Abstract
Background
Resistance commonly arises in infants exposed to single-dose nevirapine (sdNVP) for prevention of mother to child transmission (PMTCT). While K103N and Y181C are common following sdNVP, multiple other mutations also confer NVP-resistance. It remains unclear whether specific NVP-resistance mutations or combinations of mutations predict virologic failure in infants when present at low frequencies prior to NVP-based treatment.
Methods
Twenty sdNVP-exposed infants who were subsequently treated with NVP-based highly active antiretroviral therapy (HAART) were examined. Pre-treatment plasma samples were tested for the presence of NVP-resistance mutations by allele-specific PCR (ASPCR) for K103N and Y181C and ultra-deep pyrosequencing (UDPS) for all primary NVP mutations. Viral levels were determined every 3 months for up to 24months on NVP-HAART. Cox proportional hazard models were used to determine correlates of viral failure.
Results
The NVP resistance mutations K103N or Y181C were detected in pre-treatment plasma samples in 6 infants by ASPCR. NVP resistance at these or other sites was detectable by UDPS in 10 out of 20 infants tested. Virologic failure occurred in 50% of infants with any NVP resistance mutations detected, while only 20% of infants without resistance experienced viral failure, but the difference was not significant (p=0.19). An increase in the number of NVP resistance mutations detectable by UDPS in an infant was significantly associated with an increased risk of virologic failure (HR=1.79 (95%CI: 1.07, 2.99), p=0.027).
Conclusions
Low frequencies of multiple NVP resistance mutations, in addition to K103N and Y181C, present in infants before NVP-based treatment may predict treatment outcome.
Keywords: HIV, infants, nevirapine, resistance, HAART, treatment failure
Introduction
A single-dose of nevirapine (sdNVP), a non-nucleoside reverse transcriptase inhibitor (NNRTI) with a relatively long half-life, can reduce HIV-1 mother-to-child transmission (MTCT) by ~50%.1–3 This simple and effective intervention is an important component of HIV-1 prevention methods in resource-limited settings.4,5 Unfortunately, the slow decay of NVP often causes drug resistance to develop in both sdNVP-exposed HIV-positive mothers and their infants who become infected despite its use.6–9 NVP resistance is most often caused by amino acid changes K103N and Y181C in reverse transcriptase (RT) and is typically measured using population-based sequencing assays that detect mutations that comprise >20% of an individual’s viral population. Recent studies have shown that following sdNVP, virologic failure during NVP-based HAART is increased in both women and infants, even when resistance is undetectable by standard population sequencing.10,11 These findings suggest that the current standard HIV-1 sequencing assays are not sensitive enough to detect all clinically meaningful resistance.
Because NVP-resistant virus can persist as low-frequency variants for years following sdNVP12,13 and may compromise responses to NVP-containing treatment, current WHO guidelines recommend protease inhibitor (PI)-based HAART in children with any previous NVP exposure, regardless of standard HIV-1 sequencing assay results.14 However, PI-HAART is not always available as a first-line pediatric option and limits future treatment options if resistance arises, as good alternatives are not available in resource-limited settings. Ideally, it would be desirable to preserve the use of NVP-HAART as a first-line regimen, despite sdNVP-exposure if resistance at low frequencies could be ruled out.15
The suggestion that low-frequency resistant variants may have a clinical consequence has inspired the development of high-sensitivity resistance assays. These include nucleotide-specific methods such as allele-specific PCR (ASPCR) and ultra-deep pyrosequencing (UDPS) methods such as the 454 GS-FLX Titanium system (Roche). While ASPCR is relatively inexpensive and high-throughput, the reliability of ASPCR can be compromised by polymorphisms in the primer binding sites, a situation that can contribute to false positive results.16,17 In addition, ASPCR detects only one mutation at a time and has been developed primarily for select NVP-resistance mutations such as K103N and Y181C. There are other NVP-resistance mutations,18 but whether these are clinically relevant when present at low frequencies has not yet been defined. UDPS allows detection of all mutations within a region of interest, but is labor intensive, costly, and requires expertise in computational methods.16,17
Studies that have examined low-frequency NVP-resistant variants and their impact on subsequent treatment have predominately relied on ASPCR. The largest study on clinical outcome and low-frequency NVP resistance was in women. The findings suggested that levels of K103N, Y181C or G190A between 1% and 20% detected by ASPCR, which are missed by standard population sequencing assays, can reduce the efficacy of NVP-based treatment of sdNVP-exposed women.19 It is difficult to directly extrapolate findings in HIV-1-infected adults to infants because the pharmacokinetics of NVP are different in adults and children, as are viral loads and disease progression.20–22 In particular, viral loads are typically much higher in children,21 resulting in more viral replication that could contribute to increased viral diversity. One study of infants that examined K103N and Y181C using ASPCR showed a trend for an association between pre-HAART low-frequency resistance and virologic failure.23 However, low-frequency NVP resistance was not associated with failure in infants who received PI-HAART first and then switched to NVP-HAART.24 Thus, at present, the contribution of low frequencies of specific NVP resistance mutations, particularly mutations other than K103N and Y181C, to virologic failure during NVP-HAART is not well understood in children. Here we measure resistance using both ASPCR and UDPS in sdNVP-exposed infants who subsequently received NVP-HAART.
Methods
Study Population and Sample Collection
Participants were from two ongoing randomized clinical trials initiated in September 2007in Nairobi, Kenya (Optimizing Pediatric HAART 03 & 04: NCT00428116). Ethical approval was obtained from the University of Washington and Kenyatta National Hospital Institutional Review Boards. Mother-infant pairs were approached at antenatal and postnatal Nairobi city council clinics following screening for HIV-1.In addition, HIV-infected infants identified following admission to Kenyatta National hospital were recruited. Following informed consent, a filter paper blood sample was obtained and tested to determine infant HIV-1-status by HIV-1 DNA PCR. Inclusion in this laboratory sub-study was limited to HIV-positive infants with sdNVP administered either to the infant, their mother, or both, as reported by the caregiver or by clinic record. Two cases(designated in Table 1) had probable NVP exposure that could not be confirmed. Additional eligibility criteria were:1) caregiver planned to reside in Nairobi for >36 months, and2) age<12 months. Infants included were those who initiated NVP-HAART: usually zidovudine, lamivudine, plus nevirapine. A subset of the infants included(n=6) was started on NVP-HAART after standard population sequencing was used to rule out high-frequency NVP resistance. In addition, a group of infants (n=14) was treated with NVP-HAART prior to resistance testing and switched to PI-HAART after sdNVP-exposure was discovered during record review.
Table 1.
Baseline characteristics
| ID | age at NVP-HAART initiation | age at change to PI-HAART | months of follow-up on NVP-HAART | baseline VL (log copies/ml) | HIV subtype | population sequencing | treatment outcome |
|---|---|---|---|---|---|---|---|
| 7a | 3.5 | - | 21 | 7.2 | C | WT | VF |
| 9 | 11.5 | died | 6 | 6.7 | A | WT | VF |
| 11 | 12.6 | 33.6 | 21 | 7.4 | A/C | WT | VF |
| 17 | 4.2 | 9.6 | 3 | 7.2 | D | WT | VF |
| 19 | 3.6 | 8.1 | 3 | 5.4 | A | WT | VF |
| 22 | 3 | 15.6 | 12 | 6.2 | A | K103KN | VF |
| 24 | 2.1 | 14.1 | 12 | 6.6 | C/D | WT | VF |
| 2b | 4.8 | - | 24 | 6.0 | A | WT | int |
| 3 | 7 | 21 | 12 | 6.5 | A/C | WT | nf |
| 4 | 2.4 | 10.5 | 6 | 6.6 | A | WT | nf |
| 8c | 4.6 | - | 21 | 6.9 | A | 1AIT | nf |
| 12 | 4.8 | 9.9 | 3 | 7.3 | A | K103N | nf |
| 14 | 4.2 | 8.1 | 3 | 7.4 | A | G190A | nf |
| 15 | 2.4 | 7.5 | 3 | 6.0 | A | K103N | nf |
| 18 | 7.4 | 20.4 | 12 | 6.4 | A | WT | nf |
| 20 | 5.1 | 9.9 | 3 | 6.3 | D | WT | nf |
| 28 | 4.5 | 8.1 | 3 | 6.9 | C | WT | nf |
| 29 | 7 | 17 | 9 | 5.4 | A | WT | nf |
| 30 | 6.9 | 14.9 | 6 | 6.1 | A/D | WT | nf |
| 32 | 5.1 | 9 | 3 | 7.1 | A | WT | nf |
|
| |||||||
| Median | 4.7 | 10.2 | 6 | 6.6 | |||
VL = viral load, age in months. VF = viral failure, int = intermittent viremia. nf = no failure
this mother reported NVP-HAART prior to pregnancy
sdNVP exposure unkown due to maternal death prior to enrollment
conflicting data on sdNVP exposure
At enrollment, blood was collected for CD4 and viral load testing. Caregivers provided demographic information. Three adherence-counseling sessions were provided to all caregivers before HAART initiation. After HAART initiation, blood samples were collected for HIV-1 viral load testing at month 1, 3 and then every 3 months for up to 2 years.
Laboratory Assays
HIV-1 DNA filter paper assays were performed in Nairobi using a single-round PCR specific to pol, which is >98% sensitive and >98% specific.25 Plasma was separated from whole blood, aliquoted, and stored at −80°C until use. One aliquot was shipped to Seattle in liquid nitrogen. Plasma HIV-1 RNA levels were determined using the Gen-Probe HIV-1 Viral Load Assay (San Diego, CA), which has been validated for detection of HIV-1 subtypes prevalent in Kenya.26 Virologic failure was defined by ≥2 viral loads of >1000 copies/ml, or if the last sample available was >1000 copies/ml.
For all resistance assays, viral RNA was extracted from 140μl of plasma using a QIAmp viral RNA kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. Standard resistance testing at “baseline” (after sdNVP and before NVP-HAART)was performed in Nairobi using an in-house population-based sequencing method as described previously.27 HIV-1 subtypes were determined from the pol sequences with PAUP* version 4.0beta10 by creating a neighbor-joining phylogenetic tree with reference sequences from the Los Alamos National Laboratory HIV Database (http://www.hiv.lanl.gov/).
ASPCR to detect K103N and Y181C
ASPCR to detect K103N and Y181C in pre-HAART plasma samples was performed as described previously27 with modifications noted here. HIV-1 RNA was normalized to 10,000 RNA copies per 5μlas defined by the Gen-Probe assay results (targeting >2,000 RNA copies per 5μlassuming an average20% recovery after RNA extraction and cDNA synthesis efficiency as shown in similar studies)31 and 5μlwas added to each of duplicate RT-PCRs. All PCR conditions and primer sequences are described in Supplementary Table S1. To minimize errors in quantification due to polymorphisms in the ASPCR primer binding sites, a”cure” step, which uses a low annealing temperature and long primer to incorporate an invariant sequence proximal to the mutation site, 28 was performed following RT-PCR and prior to ASPCR (Teri Leigler, personal communication). The RT-PCR product was diluted 1:1000 and 5μl were added to the cure-step. The cure product was then diluted 1:50 and 5μl added to parallel reactions of allele-specific or total-copy real-time PCR performed on an ABI prism 7900HT.
To determine the error rate and lower limit of detection of ASPCR, controls were performed with mixtures of wild-type and K103N/Y181C-containing HIV-1 in-vitro transcripts. RNA was in-vitro transcribed from plasmids containing the pol region of Q23-17, a subtype A HIV-1 clone,29 and quantified by spectrophotometer before diluting and mixing in estimated ratios of 100%, 10%, 5%, 3%, 1.2%, 0.4%, and 0% mutant. ASPCR results from these mixtures were used in a receiver operator characteristic analysis and the cut-off with 100% specificity and maximum sensitivity (90.9% and 91.7% for K103N and Y181C, respectively) was used to define the lower limit.
Samples were run in duplicate and gel electrophoresis was performed on the ASPCR product. For each independent allele-specific assay run on a sample, if the measured drug-resistant copy number was between zero and one, the result of that replicate was set to zero. The proportion of drug-resistant virus was calculated for each reaction, and if it was below the lower limit or the correct size band was not visible on the gel, the proportion for that replicate was set to zero. The average proportion of drug-resistant to wild-type was calculated by averaging the replicate results from each sample.
UDPS using the 454 GS-FLX titanium system
We performed 10 parallel, high-fidelity RT-PCR reactions in the pre-HAART plasma samples, each with 10,000 viral copies based on the viral load data. Second round PCR to amplify a 555bp product spanning RT codons 50 to 234 was performed with primers linked to 454 sequencing adapters and a unique 8bp barcode (Table S1).The 10 parallel PCR products from each sample were pooled, gel-isolated and the resulting DNA concentration determined using the Qubit dsDNA HS kit (Invitrogen). The bar coded PCR amplicons were then mixed in equimolar concentrations and pyrosequenced in both forward and reverse directions on the 454 GS-FLX titanium plat format the University of Pennsylvania.
UDPS data processing
454 Sequences were grouped by sample according to an exact barcode-primer match. A quality-filtering step truncated the sequences at the first ambiguous base, or when the mean quality score in a 50bp sliding window dropped below 25. Sequences <100bp after quality filtering were discarded. Primers were trimmed prior to analysis. Cleaned sequences were pair-wise aligned to a subject-specific population-based sequence using the Needleman-Wunch algorithm (gap opening penalty: 20.0, gap extension penalty: 0.5, EDNA 4.4 scoring matrix).30 Deletions relative to the subject-specific reference sequence at a resistance site were treated as missing data. Insertions were accommodated by adding gaps to preserve the alignment, which was then confirmed by visual inspection near sites of resistance. Each sample was evaluated for the presence of any NVP-resistance mutations that confer at least moderate resistance (score ≥30) according to the Stanford resistance database.18 Forward sequence reads were used to assess RT amino acids 100 to 106 and reverse reads were used for amino acids 151to 215. Resistance sites were located on the reference population-based sequence and the aligned UDPS sequences were categorized as wild-type or mutant at each resistance site by a computer program developed in-house. Alignments and mutant categories were confirmed by visual inspection.
A Fisher’s Exact Test was used to test samples for elevated mutation rates relative to false positive rates in the pooled results of non-mutant controls. P-values were Bonferroni corrected (alpha=0.10). A lower limit of detection was determined for the minimum, median, and maximum read count at each position examined (Table S2) by testing increasing mutation counts from zero until the Bonferroni-corrected p-value was statistically significant.
Statistical Analysis
Kaplan-Meier survival curves and Cox proportional hazards models with robust standard errors were used to determine pre-HAART correlates of virologic failure. Infants without viral failure were censored at switch to PI-HAART or after 21–24 months of follow-up. In the Cox models, changes to different amino acids at the same position were counted as a single site of resistance. All analyses were performed using Stata version 9.2 (College Station, Texas, USA).
Results
Study population
The study population included 20 infants who had been exposed to sdNVP directly, through administration to their mothers during labor, or both, and who were subsequently treated with NVP-HAART. The infants ranged in age from 2.1to 12.6 months at HAART initiation(Table 1). The median pre-HAART baseline viral load was 6.6 log10 copies/ml. Phylogenetic analysis indicates that 60% of infants were infected with subtype A, 10%with subtype C,10%with subtype D, and 20% with inter-subtype recombinants.
Four infants had resistance detectable by population-based sequencing prior to HAART initiation: 3 had the K103N mutation and 1 had G190A, both of which confer high levels of NVP resistance (Table 1). No other NNRTI or NRTI resistance mutations were detectable by population sequencing in these pre-HAART samples. Seven infants experienced virologic failure while on NVP-HAART, one infant had intermittent viremia in which viral levels rose above 1000 copies/ml but then dropped below detection on subsequent measures (ID# 2) and was not categorized as virologic failure, while 12 maintained virologic suppression throughout follow-up(Table 1).
Detection limits of high-sensitivity ASPCR and UDPS resistance testing
To compare the sensitivity of our high-sensitivity resistance assays, we performed ASPCR and UDPS on duplicate low-frequency mixtures of RNA transcripts ranging from 0.1% to 10% K103N/Y181C, as well as 0% and 100% mutant controls. Both ASPCR and UDPS were able to quantify K103N and Y181C in mixtures with as little as 0.4% mutant at levels above the error rate seen in the 0% controls (Table 2). The quantification of all the low-frequency mixtures was consistently 2–5 fold below the expected mutant frequencies. Given the consistency across samples and using both methods, this difference was likely due to inaccuracies in the initial quantification of the in-vitro transcripts used and/or pipetting during the mixture preparations. Data from the 0% mutant controls reveal the error rates (false positives) at K103N and Y181C, which ranged from 0.05% to 0.55% depending on the assay used (Table 2).These error rates were used to define the lower limits of detection for the two assays: 0.97% and 0.54% for K103N and Y181C by ASPCR, and ranged from 0.16–0.27% and 0.26–0.37% for K103N and Y181C by UDPS, which varies depending on the number of sequence reads.
Table 2.
ASPCR and UDPS on low-frequency K103N/Y181C controls
| % K103N controls
|
% Y181C controls
|
|||
|---|---|---|---|---|
| expected | ASPCR | UDPS | ASPCR | UDPS |
| 100% | 94.70% | 99.86% | 95.40% | 99.94% |
| 10% | 6.55% | nt | 4.85% | nt |
| 5% | 2.95% | 1.08% | 1.55% | 1.08% |
| 3% | 1.53% | nt | 1.01% | nt |
| 1.2% | 0.95% | 0.22% | 1.07% | 0.27% |
| 0.4% | 0.98% | 0.18% | 0.47% | 0.25% |
| 0.1% | nt | 0.03% | nt | 0.06% |
| 0% | 0.55% | 0.05% | 0.16% | 0.07% |
|
| ||||
| lower limit | 0.97% | 0.16–0.27% | 0.54% | 0.26–0.37% |
nt=not tested
UDPS detection limits at other sites of resistance are shown in Supplementary Table S2, vary both by site and sequence depth, and are comparable to those published in studies using similar methods.31 These detection limits were derived from the false positive error rates of the UDPS data on the RNA transcripts (wild-type at these positions), which varied from 0.01% to 2.55%(Table S3).
Frequency of K103N and Y181C by ASPCR in pre-HAART infant plasma
K103N was detected by ASPCR in 4 infants at levels ranging from 18.7% to ~100% and Y181C in 3 infants at 0.9–3.1% (Table 3). The 3 infants in which K103N was detected by population sequencing (ID# 12,15,22) had K103N levels >20% as quantified by ASPCR. In addition, ASPCR detected K103N in 1 infant and Y181C in 3 infants at <20% (Table 3). In 1 case, both K103N and Y181C were detectable by ASPCR, while in 5 cases only one or the other mutation was detected (Table 3).
Table 3.
K103N and Y181C detected by ASPCR versus UDPS
| ASPCR
|
UDPS
|
|||
|---|---|---|---|---|
| ID | %K103N | %Y181C | %K103N | %Y181C |
| 7 | bd | 0.9% | bd | bd |
| 9 | bd | bd | bd | bd |
| 11 | bd | bd | bd | bd |
| 17 | bd | bd | bd | bd |
| 19 | bd | 1.0% | bd | bd |
| 22 | 48.9% | 3.1% | 49.90% | 0.49% |
| 24 | 18.7% | bd | 10.62% | bd |
| 2 | bd | bd | bd | bd |
| 3 | bd | bd | bd | bd |
| 4 | bd | bd | bd | bd |
| 8 | bd | bd | bd | bd |
| 12 | 106.0% | bd | 99.83% | bd |
| 14 | bd | bd | bd | bd |
| 15 | 63.5% | bd | 99.71% | bd |
| 18 | bd | bd | bd | bd |
| 20 | bd | bd | 0.18% | 0.36% |
| 28 | bd | bd | bd | bd |
| 29 | bd | bd | bd | bd |
| 30 | bd | bd | bd | bd |
| 32 | bd | bd | bd | bd |
bd = below detection
Additional low-frequency NVP resistant variants detected by UDPS
Resistance mutations K103N and Y181C were detected by UDPS in 5 and2 infants, respectively (Table 3). The level of K103N by UDPS was within two-fold of that quantified by ASPCR in samples with >10% K103N. For example, levels of K103N were 18.7% versus 10.6% (ID# 24), and 63.5% versus 99.7% (ID# 15) by each method (Table 3). Quantification below 10% was less concordant between assays. Only one sample had low levels of Y181C measured by both assays (#22) while 4 low-frequency measurements were discordant between UDPS and ASPCR, 2 each detected by one method and not the other (Table 3).
We analyzed the UDPS data at other sites known to cause intermediate to high-level NVP resistance.18 G190A, was present at a high frequency(>20%) in one infant (#14), which is consistent with results of population sequencing. Other resistance mutations detected by UDPS, but not by population sequencing, include K101E, K103S/T, V106A/M, G190T and F227C, and ranged from 0.06% to 9.97% (Table 4). Mutations that were assayed by UDPS but did not have detectable levels in any samples tested included L100I, K101P, V179F, Y181I/S/V, Y188C/H/L, and G190C/Q/S/V (data not shown).The most common mutation detected was K103N in 5 infants, followed by G190A in4 of 20 infants. Of the 10 infants that had detectable resistance by UDPS, 4 had resistance at more than one site (Table 4).
Table 4.
Resistance by UDPS in pre-HAART infant plasma
| ID | min # of 454 reads | NVP resistance mutations detected by UDPS1
|
NRTI2
|
total % reistance | # of sites with NVP resistance3 | # of sites NVP + NRTI resistance3 | months of follow-up | time of VF | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| K101
|
K103
|
V106
|
Y181
|
G190
|
F227
|
M184
|
||||||||||||
| E | N | S | T | A | M | C | A | E | T | C | 1 | |||||||
| 7 | 11134 | 0.21% | bd | bd | bd | bd | bd | bd | bd | bd | bd | bd | bd | 0.21% | 1 | 1 | 21 | month 6 |
| 9 | 8598 | bd | bd | bd | bd | bd | bd | bd | bd | bd | bd | bd | 0.13% | 0.13%* | 0 | 1 | 6 | month 3 |
| 11 | 12999 | bd | bd | bd | bd | bd | bd | bd | bd | bd | bd | 0.07% | bd | 0.07% | 1 | 1 | 21 | month 21 |
| 17 | 11029 | bd | bd | bd | bd | bd | bd | bd | bd | bd | bd | bd | 0.23% | 0.23%* | 0 | 1 | 3 | month 3 |
| 19 | 6203 | 0.20% | bd | bd | bd | bd | bd | bd | 3.76% | bd | bd | bd | bd | 3.96% | 2 | 2 | 3 | month 3 |
| 22 | 9529 | bd | 49.90% | bd | 0.32% | bd | bd | 0.49% | 0.34% | bd | bd | bd | bd | 51.05% | 3 | 3 | 12 | month 6 |
| 24 | 4212 | bd | 10.62% | bd | 0.80% | 9.97% | 0.19% | bd | 0.65% | bd | bd | bd | bd | 22.24% | 3 | 3 | 12 | month 9 |
| 2 | 5374 | bd | bd | bd | bd | bd | bd | bd | bd | bd | bd | bd | bd | 0% | 0 | 0 | 24 | int |
| 3 | 15058 | bd | bd | bd | bd | bd | bd | bd | bd | bd | bd | bd | bd | 0% | 0 | 0 | 12 | nf |
| 4 | 14402 | bd | bd | bd | bd | bd | bd | bd | bd | bd | bd | bd | bd | 0% | 0 | 0 | 6 | nf |
| 8 | 10382 | bd | bd | bd | bd | bd | bd | bd | bd | bd | bd | bd | bd | 0% | 0 | 0 | 21 | nf |
| 12 | 14318 | bd | 99.83% | 0.18% | bd | bd | bd | bd | bd | bd | bd | bd | bd | 100.01% | 1 | 1 | 3 | nf |
| 14 | 4497 | bd | bd | bd | bd | bd | bd | bd | 99.95% | bd | 0.06% | bd | bd | 100.01% | 1 | 1 | 3 | nf |
| 15 | 13633 | bd | 99.71% | 0.13% | bd | bd | bd | bd | bd | bd | bd | bd | bd | 99.83% | 1 | 1 | 3 | nf |
| 18 | 15305 | bd | bd | bd | bd | bd | bd | bd | bd | 0.64% | bd | bd | bd | 0.64% | 1 | 1 | 12 | nf |
| 20 | 14478 | bd | 0.18% | bd | bd | bd | bd | 0.36% | bd | bd | bd | bd | bd | 0.54% | 2 | 2 | 3 | nf |
| 28 | 13964 | bd | bd | bd | bd | bd | bd | bd | bd | bd | bd | bd | bd | 0% | 0 | 0 | 3 | nf |
| 29 | 4923 | bd | bd | bd | bd | bd | bd | bd | bd | bd | bd | bd | bd | 0% | 0 | 0 | 9 | nf |
| 30 | 7783 | bd | bd | bd | bd | bd | bd | bd | bd | bd | bd | bd | bd | 0% | 0 | 0 | 6 | nf |
| 32 | 13646 | bd | bd | bd | bd | bd | bd | bd | bd | bd | bd | bd | bd | 0% | 0 | 0 | 3 | nf |
bd = below detection, VF = viral failure, int = intermittent viremia, nf = no failure
NVP resistance mutations assayed by UDPS with no detectable resistance in any samples are not included in the table: L100I, K101P, V179F, Y181I/S/V, Y188C/H/L, G190C/Q/S/V
NRTI resistance mutations assayed by UDPS with no detectable resistance in any samples are not included in table: Q151M, M184V, T215F/Y
Changes to different amino acids at the same position were counted as a single site of resistance
For these cases, resistance would be 0% if NRTI sites were excluded.
In addition, we analyzed the UDPS data at RT amino acids 151, 184 and 215, known to confer high-level resistance to NRTIs. Only M184I was present at detectable levels: 0.13% and 0.23% in infants 9 and 17 respectively (Table 4).
Number of sites with NVP resistance is associated with virologic failure
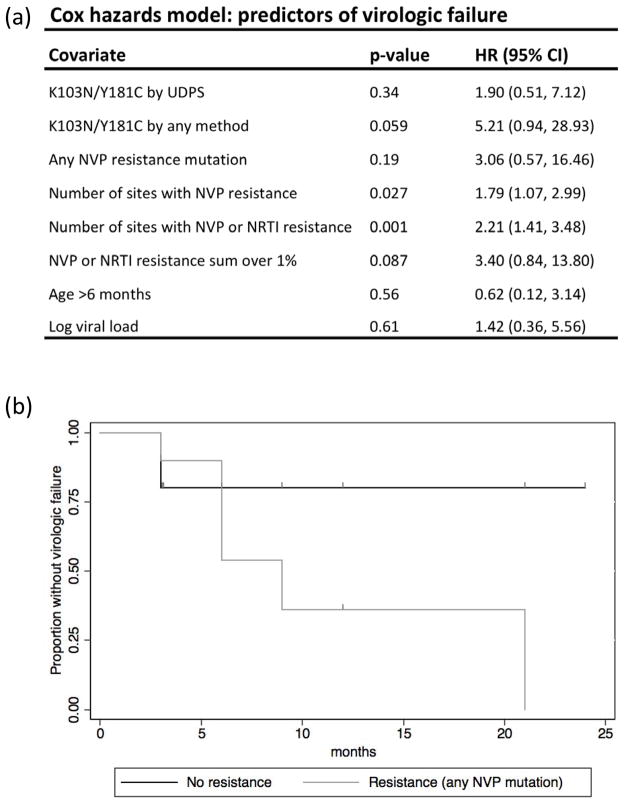
We used a Cox proportional hazards model to determine the predictors of virologic failure (Figure 1a). First we tested whether the presence, prior to HAART, of the two most common high-level NVP resistance mutations, K103N and Y181C, were associated with failure during NVP-HAART. Among the 5 infants that had detectable levels of K103N or Y181C by UDPS, 40% experienced virologic failure while failure was observed in 33% of infants without detectable K103N/Y181C,but the difference was not statistically significant (HR=1.90 (95%CI: 0.51, 7.12); P=0.34). When we included the presence of K103N or Y181C detected by any method, there was a trend towards an association (HR=5.21 (95%CI 0.94, 28.93) p=0.059).
Figure 1. Predictors of virologic failure.
(a) Univariate Cox proportional hazards models used to determine associations between pre-HAART characteristics and virologic failure. (b) Kaplan-Meier curve of the proportion of infants without virologic failure comparing infants with no detectable pre-HAART NVP resistance by UDPS (black line) to infants with any level of any NVP resistance mutation at baseline (grey line).
Next we tested whether the presence of any NVP resistance mutation at any level was associated with failure. Virologic failure occurred in 50% of infants with any NVP resistance mutations detected, while only 20% of infants without resistance experienced viral failure. This difference is shown in the Kaplan-Meier curve in Figure 1b, but the difference seen here did not reach statistical significance (HR=3.06 (95%CI: 0.57, 16.46); P=0.19; Figure 1a). A significant association was observed between the number of sites with NVP resistance detectable by UDPS and virologic failure (HR=1.79 (95%CI: 1.07, 2.99); P=0.027), suggesting an increased risk of failure with each additional site of resistance. Furthermore, when we included both NNRTI and NRTI mutations, the association between the number of sites with resistance detectable by UDPS and viral failure was highly significant(HR=2.21 (95%CI: 1.41, 3.48), p=0.001; Figure 1a). In addition, the infants with a sum of over 1% of viruses with any resistance mutations, which is a predictor of viral failure in adults,19 had a trend towards an increased risk of viral failure compared to infants with <1%resistance (HR=3.40 (95%CI: 0.84, 13.80);P=0.087). When we excluded infants with high-frequency resistance detectable by population-based sequencing(Table 1), low-frequency resistance of >1% was significantly associated with viral failure (HR=4.7 (95%CI: 1.3, 16.8), p=0.016).
No other baseline characteristics were associated with virologic failure, including age at HAART initiation or log viral load.
Discussion
A recent randomized trial showed that sdNVP-exposure increases the risk of virologic failure in children subsequently treated with NVP-HAART.10 These results prompted the WHO to recommend that all NVP-exposed children receive PI-HAART.14 However, reliance on PI-HAART for all NVP-exposed children is problematic as PI-HAART is toxic, heat-labile and costly, and alternative regimens are limited in resource-poor settings when resistance arises. Therefore, defining the contribution of low-frequency NVP-resistant variants to treatment failure in children may help define the optimal long-term treatment of NVP-exposed infants. Here we utilized two highly sensitive assays to detect low-frequency resistant variants in children subsequently treated with NVP-HAART. We observed an increased risk of virologic failure with increasing numbers of detectable NVP resistance mutations, even when present at low frequencies.
To our knowledge, this is the first study to utilize UDPS to detect multiple NVP resistance mutations in a cohort of NVP-exposed children. The most common mutations detected were K103N and G190A, while Y181C was rare. This was surprising given that Y181C is the most common mutation detected in infants in other studies.7,24,32,33 While10 infants in this study had no detectable resistance mutations, 6 had a NVP resistance mutation at a single amino acid position, 2 infants had 2 sites of NVP resistance, and 2 had NVP resistance at 3 sites. An increasing number of sites of resistance was associated with an increased risk of viral failure, suggesting that assays such as UDPS that capture all relevant NVP-resistance mutations may provide a more comprehensive view of the role of low-frequency resistance in treatment outcome than assays limited to select changes. We have very few clear cases of NRTI-based resistance, but interestingly, the two infants (ID# 9, 17) that experienced viral failure despite a lack of detectable NVP resistance did have an NRTI mutation (M184I) present at low frequency. When we include both the NVP and NRTI resistance data in our analysis, the association between the number of sites with resistance and viral failure was highly significant (p=0.001).Furthermore, of the 10 infants without detectable NVP resistance at any of the sites tested, 8 did not experience virologic failure during follow-up. This further supports the idea that mutations in addition to K103N or Y181C could be used to screen for resistance prior to NVP-based treatment.
Here we showed that the results of UDPS and ASPCR are concordant when resistance is present at high frequencies. However, there were cases in which low-frequency resistance was detected by one method and not the other, and neither method was clearly superior for detecting K103N orY181C. The one other study that has compared these assays to quantify low levels of K103N while on intermittent NNRTI-based HAART also had some discrepant results when quantifying resistance at low frequencies.34 Given that ASPCR can produce false-positives in diverse clinical samples17 and that our control data suggests UDPS is more sensitive than ASPCR(Table 2), the UDPS data may be a more accurate measure of low-frequency resistance.
The limitations of our study include a small sample size and short follow-up on NVP-HAART that resulted from study protocol changes due to the change in the WHO recommendation for treating NVP-exposed children.10,14 The lack of association seen betweenK103N or Y181C and viral failure could reflect the small sample size and limited follow-up, as other larger studies have found this association.19 Six of the8infants that were followed for only 3 months are in the no failure group, of which 4 had detectable resistance. Given the short duration of follow-up, it is possible that some of these infants are misclassified. Indeed, three infants with high frequencies of resistance were followed on NVP-HAART for only 3 months prior to switching to PI-HAART and might have experienced viral failure with longer follow-up (ID# 12, 14, 15).This would bias our analysis towards the null hypothesis that there is no difference in resistance between the two groups. Despite these limitations, we detected an association between the number of NVP resistance mutations and NVP-based treatment failure. The limited follow-up may also have contributed to the lack of association between age at HAART initiation (analogous to time since sdNVP) and treatment failure. Other studies suggest that NVP resistance and the clinical consequence of sdNVP fade with time.11,35,36
Our results are consistent with other studies that showed that low frequencies of NVP resistance have clinical consequence,16,19,23 and suggest that mutations other than K103N and Y181C may be relevant. Thus, to fully understand the contribution of low-frequency resistance to treatment failure, a comprehensive survey of the potential contributing resistance mutations is needed. While UDPS is presently not amenable to wide-scale application in resource-limited settings, it provides a potentially useful research tool for understanding the interplay between baseline resistance and treatment outcome. In the longer term, more cost-effective, lower technology methods to detect a range of resistance mutations could prove valuable for optimizing HIV treatment.
Supplementary Material
Acknowledgments
The authors thank the research personnel, laboratory staff, and data management teams in Nairobi, Kenya and Seattle, Washington; the Nairobi City Council Clinics for their participation and cooperation; the Divisions of Obstetrics and Gynaecology and Paediatrics at Kenyatta National Hospital for providing facilities for laboratory and data analysis; Maxwel Majiwa and MusaNgayo for population-based sequencing; Barbra Richardson and Katie Odem-Davis for advice on statistical analysis; Keshet Ronen for helpful discussions and scientific input. Most of all we thank the infants and their caregivers who participated in the study.
Sources of Funding
This work was supported by grants from the National Institutes of Health: HD23412 and AI076105. The Nairobi field site is supported in part by the University of Washington Center for AIDS Research (CFAR), an NIH funded program (P30 AI027757), which is supported by the following NIH Institutes and Centers (NIAID, NCI, NIMH, NIDA, NICHD, NHLBI, NIA).
Footnotes
Conflicts of Interest
The authors have no conflicts of interest.
References
- 1.Guay LA, Musoke P, Fleming T, et al. Intrapartum and neonatal single-dose nevirapine compared with zidovudine for prevention of mother-to-child transmission of HIV-1 in Kampala, Uganda: HIVNET 012 randomised trial. Lancet. 1999;354(9181):795–802. doi: 10.1016/S0140-6736(99)80008-7. [DOI] [PubMed] [Google Scholar]
- 2.Lallemant M, Jourdain G, Le Coeur S, et al. Single-dose perinatal nevirapine plus standard zidovudine to prevent mother-to-child transmission of HIV-1 in Thailand. N Engl J Med. 2004;351(3):217–228. doi: 10.1056/NEJMoa033500. [DOI] [PubMed] [Google Scholar]
- 3.Jackson JB, Musoke P, Fleming T, et al. Intrapartum and neonatal single-dose nevirapine compared with zidovudine for prevention of mother-to-child transmission of HIV-1 in Kampala, Uganda: 18-month follow-up of the HIVNET 012 randomised trial. Lancet. 2003;362(9387):859–868. doi: 10.1016/S0140-6736(03)14341-3. [DOI] [PubMed] [Google Scholar]
- 4.World Health Organization. Anitretroviral drugs for treating pregnant women and preventing HIV infection in infants, 2010 version. [Accessed August 2011]; from http://www.who.int/hiv/pub/mtct/antiretroviral2010/en/index.html. [PubMed]
- 5.McIntyre J. Use of antiretrovirals during pregnancy and breastfeeding in low-income and middle-income countries. Curr Opin HIV AIDS. 2010;5(1):48–53. doi: 10.1097/COH.0b013e328333b8ab. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackson JB, Becker-Pergola G, Guay LA, et al. Identification of the K103N resistance mutation in Ugandan women receiving nevirapine to prevent HIV-1 vertical transmission. AIDS. 2000;14(11):F111–115. doi: 10.1097/00002030-200007280-00001. [DOI] [PubMed] [Google Scholar]
- 7.Eshleman SH, Mracna M, Guay LA, et al. Selection and fading of resistance mutations in women and infants receiving nevirapine to prevent HIV-1 vertical transmission (HIVNET 012) AIDS. 2001;15(15):1951–1957. doi: 10.1097/00002030-200110190-00006. [DOI] [PubMed] [Google Scholar]
- 8.Eshleman SH, Jackson JB. Nevirapine resistance after single dose prophylaxis. AIDS Rev. 2002;4(2):59–63. [PubMed] [Google Scholar]
- 9.Johnson JA, Li J-F, Morris L, et al. Emergence of drug-resistant HIV-1 after intrapartum administration of single-dose nevirapine is substantially underestimated. J Infect Dis. 2005;192(1):16–23. doi: 10.1086/430741. [DOI] [PubMed] [Google Scholar]
- 10.Palumbo P, Lindsey JC, Hughes MD, et al. Antiretroviral treatment for children with peripartum nevirapine exposure. N Engl J Med. 2010;363(16):1510–1520. doi: 10.1056/NEJMoa1000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lockman S, Shapiro RL, Smeaton LM, et al. Response to antiretroviral therapy after a single, peripartum dose of nevirapine. N Engl J Med. 2007;356(2):135–147. doi: 10.1056/NEJMoa062876. [DOI] [PubMed] [Google Scholar]
- 12.Flys Tamara S, Donnell D, Mwatha A, et al. Persistence of K103N-Containing HIV-1 Variants after Single-Dose Nevirapine for Prevention of HIV-1 Mother-to-Child Transmission. J Infect Dis. 2007;195(5):711–715. doi: 10.1086/511433. [DOI] [PubMed] [Google Scholar]
- 13.Flys T, Nissley DV, Claasen CW, et al. Sensitive drug-resistance assays reveal long-term persistence of HIV-1 variants with the K103N nevirapine (NVP) resistance mutation in some women and infants after the administration of single-dose NVP: HIVNET 012. J Infect Dis. 2005;192(1):24–29. doi: 10.1086/430742. [DOI] [PubMed] [Google Scholar]
- 14.World Health Organization. Antiretroviral therapy for HIV infection in infants and children: Towards universal access, 2010 version. [Accessed August 2011]; from http://www.who.int/hiv/pub/paediatric/infants2010/en/index.html. [PubMed]
- 15.Coovadia A, Abrams EJ, Stehlau R, et al. Reuse of nevirapine in exposed HIV-infected children after protease inhibitor-based viral suppression: a randomized controlled trial. JAMA. 2010;304(10):1082–1090. doi: 10.1001/jama.2010.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li JZ, Paredes R, Ribaudo HJ, et al. Low-frequency HIV-1 drug resistance mutations and risk of NNRTI-based antiretroviral treatment failure: a systematic review and pooled analysis. JAMA. 2011;305(13):1327–1335. doi: 10.1001/jama.2011.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gianella S, Richman DD. Minority Variants of Drug-Resistant HIV. J Infect Dis. 2010 Jul 15; doi: 10.1086/655397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Standford University. [Accessed August, 2011];HIV drug resistance database. from: http://hivdb.stanford.edu/index.html.
- 19.Boltz VF, Zheng Y, Lockman S, et al. Role of low-frequency HIV-1 variants in failure of nevirapine-containing antiviral therapy in women previously exposed to single-dose nevirapine. Proceedings of the National Academy of Sciences. 2011 May 16;:1–6. doi: 10.1073/pnas.1105688108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newell M-L, Coovadia H, Cortina-Borja M, et al. Mortality of infected and uninfected infants born to HIV-infected mothers in Africa: a pooled analysis. Lancet. 2004;364(9441):1236–1243. doi: 10.1016/S0140-6736(04)17140-7. [DOI] [PubMed] [Google Scholar]
- 21.Richardson BA, Mbori-Ngacha D, Lavreys L, et al. Comparison of human immunodeficiency virus type 1 viral loads in Kenyan women, men, and infants during primary and early infection. J Virol. 2003;77(12):7120–7123. doi: 10.1128/JVI.77.12.7120-7123.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Musoke P, Guay LA, Bagenda D, et al. A phase I/II study of the safety and pharmacokinetics of nevirapine in HIV-1-infected pregnant Ugandan women and their neonates (HIVNET 006) AIDS. 1999;13(4):479–486. doi: 10.1097/00002030-199903110-00006. [DOI] [PubMed] [Google Scholar]
- 23.Macleod IJ, Rowley CF, Thior I, et al. Minor resistant variants in nevirapine-exposed infants may predict virologic failure on nevirapine-containing ART. J Clin Virol. 2010 doi: 10.1016/j.jcv.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunt GM, Coovadia A, Abrams EJ, et al. HIV-1 drug resistance at antiretroviral treatment initiation in children previously exposed to single-dose nevirapine. AIDS. 2011;25(12):1461–9. doi: 10.1097/QAD.0b013e3283492180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chohan BH, Emery S, Wamalwa D, John-Stewart G, Overbaugh J. Evaluation of a single round polymerase chain reaction assay using dried blood spots for diagnosis of HIV-1 infection in infants in an African setting. BMC Pediatrics. 2011;11:18. doi: 10.1186/1471-2431-11-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Emery S, Bodrug S, Richardson BA, et al. Evaluation of performance of the Gen-Probe human immunodeficiency virus type 1 viral load assay using primary subtype A, C, and D isolates from Kenya. J Clin Microbiol. 2000;38(7):2688–2695. doi: 10.1128/jcm.38.7.2688-2695.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lehman DA, Chung MH, Mabuka JM, et al. Lower risk of resistance after short-course HAART compared to zidovudine/single-dose nevirapine used for prevention of HIV-1 mother-to-child transmission. J Acquir Immune Defic Syndr. 2009;51(5):522–529. doi: 10.1097/QAI.0b013e3181aa8a22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Svarovskaia ES, Moser MJ, Bae AS, Prudent JR, Miller MD, Borroto-Esoda K. MultiCode-RTx Real-Time PCR System for Detection of Subpopulations of K65R Human Immunodeficiency Virus Type 1 Reverse Transcriptase Mutant Viruses in Clinical Samples. J Clin Microbiol. 2006;44(11):4237–4241. doi: 10.1128/JCM.01512-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poss M, Overbaugh J. Variants from the diverse virus population identified at seroconversion of a clade A human immunodeficiency virus type 1-infected woman have distinct biological properties. J Virol. 1999;73(7):5255–2564. doi: 10.1128/jvi.73.7.5255-5264.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Software Suite. Trends Genet. 2000;16(6):276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 31.Hedskog C, Mild M, Jernberg J, et al. Dynamics ofHIV-1 quasispecies during antiviral treatment dissected using ultra-deep pyrosequencing. PLoS ONE. 2010;5(7):e11345. doi: 10.1371/journal.pone.0011345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moorthy A, Gupta A, Bhosale R, et al. Nevirapine resistance and breast-milk HIV transmission: effects of single and extended-dose nevirapine prophylaxis in subtype C HIV-infected infants. PLoS ONE. 2009;4(1):e4096. doi: 10.1371/journal.pone.0004096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinson NA, Morris L, Gray G, et al. Selection and persistence of viral resistance in HIV-infected children after exposure to single-dose nevirapine. J Acquir Immune Defic Syndr. 2007;44(2):148–153. doi: 10.1097/QAI.0b013e31802b920e. [DOI] [PubMed] [Google Scholar]
- 34.Delobel P, Saliou A, Nicot F, et al. Minor HIV-1 Variants with the K103N Resistance Mutation during Intermittent Efavirenz-Containing Antiretroviral Therapy and Virological Failure. PLoS ONE. 2011;6(6):e21655. doi: 10.1371/journal.pone.0021655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jourdain G, Ngo-Giang-Huong N, Le Coeur S, et al. Intrapartum exposure to nevirapine and subsequent maternal responses to nevirapine-based antiretroviral therapy. N Engl J Med. 2004;351(3):229–240. doi: 10.1056/NEJMoa041305. [DOI] [PubMed] [Google Scholar]
- 36.Eshleman SH, Guay LA, Wang J, et al. Distinct patterns of emergence and fading of K103N and Y181C in women with subtype A vs. D after single-dose nevirapine: HIVNET 012. J Acquir Immune Defic Syndr. 2005;40(1):24–29. doi: 10.1097/01.qai.0000174656.71276.d6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.