A phase 1/2 study of a multiclade HIV-1 DNA plasmid prime and recombinant adenovirus serotype 5 boost vaccine in HIV-Uninfected East Africans (RV 172) J Infect Dis. 201:600–607. doi: 10.1086/650299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Catanzaro AT, Koup RA, Roederer M, Bailer RT, Enama ME, Moodie Z, Gu L, Martin JE, Novik L, Chakrabarti BK, Butman BT, Gall JG, King CR, Andrews CA, Sheets R, Gomez PL, Mascola JR, Nabel GJ, Graham BS. Phase 1 safety and immunogenicity evaluation of a multiclade HIV-1 candidate vaccine delivered by a replication-defective recombinant adenovirus vector. J Infect Dis. 2006;194:1638–1649. doi: 10.1086/509258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Van Rompay KK, Abel K, Lawson JR, Singh RP, Schmidt KA, Evans T, Earl P, Harvey D, Franchini G, Tartaglia J, Montefiori D, Hattangadi S, Moss B, Marthas ML. Attenuated poxvirus-based simian immunodeficiency virus (SIV) vaccines given in infancy partially protect infant and juvenile macaques against repeated oral challenge with virulent SIV. J Acquir Immune Defic Syndr. 2005;38:124–134. doi: 10.1097/00126334-200502010-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haynes BF. Case Control Study of the RV144 Trial for Immune Correlates: The Analysis and Way Forward; Presented at: AIDS Vaccine Conference, 2011; Bangkok, Thailand. 2011. PL01.04. [Google Scholar]
- 58.Vaccari M, Poonam P, Franchini G. Phase III HIV vaccine trial in Thailand: a step toward a protective vaccine for HIV. Expert Rev Vaccines. 2010;9:997–1005. doi: 10.1586/erv.10.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.