Abstract
We used the second-generation mining minima method (M2) to compute the binding affinities of the novel host-guest complexes in the SAMPL3 blind prediction challenge. The predictions were in poor agreement with experiment, and we conjectured that much of the error might derive from the force field, CHARMm with Vcharge charges. Repeating the calculations with other generalized force-fields led to no significant improvement, and we observed that the predicted affinities were highly sensitive to the choice of force-field. We therefore embarked on a systematic evaluation of a set of generalized force fields, based upon comparisons with PM6-DH2, a fast yet accurate semi-empirical quantum mechanics method. In particular, we compared gas-phase interaction energies and entropies for the host-guest complexes themselves, as well as for smaller chemical fragments derived from the same molecules. The mean deviations of the force field interaction energies from the quantum results were greater than 3 kcal/mol and 9 kcal/mol, for the fragments and host-guest systems respectively. We further evaluated the accuracy of force-fields for computing the vibrational entropies and found the mean errors to be greater than 4 kcal/mol. Given these errors in energy and entropy, it is not surprising in retrospect that the predicted binding affinities deviated from the experiment by several kcal/mol. These results emphasize the need for improvements in generalized force-fields and also highlight the importance of systematic evaluation of force-field parameters prior to evaluating different free-energy methods.
Keywords: SAMPL3, binding affinity, supramolecular, host-guest, force-field, and mining minima, semi-empirical
INTRODUCTION
Accurate computational models of biomolecular recognition are needed to speed the discovery and design of new drugs.1 However, it is difficult to test computational models for biomolecules, because of their size and complexity. Many of the physical forces involved in protein-ligand binding, including non-covalent interactions (van der Waals and electrostatic), hydrogen bonding, and solvation (or desolvation) of the ligand and receptor binding site, are also operational in host-guest systems, and the binding of these small systems are governed by the same laws of statistical mechanics.2–4 Unlike protein-ligand systems, host-guest systems are small in size (a few hundred atoms, typically) and have fewer soft degrees of freedom, so it is easier to achieve adequate sampling of the conformational space and hence well-converged thermodynamic properties.5 Owing to their computational tractability, host-guest systems make excellent model systems for validating the accuracy of different energy models, or force-fields, solvation models, and free-energy methods used in computing binding affinities. Furthermore, they are valuable in understanding the consequences of simplifying assumptions used in different free-energy methods, such as single vs. multiple conformations, harmonic approximation of potential energy wells, and implicit vs. explicit solvent. Studies of host-guest binding therefore have potential to deepen our understanding of the physical chemistry of binding.
The SAMPL3 blind prediction challenge of host-guest binding affinities presents an excellent opportunity to test the current state of the field. We used our second-generation mining minima (M2) method to predict the binding affinities for a series of novel host-guest pairs. Significant deviations between the predicted and experimental affinities prompted us to systematically investigate the source of these errors. Since the predicted affinities proved highly sensitive to the choice of force-field, we hypothesized that the errors relative to experiment stemmed primarily from the inaccuracy of the available force-field energy models. This seemed particularly plausible because the force fields for molecules other than proteins and nucleic acids -- so-called generalized force fields – are, arguably, not yet as well-optimized as those for biomolecules.
We tested this hypothesis by comparing the interaction energies computed with various force-fields with those obtained from a reference semi-empirical quantum mechanical (QM) model, for complexes constructed from small fragments that were derived from the host and 3 guest molecules in the SAMPL3 dataset. In addition, we compared the interaction energies of the various host-guest complexes in representative conformations. Furthermore, to assess the range of errors to be expected from the entropic component of the free energy, we compared the vibrational entropies of the hosts, guests, and complexes obtained from classical and QM calculations. This study was made possible in large part by the efficiency and demonstrated accuracy of the PM6-DH2 semi-empirical approach.6 The results shed light on the accuracy of several commonly used generalized force-fields, and emphasize the need for a systematic investigation of a force-field prior to its use for a molecule of interest.
MATERIALS AND METHODS
Mining minima
Host-guest binding affinities were computed with the second-generation mining minima (M2) method,7 available for download from http://pharmacy.ucsd.edu/labs/gilson/software1a.html. The M2 method is described in detail elsewhere,7–9 and so is only briefly summarized here. Binding affinities are determined from the standard chemical potential (μo) of host, guest, and their complex, using the equation,
The standard chemical potential of a molecule in solution is approximated as a sum over many local energy minima, based on the predominant states approximation,10 given by the equation
where R, T, Co, E(r), and Zi are, respectively, the gas constant, the absolute temperature, the standard concentration, the potential energy as a function of the internal coordinates r, and the configuration integral over the internal coordinates r in energy well i. The potential energy of the molecule is calculated using an empirical force-field energy model, while solvent effects are accounted for using a continuum solvation model. The Generalized-Born (GB) solvation model is used during conformational search due to its efficiency,11 and the final solvation free energies are corrected towards Poisson-Boltzmann/surface-area (PBSA) solvation model,12, 13 computed for a single representative conformation for each potential energy well using the UHBD program.14 Local energy minima conformations are identified with the Tork search algorithm,15 and duplicate conformations are eliminated based on root mean squared distance and using a symmetry-aware algorithm, to prevent double counting.16 Local configuration integrals are computed using the Harmonic Approximation/Mode Scanning (HA/MS) method.9 All free-energy calculations used the same default M2 parameters.
Starting structures of the host and guest molecules were those provided in the SAMPL3 dataset. The molecules were protonated at pH 7.4 using Avogadro software (http://avogadro.openmolecules.net/), without allowing for possible pKa shifts. Initial parameters for bonded and van der Waals interactions were assigned according to the CHARMm force-field 17 using Accelrys Discovery Studio Visualizer, while atomic partial charges were assigned using Vcharge (VC) software.18 Starting structures of host-guest complexes were constructed by docking the guest molecule in host binding pocket using Autodock Vina software.19
Gas-phase interaction energies and vibrational entropies
Interaction energies, ΔE = E(complex) − E(molecule1) − E(molecule2), of fragment and host-guest complexes in the gas phase were computed using the softwares, Macromodel, version 9.920 (Schrodinger, LLC, NY) (classical force-field), GROMACS 4.021 (classical force-field) or MOPAC 200922 (semi-empirical QM). Five different classical force-fields, CHARMm,17 OPLS-2005,23, 24 AMBER*,25, 26 MMFFs,27–29 and MM3*30 were studied, with the semi-empirical QM model PM6-DH26 serving as the reference model. A dielectric constant of 1.0 was used for Coulomb interactions, and no distance cut-offs were imposed for non-bonded interactions, for the force field calculations. Geometries of the complexes were generated for the fragments shown in Figure 1, which are derived from host and guest molecules in the SAMPL3 dataset. For each pair of SAMPL3 host-guest fragments, we performed 1000 steps of low-mode conformational search (LMOD),31 using the OPLS-2005 force-field and Macromodel. We also studied the interaction energies of the most stable conformations of host-guest complexes obtained during the M2 free-energy calculations. These lowest energy conformations of fragment and host-guest complexes were further energy minimized using the PM6-DH2 model, until the normalized energy gradient converged to 0.001 kcal/mol.Å. The optimized structures were used for computing the interaction energies at both classical and quantum levels, without any further geometry optimization, unless noted otherwise.
Figure 1.
Chemical fragments derived from the host and guest molecules in SAMPL3 dataset.
Vibrational entropies were computed using the rigid rotor harmonic-oscillator (RRHO) approximation. (See Zhou et al.32 for a detailed discussion of configurational entropy and RRHO approximation.) Briefly, the starting structures were energy minimized with a tight convergence criterion and normal mode frequencies of the 3N-6 vibrational degrees of freedom were computed from the eigenvalues of the Hessian. Vibrational entropy was then computed from the normal mode frequencies using the equation,
where R is the gas constant, β = 1/RT, T is the absolute temperature, and ωi is the angular frequency of ith mode. Normal mode frequencies were computed using Macromodel or GROMACS 4.0 for classical force-field and MOPAC 2009 for semi-empirical QM model. All interaction energy and entropy calculations were performed in the gas phase, to avoid additional complications with the different solvation models used in quantum and classical calculations.
RESULTS
M2 binding affinity predictions
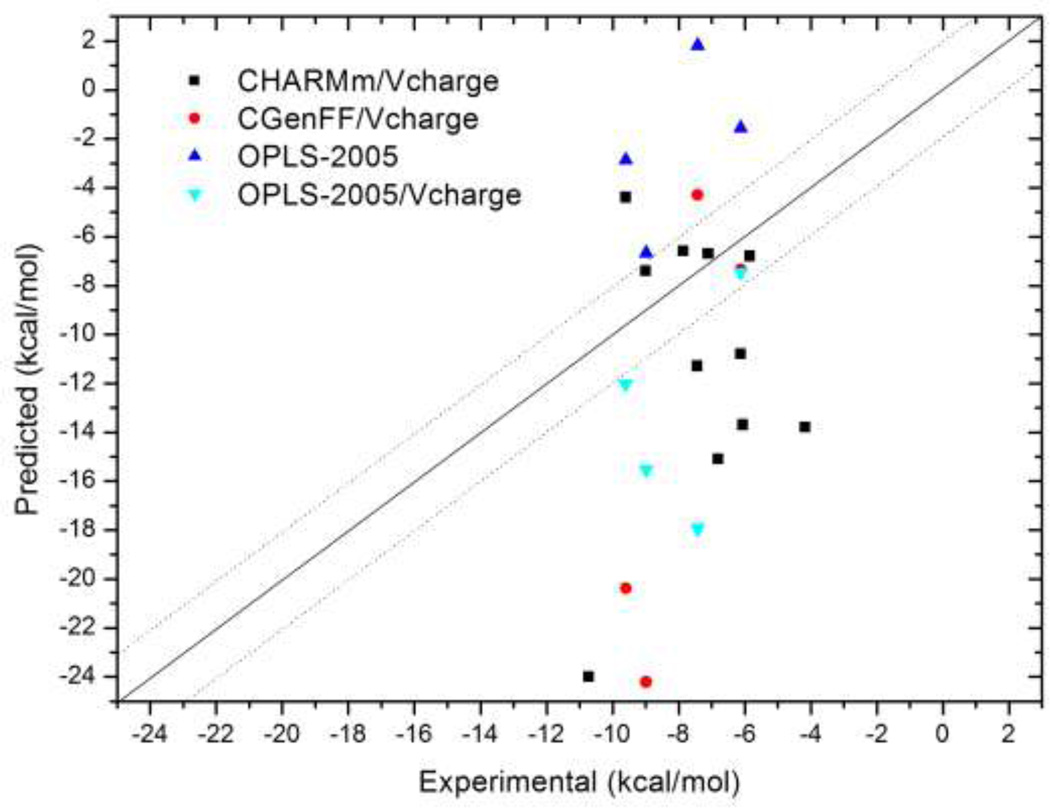
Binding affinities of SAMPL3 host-guest complexes computed using various empirical force-fields are shown in Figure 2. The corresponding error statistics are summarized in Table 1. The submitted predictions based on CHARMm force-field with Vcharge charges showed disappointing agreement with experiments. While 4 out of 11 complexes were predicted within kcal/mol of the experiment, the mean and root-mean-square (RMS) errors of the predictions were 5.2 and 6.5 kcal/mol, respectively, with a maximum error of 13.3 kcal/mol for host1/guest6 complex. Moreover, linear regression of calculation against experiment showed very poor correlation.
Figure 2.
Predicted binding affinities of SAMPL3 host-guest complexes using different force-field parameters. Dotted line represents ± 2 kcal/mol range.
Table 1.
Error statistics of computed binding affinities
| Linear Fitb |
||||
|---|---|---|---|---|
| MUE (kcal/mol) |
RMSE (kcal/mol) |
m | R2 | |
| CHARMm/Vcharge | 5.2 | 6.5 | 0.39 | 0.02 |
| host1 | 5.9 | 7.5 | 1.55 | 0.24 |
| host2/host3 | 3.8 | 4.1 | −1.8 | 0.80 |
| CGenFF/Vchargea | 7.6 | 9.5 | 5.19 | 0.70 |
| OPLS-05a | 5.7 | 6.3 | 1.22 | 0.30 |
| OPLS-05/Vchargea | 5.2 | 6.4 | 1.15 | 0.16 |
Binding affinities were computed for host2 and host3 systems only.
m and R2 are slope and correlation coefficient respectively, obtained from linear regression fitting.
One potential source of error is uncertainty in the protonation states of some of these molecules. Although we performed all our calculations with standard protonation states for the ionizable groups involved at pH 7, there may in reality be significant pKa shifts relative to the standard values. This is particularly true for host1, which has 4 carboxylic groups in close proximity to each other; and several of guests have ionizable groups whose protonation states might conceivably change on binding. The importance of ionization on the computed results is exemplified by the large difference in binding affinity of the host1/guest4 complex when computed with different charge states for the guest: the predicted affinity of this guest with the aniline group deprotonated (the preferred state at pH 7 based on the standard pka of the aniline group) was +9.4 kcal/mol, whereas in the protonated state, it was −13.7 kcal/mol. On the other hand, there is little or no uncertainty in the charge states of host2 (Cucurbit[7]uril) and host3 (Cucurbit[8]uril) and their guests studied here, but the corresponding predicted affinities still deviated significantly from experiment, and indeed were negatively correlated.
We therefore conjectured that the computational errors might result in large part from inaccuracies of the force-field. To examine this hypothesis, we computed the binding affinities of all the host-guest systems using three additional energy models: CGenFF33 (academic CHARMM) with Vcharge charges, OPLS-2005 with default charges provided by the program Macromodel, and OPLS-2005 with Vcharge charges. As shown in Figure 2 and Table 1, these calculations did not show significant improvement. Although the predicted affinities using CGenFF and OPLS force-fields were positively correlated with experiment, the correlation coefficients were still low, and the mean errors increased to the 5 – 8 kcal/mol range, compared with 3.9 kcal/mol in our original submission (Table 1). Interestingly, binding affinities computed using the OPLS-2005 force-field with Vcharge charges deviated significantly from those computed using the same force-field with default charges assigned by Macromodel, indicating that the choice of charge model can significantly influence the predicted binding affinities.
It is also worth noting that the various force fields provided quite different predictions. Thus, standard deviation of the computed affinities among the different force-fields was 7.5 kcal/mol, indicating high sensitivity of the predictions to the choice of energy model. We examined the origins of these differences by using the ability of the M2 method to provide a breakdown of the binding free energy into contributions from van der Waals (vdW), Coulomb, valence energy (bonds, angles and dihedrals), solvation, and configurational entropy. We found that, although the Coulomb energy and polar solvation energy of each host-guest system showed high standard deviations, 24 and 27 kcal/mol, respectively, across the various force fields, the standard deviation of the total electrostatic energy was only 5.2 kcal/mol, indicating significant cancellation between the Coulomb and polar solvation terms. On the other hand, the standard deviations of vdW and valence energies were 7.1 and 5.2 kcal/mol, respectively. Together, the deviation of total potential energy and entropy were 6.2 kcal/mol and 5.6 kcal/mol, respectively, contributing to the significant spread of the binding affinities predicted with different force-fields. We also observed that host2 preferred inclusion complexes with CHARMm and CGenFF force-fields, but exclusion complexes with OPLS-2005; these different conformational preferences presumably contribute to the overall sensitivity of the computed binding free energies to the choice of force field.
Force-fields vs. quantum mechanical reference calculations
Given the high sensitivity of the binding affinity predictions to the force-field and a lack of clarity whether other generalized force-fields (e.g. AMBER, MMFF) would prove any better, we sought a basis for more reliable reference energy calculations. Based on recent improvements in semi-empirical QM approach, we identified the PM6-DH26 model as a fast and accurate model to validate the accuracy of force-fields, and compared interaction energies and vibrational entropies with various force fields to those obtained with PM6-DH2 for molecules and representative fragments drawn from the host-guest systems in this SAMPL3 dataset. Here we include a wider range of force fields than considered above, now including AMBER*, MMFFs, and MM3*, with Macromodel assigned partial charges. Overall, as summarized in Figure 3 and Tables 2 & 3, the interaction energies of fragment and host-guest complexes in gas phase computed with the force-fields considered here show significant deviations from those obtained with the reference PM6-DH2 model. The following subsections discuss a first set of comparisons in which force field energies were computed for conformations energy-minimized in the quantum potential energy surface, followed by a second set of comparisons in which the quantum energy-minimized structures were reminimized with the respective forcefields before comparing. A third subsection compares entropies computed on the quantum and force field energy surfaces.
Figure 3.
Interaction energies of fragment (A) and host-guest complexes (B) with quantum optimized conformations. (C) Interaction energies of host-guest complexes with the force-field optimized conformations.
Table 2.
Error statistics for interaction energies of fragment complexes
| Linear Fita |
||||
|---|---|---|---|---|
| MUE (kcal/mol) |
RMSE (kcal/mol) |
m | R2 | |
| OPLS-2005 | 5.1 | 6.8 | 0.27 | 0.18 |
| AMBER* | 3.9 | 5.2 | 0.43 | 0.47 |
| MMFFs | 6.5 | 7.0 | 0.81 | 0.70 |
| MM3*b | 5.3 | 5.8 | 0.40 | 0.52 |
| CHARMm/VC | 3.4 | 5.2 | 0.29 | 0.30 |
m and R2 are slope and correlation coefficient respectively, obtained from linear regression fitting.
MM3* calculations do not include imidazolium containing complexes. MUE: mean unsigned error. RMSE: root-mean-squared error.
Table 3.
Error statistics for interaction energies of host-guest complexes
| Linear Fita |
RMSD (Å)c |
|||||
|---|---|---|---|---|---|---|
| MUE (kcal/mol) |
RMSE (kcal/mol) |
m | R2 | host1 | host2/3 | |
| Quantum optimized structures | ||||||
| OPLS-2005 | 59.1 | 70.9 | 0.96 | 0.90 | -- | -- |
| AMBER* | 35.2 | 53.0 | 0.91 | 0.90 | -- | -- |
| MMFFs | 25.2 | 33.5 | 0.94 | 0.94 | -- | -- |
| MM3*b | 17.2 | 21.7 | 1.07 | 0.99 | -- | -- |
| CHARMm/VC | 9.9 | 17.3 | 0.96 | 0.99 | -- | -- |
| Force-field optimized structures | ||||||
| OPLS-2005 | 32.0 | 43.3 | 1.26 | 0.97 | 2.0 | 0.4 |
| AMBER* | 16.8 | 18.4 | 0.97 | 0.98 | 2.7 | 1.2 |
| MMFFs | 17.2 | 19.3 | 1.02 | 0.98 | 3.3 | 0.5 |
| MM3*b | 20.2 | 28.2 | 1.11 | 0.98 | 2.8 | -- |
| CHARMm/VC | 24.0 | 27.4 | 0.96 | 0.97 | 2.0 | 0.5 |
m and R2 are slope and correlation coefficient respectively, obtained from linear regression fitting.
MM3* calculations do not include imidazolium containing complexes.
Root-mean-squared deviation (RMSD) was computed with respect to the quantum optimized structures.
Structures energy-minimized on the quantum-mechanical energy surface
When we compare quantum and force field energies for conformations energy-minimized on the quantum mechanical energy surface, the mean errors in interaction energies for the fragment complexes are consistently greater than 3.0 kcal/mol, and the deviations are generally larger for more tightly-bound complexes (ΔE < −10 kcal/mol), much as observed in previous studies.34, 35 Interestingly, the force fields underestimate the interaction energies in nearly all cases. The closest agreement with the reference quantum calculations (mean error 3.4 kcal/mol) are for the CHARMm/VC model used in our SAMPL3 submission. AMBER* performed similarly, with a mean error of 3.9 kcal/mol. On the other hand, OPLS-2005, MMFFs, and MM3* show slightly larger errors, on average. Complexes involving imidazolium incur some of the largest errors, always exceeding 9 kcal/mol. (Imidazolium could not be parameterized with MM3* and so we could not compute the interaction energies of imidazolium complexes with this force field and the statistics provide here should be interpreted with this in mind). Other complexes that show large errors (> 5 kcal/mol) include pyridine, methyl benzoate, acetamide, and aniline.
Interaction energies computed for bound complexes of the present host-guest complexes deviate from the reference quantum calculations by several tens of kcal/mol, but still correlate well. Much as for the fragment complexes, the force fields tend to underestimate the interaction energies. However, the errors vary drastically between force fields for these host-guest complexes, in contrast with the fragment complexes. The CHARMm/VC model shows the lowest mean error, along with a very high correlation, while the mean errors are particularly high for OPLS-2005 and AMBER*, with the host1/guest3 interaction energies provided by these models deviating from the quantum result by more than 160 kcal/mol. These were somewhat surprising results, because both MMFF and MM3* yielded larger errors than AMBER* for the fragment complexes.
Structures re-minimized on the force-field energy surfaces
We then tested whether energy-minimizing the host-guest conformations on the various force field potential energy surfaces would improve the agreement of the respective force field energies with the quantum energies minimized on the quantum energy surface. We first compared the geometries of the quantum-minimized structures to the respective force-field optimized structures. For the host1 systems, the root-mean-square deviations (RMSD) of the force-field optimized structures with respect to the quantum-optimized structures are greater than 2.0 Å (Table 3), indicating significant structural deviations (see example in Figure 4.). However, the RMSDs of the host2/3 systems are less than 1.0 Å for most force-fields (Table 3). We further compared the interaction energies of the host-guest complexes in their respective force-field optimized geometries to the interaction energies computed with the quantum model. Although the deviations from quantum are lower than they were for the quantum-minimized conformers (see prior paragraph), the mean unsigned error is still greater than 16 kcal/mol. If we exclude the host1 systems from this comparison, because of their relatively large structural deviations, we still obtain MUE and RMSE values of over 12 kcal/mol and 13 kcal/mol, respectively, for the host 2 and host3 systems, despite the fact that their force-field minimized conformers are quite similar to their quantum minimized conformers. Interestingly, whereas the interaction energies computed using force-fields for the quantum-minimized structures were consistently unfavorable compared to the interaction energies computed using the quantum model (Figures 3A,B), no such trend was observed when using the force-field minimized conformations (Figure 3C).
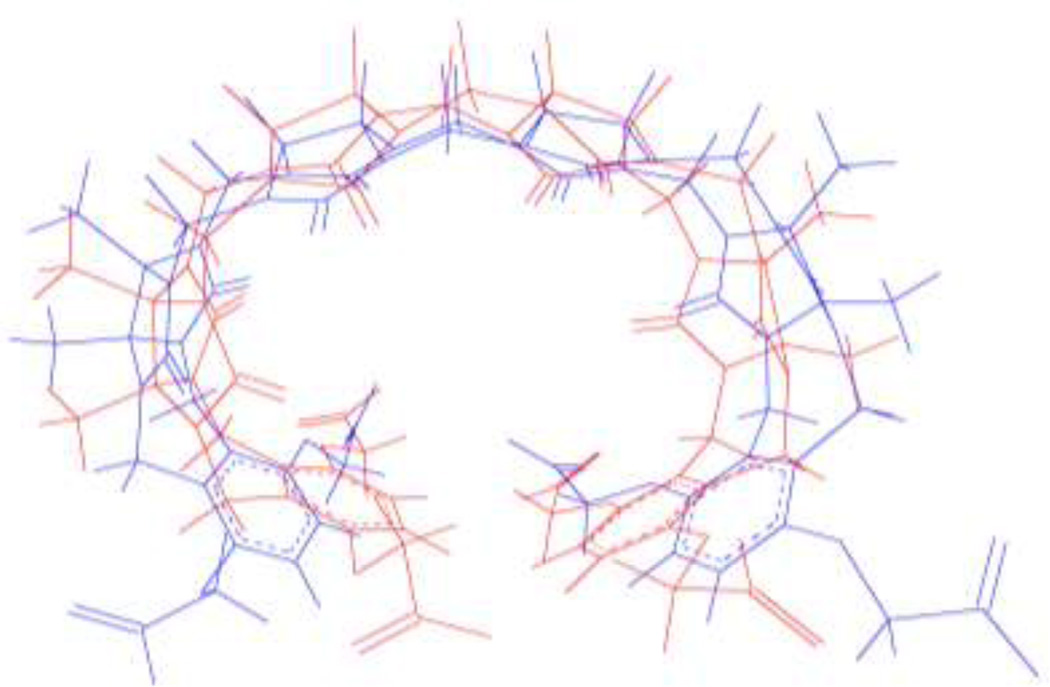
Figure 4.
Superposition of host1 in quantum-minimized conformation (red) and OPLS-2005 minimized conformation (blue), taken from host1/guest7 complex. RMSD between the two structures is 2.2 Å.
Thus, optimization on the quantum energy surface places the system in conformations that are relatively unfavorable on the force-field energy surfaces. Re-minimizing these conformers on the respective force field energy surfaces significantly lowers their force-field energies and, for the relatively flexible host 1, shifts their conformations. The deviations between force field and quantum interaction energies improves thereby, but remains substantial.
Comparison of quantum and force field entropies
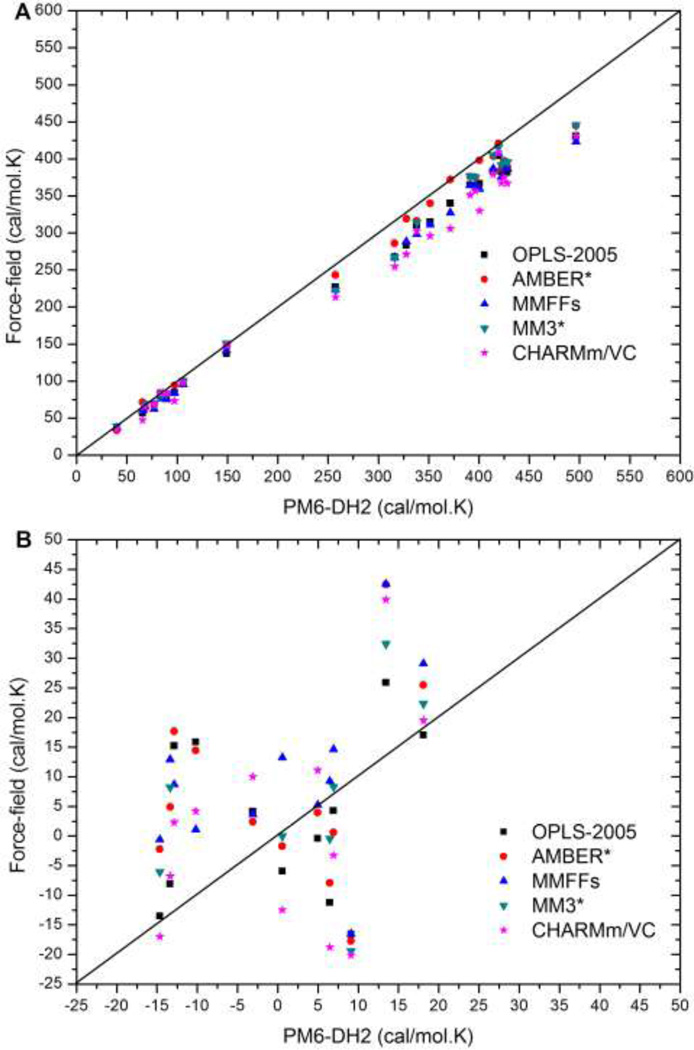
To assess the errors arising from the entropic component of the free energy, we computed the absolute vibrational entropies and binding entropies (difference in absolute entropies) of SAMPL3 molecules and their complexes, each in a representative conformation energy-minimized with the respective energy model. These results are shown in Figure 5, and the corresponding error statistics are summarized in Table 4. On average, vibrational entropies computed with force-fields deviated from the QM calculations by 20.7 cal/mol.K, or 6.2 kcal/mol at 300 K. In general, the errors increased with the size of the system. Among the different force-fields, AMBER* showed the lowest mean error, 14.0 cal/mol.K (4.2 kcal/mol at 300K), and MMFFs showed the highest mean error, 26.9 cal/mol.K (8.1 kcal/mol at 300K). However, the errors in the binding entropies are lower than those in the absolute entropies, indicating significant cancellation of error. On average, the binding entropy differences computed with the force fields deviated from the quantum results by 13.0 cal/mol.K (3.9 kcal/mol at 300K). All the force-fields performed rather similarly in this regard, with mean errors between 11 and 15 cal/mol.K.
Figure 5.
(A) Absolute vibrational entropies of hosts, guests, and complexes in SAMPL3 dataset. (B) Binding entropy differences of host-guest complexes.
Table 4.
Error statistics of absolute and binding vibrational entropies
| Linear Fita |
||||
|---|---|---|---|---|
| MUE (cal/mol.K) |
RMSE (cal/mol.K) |
m | R2 | |
| Vib. entropy | ||||
| OPLS-2005 | 24.7 | 30.2 | 0.90 | 0.99 |
| AMBER* | 14.0 | 19.2 | 0.95 | 0.99 |
| MMFFs | 26.9 | 32.4 | 0.90 | 0.99 |
| MM3*b | 17.4 | 23.7 | 0.93 | 0.99 |
| CHARMm/VC | 33.0 | 41.1 | 0.87 | 0.99 |
| Binding entropy | ||||
| OPLS-2005 | 11.6 | 15.2 | 0.28 | 0.05 |
| AMBER* | 14.9 | 18.1 | 0.28 | 0.04 |
| MMFFs | 14.1 | 16.8 | 0.57 | 0.19 |
| MM3*b | 11.3 | 14.9 | 0.55 | 0.16 |
| CHARMm/VC | 13.6 | 16.2 | 0.61 | 0.15 |
m and R2 are slope and correlation coefficient respectively, obtained from linear regression fitting.
MM3* calculations do not include imidazolium containing complexes.
DISCUSSION
Despite encouraging agreement with experiment in prior applications of M2 to host-guest systems, including one blinded prediction set,7, 36–38 our predictions for the SAMPL3 cases were quite inaccurate. Some of the error may result from incorrect assumptions about the protonation states for certain systems, especially those involving host1, with its cluster of carboxylic acid groups. However, protonation issues are highly unlikely to be the problem for the host2 and host3 cases. We therefore asked whether the problem might derive from problems with the force field, and discovered that the results could not be improved by using several other generalized force-fields. Moreover, the computed binding affinities proved to vary significantly among the different force-fields tested.
This unclear picture motivated us to use the SAMPL3 host-guest systems, and fragments thereof, as a basis for evaluating the accuracy of the force field used in our predictions, as well as several other generalized force fields available for application to these systems. Such comparisons are facilitated by the recent development of PM6-DH2, a semi-empirical QM method that has been shown to yield interaction energies within 0.4 kcal/mol of those obtained with high-level quantum calculations based on coupled-cluster theory,6 on a set of small-molecule complexes that make hydrogen bonding and dispersion interactions. The present comparisons revealed, first, that the force field we used for our predictions yields gas-phase intermolecular interaction energies which differ substantially from the reference quantum calculations. Therefore, the force field error may contribute significantly to the prediction error. Approximations inherent in the implicit solvent model and in the partitioning of the configurational integral into a sum over local energy wells may also have made a significant contribution.
Interaction energies computed with the other force fields tested also diverged substantially from the reference quantum calculations. Across all force fields tested, the errors in the gas-phase interaction energies ranged from 3.2 kcal/mol up to 160 kcal/mol, for the full host-guest complexes. Errors in interaction entropies, i.e., changes in entropy on binding, were considerably lower, amounting to about 3–4 kcal/mol at room temperature. The fact that we found larger energy errors for larger molecules is consistent with concerns recently expressed by Merz and coworkers regarding the additivity of small errors in interaction energies.39, 40 Although errors may cancel to favorable effect in some cases, in general, it clearly is important to use the most accurate possible interaction model.
The errors in interaction energy reported here are significantly larger than those of about 2 kcal/mol previously reported for some of the same force fields, with the S22 and JSCH-2005 benchmarking datasets.34 The latter contain a mix of small-molecule complexes making hydrogen bonding, π-stacking, and dispersion interactions, that are representative of a variety of chemical and biomolecular interactions. The fragment complexes studied here are predominantly dispersion complexes and are slightly larger, which may have contributed to the larger errors. Interestingly, previous validation studies of the OPLS-AA and AMBER ff03 protein force-fields, rather than of the generalized force fields used here perforce, reported mean errors less than 1 kcal/mol relative to reference quantum calculations.35 It thus appears that, although protein force fields have been quite well optimized, there is considerable room for improvement of generalized force fields. This may be particularly true for the chemistries represented in the SAMPL3 host-guest systems. It is important to recognize that there are other force-fields, including GAFF41 and CGenFF33, that we did not evaluate. On the other hand, the errors observed in SAMPL3 predictions by other groups using these force-fields hint that they may have similar issues. Overall, the present results raise concerns regarding the accuracy of generalized force fields in current use for computer-aided drug design.
It is perhaps not surprising that the problem of force field accuracy appears to be particularly acute for the many molecules that are not proteins, given that less effort has been put into such generalized force fields, and that we use them to handle a tremendous variety of chemistries. On the other hand, it is a nontrivial observation that the simplistic functional form of today’s standard force fields can yield rather good accuracy when carefully optimized for proteins; perhaps generalized force fields, too, could be greatly improved by careful optimization.
Another way forward, at least for relatively small systems like the host-guest complexes studied here, may be to directly employ semi-empirical quantum methods, like PM6-DH2, rather than using a force field at all. We are currently investigating this approach for cucurbituril-guest systems, and will report the results in a separate publication.
ACKNOWLEDGEMENTS
This publication was made possible by grant no.GM061300 from NIH. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
REFERENCES
- 1.Jorgensen WL. The many roles of computation in drug discovery. Science. 2004;303:1813–1818. doi: 10.1126/science.1096361. [DOI] [PubMed] [Google Scholar]
- 2.Gilson MK, Zhou HX. Calculation of protein-ligand binding affinities. Annual Review of Biophysics and Biomolecular Structure. 2007;36:21–42. doi: 10.1146/annurev.biophys.36.040306.132550. [DOI] [PubMed] [Google Scholar]
- 3.Houk KN, Leach AG, Kim SP, Zhang XY. Binding affinities of host-guest, protein-ligand, and protein-transition-state complexes. Angewandte Chemie-International Edition. 2003;42:4872–4897. doi: 10.1002/anie.200200565. [DOI] [PubMed] [Google Scholar]
- 4.Gohlke H, Klebe G. Approaches to the description and prediction of the binding affinity of small-molecule ligands to macromolecular receptors. Angewandte Chemie-International Edition. 2002;41:2645–2676. doi: 10.1002/1521-3773(20020802)41:15<2644::AID-ANIE2644>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 5.Rodinger T, Pomes R. Enhancing the accuracy, the efficiency and the scope of free energy simulations. Current Opinion in Structural Biology. 2005;15:164–170. doi: 10.1016/j.sbi.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 6.Rezac J, Fanfrlik J, Salahub D, Hobza P. Semiempirical Quantum Chemical PM6 Method Augmented by Dispersion and H-Bonding Correction Terms Reliably Describes Various Types of Noncovalent Complexes. Journal of Chemical Theory and Computation. 2009;5:1749–1760. doi: 10.1021/ct9000922. [DOI] [PubMed] [Google Scholar]
- 7.Chang CE, Gilson MK. Free energy, entropy, and induced fit in host-guest recognition: Calculations with the second-generation mining minima algorithm. Journal of the American Chemical Society. 2004;126:13156–13164. doi: 10.1021/ja047115d. [DOI] [PubMed] [Google Scholar]
- 8.Head MS, Given JA, Gilson MK. "Mining minima": Direct computation of conformational free energy. Journal of Physical Chemistry A. 1997;101:1609–1618. [Google Scholar]
- 9.Chang CE, Potter MJ, Gilson MK. Calculation of molecular configuration integrals. Journal of Physical Chemistry B. 2003;107:1048–1055. [Google Scholar]
- 10.Gilson MK. Multiple-Site Titration and Molecular Modeling - 2 Rapid Methods for Computing Energies and Forces for Ionizable Groups in Proteins. Proteins-Structure Function and Genetics. 1993;15:266–282. doi: 10.1002/prot.340150305. [DOI] [PubMed] [Google Scholar]
- 11.Qiu D, Shenkin PS, Hollinger FP, Still WC. The GB/SA continuum model for solvation. A fast analytical method for the calculation of approximate Born radii. Journal of Physical Chemistry A. 1997;101:3005–3014. [Google Scholar]
- 12.Gilson MK, Honig B. Calculation of the Total Electrostatic Energy of A Macromolecular System - Solvation Energies, Binding-Energies, and Conformational-Analysis. Proteins-Structure Function and Genetics. 1988;4:7–18. doi: 10.1002/prot.340040104. [DOI] [PubMed] [Google Scholar]
- 13.Sitkoff D, Sharp KA, Honig B. Accurate Calculation of Hydration Free-Energies Using Macroscopic Solvent Models. Journal of Physical Chemistry. 1994;98:1978–1988. [Google Scholar]
- 14.Madura JD, Briggs JM, Wade RC, Davis ME, Luty BA, Ilin A, Antosiewicz J, Gilson MK, Bagheri B, Scott LR, Mccammon JA. Electrostatics and Diffusion of Molecules in Solution - Simulations with the University-Of-Houston Brownian Dynamics Program. Computer Physics Communications. 1995;91:57–95. [Google Scholar]
- 15.Chang CE, Gilson MK. Tork: Conformational analysis method for molecules and complexes. Journal of Computational Chemistry. 2003;24:1987–1998. doi: 10.1002/jcc.10325. [DOI] [PubMed] [Google Scholar]
- 16.Chen W, Huang J, Gilson MK. Identification of symmetries in molecules and complexes. Journal of Chemical Information and Computer Sciences. 2004;44:1301–1313. doi: 10.1021/ci049966a. [DOI] [PubMed] [Google Scholar]
- 17.Momany FA, Rone R. Validation of the General-Purpose Quanta(R)3.2/Charmm(R) Force-Field. Journal of Computational Chemistry. 1992;13:888–900. [Google Scholar]
- 18.Gilson MK, Gilson HSR, Potter MJ. Fast assignment of accurate partial atomic charges: An electronegativity equalization method that accounts for alternate resonance forms. Journal of Chemical Information and Computer Sciences. 2003;43:1982–1997. doi: 10.1021/ci034148o. [DOI] [PubMed] [Google Scholar]
- 19.Trott O, Olson AJ. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. Journal of Computational Chemistry. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohamadi F, Richards NGJ, Guida WC, Liskamp R, Lipton M, Caufield C, Chang G, Hendrickson T, Still WC. Macromodel - An Integrated Software System for Modeling Organic and Bioorganic Molecules Using Molecular Mechanics. Journal of Computational Chemistry. 1990;11:440–467. [Google Scholar]
- 21.Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. Journal of Chemical Theory and Computation. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 22.Stewart JJP. Mopac - A Semiempirical Molecular-Orbital Program. Journal of Computer-Aided Molecular Design. 1990;4:1–45. doi: 10.1007/BF00128336. [DOI] [PubMed] [Google Scholar]
- 23.Jorgensen WL, Maxwell DS, TiradoRives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. Journal of the American Chemical Society. 1996;118:11225–11236. [Google Scholar]
- 24.Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. Journal of Physical Chemistry B. 2001;105:6474–6487. [Google Scholar]
- 25.Weiner SJ, Kollman PA, Nguyen DT, Case DA. An All Atom Force-Field for Simulations of Proteins and Nucleic-Acids. Journal of Computational Chemistry. 1986;7:230–252. doi: 10.1002/jcc.540070216. [DOI] [PubMed] [Google Scholar]
- 26.Weiner SJ, Kollman PA, Case DA, Singh UC, Ghio C, Alagona G, Profeta S, Weiner P. A New Force-Field for Molecular Mechanical Simulation of Nucleic-Acids and Proteins. Journal of the American Chemical Society. 1984;106:765–784. [Google Scholar]
- 27.Halgren TA. Merck molecular force field .1. Basis, form, scope, parameterization, and performance of MMFF94. Journal of Computational Chemistry. 1996;17:490–519. [Google Scholar]
- 28.Halgren TA. MMFF VI. MMFF94s option for energy minimization studies. Journal of Computational Chemistry. 1999;20:720–729. doi: 10.1002/(SICI)1096-987X(199905)20:7<720::AID-JCC7>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 29.Halgren TA. MMFF VII. Characterization of MMFF94, MMFF94s, and other widely available force fields for conformational energies and for intermolecular-interaction energies and geometries. Journal of Computational Chemistry. 1999;20:730–748. doi: 10.1002/(SICI)1096-987X(199905)20:7<730::AID-JCC8>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 30.Allinger NL, Yuh YH, Lii JH. Molecular Mechanics - the Mm3 Force-Field for Hydrocarbons .1. Journal of the American Chemical Society. 1989;111:8551–8566. [Google Scholar]
- 31.Kolossvary I, Guida WC. Low mode search. An efficient, automated computational method for conformational analysis: Application to cyclic and acyclic alkanes and cyclic peptides. Journal of the American Chemical Society. 1996;118:5011–5019. [Google Scholar]
- 32.Zhou HX, Gilson MK. Theory of Free Energy and Entropy in Noncovalent Binding. Chemical Reviews. 2009;109:4092–4107. doi: 10.1021/cr800551w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, MacKerell AD. CHARMM General Force Field: A Force Field for Drug-Like Molecules Compatible with the CHARMM All-Atom Additive Biological Force Fields. Journal of Computational Chemistry. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paton RS, Goodman JM. Hydrogen Bonding and pi-Stacking: How Reliable are Force Fields? A Critical Evaluation of Force Field Descriptions of Nonbonded Interactions. Journal of Chemical Information and Modeling. 2009;49:944–955. doi: 10.1021/ci900009f. [DOI] [PubMed] [Google Scholar]
- 35.Berka K, Laskowski R, Riley KE, Hobza P, Vondrasek J. Representative Amino Acid Side Chain Interactions in Proteins. A Comparison of Highly Accurate Correlated ab Initio Quantum Chemical and Empirical Potential Procedures. Journal of Chemical Theory and Computation. 2009;5:982–992. doi: 10.1021/ct800508v. [DOI] [PubMed] [Google Scholar]
- 36.Moghaddam S, Inoue Y, Gilson MK. Host-Guest Complexes with Protein-Ligand-like Affinities: Computational Analysis and Design. Journal of the American Chemical Society. 2009;131:4012–4021. doi: 10.1021/ja808175m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moghaddam S, Yang C, Rekharsky M, Ko YH, Kim K, Inoue Y, Gilson MK. New Ultrahigh Affinity Host-Guest Complexes of Cucurbit[7]uril with Bicyclo[2.2.2]octane and Adamantane Guests: Thermodynamic Analysis and Evaluation of M2 Affinity Calculations. Journal of the American Chemical Society. 2011;133:3570–3581. doi: 10.1021/ja109904u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen W, Chang CE, Gilson MK. Calculation of cyclodextrin binding affinities: Energy, entropy, and implications for drug design. Biophysical Journal. 2004;87:3035–3049. doi: 10.1529/biophysj.104.049494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Merz KM. Limits of Free Energy Computation for Protein-Ligand Interactions. Journal of Chemical Theory and Computation. 2010;6:1769–1776. doi: 10.1021/ct100102q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Faver JC, Benson ML, He X, Roberts BP, Wang B, Marshall MS, Sherrill CD, Merz KM. The Energy Computation Paradox and ab initio Protein Folding. Plos One. 2011;6 doi: 10.1371/journal.pone.0018868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang JM, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. Journal of Computational Chemistry. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]