Abstract
Tissues of the mucosa are lined by an epithelium that provides barrier and transport functions. It is now appreciated that inflammatory responses in IBD are accompanied by striking shifts in tissue metabolism. Here we examined global metabolic consequences of mucosal inflammation employing both in vitro and in vivo models of disease. Initial analysis of the metabolic signature elicited by inflammation in epithelial models and in colonic tissue isolated from murine colitis demonstrated that levels of specific metabolites associated with cellular methylation reactions are significantly altered by model inflammatory systems. Furthermore, expression of enzymes central to all cellular methylation, SAM synthetase and SAH hydrolase, are increased in response to inflammation. Subsequent studies showed that DNA methylation is substantially increased during inflammation and that epithelial NF-κB activity is significantly inhibited following treatment with a reversible SAH hydrolase inhibitor, DZ2002. Finally, these studies demonstrated that inhibition of cellular methylation in a murine model of colitis results in disease exacerbation while folate supplementation to promote methylation partially ameliorates the severity of murine colitis. Together, these results identify a global change in methylation, which during inflammation, translates to an overall protective role in mucosal epithelia.
Introduction
The inflammatory bowel diseases (IBD), including both Crohn’s disease and ulcerative colitis, are debilitating diseases of unknown etiology (1) but a combination of genetic and environmental factors are thought to be involved in disease pathology. Recent evidence suggests that IBD likely results from dysregulated immune responses to luminal triggers. Intestinal epithelial cells are a crucial cell type in the maintenance of colonic tissue homeostasis and IBD is characterized by a breakdown of the intestinal epithelial barrier leading to increased exposure of the mucosal immune system to antigenic luminal material. This exposure leads to inflammation and increased breakdown of the epithelial barrier (1–3). It is now appreciated that ongoing inflammation leads to significant shifts in tissue metabolism. These shifts result, at least in part, from perturbation of the vasculature (4, 5) leading to the decreased supply of oxygen and nutrients (6–8). The expression of increased levels of cytokines and chemokines initiates the migration of innate immune cells such as neutrophils, macrophages, and dendritic cells into the mucosal tissues (9). The recruitment of inflammatory cells increases the local immune response, as well as the oxygen and nutrient demand of the tissue, exacerbating the metabolic changes associated with IBD. IBD almost certainly elicits changes in a number of cellular metabolic pathways, however, the metabolic shifts induced in the colonic epithelia during the inflammatory process have not been elucidated in a systematic manner.
Studies to date indicate that alterations of cellular methylation have important implications in inflammation and immune responses (10). These studies have shown that methylation, through epigenetic mechanisms and protein modification, has important consequences for these processes, yet the molecular mechanisms of such changes remain poorly understood. Cellular methylation reactions include modification of DNA, RNA, proteins and lipids. These reactions all require a methyl donor for the modification of the target. The methyl donor for the majority of these reactions is S-adenosylmethionine (SAM) (11). SAM is distributed throughout the cell to act as donor for the various methyltransferases. The donation of its methyl group produces S-adenosylhomocysteine (SAH) from SAM. SAH is a potent inhibitor of methyltransferase enzymes because these enzymes have a higher affinity for SAH (12–14) and SAH is rapidly converted to homocysteine and adenosine by SAH hydrolase. Therefore, inhibition of SAH hydrolase represents a powerful means of inhibiting cellular methylation reactions (15). It has been known for a number of years that inhibition of methylation had immunomodulatory properties (16), but mechanisms remain poorly understood. This led to the development of more specific and less toxic SAH inhibitors for use in animal models. The SAH hydrolase inhibitor DZ2002 [methyl 4-(adenin-9-yl)-2-hydroxybutanoate] has been found to have immunosuppressive actions and ameliorate disease in a number of animal models (17–21). One mechanism for the immuno-suppressive action of DZ2002 was shown to be the cell type-specific inhibition of NF-κB activation (19). These studies demonstrate that cellular methylation reactions play an important role in chronic inflammatory disease, such as IBD.
The hypothesis of the present study was that ongoing mucosal inflammatory responses are characterized by specific shifts in metabolic profiles. To define these principles, an NMR-based metabolomics approach was utilized to elucidate the metabolic changes associated with murine models of IBD and in cellular models of mucosal inflammation. This analysis revealed that a number of metabolites associated with cellular methylation reactions are significantly impacted by ongoing inflammation. Dovetailing these findings with microarray analysis of epithelial inflammation models, we found that the expression of enzymes central to cellular methylation are significantly altered during mucosal inflammation. We confirmed these findings at the molecular level and extended these studies to a murine model of colitis. These results indicate that methylation reactions play an important role in mucosal inflammation.
Materials and Methods
Cell culture
Human intestinal epithelial cells (T84) and HeLa epithelial cells were grown and maintained in T175 cell culture flasks (Costar Corp., Cambridge MA) as described previously (22). DZ2002 (Diazyme, San Diego, CA) was prepared as described (19). Where indicated, DZ2002 was added at the indicated concentrations by pre-incubation for1 hr prior to indicated treatments.
Western Blot Analysis
Cells were harvested by scraping, pelleted by centrifugation, washed with ice-cold phosphate-buffered saline, and lysed by sonication in Tris lysis buffer (150 mM NaCl; 20 mM Tris, pH 5.5; 1 mM EDTA; 1 mM EGTA; 1% Triton X-100). Similarly, snap frozen colon tissues samples were sonicated in Tris buffer. Protein concentration was assessed by the bicinchoninic acid (BCA) assay following the manufacturer’s instructions (Thermo Scientific) in order to ensure equal protein loading of each preparation. Proteins were separated by SDS-PAGE electrophoresis and transferred to PVDF membrane (Bio-Rad Laboratories, Hercules, CA) for immunoblotting. Antibodies utilized for this study were: anti-SAM synthetase (1:1000, Abcam), anti-SAH hydrolase (1:1000, gift of Dr. Doris Kloor), anti-IκBα (1:2000, Cell Signaling Technologies), anti-β–actin (1:10000, Abcam), anti-β–tubulin (1:10000, Abcam). Proteins were visualized using the SuperSignal detection substrate (Thermo Scientific).
RNA isolation and transcriptional analysis
RNA was isolated using Trizol (Invitrogen) and cDNA synthesized as described previously(10). Potential contaminating genomic DNA was digested using Turbo DNA-free (Ambion, Austin, TX, USA). The mRNA profile of intestinal epithelial cells (T84) subjected to modeled inflammation (IFN-γ at 10ng/ml for 6 or 18h) was compared to that of control cells exposed to vehicle only using genechip expression arrays (Affymetrix, Inc.) as described previously (23). Semi-quantitative and real-time PCR were performed using increasing numbers of cycles of 95°C for 45 sec, 59°C for 35 sec and 72°C for 45 sec and a final extension time of 7 min. The following primers were used to quantify expression in intestinal epithelial cells: MAT2A (SAM synthetase): forward 5'-ATACAATCTACCACCTACAGCC-3' and reverse 5'-CATAAGAGACCTGAACAAGAACC-3'; AHCY (SAH hydrolase): forward 5'-GGTATCGGTTGAAGAATGG-3' and reverse 5'- GGTACTTGTCTGGATGGGTC-3'; DNMT3B: forward 5’-ATAAGACACCCCCTCAAACC-3’ and reverse 5’-TTCCCGTTCTCCCTAAAAAC-3’; IL-8: forward 5'-ATGACTTCCAAGCTGGCCGTGG-3' and reverse 5'-CATAATTTCTGTGTTGGCGCAGTGTGG-3'; TNF-α: forward 5'- TCCTTCAGACACCCTCAACC-3' reverse 5'- AGGCCCCAGTTTGAATTCTT -3'; β-actin: forward 5’-GCACTCTTCCAGCCTTCCTTCC-3’ and reverse 5’-CAGGTCTTTGCGGATGTCCACG-3’. Transcript levels and fold change in mRNA were determined as described previously (24). Intron-spanning primer pairs for were designed by using primer3 software (http://frodo.wi.mit.edu/). Primer properties and secondary structures including hairpins, self-dimers, and cross-dimers were evaluated in a second step using Netprimer software (http://www.premierbiosoft.com/netprimer). Samples were controlled for β-actin.
pNFκB-Luc plasmid transfection and luciferase reporter assay
HeLa cells were passaged into 24 well plates and allowed to attach for 24 hours. Transient transfection of Hela cells and assessment of luciferase activity was carried out as described previously (22).
DSS colitis model
DSS colitis was induced with a modification of the technique of Okayasu, et al. (25). Colitis was induced on day 0 by the addition of 3% DSS (MW = 36,000–50,000, MP Biomedicals, Illkirch, France) solution in drinking water. Control animals received water alone. DZ2002 was delivered i.p. at a concentration of 50 mg/kg/day (19) beginning one day prior (day -1) to DSS administration. Folic acid (Sigma Aldrich) was delivered i.p. at a concentration of 50 mg/kg beginning one day prior (day -1) to DSS administration and then every other day for the course of the experiment. BAY 11-7082 (Cayman Chemical) was administered i.p. at a concentration of 5mg/kg beginning one day prior (day -1) to DSS administration and then every other day for the course of the experiment, as previously described (26).
Quantification of IFN-γ in murine colonic tissue
For cytokine analysis, colonic tissue was extracted in Tris lysis buffer by sonication and protein homogenates were stored at −80°C until use. Tissue concentrations of IFN-γ were measured in colonic protein extracts using a pro-inflammatory cytokine screen (Meso Scale Discovery). Assays were performed per manufacturer’s instructions and analyzed using a Sector Imager 2400 (Meso Scale Discovery). IFN-γ concentrations were normalized to total protein concentration.
Flow Cytometry
5-methyl cytidine staining was quantified using flow cytometry essentially as described by Watson et al (27). Briefly, cells were trypsinised and fixed in 70% MeOH. Following fixation, cells were incubated in 1M HCl for 1 hr at 37°C. Cells were incubated with anti-5-methyl cytidine Ab (Abcam) or IgG control at identical concentrations followed by application of AlexaFluor 555 (Invitrogen) secondary antibody. Cells were analyzed using the FACS® Canto system (Beckton-Dickinson Immunocytometry Systems, San José, CA). Post-analyses were performed using FLOWJo software (Tree Star Inc, Ashland, OR).
Immunocytochemistry
T84 human epithelial cells were cultured on glass coverslips and fixed in ice-cold methanol followed by permeablization in 0.2% Triton X-100. Cells were incubated in 1M HCl for 1 hr at 37°C. Cells were incubated with blocking buffer (1% BSA) then incubated with anti-5-methyl cytidine Ab (Abcam) or IgG negative control followed by AlexaFluor 555 secondary Ab. Coverslips were mounted using ProLong Gold antifade (Invitrogen). Staining was visualized using Axioplan Zeiss microscope equipped with an AxioCam MRc5 system. Imaging software for data capture was Axiovision 4.6.
Immunohistochemistry
Blank sections were cut from formalin fixed, paraffin embedded samples. Following deparaffinization, slides were subjected to antigen retrieval utilizing a sodium citrate buffer. Sections were permeabilized using 2% Triton X-100 followed by blocking in 1% BSA. Sections were incubated with anti-5-methyl cytidine Ab followed by incubation with AexaFluor 555 secondary Ab. Sections were counterstained with DAPI (Invitrogen). Images were visualized and captured as described above.
Extraction Protocols for Metabolic NMR
Collected cell pellets or frozen tumor specimens were homogenated in ice-cold 8% perchloric acid (PCA) as described previously (28, 29). Briefly, after centrifugation, the supernatants (containing hydrophilic metabolites) were collected and pH was adjusted to pH=7 using KOH. The potassium perchlorate was removed by centrifugation, and the hydrophilic fraction was lyophilized overnight. The cell and tissue pellets (after the first centrifugation), which contained the lipophilic metabolites, were re-dissolved in water and pH was adjusted (7.0). The lipophilic fraction was lyophilized overnight. For the cell experiments only, previously collected media were lyophilized overnight after adjusting pH. The dried hydrophilic cell and tissue extracts were re-dissolved in 0.5 mL of deuterium oxide (D2O), transferred into 5-mm NMR tubes and used for 1H-, 31P- and 13C-NMR analysis. The medium extracts were re-dissolved in 1 mL of D2O, transferred to 5-mm NMR tubes and underwent 1H- and 13C-NMR analysis. The cell and tissue lipid extracts were re-dissolved in 1.2 ml of deuterated chloroform/deuterated methanol mixture (2:1 vol/vol).
NMR Analysis on Cell and Tissue Extracts
All 1H- and 13C-NMR spectra were obtained at the Bruker 500 MHz DRX NMR spectrometer using an inverse Bruker 5-mm TXI probe. To assist 1H-NMR peak assignment and metabolite identification in cell, media and biopsy extracts, two-dimensional (2D)-H/C-HSQC (heteronuclear single quantum correlation) NMR techniques were used. All spectra were Fourier transformed and lactate (Lac3, CH3) was used as an internal chemical reference for both carbon (21 ppm) and proton (1.32 ppm) axes. For metabolite quantification, one dimensional 1H-NMR spectra were obtained from each sample, with a standard water pre-saturation pulse program “zgpr”. A thin sealed glass capillary, containing TSP, was placed in each 5-m m tube prior to 1H-NMR experiments. The total number of acquisitions varied from 40 to 128. Conventional 1H acquisition parameters were: power level pl1=20dB; power angle p1=6.3 msec (90 degree pulse); power level for water pre-saturation pl9=77 dB; water suppression at O1P=4.76 ppm; spectral width SW=5000 MHz; and the pulse delay of 12.75 s (calculated as 5*T1) was applied between acquisitions. The TSP from the reference capillary served as a chemical shift (0 ppm) and proton metabolite concentration reference. The cell and media extracts subsequently underwent 13C-NMR analysis (with proton decoupling). The total number of scans was 24,000 and 800 for each cell and medium extract, respectively. The concentration of [3-13C] lactate at 21 ppm (calculated from two satellite peaks at the 1H-NMR spectra) served as an internal carbon concentration reference.
Before 31P-NMR analysis, 100 mmol/L EDTA was added to the extracts to complex divalent cations (for ATP and ADP quantification). All 31P-NMR spectra (with proton decoupling) on cell and tissue extracts were obtained at the Bruker 300 MHz Avance NMR spectrometer using a Bruker QNP probe. The total number of scans was 8,000–16,000 per extract. A thin capillary glass containing 2.3 mmol/L methyl-diphosphoric acid (MDPA) served as a chemical shift (18.6 ppm) and phosphor metabolite concentration reference.
Histological scoring
Histological examination was performed on three samples of the distal colon. Samples were fixed in 4% formalin before staining with hematoxylin and eosin. All histological quantitation was performed in a blinded fashion, using a previously described scoring system (30). Severity of inflammation: rare inflammatory cells in the lamina propria (0); increased numbers of inflammatory cells (1); confluence of inflammatory cells extending into the submucosa (2); transmural extension of the inflammatory cell infiltrate (3). Extend of injury: nil mucosal damage (0); discrete lymphoepithelial lesions (1); surface mucosal erosion (2); and widespread mucosal ulceration and extension through deeper bowel wall structures (3). The scores of the two parameters were added and the mean was calculated. Maximum possible score was 3.
Statistics
Unpaired T-test and/or one-way analysis of variance (ANOVA) test were used to determine differences between groups, as indicated. The significance level was set at p<0.05 for all tests (Prism 4, GraphPad Software, San Diego, CA).
Results
Metabolic Profiling of Modeled Inflammation Identifies a “Methylation Signature”
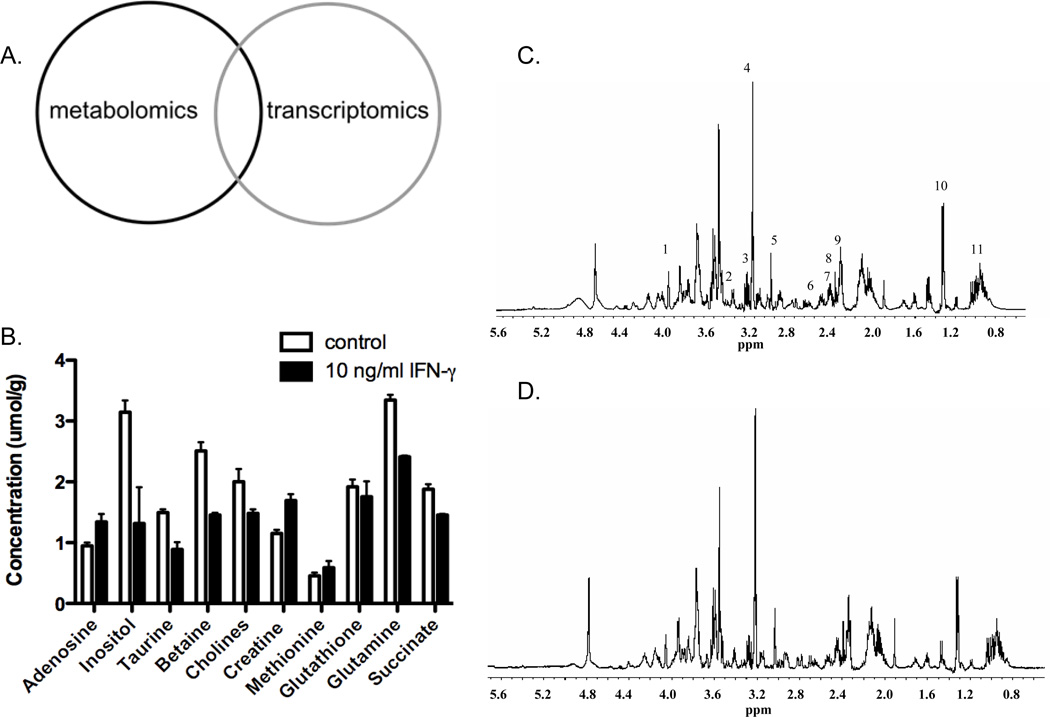
Previous studies have shown that inflammatory lesions associated with IBD are the site of significant metabolic changes (31–33). These changes in metabolism, however, have not been studied in a systematic manner. Here, we sought to define the metabolic changes associated with modeled inflammation. Since previous reports strongly implicated intestinal epithelial cells (23, 32), we utilized T84 cells, a well characterized human intestinal epithelial cell line. Initial experiments involved subjecting T84 human colonic epithelial cells to IFN-γ for 48 hrs. Additionally, cells were cultured in the presence of [1-13C]-glucose for the final 4 hours of culture in order to obtain data on the fate of glucose during inflammation. Following the incubation period, cells were immediately harvested and prepared for magnetic resonance spectroscopy (MRS) analysis. The media was also preserved and subjected to metabolite quantification. It has been previously shown that intestinal inflammation results in a state termed ‘inflammatory hypoxia’ (32). Additionally, it has been previously shown that treatment of human epithelial cells with pro-inflammatory cytokines leads to an increase in glycolytic rate (34, 35). The present study corroborates these studies with the finding that treatment of T84 cells with IFN-γ results in an increase in glucose uptake and lactate production, as well as intracellular adenosine levels, all of which are indicative of hypoxia (Figure 1 A; Figure 1 and Table 1, supplementary data). Importantly, we also found changes in a number of metabolites that participate in (betaine, cholines) or are products of (creatine, taurine, adenosine) cellular methylation reactions (Figure 1 A; Figure 1 and Table 1, supplementary data). These results strongly implicate methylation reactions as a contributing factor to mucosal inflammatory responses. Moving forward, we compared these results with those obtained from microarray analysis (Figure 1B).
Figure 1. Induction of SAM synthetase and SAH hydrolase during modeled inflammation.
T84 human epithelial cells were cultured in the presence of IFN-γ (10 ng/ml) for the indicated times. Panel A displays absolute metabolite concentrations for a subset of the measured metabolites from all treatment groups. Panel B displays a heat map depiction of gene expression data from microarray analysis showing both control (IFN-γ inducible) and methylation-related genes. Panel C displays real time PCR and western blot analysis for SAM synthetase and panel D shows same analysis for SAH hydrolase. PCR data (n=3, *p<0.05, ***p<0.005). Western blot (n=2).
The Central Enzymes of Cellular Methylation Reactions are Induced by IFN-γ
After finding that key metabolites of methylation reactions were changed in our in vitro model of mucosal inflammation, we compared these metabolomic endpoints with microarray data generated in T84 intestinal epithelial cells following exposure to IFN-γ for either 6 or 18 hrs. As shown in Figure 1B, a number of genes strongly implicated in IFN-γ signaling (e.g. CXL11, CXCL10 and IDO1) were highly induced by IFN-γ and thus served as positive controls for this microarray. More relevant for the present study, this microarray also revealed strong induction of several methylation-related gene transcripts, including S-adenosylmethionine (SAM) synthetase (MAT2A), S-adenosylhomocysteine hydrolase (AHCY), O-6-methylguanine-DNA methyltransferase (MGMT), and guanidinoacetate N-methyltransferase (GAMT). MAT2A, the enzyme responsible for SAM production, was among the most highly up-regulated genes in the array (Fig. 1B). Given its central role in all methylation reactions (14), we pursued the induction of MAT2A by IFN-γ. To confirm this microarray result, we performed real time PCR using primers specific for the MAT2A and examined SAM synthetase protein levels by immuno-blot analysis. These results indicate that both MAT2A expression and SAM synthetase protein are increased in epithelial cells following exposure to IFN-γ (Fig. 1C, for both p<0.05 by ANOVA). In a similar manner, we also examined the expression of SAH hydrolase. This enzyme plays a key role in methylation reactions by catabolizing SAH, a potent methyltransferase inhibitor, to homocysteine and adenosine (14). These studies revealed that SAH hydrolase expression and protein levels are also up-regulated during inflammation (Fig. 1D). Taken together, these results and that of metabolomic analysis provide strong evidence that cellular methylation pathways are important in the inflammatory process.
Increased DNA Methylation in a Model of Epithelial Inflammation
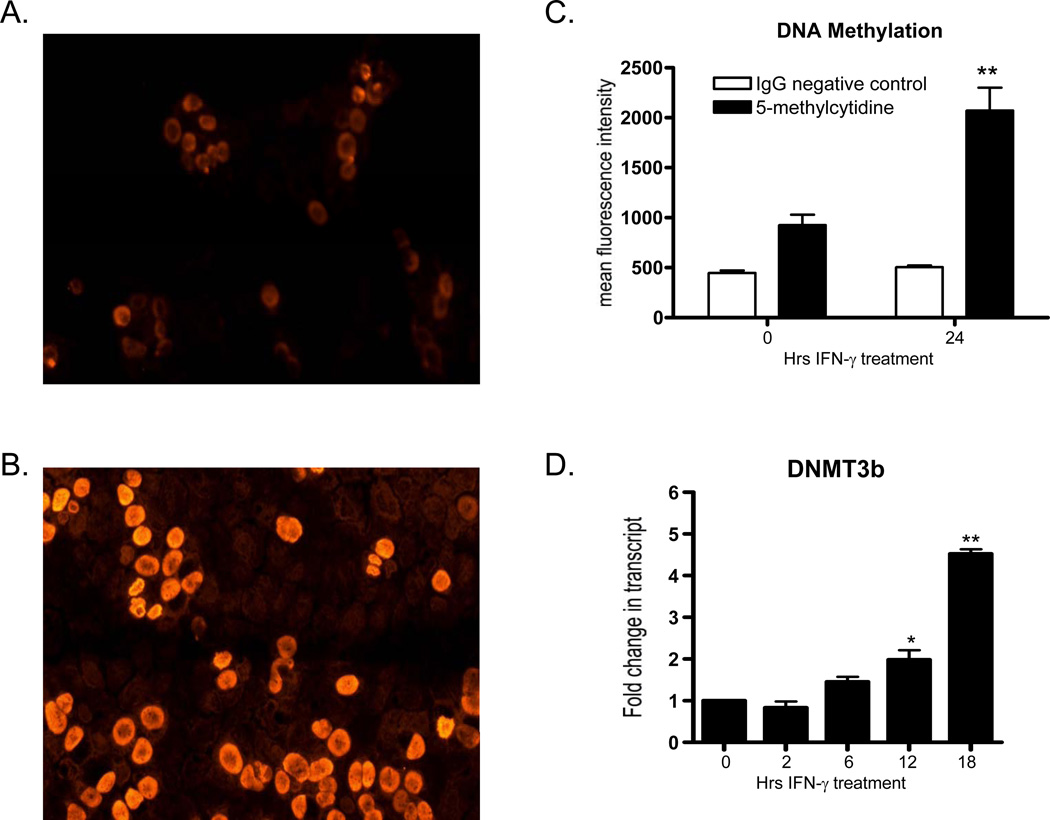
Having determined that key enzymes of cellular methylation are up-regulated during inflammation, we next examined functional endpoints of these reactions, namely DNA methylation. For these studies, T84 epithelial cells were exposed to IFN-γ (10ng/ml) for the indicated times and analyzed levels of 5’-methylcytidine, which is indicative of DNA methylation, utilizing fluorescent microscopy and flow cytometry. As depicted in Figure 2A, untreated T84 cells exhibit a small amount of nuclear 5’-methylcytidine staining. In stark contrast, as shown in Figure 2B, nuclear staining for 5’-methylcytidine is substantially increased following exposure to IFN-γ for 24 hr. This result was corroborated using flow cytometry. As depicted in Figure 2C, incubation of T84 cells for 24 hr with IFN-γ resulted in a 2.23±0.02) increase in 5’-methylcytidine (p<0.01). An insight into a potential mechanism for the increase in DNA methylation came when the expression of DNMT3b, a DNA methyltransferase responsible for de novo DNA methylation (14), was examined in cells exposed to IFN-γ. As shown in Figure 2D, DNMT3b expression is significantly induced in T84 cells in an inflammatory setting (p<0.01 by ANOVA). These data indicate that DNA methylation is increased in an in vitro model of inflammation.
Figure 2. Increase in DNA methylation during inflammation.
T84 human epithelial cells were cultured in the presence of IFN-γ (10 ng/ml) for the indicated times. Panel A shows nuclear staining using fluorescent microscopy utilizing an antibody specific for 5’-methylcytidine. Panel B displays nuclear staining for 5’-methylcytidine in T84 cells incubated with IFN-γ for 24 hr. Panel C shows flow cytometry data for control or IFN-γ exposed T84 cells using a anti-5’-methylcytidine antibody or an isotype matched negative control (n=3). Panel D depicts real time PCR data for DNMT3b in control and IFN-γ exposed T84 cells for 2, 6, 12, and 18 hr (n=3, *p<0.05, **p<0.01).
NF-κB Activity is Modulated in a Methylation-dependent Manner
NF-κB is one of the master regulators of pro-inflammatory gene expression (for review see (36)). NF-κB upregulates a number of genes including pro-inflammatory cytokines, chemokines, and adhesion molecules, and can induce specific sets of genes in response to a particular triggers (37). NF-κB has been shown by EMSA and IHC studies to be highly activated at the site of inflammation in a number of diseases, including IBD (36), as wells as activated under conditions of hypoxia (38, 39). Additionally, NF-κB has been found to be protective of the mucosal epithelium in murine disease models (40, 41). Interestingly, NF-κB has been shown to elicit epigenetic modifications of its inducible genes (42). Based on these studies and our preliminary data suggesting a link between methylation and the inflammatory response in epithelial cell models, we examined NF-κB activity using a luciferase reporter assay following inhibition of methylation by DZ2002, a potent reversible SAH hydrolase inhibitor (21). As shown in Figure 3A, IFN-γ elicited a greater than 8-fold increase in NF-κB activity at the highest concentrations (p<0.01). Pre-treatment with DZ2002 effectively inhibited NF-κB activity in a concentration-dependent manner (Figure 3A). As IFN-γ does not represent a canonical activator of NF-κB, we also studied the impact of DZ2002 on NF-κB following treatment with TNF-α. As shown in Figure 3B, while TNF-α is a much more potent NF-κB activator (maximal 58.02±9.66, p<0.001), inhibition of methylation significantly repressed such induction, likely indicating that this inhibition of NF-κB by DZ2002 is not stimulus dependent.
Figure 3. NF-κB activity is inhibited by DZ2002.
Panels A and B, cells were cultured in 24 well plates for 24 hr then transfected with an NF-κB luciferase reporter construct. Cells were then treated with DZ2002 or vehicle for 1 hr at the indicated concentrations followed by addition of IFN-γ at the indicated concentrations (Panel A) or TNF-α (10 ng/ml) (Panel B). Cells were incubated for 12hr and immediately prepared for analysis of luciferase expression (n=3, data are expressed as mean ± SD, *p<0.05, **p<0.01, ***p<0.005). Panel C, western blot analysis of IKBα following exposure to IFN-γ (10 ng/ml) or DZ2002 (100 µM) and IFN-γ for the indicated time periods (n=3). Panel D–E, real time PCR analysis of TNF-α (D) and IL-8 (E) expression in T84 cells following IFN-γ treatment (10 ng/ml) or DZ2002 and IFN-γ for indicated time periods. Values are expressed as fold increase over vehicle treated control (n=3; *p<0.05, **p<0.01, ***p<0.001).
To confirm that DZ2002 directly impacts NF-κB, we next examined the fate of the NF-κB inhibitory protein, IκBα, in response to DZ2002. As shown in Figure 3C, IκBα levels are decreased in response to IFN-γ treatment. This response is abrogated with pretreatment with DZ2002 indicating that the inhibition of NF-κB is mediated, at least in part, through modulation of the inhibitory protein IκBα. To confirm these results at a functional level, we examined the expression of TNF-α and IL-8, two known NF-κB target genes. As depicted in Figure 3D and 3E, expression of both TNF-α and IL-8 are increased in response to IFN-γ treatment. In support of our hypothesis, expression of both of these targets was diminished upon pretreatment with DZ2002 (p<0.05). These results strongly implicate methylation in NF-κB activation, at least in part, through the regulation of IκBα.
Metabolic Profiling of Experimental Colitis Identifies a “Methylation Signature”
Previous studies have shown that inflammatory lesions in mucosal inflammation (e.g. IBD) result in substantial metabolic changes and major shifts in metabolite supply and demand (10). Here, we sought to define the metabolic changes induced during inflammation in an epithelial-driven in vivo model of experimental colitis. For these purposes, we selected the DSS model. C57B6 mice were administered dextran sodium sulfate (DSS) or vehicle (water) alone for 5 days, after which mice were euthanized and colon tissue was immediately removed and frozen. Colons were extracted and prepared for MRS analysis. Figure 4 displays representative proton spectra for both control (panel A) and DSS treated (panel B) colonic tissue. These experiments revealed alterations in a number of metabolites associated with methylation reactions, including adenosine, betaine, choline, creatine, glutathione (p<0.05, n=6, Figure 4C, Table 2, supplementary data). In addition to an impact on metabolites involved in methylation, changes in several metabolites indicative of hypoxia (glucose, lactate, adenosine), consistent with previous work in experimental colitis models (23, 32, 33, 43). To confirm our in vitro experiments, we examined the parallel expression of SAM synthetase/SAH hydrolase and IFN-γ in colonic tissue extracts from control and DSS-treated mice harvested at two and four days of treatment. As shown in figure 4D, both SAM synthetase and SAH hydrolase expression were increased after 2 and 4 days of DSS administration. Likewise, tissue concentrations of IFN-γ were increased at day 4 of DSS treatment (Fig. 4E).
Figure 4. Metabolic analysis of murine colonic inflammation.
Wild type mice received DSS for 5 days. Tissue was extracted and used for MRS analysis of endogenous metabolites. Panel A–B representative proton spectra from control (A) and DSS-treated (3%, 6 days treatment) colonic tissue extracts. Numbered peaks are: 1, glucose; 2, inositol; 3, betaine; 4, total cholines; 5, total creatine; 6, total glutathione; 7, succinate; 8, lactate; 9, leucine, isoleucine, and valine. Panel C displays absolute metabolite concentrations for a subset of the measured metabolites for control and DSS-treated tissue. Panel D, western blot analysis of SAM synthetase and SAH hydrolase in colonic tissue extracts from control and DSS treated animals (3 animals per treatment). Panel E, MesoScale analysis of IFN-γ concentrations in colonic tissue from control (day 0) and DSS-treated animals (2 and 4 days of DSS) (n=4 animals per group).
Inhibition of Methylation Exacerbates Disease in a Mouse Model of Colitis
We next examined the impact of methylation-inhibition on DSS colitis outcomes. Given that the SAH hydrolase (AHCY)-null mouse is embryonic lethal (44) and that SAM synthetase is not easily inhibited pharmacologically (15), we reverted to a pharmacological approach using DZ2002. Our in vitro data revealed that modeled inflammation results in an increase in DNA methylation in epithelial models, as evidenced by an increase in 5-methylcytidine staining detected both by immunocytochemistry and flow cytometry (Figure 2). Based on these findings, we initially examined the extent of DNA methylation in response to DSS in the presence and absence of DZ2002 in vivo. As shown in Figure 5, treatment with DSS resulted in a significant increase in colonic DNA methylation. Colonic tissue from vehicle-treated animals displayed low levels of basal 5-methyl cytidine staining (Figure 5, column 1). By contrast, DSS induction resulted in a substantial increase in DNA methylation as indicated by the increase in 5-methyl cytidine and DAPI/5-methyl cytidine co-staining (see arrows in merged image, column 2). Treatment of animals with DZ2002 in combination with DSS significantly dampened DNA methylation as indicated by the decrease in both 5-methyl cytidine staining as well as DAPI/5-methyl cytidine co-localization (see arrows in merged image, column 3).
Figure 5. DSS treatment increases DNA methylation in vivo.
12-week old C57-BL6 mice were treated as described above. Tissue sections were prepared as previously described(32) followed by labeling with anti-5-methyl cytidine antibody (Abcam) and detection with anti-rabbit Alexafluor 555 secondary Ab. Tissues were counterstained with DAPI. Arrows in merged image indicate representative cells displaying staining for both 5-methyl cytidine and DAPI.
We next demonstrated that DZ2002, administered in combination with administration of DSS, exacerbated disease progression. As shown in Figure 6 A–C, inhibition of methylation in combination with DSS treatment resulted in significant decrease in weight (Figure 6A, p<.05), increased colon shortening (Figure 6B, p<0.001), and increase in disease activity as indicated by histological score (Figure 6C, p<0.001). Importantly, DZ2002 treatment alone resulted in no weight loss or colon shortening (Figure 6A and 6B), indicating that this inhibitor is not toxic at the doses administered. Additionally, these indicators of disease severity were confirmed by histology. As shown in Figure 6D, vehicle treated mice display normal tissue morphology, including intact epithelium and crypt structure. DSS-treated animals exhibited a deterioration in normal crypt architecture and epithelial cell depletion (Figure 6E) and animals treated with DZ2002 and DSS display an almost complete lack of normal tissue architecture and loss of the epithelium (Figure 6F). These results confirm that inhibition of methylation worsens disease progression in a murine model of colitis and strongly implicate that inflammation-associated methylation functions as an endogenous protective mechanism. Importantly, the finding that NF-κB inhibition exacerbates epithelial-driven colitis is consistent with recent literature (45, 46).
Figure 6. DSS colitis is enhanced by DZ2002.
12–18 week old C57-B6 mice were IP injected daily with vehicle (0.4% DMSO in PBS) or DZ2002 (50 mg/kg) beginning at day 0. From day 1, mice were administered water (control) or 3% DSS ad libitum for 6 days. Panel A displays percent weight loss; panel B displays colon length; panel C represents histological score (n=5, data are expressed as mean ± SD, *p<0.05, **p<0.01, ***p<0.005). Histological score determined as previously described (33). Panel D–E, H&E staining of tissue isolated from vehicle (D), DSS (E), and DSS + DZ2002 (F) treated animals.
In an attempt to clarify the role of the NF-κB pathway in the context of our findings, we administered the NF-κB inhibitor BAY 11-7082 to mice, alone or in combination with DSS. This inhibitor selectively and irreversibly inhibits NF-κB activation by blocking cytokine-induced phosphorylation of IκB-α without influencing constitutive IκB-α phosphorylation (47). While animals receiving BAY 11-7082 alone showed no symptomology compared to vehicle controls, mice receiving both BAY 11-7082 and DSS developed significantly more severe disease, to the extent that it was necessary that we humanely sacrifice animals prior to completion of the DSS time course (data not shown). These experiments do not provide direct evidence of a role for NF-κB activation in the protection of the mucosa in a DSS model of colitis. However, the results do corroborate our findings and those of others indicating that NF-κB is protective within the intestinal epithelium (40, 41, 48), and suggest that the endogenously protective effect of methylation may be due, at least in part, to epithelial NF-κB activation.
Augmentation with the Methyl Donor Folate Ameliorates Disease in a DSS Mouse Model of Colitis
Having shown that inhibition of methylation using DZ2002 in combination with administration of DSS worsens colitis in mice, we next determined if augmentation of methylation would be protective in the DSS model. For these experiments we administered folate in the same manner as the methylation inhibitor (i.e. systemic via i.p. injection) in order to make the experiments as comparable as possible. As shown in Figure 7 A–C, administration of folate in combination with DSS treatment resulted in an increase in weight (Figure 7A, p<.05), significantly decreased colon shortening (Figure 7B, p<0.05), and a significant decrease in disease activity as indicated by histological score (Figure 7C, p<0.01). Administration of folate alone had no effect on weight loss or colon length (Figure 7A and 7B). Once again, these indicators of disease severity were confirmed by histology. As shown in Figure 7D, vehicle treated mice display normal tissue morphology, including intact epithelium and crypt structure. DSS-treated animals exhibited a deterioration in normal crypt architecture and epithelial cell depletion (Figure 7E). Importantly, animals administered folate in combination with DSS display improved tissue architecture, diminished loss of the epithelium, and less inflammatory cell migration than DSS alone (Figure 6F). These results confirm that augmentation of methylation through the administration of folic acid ameliorates disease progression and provides further evidence that inflammation-associated methylation is protective in a murine colitic model. These data are in agreement with clinical studies demonstrating vitamin B12 and folate deficiencies are common in IBD patients (49–51), and suggest folate supplementation in IBD treatment. Additionally, a recent study demonstrated that a methyl-deficient diet exacerbates disease in an animal model of colitis (52).
Figure 7. DSS colitis is reduced in folic acid treated animals.
8–10 week old C57-B6 mice were IP injected every other day with vehicle (PBS) or folic acid (50 mg/kg) beginning at day 0. From day 1, mice were administered water (control) or 3% DSS ad libitum for 6 days. Panel A displays percent weight loss; panel B displays colon length; panel C represents histological score (n=4, data are expressed as mean ± SD, *p<0.05, **p<0.01, ***p<0.005). Histological score determined as previously described (33). Panel D–E, H&E staining of tissue isolated from vehicle (D), DSS (E), and DSS + folic acid (F) treated animals.
Discussion
This study aimed to identify metabolic changes associated with modeled intestinal inflammation, particularly related to the epithelium. As previous work had demonstrated specific shifts in metabolism during inflammation, we reasoned that such changes could be reflected on a more global basis. An amalgamated approach using NMR-based metabolomics and transcriptional arrays identified shifts in methylation-dependent pathways as a major metabolic fingerprint. These studies significantly extend previous work related to inflammation-associated metabolism and identify changes in methylation as a target signature within the epithelium during mucosal inflammation. Evidence is provided that shifts in methylation associated with inflammation serve an endogenously protective role in murine colitis.
Ongoing mucosal inflammatory responses are characterized by significant shifts in tissue metabolism (10). These changes include a shift toward a glycolytic phenotype and careful analysis has shown the development of significant hypoxia, termed “inflammatory hypoxia”, particularly prominent within the epithelium (53). Coinciding with inflammation-associated hypoxia is the stabilization of HIF (32, 53), and given the central role of HIF in most metabolic processes (54, 55), we reasoned that overall metabolism would be changed. In support of this hypothesis, global analysis of metabolism using MRS identified significant changes in a number of cellular metabolites. Many of these alterations are in agreement with results from our lab indicating the presence of “inflammatory hypoxia”. These include increased intracellular adenosine, increased glucose uptake and intracellular glucose, increased intracellular lactate and lactate export, and increased turnover of ATP (Figures 1 and 4; Supplemental Figure 1 and Supplemental Tables 1 and 2). Additionally, significant changes were observed in a number of lipid metabolites including monounsaturated fatty acids (MUFA), triacylglycerol (TAG), polyunsaturated fatty acids (PUFA), phosphatidyl choline, and phosphatidyl ethanolamine (Supplemental Tables 1 and 2), which may be indicative of dynamic alterations in cellular membranes in response to inflammation. Importantly, this analysis revealed a distinct and specific methylation fingerprint. Cellular methylation reactions include modification of DNA, RNA, proteins and lipids (56, 57). These reactions all require a methyl donor for the modification of the target. The methyl donor for the majority of these reactions is S-adenosylmethionine (SAM) (11). SAM is distributed within all cell and tissue compartments and functions as a methyl-donor for a number of different methyltransferases. The donation of methyl groups results in the generation of S-adenosylhomocysteine (SAH). Methyltransferase enzymes have a higher affinity for SAH than SAM, and thus, SAH functions as a potent feedback inhibitor (14). SAH is rapidly converted to homocysteine and adenosine by SAH hydrolase. Based on this methylation fingerprint, we performed parallel microarray analysis of modeled inflammation using a Th1 cytokine (IFN-γ) as a stimulus and discovered that SAM synthetase (MAT2A) and SAH hydrolase (AHCY) were among the top twenty highest IFN-γ-induced transcripts in T84 intestinal epithelia. The mammalian MAT2A gene promoter is relatively well-characterized. Previous studies have shown that the human MAT2A promoter is basally controlled by three tandem Sp1 sites (58). Under modeled inflammatory conditions (TNF-α activation), MAT2A is strongly induced through activation of AP-1 and NFκB (59). These studies support our findings of MAT2A induction by IFN-γ, a known activator of both NFκB and AP-1 (60). At present, essentially nothing is known about the regulation of the mammalian AHCY gene.
As a proof of principle, we extended these findings to a mucosal inflammation model. Given our understanding of inflammation-associated metabolic changes within the epithelium, we selected DSS colitis as an appropriate animal model to study colitis. DSS functions primarily as an epithelial irritant to drive permeability-induced colonic inflammation (61). Therefore, inhibition of SAH hydrolase represents a powerful means of inhibiting cellular methylation reactions (15). It has been known for several years that inhibition of methylation had immunosuppressive influences (16). This led to the development of more specific, reversible, and less toxic SAH inhibitors for use in animal models of inflammation. Utilizing these compounds, it was demonstrated that SAH hydrolase inhibition particularly down-regulates T cell activation and adaptive immune responses. One of these SAH hydrolase inhibitors, DZ2002, has been found to have potent immunosuppressive effects and ameliorates disease in a number of animal models including delayed-type hypersensitivity (20), arthritis (20), and EAE (19). While we do not know the exact mechanism by which DZ2002, we have shown that inhibition of methylation by DZ2002 inhibits NF-κB in vitro. NF-κB is a master regulator of the pro-inflammatory response, upregulating a number of genes including cytokines, chemokines, and adhesion molecules, as well as activated under conditions of hypoxia (62). A number of studies have shown that inhibition of NF-κB may be protective in animal models of IBD ((63–65)). Paradoxically, however, NF-κB activation has also been found to be protective in the context of the mucosal epithelium in murine disease models (40, 41). Our findings with DZ20002 support this hypothesis and demonstrate that parenteral administration of DZ2002 significantly inhibits tissue methylation and that such inhibition is associated with exacerbated DSS colitic responses. Additionally, augmentation of methylation through the administration of folate, ameliorates disease in this colitic model. Whether methylation inhibition would impact other models of intestinal inflammation in the same manner is not currently known.
Taken together, these studies provide a new and compelling role for methylation as an endogenously protective mechanism for mucosal inflammation. Additionally, the findings herein provide a tractable and potentially new therapeutic opportunities for methylation-dependent targets in mucosal diseases such as IBD.
Supplementary Material
Footnotes
The authors declare no financial interests in any of the work submitted here.
References
- 1.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 2.Baumgart DC, Carding SR. Inflammatory bowel disease: cause and immunobiology. Lancet. 2007;369:1627–1640. doi: 10.1016/S0140-6736(07)60750-8. [DOI] [PubMed] [Google Scholar]
- 3.Sartor RB. Current concepts of the etiology and pathogenesis of ulcerative colitis and Crohn's disease. Gastroenterol Clin North Am. 1995;24:475–507. [PubMed] [Google Scholar]
- 4.Danese S, Dejana E, Fiocchi C. Immune regulation by microvascular endothelial cells: directing innate and adaptive immunity, coagulation, and inflammation. J Immunol. 2007;178:6017–6022. doi: 10.4049/jimmunol.178.10.6017. [DOI] [PubMed] [Google Scholar]
- 5.Hatoum OA, Binion DG, Gutterman DD. Paradox of simultaneous intestinal ischaemia and hyperaemia in inflammatory bowel disease. Eur J Clin Invest. 2005;35:599–609. doi: 10.1111/j.1365-2362.2005.01567.x. [DOI] [PubMed] [Google Scholar]
- 6.Haddad JJ. Science review: redox and oxygen-sensitive transcription factors in the regulation of oxidant-mediated lung injury: role for hypoxia-inducible factor-1alpha. Crit Care. 2003;7:47–54. doi: 10.1186/cc1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kokura S, Yoshida N, Yoshikawa T. Anoxia/reoxygenation-induced leukocyte-endothelial cell interactions. Free Radic Biol Med. 2002;33:427–432. doi: 10.1016/s0891-5849(02)00852-3. [DOI] [PubMed] [Google Scholar]
- 8.Saadi S, Wrenshall LE, Platt JL. Regional manifestations and control of the immune system. Faseb J. 2002;16:849–856. doi: 10.1096/fj.01-0690hyp. [DOI] [PubMed] [Google Scholar]
- 9.Lewis JS, Lee JA, Underwood JC, Harris AL, Lewis CE. Macrophage responses to hypoxia: relevance to disease mechanisms. J Leukoc Biol. 1999;66:889–900. doi: 10.1002/jlb.66.6.889. [DOI] [PubMed] [Google Scholar]
- 10.Kominsky DJ, Campbell EL, Colgan SP. Metabolic shifts in immunity and inflammation. J Immunol. 184:4062–4068. doi: 10.4049/jimmunol.0903002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baldessarini RJ. Neuropharmacology of S-adenosyl-L-methionine. Am J Med. 1987;83:95–103. doi: 10.1016/0002-9343(87)90860-6. [DOI] [PubMed] [Google Scholar]
- 12.Cantoni G, Chiang P. The role of S-adenosylhomocysteine and S-adenosylhomocysteine hydrolase in the control of biological methylations. 1980:67–80. [Google Scholar]
- 13.Cantoni GL, Scarano E. The formation of S-adenosylhomocysteine in enzymatic transmethylation reactions. J. Am. Chem. Soc. 1954;76:4744. [Google Scholar]
- 14.Clarke S, Banfield K. S-adenosylmethionine-dependent methyltransferases. 2001:63–78. [Google Scholar]
- 15.Chiang PK. Biological effects of inhibitors of S-adenosylhomocysteine hydrolase. Pharmacol Ther. 1998;77:115–134. doi: 10.1016/s0163-7258(97)00089-2. [DOI] [PubMed] [Google Scholar]
- 16.Wolos JA, Frondorf KA, Babcock GF, Stripp SA, Bowlin TL. Immunomodulation by an inhibitor of S-adenosyl-L-homocysteine hydrolase: inhibition of in vitro and in vivo allogeneic responses. Cell Immunol. 1993;149:402–408. doi: 10.1006/cimm.1993.1165. [DOI] [PubMed] [Google Scholar]
- 17.Fu YF, Wang JX, Zhao Y, Yang Y, Tang W, Ni J, Zhu YN, Zhou R, He PL, Li C, Li XY, Yang YF, Lawson BR, Zuo JP. S-adenosyl-L-homocysteine hydrolase inactivation curtails ovalbumin-induced immune responses. J Pharmacol Exp Ther. 2006;316:1229–1237. doi: 10.1124/jpet.105.093369. [DOI] [PubMed] [Google Scholar]
- 18.Fu YF, Zhu YN, Ni J, Zhong XG, Tang W, Re YD, Shi LP, Wan J, Yang YF, Yuan C, Nan FJ, Lawson BR, Zuo JP. A reversible S-adenosyl-L-homocysteine hydrolase inhibitor ameliorates experimental autoimmune encephalomyelitis by inhibiting T cell activation. J Pharmacol Exp Ther. 2006;319:799–808. doi: 10.1124/jpet.106.107185. [DOI] [PubMed] [Google Scholar]
- 19.Lawson BR, Manenkova Y, Ahamed J, Chen X, Zou JP, Baccala R, Theofilopoulos AN, Yuan C. Inhibition of transmethylation down-regulates CD4 T cell activation and curtails development of autoimmunity in a model system. J Immunol. 2007;178:5366–5374. doi: 10.4049/jimmunol.178.8.5366. [DOI] [PubMed] [Google Scholar]
- 20.Saso Y, Conner EM, Teegarden BR, Yuan CS. S-Adenosyl-L-homocysteine hydrolase inhibitor mediates immunosuppressive effects in vivo: suppression of delayed type hypersensitivity ear swelling and peptidoglycan polysaccharide-induced arthritis. J Pharmacol Exp Ther. 2001;296:106–112. [PubMed] [Google Scholar]
- 21.Wu QL, Fu YF, Zhou WL, Wang JX, Feng YH, Liu J, Xu JY, He PL, Zhou R, Tang W, Wang GF, Zhou Y, Yang YF, Ding J, Li XY, Chen XR, Yuan C, Lawson BR, Zuo JP. Inhibition of S-adenosyl-L-homocysteine hydrolase induces immunosuppression. J Pharmacol Exp Ther. 2005;313:705–711. doi: 10.1124/jpet.104.080416. [DOI] [PubMed] [Google Scholar]
- 22.Khoury J, Ibla JC, Neish AS, Colgan SP. Antiinflammatory adaptation to hypoxia through adenosine-mediated cullin-1 deneddylation. J Clin Invest. 2007;117:703–711. doi: 10.1172/JCI30049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Synnestvedt K, Furuta GT, Comerford KM, Louis N, Karhausen J, Eltzschig HK, Hansen KR, Thompson LF, Colgan SP. Ecto-5'-nucleotidase (CD73) regulation by hypoxia-inducible factor-1 mediates permeability changes in intestinal epithelia. J Clin Invest. 2002;110:993–1002. doi: 10.1172/JCI15337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eltzschig HK, Ibla JC, Furuta GT, Leonard MO, Jacobson KA, Enjyoji K, Robson SC, Colgan SP. Coordinated adenine nucleotide phosphohydrolysis and nucleoside signaling in posthypoxic endothelium: role of ectonucleotidases and adenosine A2B receptors. J Exp Med. 2003;198:783–796. doi: 10.1084/jem.20030891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okayasu I, Hatakeyama S, Yamada M, Ohkusa T, Inagaki Y, Nakaya R. A novel method in the induction of reliable experimental acute and chronic ulcerative colitis in mice. Gastroenterology. 1990;98:694–702. doi: 10.1016/0016-5085(90)90290-h. [DOI] [PubMed] [Google Scholar]
- 26.Magness ST, Jijon H, Van Houten Fisher N, Sharpless NE, Brenner DA, Jobin C. In vivo pattern of lipopolysaccharide and anti-CD3-induced NF-kappa B activation using a novel gene-targeted enhanced GFP reporter gene mouse. J Immunol. 2004;173:1561–1570. doi: 10.4049/jimmunol.173.3.1561. [DOI] [PubMed] [Google Scholar]
- 27.Watson JA, Watson CJ, McCrohan AM, Woodfine K, Tosetto M, McDaid J, Gallagher E, Betts D, Baugh J, O'Sullivan J, Murrell A, Watson RW, McCann A. Generation of an epigenetic signature by chronic hypoxia in prostate cells. Hum Mol Genet. 2009;18:3594–3604. doi: 10.1093/hmg/ddp307. [DOI] [PubMed] [Google Scholar]
- 28.Gottschalk S, Anderson N, Hainz C, Eckhardt SG, Serkova NJ. Imatinib (STI571)-mediated changes in glucose metabolism in human leukemia BCR-ABL-positive cells. Clin Cancer Res. 2004;10:6661–6668. doi: 10.1158/1078-0432.CCR-04-0039. [DOI] [PubMed] [Google Scholar]
- 29.Serkova N, Fuller TF, Klawitter J, Freise CE, Niemann CU. H-NMR-based metabolic signatures of mild and severe ischemia/reperfusion injury in rat kidney transplants. Kidney Int. 2005;67:1142–1151. doi: 10.1111/j.1523-1755.2005.00181.x. [DOI] [PubMed] [Google Scholar]
- 30.Hartmann G, Bidlingmaier C, Siegmund B, Albrich S, Schulze J, Tschoep K, Eigler A, Lehr HA, Endres S. Specific type IV phosphodiesterase inhibitor rolipram mitigates experimental colitis in mice. J Pharmacol Exp Ther. 2000;292:22–30. [PubMed] [Google Scholar]
- 31.Furuta GT, Turner JR, Taylor CT, Hershberg RM, Comerford K, Narravula S, Podolsky DK, Colgan SP. Hypoxia-inducible factor 1-dependent induction of intestinal trefoil factor protects barrier function during hypoxia. J Exp Med. 2001;193:1027–1034. doi: 10.1084/jem.193.9.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karhausen J, Furuta GT, Tomaszewski JE, Johnson RS, Colgan SP, Haase VH. Epithelial hypoxia-inducible factor-1 is protective in murine experimental colitis. J Clin Invest. 2004;114:1098–1106. doi: 10.1172/JCI21086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson A, Keely S, Karhausen J, Gerich ME, Furuta GT, Colgan SP. Mucosal protection by hypoxia-inducible factor prolyl hydroxylase inhibition. Gastroenterology. 2008;134:145–155. doi: 10.1053/j.gastro.2007.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Berg S, Sappington PL, Guzik LJ, Delude RL, Fink MP. Proinflammatory cytokines increase the rate of glycolysis and adenosine-5'-triphosphate turnover in cultured rat enterocytes. Crit Care Med. 2003;31:1203–1212. doi: 10.1097/01.CCM.0000059647.92390.92. [DOI] [PubMed] [Google Scholar]
- 35.Scharte M, Han X, Bertges DJ, Fink MP, Delude RL. Cytokines induce HIF-1 DNA binding and the expression of HIF-1-dependent genes in cultured rat enterocytes. Am J Physiol Gastrointest Liver Physiol. 2003;284:G373–G384. doi: 10.1152/ajpgi.00076.2002. [DOI] [PubMed] [Google Scholar]
- 36.Li X, Stark GR. NFkappaB-dependent signaling pathways. Exp Hematol. 2002;30:285–296. doi: 10.1016/s0301-472x(02)00777-4. [DOI] [PubMed] [Google Scholar]
- 37.Werner SL, Barken D, Hoffmann A. Stimulus specificity of gene expression programs determined by temporal control of IKK activity. Science. 2005;309:1857–1861. doi: 10.1126/science.1113319. [DOI] [PubMed] [Google Scholar]
- 38.Cockman ME, Lancaster DE, Stolze IP, Hewitson KS, McDonough MA, Coleman ML, Coles CH, Yu X, Hay RT, Ley SC, Pugh CW, Oldham NJ, Masson N, Schofield CJ, Ratcliffe PJ. Posttranslational hydroxylation of ankyrin repeats in IkappaB proteins by the hypoxia-inducible factor (HIF) asparaginyl hydroxylase, factor inhibiting HIF (FIH) Proc Natl Acad Sci U S A. 2006;103:14767–14772. doi: 10.1073/pnas.0606877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cummins EP, Berra E, Comerford KM, Ginouves A, Fitzgerald KT, Seeballuck F, Godson C, Nielsen JE, Moynagh P, Pouyssegur J, Taylor CT. Prolyl hydroxylase-1 negatively regulates IkappaB kinase-beta, giving insight into hypoxia-induced NFkappaB activity. Proc Natl Acad Sci U S A. 2006;103:18154–18159. doi: 10.1073/pnas.0602235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen LW, Egan L, Li ZW, Greten FR, Kagnoff MF, Karin M. The two faces of IKK and NF-kappaB inhibition: prevention of systemic inflammation but increased local injury following intestinal ischemia-reperfusion. Nat Med. 2003;9:575–581. doi: 10.1038/nm849. [DOI] [PubMed] [Google Scholar]
- 41.Greten FR, Eckmann L, Greten TF, Park JM, Li ZW, Egan LJ, Kagnoff MF, Karin M. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118:285–296. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 42.Vanden Berghe W, Ndlovu MN, Hoya-Arias R, Dijsselbloem N, Gerlo S, Haegeman G. Keeping up NF-kappaB appearances: epigenetic control of immunity or inflammation-triggered epigenetics. Biochem Pharmacol. 2006;72:1114–1131. doi: 10.1016/j.bcp.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 43.Cummins EP, Seeballuck F, Keely SJ, Mangan NE, Callanan JJ, Fallon PG, Taylor CT. The hydroxylase inhibitor dimethyloxalylglycine is protective in a murine model of colitis. Gastroenterology. 2008;134:156–165. doi: 10.1053/j.gastro.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 44.Miller MW, Duhl DM, Winkes BM, Arredondo-Vega F, Saxon PJ, Wolff GL, Epstein CJ, Hershfield MS, Barsh GS. The mouse lethal nonagouti (a(x)) mutation deletes the S-adenosylhomocysteine hydrolase (Ahcy) gene. EMBO J. 1994;13:1806–1816. doi: 10.1002/j.1460-2075.1994.tb06449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eckmann L, Nebelsiek T, Fingerle AA, Dann SM, Mages J, Lang R, Robine S, Kagnoff MF, Schmid RM, Karin M, Arkan MC, Greten FR. Opposing functions of IKKbeta during acute and chronic intestinal inflammation. Proc Natl Acad Sci U S A. 2008;105:15058–15063. doi: 10.1073/pnas.0808216105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zaph C, Troy AE, Taylor BC, Berman-Booty LD, Guild KJ, Du Y, Yost EA, Gruber AD, May MJ, Greten FR, Eckmann L, Karin M, Artis D. Epithelial-cell-intrinsic IKK-beta expression regulates intestinal immune homeostasis. Nature. 2007;446:552–556. doi: 10.1038/nature05590. [DOI] [PubMed] [Google Scholar]
- 47.Pierce JW, Schoenleber R, Jesmok G, Best J, Moore SA, Collins T, Gerritsen ME. Novel inhibitors of cytokine-induced IkappaBalpha phosphorylation and endothelial cell adhesion molecule expression show anti-inflammatory effects in vivo. J Biol Chem. 1997;272:21096–21103. doi: 10.1074/jbc.272.34.21096. [DOI] [PubMed] [Google Scholar]
- 48.Cummins EP, Seeballuck F, Keely SJ, Mangan NE, Callanan JJ, Fallon PG, Taylor CT. The hydroxylase inhibitor dimethyloxalylglycine is protective in a murine model of colitis. Gastroenterology. 2008;134:156–165. doi: 10.1053/j.gastro.2007.10.012. Epub 2007 Oct 2010. [DOI] [PubMed] [Google Scholar]
- 49.Erzin Y, Uzun H, Celik AF, Aydin S, Dirican A, Uzunismail H. Hyperhomocysteinemia in inflammatory bowel disease patients without past intestinal resections: correlations with cobalamin, pyridoxine, folate concentrations, acute phase reactants, disease activity, and prior thromboembolic complications. J Clin Gastroenterol. 2008;42:481–486. doi: 10.1097/MCG.0b013e318046eab0. [DOI] [PubMed] [Google Scholar]
- 50.Yakut M, Ustun Y, Kabacam G, Soykan I. Serum vitamin B12 and folate status in patients with inflammatory bowel diseases. Eur J Intern Med. 21:320–323. doi: 10.1016/j.ejim.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 51.Zezos P, Papaioannou G, Nikolaidis N, Vasiliadis T, Giouleme O, Evgenidis N. Hyperhomocysteinemia in ulcerative colitis is related to folate levels. World J Gastroenterol. 2005;11:6038–6042. doi: 10.3748/wjg.v11.i38.6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen M, Peyrin-Biroulet L, George A, Coste F, Bressenot A, Bossenmeyer-Pourie C, Alberto JM, Xia B, Namour B, Gueant JL. Methyl deficient diet aggravates experimental colitis in rats. J Cell Mol Med. doi: 10.1111/j.1582-4934.2010.01252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taylor CT, Colgan SP. Hypoxia and gastrointestinal disease. J Mol Med. 2007;85:1295–1300. doi: 10.1007/s00109-007-0277-z. [DOI] [PubMed] [Google Scholar]
- 54.Semenza GL. Regulation of oxygen homeostasis by hypoxia-inducible factor 1. Physiology (Bethesda) 2009;24:97–106. doi: 10.1152/physiol.00045.2008. [DOI] [PubMed] [Google Scholar]
- 55.Tormos KV, Chandel NS. Inter-connection between mitochondria and HIFs. J Cell Mol Med. 14:795–804. doi: 10.1111/j.1582-4934.2010.01031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet. 2005;14(Spec No 1):R47–R58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- 57.Trojer P, Reinberg D. Histone lysine demethylases and their impact on epigenetics. Cell. 2006;125:213–217. doi: 10.1016/j.cell.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 58.Halim AB, LeGros L, Chamberlin ME, Geller A, Kotb M. Regulation of the human MAT2A gene encoding the catalytic alpha 2 subunit of methionine adenosyltransferase, MAT II: gene organization, promoter characterization, and identification of a site in the proximal promoter that is essential for its activity. J Biol Chem. 2001;276:9784–9791. doi: 10.1074/jbc.M002347200. [DOI] [PubMed] [Google Scholar]
- 59.Yang H, Sadda MR, Yu V, Zeng Y, Lee TD, Ou X, Chen L, Lu SC. Induction of human methionine adenosyltransferase 2A expression by tumor necrosis factor alpha. Role of NF-kappa B and AP-1. J Biol Chem. 2003;278:50887–50896. doi: 10.1074/jbc.M307600200. [DOI] [PubMed] [Google Scholar]
- 60.Gough DJ, Levy DE, Johnstone RW, Clarke CJ. IFNgamma signaling-does it mean JAK-STAT? Cytokine Growth Factor Rev. 2008;19:383–394. doi: 10.1016/j.cytogfr.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 61.Strober W, Fuss IJ, Blumberg RS. The immunology of mucosal models of inflammation. Annu Rev Immunol. 2002;20:495–549. doi: 10.1146/annurev.immunol.20.100301.064816. [DOI] [PubMed] [Google Scholar]
- 62.Taylor CT, Cummins EP. The role of NF-kappaB in hypoxia-induced gene expression. Ann N Y Acad Sci. 2009;1177:178–184. doi: 10.1111/j.1749-6632.2009.05024.x. [DOI] [PubMed] [Google Scholar]
- 63.Hirata I, Yasumoto S, Toshina K, Inoue T, Nishikawa T, Murano N, Murano M, Wang FY, Katsu K. Evaluation of the effect of pyrrolidine dithiocarbamate in suppressing inflammation in mice with dextran sodium sulfate-induced colitis. World J Gastroenterol. 2007;13:1666–1671. doi: 10.3748/wjg.v13.i11.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schmidt N, Gonzalez E, Visekruna A, Kuhl AA, Loddenkemper C, Mollenkopf H, Kaufmann SH, Steinhoff U, Joeris T. Targeting the proteasome: partial inhibition of the proteasome by bortezomib or deletion of the immunosubunit LMP7 attenuates experimental colitis. Gut. 59:896–906. doi: 10.1136/gut.2009.203554. [DOI] [PubMed] [Google Scholar]
- 65.Zhang DK, Cheng LN, Huang XL, Shi W, Xiang JY, Gan HT. Tetrandrine ameliorates dextran-sulfate-sodium-induced colitis in mice through inhibition of nuclear factor -kappaB activation. Int J Colorectal Dis. 2009;24:5–12. doi: 10.1007/s00384-008-0544-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.