Abstract
The kinetochore is the proteinaceous complex that governs the movement of duplicated chromosomes by interacting with spindle microtubules during mitosis and meiosis. Faithful chromosome segregation requires that kinetochores form robust load-bearing attachments to the tips of dynamic spindle microtubules, correct microtubule attachment errors, and delay the onset of anaphase until all chromosomes have made proper attachments. To understand how this macromolecular machine operates to segregate duplicated chromosomes with exquisite accuracy, it is critical to reconstitute and study kinetochore-microtubule interactions in vitro using defined components. Here, we review the current status of reconstitution as well as recent progress in understanding the microtubule binding functions of kinetochores in vivo.
Keywords: Chromosome segregation, mitosis, kinetochore, microtubules, in vitro reconstitution
Introduction
Accurate partitioning of genetic material during mitosis and meiosis is essential for all organisms to proliferate. Defects in this process result in aneuploidy that can lead to cell death, cancer or birth defects (Holland and Cleveland 2009; Schvartzman et al. 2010). In eukaryotes, chromosome segregation is directed by the kinetochore that assembles onto the centromeric region of each chromosome, and by dynamic microtubules that grow and shrink by incorporating or dissociating α/β tubulin subunits at the ends (Nicklas 1997; Cheeseman and Desai 2008; Santaguida and Musacchio 2009; Lampert and Westermann 2011). Microtubules continuously switch from growth to shortening (catastrophe) and from shortening to growth (rescue), a property known as dynamic instability (Mitchison and Kirschner 1984). Kinetochores govern chromosome movement by coupling to these growing and shrinking microtubule tips. Faithful chromosome segregation requires that sister kinetochores form bi-oriented attachments to spindle microtubules emanating from opposite poles. In addition, attachment errors must be corrected to avoid mis-segregation. The spindle checkpoint monitors attachment status and delays the onset of anaphase until all chromosomes form correct bi-oriented attachments.
The simplest characterized kinetochore is found in budding yeast where each kinetochore assembles onto a defined 125 bp of centromeric DNA and binds a single microtubule (Winey et al. 1995; Westermann et al. 2007). In contrast, most organisms have much larger centromeres and their kinetochores bind to multiple microtubules (Cleveland et al. 2003). Yet, the conservation of kinetochore proteins suggests that the larger kinetochores of other eukaryotes may be assembled from the repetition of the budding yeast-type kinetochores, called the repeat subunit model (Zinkowski et al. 1991; Joglekar et al. 2008). The core of the budding yeast kinetochore is composed of ~40 proteins, most of which are present in multiple copies within each kinetochore. It has been estimated that anywhere from approximately 200 or more (Joglekar et al. 2006) to as many as 500 (Coffman et al. 2011; Lawrimore et al. 2011) total proteins constitute the core of the budding yeast kinetochore. In addition, >20 additional regulatory proteins modulate kinetochore functions, such as kinases and motor proteins (Cheeseman and Desai 2008). The complexity and dynamic nature of the kinetochore makes it a daunting task to understand how it accurately segregates duplicated chromosomes. The most basic kinetochore function is to mediate the interaction between centromeric DNA and spindle microtubules. In this regard, kinetochores can be classified into two functional parts: the inner kinetochore that binds centromeric DNA and the outer kinetochore that binds microtubules. Here, we focus on the kinetochore-microtubule interface. We refer readers to recent reviews on inner kinetochores (Buscaino et al. 2010; Black and Cleveland 2011; Gascoigne and Cheeseman 2011; Perpelescu and Fukagawa 2011; Verdaasdonk and Bloom 2011).
Kinetochore constituent lists
Extensive efforts over the last 30 years have led to the identification of most kinetochore proteins in major model organisms (for detailed reviews, see (Westermann et al. 2007; Cheeseman and Desai 2008; Santaguida and Musacchio 2009; Przewloka and Glover 2009; Perpelescu and Fukagawa 2011). The following picture has emerged from these studies. Kinetochores consist of several subcomplexes that represent functional units. Overall kinetochore architecture appears conserved from yeast to humans based on the conservation of kinetochore proteins, as well as the similar stoichiometry and position of subcomplexes across species (Joglekar et al. 2006; Schittenhelm et al. 2007; Joglekar et al. 2008; Joglekar et al. 2009; Wan et al. 2009; Johnston et al. 2010). At the base of the kinetochore lies the centromere-specific histone H3 variant, CENP-A (also called CenH3) (Allshire and Karpen 2008; Mendiburo et al. 2011). Although CENP-A forms specialized chromatin that is essential for kinetochore assembly, the exact composition and nature of CENP-A containing nucleosomes in vivo remains unclear (Henikoff and Furuyama 2010; Black and Cleveland 2011). The so-called CCAN (constitutive centromere-associated network: at least 16 components in human, 13 in budding yeast) loads onto centromeric chromatin and is important for outer kinetochore assembly (Perpelescu and Fukagawa 2011). The outer kinetochore consists of the conserved KMN network (KNL1, Mis12, Ndc80 subcomplexes) that serves as a core microtubule-binding module, as well as other microtubule binding subcomplexes (e.g. Dam1 complex in fungi, Ska1 complex in mammals) (Cheeseman and Desai 2008). It is estimated that there are ~7 copies of KMN per microtubule binding unit per single CENP-A nucleosome (Joglekar et al. 2006; Furuyama and Biggins 2007; Joglekar et al. 2008; Joglekar et al. 2009; Wan et al. 2009; Johnston et al. 2010; Coffman et al. 2011; Lawrimore et al. 2011). In addition to these core kinetochore components, many regulatory proteins also localize to kinetochores, such as spindle checkpoint proteins, microtubule-associated proteins (MAPs), motor proteins, mitotic kinases and phosphatases (Cheeseman and Desai 2008). As the list of kinetochore components is becoming complete, the next challenge is to understand how these numerous proteins function to form the macromolecular machine that segregates chromosomes with exquisite fidelity. In vitro reconstitution of kinetochore functions using individual subcomplexes as well as their ensembles is critical to achieve this goal.
What are the functions of kinetochores?
Lateral and end-on attachments, error-correction and the spindle checkpoint
A fundamental kinetochore function is to interact with dynamic microtubules. Because the microtubule lattice offers a much larger contact surface than its tips, kinetochores first form lateral attachments, which are then converted to end-on attachments (Fig. 1) (Rieder and Alexander 1990; Tanaka et al. 2005). The fact that microtubule polymerization/depolymerization occurs only at the tips means that kinetochores must stay bound to dynamic microtubule tips while allowing the incorporation and dissociation of thousands of tubulin subunits at the ends. This end-on attachment allows kinetochores to translate the energy stored in dynamic microtubules to move chromosomes upon depolymerization (Mandelkow et al. 1991; Grishchuk et al. 2005; Wang and Nogales 2005). End-on attachments also allow kinetochores to modulate microtubule dynamics, which may be critical to regulate attachment stability (see below). The observation that tip-bound kinetochores appear to be under tension in vivo means that kinetochores must form load-bearing attachments (Khodjakov and Rieder 1996; Waters et al. 1996). It is estimated that 0.4 to 8 pN of force is transmitted to kinetochores per attached microtubule in vivo (Nicklas 1988; Powers et al. 2009). The interaction between kinetochores and microtubules must be stable enough to maintain attachments throughout mitosis (which can last for more than 1 hour), yet flexible enough to allow destabilization of erroneous attachments. Although a back-to-back positioning of sister kinetochores increases the chance of bi-orientation (Östergren 1951; Indjeian and Murray 2007; Sakuno et al. 2009), the formation of erroneous attachments is somewhat inevitable because kinetochores are unable to tell from which pole any given microtubules emanate. For example, both kinetochores can attach to microtubules from the same pole (called mono-oriented attachment, as shown in Fig. 2b), or one kinetochore can attach to microtubules from both poles (called merotelic attachment). Although it remains unclear how often erroneous attachments are formed during mitosis, many erroneous kinetochore-microtubule attachments are observed during prometaphase of meiosis I, suggesting that error correction is essential to achieve correct bi-orientation (Kitajima et al. 2011; Sakuno et al. 2011). How do cells distinguish bi-oriented attachments from erroneous ones? One distinct feature of bi-oriented attachment is the presence of robust tension due to microtubule pulling forces that are opposed by the linkage between sister chromatids (Fig. 2a). In contrast, less tension is produced when sister kinetochores are mono-oriented (Fig. 2b). Therefore, by destabilizing tension-less attachments and/or stabilizing attachments under full tension, all chromosomes should be able to achieve bi-orientation. Classic micromanipulation studies in insect spermatocytes as well as chromosome engineering experiments in budding yeast demonstrated that tension indeed stabilizes attachments in vivo (Nicklas and Koch 1969; Nicklas and Ward 1994; Dewar et al. 2004). However, because protein-protein interactions are typically destabilized by tension (Bell 1978; Merkel et al. 1999), it is intriguing that cells rely on tension to stabilize kinetochore-microtubule attachments. As discussed later, the prevailing model for the underlying molecular mechanism that leads to the destabilization of attachments is phospho-regulation by the Aurora B kinase. In addition, our recent reconstitution studies discovered another mechanism by which tension directly stabilizes attachments by modulating microtubule tip dynamics (Akiyoshi et al. 2010).
Fig. 1.
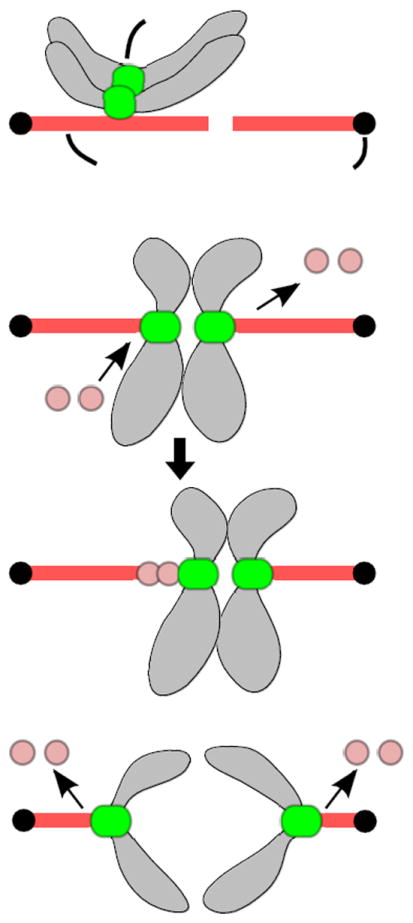
Lateral and end-on attachments. a Lateral attachments. Kinetochores initially encounter the lateral side of spindle microtubules during prometaphase. b End-on attachments during metaphase. Kinetochores interact with the tips of spindle microtubules where incorporation and dissociation of tubulin subunits occur. In this example, microtubules emanating from the left pole grow and those from the right pole shorten (top), leading to rightward movement of the attached chromosomes (bottom). c End-on attachments during anaphase. Linkage between sister chromatids is lost and microtubules from both sides shorten, resulting in the separation of sister chromatids to opposite poles.
Fig. 2.
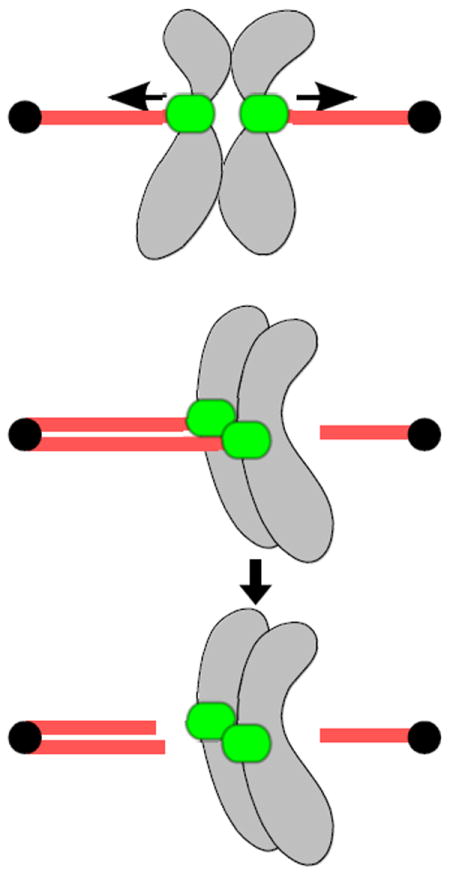
Different types of kinetochore-microtubule attachments. a Bi-oriented attachments in which sister kinetochores attach to microtubules emanating from opposite poles. Kinetochores are under robust tension due to microtubule pulling forces that are opposed by linkage between sister chromatids. Tension stabilizes these proper bi-oriented attachments. b Mono-oriented attachment in which sister kinetochores are attached to microtubules emanating from the same pole (top). Lack of tension destabilizes these improper attachments, giving cells another chance to achieve correct bi-oriented attachments (bottom).
Cells must also coordinate the timing of anaphase onset with chromosome alignment so that segregation occurs only after all chromosomes have achieved bi-orientation. A surveillance mechanism, known as the spindle checkpoint, evolved for this purpose. Spindle checkpoint proteins are recruited to kinetochores prior to bi-orientation, activating the signaling cascade that inhibits the activation of the anaphase promoting complex (APC/C) (for reviews, see (Peters 2006; Musacchio and Salmon 2007; Nezi and Musacchio 2009; Murray 2011; Pines 2011)). These proteins dissociate from kinetochores once correct attachments are formed, suggesting that they monitor attachment status and/or tension at kinetochores. However, it is still unclear how many of the spindle checkpoint proteins interact with kinetochores and how this interaction is regulated.
Regulation of microtubule attachments by mitotic kinases and phosphatases
The Aurora B kinase, a catalytic subunit of the chromosomal passenger complex (CPC), plays a critical role in destabilizing erroneous attachments (Ruchaud et al. 2007; Kelly and Funabiki 2009). Because it localizes to the inner centromere where it overlaps with kinetochores before bi-orientation but not after, it was proposed that this spatial arrangement of Aurora B allows selective destabilization of tension-less attachments (Tanaka et al. 2002). More recently, based on previous observations that Aurora B rapidly turns over at inner centromeres (Murata-Hori and Wang 2002), a diffusion-based intra-cellular phosphorylation gradient was proposed and experimentally observed in vivo (Fuller et al. 2008; Liu et al. 2009; Welburn et al. 2010; Wang et al. 2011; Tan and Kapoor 2011). However, Aurora B can efficiently phosphorylate its targets even if they localize at a micrometer rather than nanometer distance scale from the inner centromere, suggesting that counteracting phosphatases must play critical roles to balance local phosphorylation on kinetochores (Tan and Kapoor 2011; Wang et al. 2011). An important phosphatase that counteracts Aurora B is the protein phosphatase 1 (PP1) (for reviews, see (De Wulf et al. 2009; Wurzenberger and Gerlich 2011)). PP1 is found at outer kinetochores, showing enrichment at bi-oriented kinetochores (Trinkle-Mulcahy et al. 2003; Liu et al. 2010). Several kinetochore proteins recruit PP1 onto kinetochores, including KNL1, CENP-E and Fin1 (Akiyoshi et al. 2009a; Kim et al. 2010; Liu et al. 2010; Meadows et al. 2011; Rosenberg et al. 2011). It remains to be determined if PP1 is recruited by different kinetochore proteins to dephosphorylate distinct substrates. Recent work suggests that PP2A may also promote attachment stability by counteracting Aurora B and Plk1 (Foley et al. 2011). In addition to Aurora B, the Mps1 kinase is also implicated in regulating kinetochore-microtubule attachments, although its mechanism of action remains to be determined (Maure et al. 2007; Jelluma et al. 2008; Kemmler et al. 2009; Hewitt et al. 2010; Lan and Cleveland 2010; Maciejowski et al. 2010; Santaguida et al. 2010; Dou et al. 2011).
Numerous microtubule binding kinetochore proteins are phosphorylated by Aurora B. For example, the Ndc80 protein is regulated by Aurora B and the non-phosphorylatable mutant shows hyper-stable attachments and severe segregation defects in human and PtK1 cells, demonstrating the importance of phospho-regulation (DeLuca et al. 2006; DeLuca et al. 2011). In contrast, the non-phosphorylatable mutant is viable in budding yeast and chicken cells, implying that the regulation of additional targets is sufficient for faithful chromosome segregation in these organisms (Akiyoshi et al. 2009b; Welburn et al. 2010). KNL1 and Dsn1 are also phosphorylated by Aurora B, and their combinatorial phosphorylation appears important to regulate the microtubule binding activity of the KMN network (Welburn et al. 2010). Dam1 is a key Aurora target in budding yeast, and non-phosphorylatable mutants show chromosome segregation defects while phospho-mimicking mutants can suppress temperature sensitivity of a kinase mutant (Cheeseman et al. 2002). Although many kinetochore proteins are regulated by kinases, it is less clear how phosphorylation affects their activity or behavior at molecular levels. Phosphorylation can regulate attachment stability by affecting various parameters, such as detachment rate, microtubule dynamics, and interaction strength with associating kinetochore proteins. To understand how these kinases/phosphatases regulate kinetochore-microtubule attachments, it is essential to identify their targets as well as to reveal the parameters that are modulated by phosphorylation.
Why do we need to reconstitute the kinetochore-microtubule interface?
Reconstitution of the kinetochore-microtubule interface is critical to understanding aspects of kinetochore function that have not been easily answered by in vivo studies. Within the cell, numerous factors affect kinetochore-microtubule attachments either directly or indirectly. For example, inactivation of proteins important for microtubule dynamics can lead to kinetochore-microtubule attachment defects even if they have no direct roles at kinetochores. Also, kinetochores are thought to assemble in a hierarchical manner such that the localization of outer kinetochore proteins depends on inner kinetochores (De Wulf et al. 2003; Nekrasov et al. 2003; Pinsky et al. 2003). Therefore, the inactivation of one kinetochore protein can indirectly affect many others, complicating the interpretation of the results. Reconstituting kinetochore functions in vitro with pure components allows researchers to observe a system in a controlled manner as well as to interrogate specific proteins or change other parameters to see how the system responds. One of the earliest reconstitution studies using isolated chromosomes led to the important finding that depolymerizing microtubules can do mechanical work to move chromosomes in the absence of any molecular motor (Mitchison and Kirschner 1985; Koshland et al. 1988; Coue et al. 1991). This was later corroborated by the finding that minus end directed motors are not required for poleward chromosome movement in yeast (Grishchuk and McIntosh 2006; Tanaka et al. 2007).
In vitro studies are also crucial to reveal the structure of the kinetochore-microtubule interface, which is essential to ultimately understanding how kinetochores couple chromosomes to dynamic microtubule ends (Welburn and Cheeseman 2008). Although electron microscopy (EM) studies on cells have pictured overall kinetochore structure (Brinkley and Stubblefield 1966; McEwen et al. 1998; Dong et al. 2007; McIntosh et al. 2008; McEwen and Dong 2010), higher resolution images are required to reveal the nature of individual kinetochore-microtubule attachments. Another limitation of cellular EM is that it is difficult to reveal the identity of observed structures. For example, although EM tomographic reconstitution led McIntosh and colleagues to discover slender structures (called “fibrils”) that appear to connect kinetochores to the inner face of microtubule plus ends, it remains unknown which proteins make up the fibrils (McIntosh et al. 2008). Reconstituting kinetochore functions in vitro for biochemical and structural analyses is therefore key to understanding how the dynamic kinetochore-microtubule interface is formed and regulated.
Reconstitution should also be invaluable to understanding the spindle checkpoint. Although checkpoint proteins localize to unattached kinetochores, the molecular requirements to displace checkpoint proteins from kinetochores are unclear. Do they dissociate upon formation of lateral or end-on attachment? Or, if attachment is not enough, how much tension is required to displace them from kinetochores? Although the extent of inter-kinetochore stretch (distance between sister kinetochores) is often used as a read-out for tension, work from several labs suggests that it is rather the intra-kinetochore stretch that is monitored by the checkpoint (Maresca and Salmon 2009; Uchida et al. 2009; Wan et al. 2009; Maresca and Salmon 2010; Suzuki et al. 2011). Further complicating the matter, recent high-resolution 3D imaging of human cells showed that even laterally-attached kinetochores may also be under some tension (Magidson et al. 2011). Difficulties in understanding the molecular details of the spindle checkpoint derive from a lack of robust reconstitution assays that would allow researchers to directly examine the effect of microtubule binding and tension on the behavior of checkpoint proteins at kinetochores (Murray 2011).
How can the kinetochore-microtubule interface be reconstituted?
Preparation of pure kinetochores, and their functional assays
To reconstitute and study the kinetochore-microtubule interface in vitro, one needs kinetochores, microtubules and assays. There are two major approaches to prepare pure kinetochores: expression of recombinant proteins and purification from cells. Due to difficulties in purifying native kinetochores or obtaining large quantities of kinetochore proteins from cells, most studies have utilized recombinant expression to reconstitute kinetochore subcomplexes. Although many kinetochore proteins are insoluble when expressed individually, solubility can often be achieved by co-expressing interacting partners (for example (Ciferri et al. 2005; Wei et al. 2005)). Polycistronic vectors that allow the simultaneous expression of multiple genes from a single vector facilitate the formation and purification of stable subcomplexes (Tan 2001). Although whole kinetochores have not yet been reconstituted, studies of subcomplexes have led to significant progress in understanding the molecular mechanism of kinetochore functions. While recombinant expression represents a bottom-up approach, purification of native kinetochores from cells is a top-down one. Both approaches are critical to understanding the functions of individual complexes as well as their ensemble properties. Table 1 summarizes the complexes reconstituted or purified and used in microtubule binding assays in vitro to date.
Table 1.
List of characterized microtubule-binding kinetochore proteins and complexes
| Name | Assays | Comments | References |
|---|---|---|---|
| Ndc80 complex | Sedimentation TIRF Optical trap EM |
|
(Ciferri et al. 2005; Wei et al. 2005; Cheeseman et al. 2006; Wei et al. 2007; Ciferri et al. 2008; McIntosh et al. 2008; Wilson-Kubalek et al. 2008; Powers et al. 2009; Alushin et al. 2010) |
| KMN network | Sedimentation |
|
(Cheeseman et al. 2004; Cheeseman et al. 2006; Welburn et al. 2010) |
| Dam1 complex | Sedimentation TIRF Optical trap EM |
|
(Hofmann et al. 1998; Miranda et al. 2005; Westermann et al. 2005; Asbury et al. 2006; Westermann et al. 2006; Franck et al. 2007; Gestaut et al. 2008; Grishchuk et al. 2008; Gao et al. 2010; Lampert et al. 2010) |
| Ndc80 complex + Dam1 complex | Sedimentation TIRF Optical trap |
|
(Lampert et al. 2010; Tien et al. 2010) |
| Ska1 complex | Sedimentation Beads assay EM |
|
(Welburn et al. 2009) |
| Kinetochore particles purified from budding yeast | TIRF Optical trap |
|
(Akiyoshi et al. 2010) |
The following proteins show microtubule binding activity as well as localization to kinetochores: SKAP-Astrin/Kinastrin complex (Mack and Compton 2001; Manning et al. 2010; Schmidt et al. 2010; Dunsch et al. 2011), CENP-Q (Amaro et al. 2010), MCAK (Wordeman and Mitchison 1995; Desai et al. 1999; Cooper et al. 2010; Oguchi et al. 2011), CENP-E, (Kim et al. 2008; Maffini et al. 2009), Fin1 (Woodbury and Morgan 2007), Kar3 (Tanaka et al. 2005; Tytell and Sorger 2006), INCENP/Survivin (Wheatley et al. 2001; Sandall et al. 2006), XMAP215 (Garcia et al. 2001; He et al. 2001; Brouhard et al. 2008; Kitamura et al. 2010; Al-Bassam and Chang 2011), CLASP (Maiato et al. 2003; Cheeseman et al. 2005; Ortiz et al. 2009), CENP-F (Feng et al. 2006) that recruits NDE1, NDEL1 (Vergnolle and Taylor 2007), RZZ complex (Karess 2005) and Spindly (Griffis et al. 2007; Gassmann et al. 2008) that recruit dynein/dynactin to kinetochores (Howell et al. 2001; Yang et al. 2007). Other proteins that may directly bind microtubules or contribute to microtubule binding by recruiting other factors include: Cep57 (Emanuele and Stukenberg 2007), Bod1 (Porter et al. 2007), Shugoshin (Salic et al. 2004; Huang et al. 2007; Tanno et al. 2010), mDia3 (Yasuda et al. 2004; Cheng et al. 2011), Kebab (Meireles et al. 2011), and CAMP (Itoh et al. 2011).
There are a variety of microtubule-binding assays. So far, most experiments have used stabilized microtubules and conventional microtubule sedimentation assays (Fig. 3a). Although these assays are useful to determine if proteins possess microtubule binding activity, they can’t distinguish the precise type of interaction (for example, do they bind the microtubule lattice or the tip? How many contact points do they make?). Therefore, it is essential to directly study the interface between kinetochore proteins and individual microtubules. EM is a powerful technique that can visualize the kinetochore-microtubule interface at the nanometer level. It is also critical to use dynamic microtubules to understand how kinetochores couple to polymerizing and depolymerizing microtubule tips. Single-molecule techniques enable studies of individual interactions between microtubules and kinetochore proteins/subcomplexes. TIRF (Total Internal Reflection Fluorescence) microscopy allows visualization of fluorescently labeled proteins and microtubules at a single molecule sensitivity by restricting the imaging area within ~100 nm of the coverslip (Axelrod et al. 1984) (Fig. 3b). Optical trapping is a technique that can apply mechanical force to protein-protein linkages (Block et al. 1990). In this assay, polystyrene beads that are decorated with kinetochore subcomplexes (or larger assemblies) are attached to microtubules so one can assess their biophysical characteristics, such as how much force they can bear (Franck et al. 2010) (Fig. 3c). Below, we highlight some reconstituted subcomplexes and the insights gained from their study.
Fig. 3.
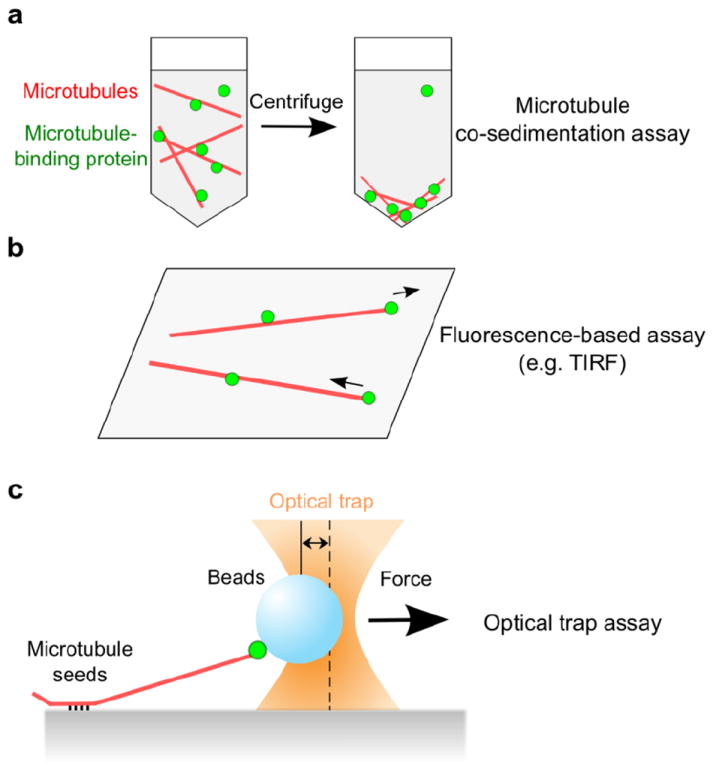
Examples of microtubule-binding assays. a A microtubule co-sedimentation assay analyzes whether proteins of interest stay associated with stabilized (non-dynamic) microtubules that are pelleted by centrifugation. The level of co-sedimentation is typically determined by immunoblots. b A fluorescence-based assay observes binding between fluorescent microtubules attached to a coverslip and fluorescently labeled proteins. TIRF microscopy enables visualization of molecules only in the evanescent field (within ~100 nm of cover slip), providing single-molecule sensitivity (i.e. observation of binding events between individual microtubules and individual kinetochore proteins or complexes). Dynamically growing and shortening microtubules can also be used, allowing studies of lateral versus end-on attachments. c Optical trap assays use a focused laser beam to apply force to the interaction between bead-bound proteins and dynamic microtubule tips. Growth and shrinkage of individual microtubules can be monitored by video-enhanced microscopy and the number of bead-bound proteins can be diluted low enough to allow studies of single molecules or complexes.
Reconstitution using recombinant individual kinetochore subcomplexes: Ndc80, Dam1 and more
Despite the identification of numerous kinetochore proteins, the identity of the factors that directly bind to microtubules remained unknown for a long time. Although unattached kinetochores were observed in mutational studies of many kinetochore proteins, one could not tell if they have direct microtubule binding activity or general structural roles that lead to defective microtubule attachments when inactivated (Pinsky et al. 2006). A breakthrough came when the conserved Ndc80 complex (composed of Ndc80, Nuf2, Spc24 and Spc25) was reconstituted and shown to have microtubule binding activity in vitro (Cheeseman et al. 2006; Wei et al. 2007). Structural studies showed that the Ndc80 complex is a 57 nm elongated structure with globular domains at both ends (Ciferri et al. 2005; Wei et al. 2005). The Ndc80-Nuf2 globular domain binds microtubules while the Spc24-Spc25 globular domain faces the inner kinetochore (Cheeseman et al. 2006; DeLuca et al. 2006). Initial EM analysis revealed that the Ndc80 complex binds microtubules at a defined angle (Cheeseman et al. 2006). X-ray crystallography studies show that both Ndc80 and Nuf2 possess a calponin-homology (CH) domain (Wei et al. 2007; Ciferri et al. 2008), which is found in the EB1 microtubule tip-tracking protein (Hayashi and Ikura 2003), although the Ndc80 complex does not exhibit high affinity to microtubule tips (Alushin et al. 2010). Furthermore, higher-resolution images of the Ndc80 complex binding to taxol-stabilized microtubules revealed that the complex contacts microtubules at the interface between tubulin monomers via the CH domain of Ndc80, not Nuf2 (Wilson-Kubalek et al. 2008; Alushin et al. 2010). Consistent with these results, mutations of the Ndc80 CH domain cause severe defects in vivo, while those of Nuf2 CH domain result in only minor phenotypes (Guimaraes et al. 2008; Miller et al. 2008; Sundin et al. 2011). Furthermore, Ndc80 complexes self-associate to form oligomeric arrays along the microtubule lattice, which are mediated by its N-terminal extension (Alushin et al. 2010). Aurora B phosphorylates this N-terminal extension at multiple sites (Cheeseman et al. 2006; DeLuca et al. 2006; Ciferri et al. 2008) and inhibits the oligomerization of Ndc80 complexes (Alushin et al. 2010), reducing its microtubule binding activity. We note that the oligomeric arrays have so far only been observed in vitro and their physiological relevance remains unclear. It is likely that phosphorylation also directly reduces attachment stability by adding negative charges to Ndc80. Therefore, Aurora B may regulate Ndc80’s microtubule-binding activity by multiple mechanisms.
In contrast to EM techniques that reveal static images of the kinetochore-microtubule interface at sub-nanometer levels, single-molecule techniques can reveal dynamic pictures. The Asbury lab carried out a detailed characterization of the Ndc80 complex using TIRF microscopy and optical trap assays to reveal biophysical insights into their functions (Powers et al. 2009). They found that individual Ndc80 complexes bind to the microtubule lattice with a weak affinity and rapidly diffuse along the lattice. Oligomers of Ndc80 complexes can track with depolymerizing tips as microtubules shorten, while individual complexes cannot. Based on these results, they proposed that a biased-diffusion mechanism (Hill 1985) underlies Ndc80’s motility such that the movement of Ndc80 complexes is biased by depolymerizing microtubule tips. Using an optical trap, they further show that Ndc80 complexes can form load-bearing attachments (up to 3 pN of force) to dynamic microtubule tips as long as 6 to 30 complexes are present (Powers et al. 2009). Although the estimates of Ndc80 complexes per microtubule binding site vary between studies (~7 (Joglekar et al. 2006; Johnston et al. 2010), ~20 (Lawrimore et al. 2011), or ~40 (Coffman et al. 2011)), the Ndc80 complexes clearly play a significant role in microtubule binding in vivo. Taken together, reconstitution of the Ndc80 complex and analyses of its mode of microtubule binding facilitated an in-depth characterization of this important kinetochore subcomplex, demonstrating the power of the approach.
Although the Ndc80 complex is essential for microtubule attachments, it is not sufficient to achieve bi-orientation. In budding yeast, cells fail to form bi-oriented attachments when any component of the 10-subunit Dam1 complex is inactivated (Hofmann et al. 1998; Jones et al. 2001; Cheeseman et al. 2002; Janke et al. 2002). To understand how the Dam1 complex binds to microtubules, the Harrison lab constructed an elegant polycistronic vector containing all 10-subunits (Miranda et al. 2005). Initial characterization showed that Dam1 complexes form rings and spirals around microtubules (Miranda et al. 2005; Westermann et al. 2005). This was an exciting finding because rings had previously been proposed to function as a coupler to track on depolymerizing microtubule tips (Margolis and Wilson 1981; Mitchison et al. 1986). Indeed, Dam1 complexes can be induced to move by depolymerizing tips (Westermann et al. 2005; Westermann et al. 2006), and they can form attachments even in the presence of external force as high as 3 pN (Asbury et al. 2006; Franck et al. 2007). The estimate of at least 16 copies per kinetochore is sufficient to form a ring (Joglekar et al. 2006; Coffman et al. 2011; Lawrimore et al. 2011), implying that this complex might form a ring or similar structure in vivo. However, this model was challenged by the finding that ring formation is not required for its processive movements in vitro (Gestaut et al. 2008; Grishchuk et al. 2008; Gao et al. 2010) and the failure so far to identify ring-like structures in vivo. Furthermore, the Dam1 complex is not essential for viability in S. pombe and is not found outside of fungi (Liu et al. 2005; Sanchez-Perez et al. 2005). One critical difference between the kinetochores of budding yeast and fission yeast is that the former bind a single microtubule while the latter bind multiple microtubules. Therefore, it is possible that budding yeast heavily relies on Dam1 to avoid complete loss of attachments, an idea supported by recent studies in Candida albicans (Burrack et al. 2011; Thakur and Sanyal 2011). The Ska1 complex has been proposed to be a functional homolog of Dam1 because it shows the most similar biophysical properties (Welburn et al. 2009). Ska1 localizes to kinetochores and spindles in vivo, possesses direct microtubule-binding activities, can track on depolymerizing microtubules, and is phosphorylated by Aurora B (Hanisch et al. 2006; Daum et al. 2009; Gaitanos et al. 2009; Raaijmakers et al. 2009; Welburn et al. 2009; McIntosh et al. 2010). Ska1 complexes also form oligomers around microtubules in vitro, although rings have not been detected (Welburn et al. 2009). Additional biophysical assays should reveal whether the Ska1 complex can also form load-bearing attachments and is a functional equivalent of the yeast Dam1 complex.
Besides these proteins, there are many others that affect kinetochore-microtubule attachment either directly or indirectly. For example, microtubule sedimentation assays showed that KNL1/Blinkin/Spc105 has weak microtubule binding activity (Cheeseman et al. 2006; Pagliuca et al. 2009). Its depletion phenotype is very severe even though Ndc80 still localizes to kinetochores (Kiyomitsu et al. 2007). This result also shows that Ndc80 is not sufficient for kinetochore-microtubule interaction, consistent with the cooperative behavior of the KMN complex (see below). Other proteins that have microtubule binding activities are shown in Table 1, but because these proteins have not been analyzed in detail in vitro, we will not discuss them further in this review.
Multiple subcomplexes
Once individual subcomplexes are reconstituted, the next goal is to reconstitute larger assemblies (ultimately whole kinetochores) to understand how the numerous kinetochore components function as a single macromolecular unit. This is also useful to infer which subcomplexes interact with each other within kinetochores. For example, the CENP-C inner kinetochore protein directly interacts with the reconstituted Mis12 complex (composed of Mis12, Dsn1, Nsl1 and Nnf1 proteins), revealing an important linkage between inner and outer kinetochores (Gascoigne et al. 2011; Przewloka et al. 2011; Screpanti et al. 2011). Interestingly, the Mis12 complex changes its shape upon CENP-C binding (Screpanti et al. 2011), which might be important for the regulation of kinetochore assembly. The Mis12 complex also directly binds to KNL1 (Maskell et al. 2010; Petrovic et al. 2010) as well as to the Ndc80 complex (Petrovic et al. 2010; Hornung et al. 2011). These results suggest that the Mis12 complex also plays a key role in linking inner and outer kinetochore components. Similarly, characterization of a mixture of Ndc80 and Dam1 complexes suggested that Dam1 interacts with Ndc80 in the presence of microtubules and that the interaction enhances the microtubule-binding activity of the Ndc80 complex (Lampert et al. 2010; Tien et al. 2010). Like Ndc80, the Dam1 complex is targeted by Aurora B (Cheeseman et al. 2002), resulting in reduced microtubule-binding activity as well as weakened interaction with the Ndc80 complex (Shang et al. 2003; Wang et al. 2007; Gestaut et al. 2008; Lampert et al. 2010; Tien et al. 2010).
Pioneering work by Cheeseman and colleagues succeeded in reconstituting a complex composed of KNL1, Mis12 and Ndc80, called the KMN network (Cheeseman et al. 2006). This study showed that KNL1 and Ndc80 complex synergize to bind to microtubules in the presence of the Mis12 complex that does not directly bind microtubules. Because of the conservation of KMN components and the severity of their knockdown phenotype, it is now widely accepted that KMN forms a core microtubule-binding module across eukaryotes. However, although a KMN complex containing 1 copy each of K, M and N can be readily reconstituted in vitro (at least using worm proteins), it is not clear if larger kinetochore assemblies can be reconstituted without additional post-translational modifications. For example, the Aurora B kinase is implicated in promoting outer kinetochore assembly onto inner kinetochore proteins by phosphorylating Dsn1 (Emanuele et al. 2008; Yang et al. 2008), and Cdk1 phosphorylates the CENP-T component of the CCAN to promote kinetochore assembly (Gascoigne et al. 2011).
Kinetochore particles isolated from cells
As an alternative approach to reconstituting kinetochores with recombinant proteins, researchers tried to purify kinetochores from cells. However, this approach was initially not very successful due to several challenges, including their low abundance and unknown biochemical properties. By utilizing artificial minichromosomes that possess microtubule-binding activity (Kingsbury and Koshland 1993) and divide faithfully during cell division (Clarke and Carbon 1980), we established a method to purify centromere-bound kinetochores from budding yeast (Akiyoshi et al. 2009a). Taking advantage of the information we obtained to maintain kinetochores throughout the purification process (e.g. buffer containing a physiological salt concentration was important to maintain kinetochore integrity), we then succeeded in isolating a complex in higher purity and quantity that contains almost all core kinetochore proteins via affinity-purification of the Dsn1 protein (a component of the Mis12 complex) (Akiyoshi et al. 2010). Using TIRF microscopy, we found that fluorescently labeled kinetochore particles stably bind to the microtubule lattice. This lateral binding depends on the Ndc80 and KNL1/Spc105 complexes, but not on the Dam1 complex, consistent with in vivo studies that showed the Dam1 complex is dispensable for initial lateral attachments (Tanaka et al. 2005). Unlike the Ndc80 complex, individual kinetochore particles do not diffuse on the lattice, yet they track on depolymerizing microtubule tips, consistent with biased-diffusion based motility. Using an optical trap, we showed that the individual kinetochore particles stay bound to the tip of dynamically growing and shrinking microtubules in the presence of external force. Compared to the attachments mediated by Ndc80 or Dam1 complexes, kinetochore particle-mediated attachments persisted for a much longer time (>20 min), comparable to the duration of mitosis in budding yeast. Purified kinetochores form robust load-bearing attachments, persisting even under 11 pN of force. Although not required for lateral attachment, Dam1 is essential to form robust tip attachments in these assays, again consistent with previous in vivo studies. Therefore, kinetochore particles purified from budding yeast reconstitute fundamental kinetochore functions.
Our in vitro reconstitution of kinetochore-microtubule attachments allowed us to directly study the effect of tip attachments and mechanical tension on the dynamics of individual microtubules, as well as the stability of the attachment depending on the state of the microtubule (Fig. 4a). It has long been known that the dynamics of kinetochore microtubules are different from non-kinetochore microtubules in vivo (Nicklas and Kubai 1985; Mitchison et al. 1986; Tanaka et al. 2007), although the significance of this to the state of kinetochore-microtubule attachments was unclear. It was also unknown whether kinetochores bind more stably to growing or shortening microtubules. We therefore measured the following four parameters in the presence of a variable level of tension: catastrophe rate, rescue rate, and the detachment rate from growing versus shortening microtubules. Strikingly, tension increases the rescue rate and decreases the catastrophe rate, causing microtubules to spend more time in the assembly phase (Fig. 4b). In addition, kinetochores bind more stably to growing microtubules than to shortening microtubules (Fig. 4b). Changes in these four parameters result in a net increase in total attachment time as tension is increased within a certain force range (0.5 – 5 pN). In the presence of higher force (> 5 pN), total attachment time then decreases. This behavior resembles the “catch bonds” that occur between receptor-ligand interactions to enhance cell adhesion in the presence of mechanical tension (Marshall et al. 2003; Thomas et al. 2008; McEver and Zhu 2010). Therefore, reconstituting kinetochore-microtubule attachments in vitro demonstrated that tension directly stabilizes kinetochore-attachments, and the properties that regulate this behavior can now be further explored.
Fig. 4.
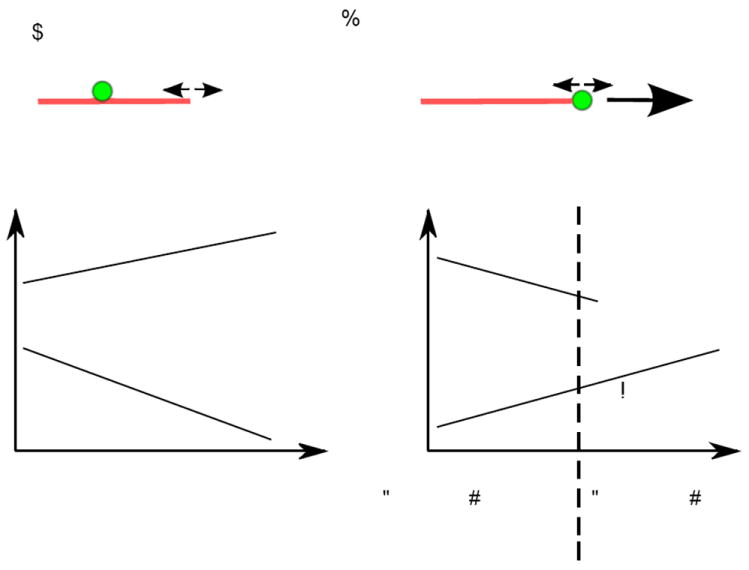
Effects of tension on microtubule tip dynamics and attachment stability. a End-on attachments allow kinetochores and tension to directly regulate microtubule tip dynamics. b Tension stabilizes reconstituted kinetochore-microtubule interactions. Left: Tension increases rescue rate and decreases catastrophe rates, resulting in a state where microtubules spend more time in the assembly phase. Right: There is also a tendency for the detachment rate from shortening microtubules to decrease in the presence of higher tension whereas that from growing microtubules increases. Changes in these four parameters result in a net increase in total attachment time as tension is increased within a certain force range (0.5 – 5 pN) (Akiyoshi et al. 2010).
One major advantage of this top-down approach is that purified native kinetochore particles may retain important post-translational modifications and structural integrity, which might be difficult to achieve from the bottom-up approach. It will be important to identify and characterize these modifications to facilitate the reconstitution of larger kinetochore assemblies from recombinant proteins. In addition, the availability of conditional mutants that can be inactivated prior to purification allows particles lacking essential proteins to be analyzed. However, a limitation of the approach is that the purified kinetochore particles are unlikely to be bound to centromeric DNA. Therefore, although kinetochore particles show robust microtubule binding activity, it is not clear if they truly represent the state of kinetochores within the cell. It will be important to determine whether kinetochore particles possess DNA-binding activity and, if so, how centromeric DNA affects their behavior. Regardless of the nature of kinetochore particles, they are the most complete kinetochore assemblies obtained thus far and provide excellent opportunities to analyze the ensemble properties of kinetochores in vitro. For example, we are actively using the system to further characterize the kinetochore-microtubule interface by EM and other techniques. These efforts should synergize with the bottom-up approach that reveals the function of individual kinetochore subcomplexes and lead to better understanding of kinetochore functions.
Other approaches to understanding kinetochores
Reconstitution of kinetochores on exogenous DNA templates in vitro
In vitro reconstitution of kinetochore proteins on exogenous DNA sequences is another approach to study kinetochore functions. Because of its simplicity and sequence specificity, the budding yeast centromere was utilized for this purpose. In the presence of yeast extracts, fluorescent beads coated with centromere DNA can bind microtubules (Sorger et al. 1994). However, this activity does not depend on CENP-A or the KMN core-microtubule binding factor (Sandall et al. 2006). Instead, the DNA binding CBF3 complex and Sli15/Bir1 complex connect centromeric DNA and microtubules (Sandall et al. 2006), suggesting that this reconstitution system is unlikely to represent a core microtubule binding event in vivo.
Recently, the Straight lab achieved the significant milestone of assembling kinetochores on reconstituted CENP-A chromatin (Guse et al. 2011). The reconstituted kinetochores contain key kinetochore components (including the KMN network) and stabilize microtubules. They can, albeit at a low efficiency, activate the spindle checkpoint when treated with microtubule drugs. To date, this is the most successful reconstitution of kinetochore assembly in vitro, and it will be important to further test functionality with biophysical and structural assays. This cell-free system also provides a unique opportunity to study how the underlying chromatin environment affects kinetochore assembly. Because there is much controversy about the composition of centromeric chromatin (Henikoff and Furuyama 2010; Black and Cleveland 2011), it will be interesting to test whether kinetochores can be assembled onto various types of CENP-A nucleosomes (e.g. right-handed nucleosomes, hemisomes) and to examine how the underlying chromatin environment affects kinetochore assembly and function.
Construction of minimal kinetochores
It is not clear why there are so many constituents in even the simplest eukaryotic kinetochore. In striking contrast, elegant reconstitution of the bacterial R1 plasmid segregation system showed that just two proteins are sufficient to segregate the plasmid DNA in vitro (Garner et al. 2007). In this system, one protein binds DNA while the other protein forms a polymer that is stabilized by the DNA-binding protein, promoting DNA segregation. To understand the design and working principles of kinetochores that contain numerous components, one approach is thus to dissect their minimal requirements. Artificially tethering the Dam1 complex to non-centromeric DNA was found to promote chromosome segregation in budding yeast (Lacefield et al. 2009; Kiermaier et al. 2009). Although the DNA-binding CBF3 complex is dispensable for its function, the majority of other kinetochore proteins appear to be recruited onto this Dam1-based machinery. Recently, tethering the CENP-C protein to the spindle pole was shown to be sufficient to assemble kinetochore proteins including KMN and spindle checkpoint proteins in the fly (Przewloka et al. 2011), and similar results were obtained by tethering CENP-C/CENP-T to the chromosome arm region in human and chicken cells (Gascoigne et al. 2011). Although CENP-A is dispensable in these systems, the CENP-T-based system cannot replace endogenous kinetochore functions (Gascoigne et al. 2011) and the Dam1-based segregation system is not as efficient as endogenous kinetochores (Lacefield et al. 2009; Kiermaier et al. 2009). It remains to be determined whether the other systems lacking CENP-A are functional. Interestingly, meiotic chromosome segregation in C. elegans appears to occur in a CENP-A-independent manner (Monen et al. 2005). It is noteworthy that trypanosomatids (such as Trypanosoma brucei that causes African sleeping sickness) lack CENP-A (Malik and Henikoff 2003; Lowell and Cross 2004; Berriman et al. 2005). Thus far, no kinetochore proteins have been identified in these organisms and their mechanism of chromosome segregation is a black box. It will be interesting to reveal how these organisms assemble DNA-segregation machinery without CENP-A, which may also reveal why CENP-A is used to assemble kinetochores in most eukaryotes.
Conclusions and Perspectives
Although reconstituting kinetochore functions in vitro has revealed key mechanistic details about kinetochore-microtubule interactions, there are a number of challenges for the future. Higher resolution structural analyses of larger assemblies such as the KMN network and purified kinetochore particles are a critical next step to understanding how individual kinetochore subcomplexes cooperate to interact with microtubules. These studies may also shed light on how kinetochores that possess multiple microtubule-binding sites are arranged, and how attachments to individual binding sites are coordinated as a whole. In vitro studies will also lead to a more complete understanding of how kinases and phosphatases control kinetochore-microtubule interactions, as well as determine whether phosphorylation solely affects attachment stability or also regulates tip dynamics and other parameters. Finally, in vitro reconstitution will be essential for fully dissecting the spindle checkpoint. Purified chromosomes have been used to recapitulate the wait-anaphase signal from kinetochores in vitro (Kulukian et al. 2009), but the efficiency of the checkpoint was low and it was never tested whether microtubule attachment silences the signal. Kinetochore particles purified from budding yeast provide a novel substrate for this approach because many checkpoint proteins co-purify (Akiyoshi et al. 2010). Addressing these and other questions should lead to better understanding of kinetochores, the intricate chromosome segregation machinery in eukaryotes.
Acknowledgments
We thank Chip Asbury for critically reading the manuscript. B.A. was supported by postdoctoral fellowships from the EMBO and Human Frontier Science Program. S.B. was supported by grants from the National Institutes of Health (GM078069 and GM064386).
Contributor Information
Bungo Akiyoshi, Sir William Dunn School of Pathology, University of Oxford, Oxford, OX1 3RE, UK.
Sue Biggins, Division of Basic Sciences, Fred Hutchinson Cancer Research Center, 1100 Fairview Ave N, PO Box 19024, Seattle, WA 98109, USA.
References
- Akiyoshi B, Nelson CR, Ranish JA, Biggins S. Quantitative proteomic analysis of purified yeast kinetochores identifies a PP1 regulatory subunit. Genes Dev. 2009a;23:2887–2899. doi: 10.1101/gad.1865909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyoshi B, Nelson CR, Ranish JA, Biggins S. Analysis of Ipl1-mediated phosphorylation of the Ndc80 kinetochore protein in Saccharomyces cerevisiae. Genetics. 2009b;183:1591–1595. doi: 10.1534/genetics.109.109041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyoshi B, Sarangapani KK, Powers AF, et al. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature. 2010;468:576–579. doi: 10.1038/nature09594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bassam J, Chang F. Regulation of microtubule dynamics by TOG-domain proteins XMAP215/Dis1 and CLASP. Trends Cell Biol. 2011;21:604–614. doi: 10.1016/j.tcb.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet. 2008;9:923–937. doi: 10.1038/nrg2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alushin GM, Ramey VH, Pasqualato S, et al. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature. 2010;467:805–810. doi: 10.1038/nature09423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaro AC, Samora CP, Holtackers R, et al. Molecular control of kinetochore-microtubule dynamics and chromosome oscillations. Nat Cell Biol. 2010;12:319–329. doi: 10.1038/ncb2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asbury CL, Gestaut DR, Powers AF, et al. The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc Natl Acad Sci USA. 2006;103:9873–9878. doi: 10.1073/pnas.0602249103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod D, Burghardt TP, Thompson NL. Total internal reflection fluorescence. Annu Rev Biophys Bioeng. 1984;13:247–268. doi: 10.1146/annurev.bb.13.060184.001335. [DOI] [PubMed] [Google Scholar]
- Bell GI. Models for the specific adhesion of cells to cells. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- Berriman M, Ghedin E, Hertz-Fowler C, et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- Black BE, Cleveland DW. Epigenetic centromere propagation and the nature of CENP-A nucleosomes. Cell. 2011;144:471–479. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block SM, Goldstein LS, Schnapp BJ. Bead movement by single kinesin molecules studied with optical tweezers. Nature. 1990;348:348–352. doi: 10.1038/348348a0. [DOI] [PubMed] [Google Scholar]
- Brinkley BR, Stubblefield E. The fine structure of the kinetochore of a mammalian cell in vitro. Chromosoma. 1966;19:28–43. doi: 10.1007/BF00332792. [DOI] [PubMed] [Google Scholar]
- Brouhard GJ, Stear JH, Noetzel TL, et al. XMAP215 is a processive microtubule polymerase. Cell. 2008;132:79–88. doi: 10.1016/j.cell.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrack LS, Applen SE, Berman J. The requirement for the Dam1 complex is dependent upon the number of kinetochore proteins and microtubules. Curr Biol. 2011;21:889–896. doi: 10.1016/j.cub.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscaino A, Allshire R, Pidoux A. Building centromeres: home sweet home or a nomadic existence? Curr Opin Genet Dev. 2010;20:118–126. doi: 10.1016/j.gde.2010.01.006. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Anderson S, Jwa M, et al. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111:163–172. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–997. doi: 10.1016/j.cell.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, MacLeod I, Yates JR, 3, et al. The CENP-F-like proteins HCP-1 and HCP-2 target CLASP to kinetochores to mediate chromosome segregation. Curr Biol. 2005;15:771–777. doi: 10.1016/j.cub.2005.03.018. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Niessen S, Anderson S, et al. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 2004;18:2255–2268. doi: 10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L, Zhang J, Ahmad S, et al. Aurora B regulates formin mDia3 in achieving metaphase chromosome alignment. Dev Cell. 2011;20:342–352. doi: 10.1016/j.devcel.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciferri C, De Luca J, Monzani S, et al. Architecture of the human ndc80-hec1 complex, a critical constituent of the outer kinetochore. J Biol Chem. 2005;280:29088–29095. doi: 10.1074/jbc.M504070200. [DOI] [PubMed] [Google Scholar]
- Ciferri C, Pasqualato S, Screpanti E, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L, Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980;287:504–509. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 2003;112:407–421. doi: 10.1016/s0092-8674(03)00115-6. [DOI] [PubMed] [Google Scholar]
- Coffman VC, Wu P, Parthun MR, Wu J-Q. CENP-A exceeds microtubule attachment sites in centromere clusters of both budding and fission yeast. J Cell Biol. 2011;195:563–572. doi: 10.1083/jcb.201106078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JR, Wagenbach M, Asbury CL, Wordeman L. Catalysis of the microtubule on-rate is the major parameter regulating the depolymerase activity of MCAK. Nat Struct Mol Biol. 2010;17:77–82. doi: 10.1038/nsmb.1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coue M, Lombillo VA, McIntosh JR. Microtubule depolymerization promotes particle and chromosome movement in vitro. J Cell Biol. 1991;112:1165–1175. doi: 10.1083/jcb.112.6.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum JR, Wren JD, Daniel JJ, et al. Ska3 is required for spindle checkpoint silencing and the maintenance of chromosome cohesion in mitosis. Curr Biol. 2009;19:1467–1472. doi: 10.1016/j.cub.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca JG, Gall WE, Ciferri C, et al. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- DeLuca KF, Lens SMA, DeLuca JG. Temporal changes in Hec1 phosphorylation control kinetochore-microtubule attachment stability during mitosis. J Cell Sci. 2011;124:622–634. doi: 10.1242/jcs.072629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A, Verma S, Mitchison TJ, Walczak CE. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 1999;96:69–78. doi: 10.1016/s0092-8674(00)80960-5. [DOI] [PubMed] [Google Scholar]
- Dewar H, Tanaka K, Nasmyth K, Tanaka TU. Tension between two kinetochores suffices for their bi-orientation on the mitotic spindle. Nature. 2004;428:93–97. doi: 10.1038/nature02328. [DOI] [PubMed] [Google Scholar]
- De Wulf P, McAinsh AD, Sorger PK. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 2003;17:2902–2921. doi: 10.1101/gad.1144403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wulf P, Montani F, Visintin R. Protein phosphatases take the mitotic stage. Curr Opin Cell Biol. 2009;21:806–815. doi: 10.1016/j.ceb.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Dong Y, Vanden Beldt KJ, Meng X, et al. The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nat Cell Biol. 2007;9:516–522. doi: 10.1038/ncb1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Z, von Schubert C, Körner R, et al. Quantitative mass spectrometry analysis reveals similar substrate consensus motif for human Mps1 kinase and Plk1. PLoS ONE. 2011;6:e18793. doi: 10.1371/journal.pone.0018793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunsch AK, Linnane E, Barr FA, Gruneberg U. The astrin-kinastrin/SKAP complex localizes to microtubule plus ends and facilitates chromosome alignment. J Cell Biol. 2011;192:959–968. doi: 10.1083/jcb.201008023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuele MJ, Lan W, Jwa M, et al. Aurora B kinase and protein phosphatase 1 have opposing roles in modulating kinetochore assembly. J Cell Biol. 2008;181:241–254. doi: 10.1083/jcb.200710019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuele MJ, Stukenberg PT. Xenopus Cep57 is a novel kinetochore component involved in microtubule attachment. Cell. 2007;130:893–905. doi: 10.1016/j.cell.2007.07.023. [DOI] [PubMed] [Google Scholar]
- Feng J, Huang H, Yen TJ. CENP-F is a novel microtubule-binding protein that is essential for kinetochore attachments and affects the duration of the mitotic checkpoint delay. Chromosoma. 2006;115:320–329. doi: 10.1007/s00412-006-0049-5. [DOI] [PubMed] [Google Scholar]
- Foley EA, Maldonado M, Kapoor TM. Formation of stable attachments between kinetochores and microtubules depends on the B56-PP2A phosphatase. Nat Cell Biol. 2011;13:1265–1271. doi: 10.1038/ncb2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck AD, Powers AF, Gestaut DR, et al. Direct physical study of kinetochore-microtubule interactions by reconstitution and interrogation with an optical force clamp. Methods. 2010;51:242–250. doi: 10.1016/j.ymeth.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck AD, Powers AF, Gestaut DR, et al. Tension applied through the Dam1 complex promotes microtubule elongation providing a direct mechanism for length control in mitosis. Nat Cell Biol. 2007;9:832–837. doi: 10.1038/ncb1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller BG, Lampson MA, Foley EA, et al. Midzone activation of aurora B in anaphase produces an intracellular phosphorylation gradient. Nature. 2008;453:1132–1136. doi: 10.1038/nature06923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama S, Biggins S. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc Natl Acad Sci USA. 2007;104:14706–14711. doi: 10.1073/pnas.0706985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaitanos TN, Santamaria A, Jeyaprakash AA, et al. Stable kinetochore-microtubule interactions depend on the Ska complex and its new component Ska3/C13Orf3. EMBO J. 2009;28:1442–1452. doi: 10.1038/emboj.2009.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, Courtheoux T, Gachet Y, et al. A non-ring-like form of the Dam1 complex modulates microtubule dynamics in fission yeast. Proc Natl Acad Sci USA. 2010;107:13330–13335. doi: 10.1073/pnas.1004887107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MA, Vardy L, Koonrugsa N, Toda T. Fission yeast ch-TOG/XMAP215 homologue Alp14 connects mitotic spindles with the kinetochore and is a component of the Mad2-dependent spindle checkpoint. EMBO J. 2001;20:3389–3401. doi: 10.1093/emboj/20.13.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner EC, Campbell CS, Weibel DB, Mullins RD. Reconstitution of DNA segregation driven by assembly of a prokaryotic actin homolog. Science. 2007;315:1270–1274. doi: 10.1126/science.1138527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne KE, Cheeseman IM. Kinetochore assembly: if you build it, they will come. Curr Opin Cell Biol. 2011;23:102–108. doi: 10.1016/j.ceb.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne KE, Takeuchi K, Suzuki A, et al. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell. 2011;145:410–422. doi: 10.1016/j.cell.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R, Essex A, Hu J-S, et al. A new mechanism controlling kinetochore-microtubule interactions revealed by comparison of two dynein-targeting components: SPDL-1 and the Rod/Zwilch/Zw10 complex. Genes Dev. 2008;22:2385–2399. doi: 10.1101/gad.1687508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gestaut DR, Graczyk B, Cooper J, et al. Phosphoregulation and depolymerization-driven movement of the Dam1 complex do not require ring formation. Nat Cell Biol. 2008;10:407–414. doi: 10.1038/ncb1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffis ER, Stuurman N, Vale RD. Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore. J Cell Biol. 2007;177:1005–1015. doi: 10.1083/jcb.200702062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk EL, McIntosh JR. Microtubule depolymerization can drive poleward chromosome motion in fission yeast. EMBO J. 2006;25:4888–4896. doi: 10.1038/sj.emboj.7601353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk EL, Molodtsov MI, Ataullakhanov FI, McIntosh JR. Force production by disassembling microtubules. Nature. 2005;438:384–388. doi: 10.1038/nature04132. [DOI] [PubMed] [Google Scholar]
- Grishchuk EL, Spiridonov IS, Volkov VA, et al. Different assemblies of the DAM1 complex follow shortening microtubules by distinct mechanisms. Proc Natl Acad Sci USA. 2008;105:6918–6923. doi: 10.1073/pnas.0801811105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes GJ, Dong Y, McEwen BF, Deluca JG. Kinetochore-microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Curr Biol. 2008;18:1778–1784. doi: 10.1016/j.cub.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guse A, Carroll CW, Moree B, et al. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature. 2011;477:354–358. doi: 10.1038/nature10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanisch A, Silljé HHW, Nigg EA. Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. EMBO J. 2006;25:5504–5515. doi: 10.1038/sj.emboj.7601426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi I, Ikura M. Crystal structure of the amino-terminal microtubule-binding domain of end-binding protein 1 (EB1) J Biol Chem. 2003;278:36430–36434. doi: 10.1074/jbc.M305773200. [DOI] [PubMed] [Google Scholar]
- He X, Rines DR, Espelin CW, Sorger PK. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell. 2001;106:195–206. doi: 10.1016/s0092-8674(01)00438-x. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Furuyama T. Epigenetic inheritance of centromeres. Cold Spring Harb Symp Quant Biol. 2010;75:51–60. doi: 10.1101/sqb.2010.75.001. [DOI] [PubMed] [Google Scholar]
- Hewitt L, Tighe A, Santaguida S, et al. Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1-C-Mad2 core complex. J Cell Biol. 2010;190:25–34. doi: 10.1083/jcb.201002133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill TL. Theoretical problems related to the attachment of microtubules to kinetochores. Proc Natl Acad Sci USA. 1985;82:4404–4408. doi: 10.1073/pnas.82.13.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann C, Cheeseman IM, Goode BL, et al. Saccharomyces cerevisiae Duo1p and Dam1p, novel proteins involved in mitotic spindle function. J Cell Biol. 1998;143:1029–1040. doi: 10.1083/jcb.143.4.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10:478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung P, Maier M, Alushin GM, et al. Molecular architecture and connectivity of the budding yeast Mtw1 kinetochore complex. J Mol Biol. 2011;405:548–559. doi: 10.1016/j.jmb.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BJ, McEwen BF, Canman JC, et al. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J Cell Biol. 2001;155:1159–1172. doi: 10.1083/jcb.200105093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Feng J, Famulski J, et al. Tripin/hSgo2 recruits MCAK to the inner centromere to correct defective kinetochore attachments. J Cell Biol. 2007;177:413–424. doi: 10.1083/jcb.200701122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indjeian VB, Murray AW. Budding yeast mitotic chromosomes have an intrinsic bias to biorient on the spindle. Curr Biol. 2007;17:1837–1846. doi: 10.1016/j.cub.2007.09.056. [DOI] [PubMed] [Google Scholar]
- Itoh G, Kanno S-ichiro, Uchida KSK, et al. CAMP (C13orf8, ZNF828) is a novel regulator of kinetochore-microtubule attachment. EMBO J. 2011;30:130–144. doi: 10.1038/emboj.2010.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke C, Ortíz J, Tanaka TU, et al. Four new subunits of the Dam1-Duo1 complex reveal novel functions in sister kinetochore biorientation. EMBO J. 2002;21:181–193. doi: 10.1093/emboj/21.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelluma N, Brenkman AB, van den Broek NJF, et al. Mps1 phosphorylates Borealin to control Aurora B activity and chromosome alignment. Cell. 2008;132:233–246. doi: 10.1016/j.cell.2007.11.046. [DOI] [PubMed] [Google Scholar]
- Joglekar AP, Bloom K, Salmon ED. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr Biol. 2009;19:694–699. doi: 10.1016/j.cub.2009.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar AP, Bouck D, Finley K, et al. Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres. J Cell Biol. 2008;181:587–594. doi: 10.1083/jcb.200803027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joglekar AP, Bouck DC, Molk JN, et al. Molecular architecture of a kinetochore-microtubule attachment site. Nat Cell Biol. 2006;8:581–585. doi: 10.1038/ncb1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston K, Joglekar A, Hori T, et al. Vertebrate kinetochore protein architecture: protein copy number. J Cell Biol. 2010;189:937–943. doi: 10.1083/jcb.200912022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MH, He X, Giddings TH, Winey M. Yeast Dam1p has a role at the kinetochore in assembly of the mitotic spindle. Proc Natl Acad Sci USA. 2001;98:13675–13680. doi: 10.1073/pnas.241417098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karess R. Rod-Zw10-Zwilch: a key player in the spindle checkpoint. Trends Cell Biol. 2005;15:386–392. doi: 10.1016/j.tcb.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Kelly AE, Funabiki H. Correcting aberrant kinetochore microtubule attachments: an Aurora B-centric view. Curr Opin Cell Biol. 2009;21:51–58. doi: 10.1016/j.ceb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmler S, Stach M, Knapp M, et al. Mimicking Ndc80 phosphorylation triggers spindle assembly checkpoint signalling. EMBO J. 2009;28:1099–1110. doi: 10.1038/emboj.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A, Rieder CL. Kinetochores moving away from their associated pole do not exert a significant pushing force on the chromosome. J Cell Biol. 1996;135:315–327. doi: 10.1083/jcb.135.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiermaier E, Woehrer S, Peng Y, et al. A Dam1-based artificial kinetochore is sufficient to promote chromosome segregation in budding yeast. Nat Cell Biol. 2009;11:1109–1115. doi: 10.1038/ncb1924. [DOI] [PubMed] [Google Scholar]
- Kim Y, Heuser JE, Waterman CM, Cleveland DW. CENP-E combines a slow, processive motor and a flexible coiled coil to produce an essential motile kinetochore tether. J Cell Biol. 2008;181:411–419. doi: 10.1083/jcb.200802189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Holland AJ, Lan W, Cleveland DW. Aurora kinases and protein phosphatase 1 mediate chromosome congression through regulation of CENP-E. Cell. 2010;142:444–455. doi: 10.1016/j.cell.2010.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury J, Koshland D. Centromere function on minichromosomes isolated from budding yeast. Mol Biol Cell. 1993;4:859–870. doi: 10.1091/mbc.4.8.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima TS, Ohsugi M, Ellenberg J. Complete kinetochore tracking reveals error-prone homologous chromosome biorientation in Mammalian oocytes. Cell. 2011;146:568–581. doi: 10.1016/j.cell.2011.07.031. [DOI] [PubMed] [Google Scholar]
- Kitamura E, Tanaka K, Komoto S, et al. Kinetochores generate microtubules with distal plus ends: their roles and limited lifetime in mitosis. Dev Cell. 2010;18:248–259. doi: 10.1016/j.devcel.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyomitsu T, Obuse C, Yanagida M. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev Cell. 2007;13:663–676. doi: 10.1016/j.devcel.2007.09.005. [DOI] [PubMed] [Google Scholar]
- Koshland DE, Mitchison TJ, Kirschner MW. Polewards chromosome movement driven by microtubule depolymerization in vitro. Nature. 1988;331:499–504. doi: 10.1038/331499a0. [DOI] [PubMed] [Google Scholar]
- Kulukian A, Han JS, Cleveland DW. Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding. Dev Cell. 2009;16:105–117. doi: 10.1016/j.devcel.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacefield S, Lau DTC, Murray AW. Recruiting a microtubule-binding complex to DNA directs chromosome segregation in budding yeast. Nat Cell Biol. 2009;11:1116–1120. doi: 10.1038/ncb1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampert F, Hornung P, Westermann S. The Dam1 complex confers microtubule plus end-tracking activity to the Ndc80 kinetochore complex. J Cell Biol. 2010;189:641–649. doi: 10.1083/jcb.200912021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampert F, Westermann S. A blueprint for kinetochores - new insights into the molecular mechanics of cell division. Nat Rev Mol Cell Biol. 2011;12:407–412. doi: 10.1038/nrm3133. [DOI] [PubMed] [Google Scholar]
- Lan W, Cleveland DW. A chemical tool box defines mitotic and interphase roles for Mps1 kinase. J Cell Biol. 2010;190:21–24. doi: 10.1083/jcb.201006080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrimore J, Bloom KS, Salmon ED. Point centromeres contain more than a single centromere-specific Cse4 (CENP-A) nucleosome. J Cell Biol. 2011;195:573–582. doi: 10.1083/jcb.201106036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Vader G, Vromans MJM, et al. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 2009;323:1350–1353. doi: 10.1126/science.1167000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Vleugel M, Backer CB, et al. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J Cell Biol. 2010;188:809–820. doi: 10.1083/jcb.201001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, McLeod I, Anderson S, et al. Molecular analysis of kinetochore architecture in fission yeast. EMBO J. 2005;24:2919–2930. doi: 10.1038/sj.emboj.7600762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowell JE, Cross GAM. A variant histone H3 is enriched at telomeres in Trypanosoma brucei. J Cell Sci. 2004;117:5937–5947. doi: 10.1242/jcs.01515. [DOI] [PubMed] [Google Scholar]
- Maciejowski J, George KA, Terret M-E, et al. Mps1 directs the assembly of Cdc20 inhibitory complexes during interphase and mitosis to control M phase timing and spindle checkpoint signaling. J Cell Biol. 2010;190:89–100. doi: 10.1083/jcb.201001050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack GJ, Compton DA. Analysis of mitotic microtubule-associated proteins using mass spectrometry identifies astrin, a spindle-associated protein. Proc Natl Acad Sci USA. 2001;98:14434–14439. doi: 10.1073/pnas.261371298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maffini S, Maia ARR, Manning AL, et al. Motor-independent targeting of CLASPs to kinetochores by CENP-E promotes microtubule turnover and poleward flux. Curr Biol. 2009;19:1566–1572. doi: 10.1016/j.cub.2009.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magidson V, O’Connell CB, Lončarek J, et al. The Spatial Arrangement of Chromosomes during Prometaphase Facilitates Spindle Assembly. Cell. 2011;146:555–567. doi: 10.1016/j.cell.2011.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H, Fairley EAL, Rieder CL, et al. Human CLASP1 is an outer kinetochore component that regulates spindle microtubule dynamics. Cell. 2003;113:891–904. doi: 10.1016/s0092-8674(03)00465-3. [DOI] [PubMed] [Google Scholar]
- Malik HS, Henikoff S. Phylogenomics of the nucleosome. Nat Struct Biol. 2003;10:882–891. doi: 10.1038/nsb996. [DOI] [PubMed] [Google Scholar]
- Mandelkow EM, Mandelkow E, Milligan RA. Microtubule dynamics and microtubule caps: a time-resolved cryo-electron microscopy study. J Cell Biol. 1991;114:977–991. doi: 10.1083/jcb.114.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning AL, Bakhoum SF, Maffini S, et al. CLASP1, astrin and Kif2b form a molecular switch that regulates kinetochore-microtubule dynamics to promote mitotic progression and fidelity. EMBO J. 2010;29:3531–3543. doi: 10.1038/emboj.2010.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca TJ, Salmon ED. Welcome to a new kind of tension: translating kinetochore mechanics into a wait-anaphase signal. J Cell Sci. 2010;123:825–835. doi: 10.1242/jcs.064790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca TJ, Salmon ED. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J Cell Biol. 2009;184:373–381. doi: 10.1083/jcb.200808130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis RL, Wilson L. Microtubule treadmills--possible molecular machinery. Nature. 1981;293:705–711. doi: 10.1038/293705a0. [DOI] [PubMed] [Google Scholar]
- Marshall BT, Long M, Piper JW, et al. Direct observation of catch bonds involving cell-adhesion molecules. Nature. 2003;423:190–193. doi: 10.1038/nature01605. [DOI] [PubMed] [Google Scholar]
- Maskell DP, Hu X-W, Singleton MR. Molecular architecture and assembly of the yeast kinetochore MIND complex. J Cell Biol. 2010;190:823–834. doi: 10.1083/jcb.201002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maure J-F, Kitamura E, Tanaka TU. Mps1 kinase promotes sister-kinetochore bi-orientation by a tension-dependent mechanism. Curr Biol. 2007;17:2175–2182. doi: 10.1016/j.cub.2007.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEver RP, Zhu C. Rolling cell adhesion. Annu Rev Cell Dev Biol. 2010;26:363–396. doi: 10.1146/annurev.cellbio.042308.113238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BF, Dong Y. Contrasting models for kinetochore microtubule attachment in mammalian cells. Cell Mol Life Sci. 2010;67:2163–2172. doi: 10.1007/s00018-010-0322-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BF, Hsieh CE, Mattheyses AL, Rieder CL. A new look at kinetochore structure in vertebrate somatic cells using high-pressure freezing and freeze substitution. Chromosoma. 1998;107:366–375. doi: 10.1007/s004120050320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JR, Grishchuk EL, Morphew MK, et al. Fibrils connect microtubule tips with kinetochores: a mechanism to couple tubulin dynamics to chromosome motion. Cell. 2008;135:322–333. doi: 10.1016/j.cell.2008.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh JR, Volkov V, Ataullakhanov FI, Grishchuk EL. Tubulin depolymerization may be an ancient biological motor. J Cell Sci. 2010;123:3425–3434. doi: 10.1242/jcs.067611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows JC, Shepperd LA, Vanoosthuyse V, et al. Spindle checkpoint silencing requires association of PP1 to both Spc7 and kinesin-8 motors. Dev Cell. 2011;20:739–750. doi: 10.1016/j.devcel.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meireles AM, Dzhindzhev NS, Ohkura H. Kebab: Kinetochore and EB1 Associated Basic Protein That Dynamically Changes Its Localisation during Drosophila Mitosis. PLoS ONE. 2011;6:e24174. doi: 10.1371/journal.pone.0024174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendiburo MJ, Padeken J, Fülöp S, et al. Drosophila CENH3 is sufficient for centromere formation. Science. 2011;334:686–690. doi: 10.1126/science.1206880. [DOI] [PubMed] [Google Scholar]
- Merkel R, Nassoy P, Leung A, et al. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature. 1999;397:50–53. doi: 10.1038/16219. [DOI] [PubMed] [Google Scholar]
- Miller SA, Johnson ML, Stukenberg PT. Kinetochore attachments require an interaction between unstructured tails on microtubules and Ndc80(Hec1) Curr Biol. 2008;18:1785–1791. doi: 10.1016/j.cub.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda JJL, De Wulf P, Sorger PK, Harrison SC. The yeast DASH complex forms closed rings on microtubules. Nat Struct Mol Biol. 2005;12:138–143. doi: 10.1038/nsmb896. [DOI] [PubMed] [Google Scholar]
- Mitchison T, Evans L, Schulze E, Kirschner M. Sites of microtubule assembly and disassembly in the mitotic spindle. Cell. 1986;45:515–527. doi: 10.1016/0092-8674(86)90283-7. [DOI] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ, Kirschner MW. Properties of the kinetochore in vitro. II. Microtubule capture and ATP-dependent translocation. J Cell Biol. 1985;101:766–777. doi: 10.1083/jcb.101.3.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monen J, Maddox PS, Hyndman F, et al. Differential role of CENP-A in the segregation of holocentric C. elegans chromosomes during meiosis and mitosis. Nat Cell Biol. 2005;7:1248–1255. doi: 10.1038/ncb1331. [DOI] [PubMed] [Google Scholar]
- Murata-Hori M, Wang Y-L. Both midzone and astral microtubules are involved in the delivery of cytokinesis signals: insights from the mobility of aurora B. J Cell Biol. 2002;159:45–53. doi: 10.1083/jcb.200207014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AW. A brief history of error. Nat Cell Biol. 2011;13:1178–1182. doi: 10.1038/ncb2348. [DOI] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Nekrasov VS, Smith MA, Peak-Chew S, Kilmartin JV. Interactions between centromere complexes in Saccharomyces cerevisiae. Mol Biol Cell. 2003;14:4931–4946. doi: 10.1091/mbc.E03-06-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezi L, Musacchio A. Sister chromatid tension and the spindle assembly checkpoint. Curr Opin Cell Biol. 2009;21:785–795. doi: 10.1016/j.ceb.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Nicklas RB. How cells get the right chromosomes. Science. 1997;275:632–637. doi: 10.1126/science.275.5300.632. [DOI] [PubMed] [Google Scholar]
- Nicklas RB. The forces that move chromosomes in mitosis. Annu Rev Biophys Biophys Chem. 1988;17:431–449. doi: 10.1146/annurev.bb.17.060188.002243. [DOI] [PubMed] [Google Scholar]
- Nicklas RB, Koch CA. Chromosome micromanipulation. 3. Spindle fiber tension and the reorientation of mal-oriented chromosomes. J Cell Biol. 1969;43:40–50. doi: 10.1083/jcb.43.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas RB, Kubai DF. Microtubules, chromosome movement, and reorientation after chromosomes are detached from the spindle by micromanipulation. Chromosoma. 1985;92:313–324. doi: 10.1007/BF00329815. [DOI] [PubMed] [Google Scholar]
- Nicklas RB, Ward SC. Elements of error correction in mitosis: microtubule capture, release, and tension. J Cell Biol. 1994;126:1241–1253. doi: 10.1083/jcb.126.5.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguchi Y, Uchimura S, Ohki T, et al. The bidirectional depolymerizer MCAK generates force by disassembling both microtubule ends. Nat Cell Biol. 2011;13:846–852. doi: 10.1038/ncb2256. [DOI] [PubMed] [Google Scholar]
- Ortiz J, Funk C, Schäfer A, Lechner J. Stu1 inversely regulates kinetochore capture and spindle stability. Genes Dev. 2009;23:2778–2791. doi: 10.1101/gad.541309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Östergren G. The Mechanism of Co-orientation in Bivalents and Multivalents. Hereditas. 1951;37:85–156. doi: 10.1111/j.1601-5223.1951.tb02891.x. [DOI] [Google Scholar]
- Pagliuca C, Draviam VM, Marco E, et al. Roles for the conserved spc105p/kre28p complex in kinetochore-microtubule binding and the spindle assembly checkpoint. PLoS ONE. 2009;4:e7640. doi: 10.1371/journal.pone.0007640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perpelescu M, Fukagawa T. The ABCs of CENPs. Chromosoma. 2011;120:425–446. doi: 10.1007/s00412-011-0330-0. [DOI] [PubMed] [Google Scholar]
- Peters J-M. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- Petrovic A, Pasqualato S, Dube P, et al. The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J Cell Biol. 2010;190:835–852. doi: 10.1083/jcb.201002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines J. Cubism and the cell cycle: the many faces of the APC/C. Nat Rev Mol Cell Biol. 2011;12:427–438. doi: 10.1038/nrm3132. [DOI] [PubMed] [Google Scholar]
- Pinsky BA, Kung C, Shokat KM, Biggins S. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat Cell Biol. 2006;8:78–83. doi: 10.1038/ncb1341. [DOI] [PubMed] [Google Scholar]
- Pinsky BA, Tatsutani SY, Collins KA, Biggins S. An Mtw1 complex promotes kinetochore biorientation that is monitored by the Ipl1/Aurora protein kinase. Dev Cell. 2003;5:735–745. doi: 10.1016/s1534-5807(03)00322-8. [DOI] [PubMed] [Google Scholar]
- Porter IM, McClelland SE, Khoudoli GA, et al. Bod1, a novel kinetochore protein required for chromosome biorientation. J Cell Biol. 2007;179:187–197. doi: 10.1083/jcb.200704098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers AF, Franck AD, Gestaut DR, et al. The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell. 2009;136:865–875. doi: 10.1016/j.cell.2008.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przewloka MR, Glover DM. The kinetochore and the centromere: a working long distance relationship. Annu Rev Genet. 2009;43:439–465. doi: 10.1146/annurev-genet-102108-134310. [DOI] [PubMed] [Google Scholar]
- Przewloka MR, Venkei Z, Bolanos-Garcia VM, et al. CENP-C is a structural platform for kinetochore assembly. Curr Biol. 2011;21:399–405. doi: 10.1016/j.cub.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Raaijmakers JA, Tanenbaum ME, Maia AF, Medema RH. RAMA1 is a novel kinetochore protein involved in kinetochore-microtubule attachment. J Cell Sci. 2009;122:2436–2445. doi: 10.1242/jcs.051912. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Alexander SP. Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. J Cell Biol. 1990;110:81–95. doi: 10.1083/jcb.110.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg JS, Cross FR, Funabiki H. KNL1/Spc105 recruits PP1 to silence the spindle assembly checkpoint. Curr Biol. 2011;21:942–947. doi: 10.1016/j.cub.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol. 2007;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- Sakuno T, Tada K, Watanabe Y. Kinetochore geometry defined by cohesion within the centromere. Nature. 2009;458:852–858. doi: 10.1038/nature07876. [DOI] [PubMed] [Google Scholar]
- Sakuno T, Tanaka K, Hauf S, Watanabe Y. Repositioning of aurora B promoted by chiasmata ensures sister chromatid mono-orientation in meiosis I. Dev Cell. 2011;21:534–545. doi: 10.1016/j.devcel.2011.08.012. [DOI] [PubMed] [Google Scholar]
- Salic A, Waters JC, Mitchison TJ. Vertebrate shugoshin links sister centromere cohesion and kinetochore microtubule stability in mitosis. Cell. 2004;118:567–578. doi: 10.1016/j.cell.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Sanchez-Perez I, Renwick SJ, Crawley K, et al. The DASH complex and Klp5/Klp6 kinesin coordinate bipolar chromosome attachment in fission yeast. EMBO J. 2005;24:2931–2943. doi: 10.1038/sj.emboj.7600761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandall S, Severin F, McLeod IX, et al. A Bir1-Sli15 complex connects centromeres to microtubules and is required to sense kinetochore tension. Cell. 2006;127:1179–1191. doi: 10.1016/j.cell.2006.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S, Musacchio A. The life and miracles of kinetochores. EMBO J. 2009;28:2511–2531. doi: 10.1038/emboj.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S, Tighe A, D’Alise AM, et al. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J Cell Biol. 2010;190:73–87. doi: 10.1083/jcb.201001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittenhelm RB, Heeger S, Althoff F, et al. Spatial organization of a ubiquitous eukaryotic kinetochore protein network in Drosophila chromosomes. Chromosoma. 2007;116:385–402. doi: 10.1007/s00412-007-0103-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JC, Kiyomitsu T, Hori T, et al. Aurora B kinase controls the targeting of the Astrin-SKAP complex to bioriented kinetochores. J Cell Biol. 2010;191:269–280. doi: 10.1083/jcb.201006129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvartzman J-M, Sotillo R, Benezra R. Mitotic chromosomal instability and cancer: mouse modelling of the human disease. Nat Rev Cancer. 2010;10:102–115. doi: 10.1038/nrc2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Screpanti E, De Antoni A, Alushin GM, et al. Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr Biol. 2011;21:391–398. doi: 10.1016/j.cub.2010.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang C, Hazbun TR, Cheeseman IM, et al. Kinetochore protein interactions and their regulation by the Aurora kinase Ipl1p. Mol Biol Cell. 2003;14:3342–3355. doi: 10.1091/mbc.E02-11-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger PK, Severin FF, Hyman AA. Factors required for the binding of reassembled yeast kinetochores to microtubules in vitro. J Cell Biol. 1994;127:995–1008. doi: 10.1083/jcb.127.4.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundin LJR, Guimaraes GJ, Deluca JG. The NDC80 complex proteins Nuf2 and Hec1 make distinct contributions to kinetochore-microtubule attachment in mitosis. Mol Biol Cell. 2011;22:759–768. doi: 10.1091/mbc.E10-08-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, Hori T, Nishino T, et al. Spindle microtubules generate tension-dependent changes in the distribution of inner kinetochore proteins. J Cell Biol. 2011;193:125–140. doi: 10.1083/jcb.201012050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L, Kapoor TM. Examining the dynamics of chromosomal passenger complex (CPC)-dependent phosphorylation during cell division. Proc Natl Acad Sci USA. 2011;108:16675–16680. doi: 10.1073/pnas.1106748108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan S. A modular polycistronic expression system for overexpressing protein complexes in Escherichia coli. Protein Expr Purif. 2001;21:224–234. doi: 10.1006/prep.2000.1363. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Kitamura E, Kitamura Y, Tanaka TU. Molecular mechanisms of microtubule-dependent kinetochore transport toward spindle poles. J Cell Biol. 2007;178:269–281. doi: 10.1083/jcb.200702141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Mukae N, Dewar H, et al. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature. 2005;434:987–994. doi: 10.1038/nature03483. [DOI] [PubMed] [Google Scholar]
- Tanaka TU, Rachidi N, Janke C, et al. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- Tanno Y, Kitajima TS, Honda T, et al. Phosphorylation of mammalian Sgo2 by Aurora B recruits PP2A and MCAK to centromeres. Genes Dev. 2010;24:2169–2179. doi: 10.1101/gad.1945310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur J, Sanyal K. The essentiality of the fungal specific Dam1 complex is correlated with one kinetochore one microtubule interaction present throughout the cell cycle, independent of the nature of a centromere. Eukaryotic Cell. 2011 doi: 10.1128/EC.05093-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas WE, Vogel V, Sokurenko E. Biophysics of catch bonds. Annu Rev Biophys. 2008;37:399–416. doi: 10.1146/annurev.biophys.37.032807.125804. [DOI] [PubMed] [Google Scholar]
- Tien JF, Umbreit NT, Gestaut DR, et al. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. J Cell Biol. 2010;189:713–723. doi: 10.1083/jcb.200910142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L, Andrews PD, Wickramasinghe S, et al. Time-lapse imaging reveals dynamic relocalization of PP1gamma throughout the mammalian cell cycle. Mol Biol Cell. 2003;14:107–117. doi: 10.1091/mbc.E02-07-0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tytell JD, Sorger PK. Analysis of kinesin motor function at budding yeast kinetochores. J Cell Biol. 2006;172:861–874. doi: 10.1083/jcb.200509101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida KSK, Takagaki K, Kumada K, et al. Kinetochore stretching inactivates the spindle assembly checkpoint. J Cell Biol. 2009;184:383–390. doi: 10.1083/jcb.200811028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdaasdonk JS, Bloom K. Centromeres: unique chromatin structures that drive chromosome segregation. Nat Rev Mol Cell Biol. 2011;12:320–332. doi: 10.1038/nrm3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergnolle MAS, Taylor SS. Cenp-F links kinetochores to Ndel1/Nde1/Lis1/dynein microtubule motor complexes. Curr Biol. 2007;17:1173–1179. doi: 10.1016/j.cub.2007.05.077. [DOI] [PubMed] [Google Scholar]
- Wan X, O’Quinn RP, Pierce HL, et al. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–684. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, Ballister ER, Lampson MA. Aurora B dynamics at centromeres create a diffusion-based phosphorylation gradient. J Cell Biol. 2011;194:539–549. doi: 10.1083/jcb.201103044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H-W, Nogales E. Nucleotide-dependent bending flexibility of tubulin regulates microtubule assembly. Nature. 2005;435:911–915. doi: 10.1038/nature03606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H-W, Ramey VH, Westermann S, et al. Architecture of the Dam1 kinetochore ring complex and implications for microtubule-driven assembly and force-coupling mechanisms. Nat Struct Mol Biol. 2007;14:721–726. doi: 10.1038/nsmb1274. [DOI] [PubMed] [Google Scholar]
- Waters JC, Skibbens RV, Salmon ED. Oscillating mitotic newt lung cell kinetochores are, on average, under tension and rarely push. J Cell Sci. 1996;109(Pt 12):2823–2831. doi: 10.1242/jcs.109.12.2823. [DOI] [PubMed] [Google Scholar]
- Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat Struct Mol Biol. 2007;14:54–59. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
- Wei RR, Sorger PK, Harrison SC. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc Natl Acad Sci USA. 2005;102:5363–5367. doi: 10.1073/pnas.0501168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn JPI, Cheeseman IM. Toward a molecular structure of the eukaryotic kinetochore. Dev Cell. 2008;15:645–655. doi: 10.1016/j.devcel.2008.10.011. [DOI] [PubMed] [Google Scholar]
- Welburn JPI, Grishchuk EL, Backer CB, et al. The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev Cell. 2009;16:374–385. doi: 10.1016/j.devcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn JPI, Vleugel M, Liu D, et al. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol Cell. 2010;38:383–392. doi: 10.1016/j.molcel.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S, Avila-Sakar A, Wang H-W, et al. Formation of a dynamic kinetochore-microtubule interface through assembly of the Dam1 ring complex. Mol Cell. 2005;17:277–290. doi: 10.1016/j.molcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- Westermann S, Drubin DG, Barnes G. Structures and functions of yeast kinetochore complexes. Annu Rev Biochem. 2007;76:563–591. doi: 10.1146/annurev.biochem.76.052705.160607. [DOI] [PubMed] [Google Scholar]
- Westermann S, Wang H-W, Avila-Sakar A, et al. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature. 2006;440:565–569. doi: 10.1038/nature04409. [DOI] [PubMed] [Google Scholar]
- Wheatley SP, Kandels-Lewis SE, Adams RR, et al. INCENP binds directly to tubulin and requires dynamic microtubules to target to the cleavage furrow. Exp Cell Res. 2001;262:122–127. doi: 10.1006/excr.2000.5088. [DOI] [PubMed] [Google Scholar]
- Wilson-Kubalek EM, Cheeseman IM, Yoshioka C, et al. Orientation and structure of the Ndc80 complex on the microtubule lattice. J Cell Biol. 2008;182:1055–1061. doi: 10.1083/jcb.200804170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M, Mamay CL, O’Toole ET, et al. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodbury EL, Morgan DO. Cdk and APC activities limit the spindle-stabilizing function of Fin1 to anaphase. Nat Cell Biol. 2007;9:106–112. doi: 10.1038/ncb1523. [DOI] [PubMed] [Google Scholar]
- Wordeman L, Mitchison TJ. Identification and partial characterization of mitotic centromere-associated kinesin, a kinesin-related protein that associates with centromeres during mitosis. J Cell Biol. 1995;128:95–104. doi: 10.1083/jcb.128.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurzenberger C, Gerlich DW. Phosphatases: providing safe passage through mitotic exit. Nat Rev Mol Cell Biol. 2011;12:469–482. doi: 10.1038/nrm3149. [DOI] [PubMed] [Google Scholar]
- Yang Y, Wu F, Ward T, et al. Phosphorylation of HsMis13 by Aurora B kinase is essential for assembly of functional kinetochore. J Biol Chem. 2008;283:26726–26736. doi: 10.1074/jbc.M804207200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Tulu US, Wadsworth P, Rieder CL. Kinetochore dynein is required for chromosome motion and congression independent of the spindle checkpoint. Curr Biol. 2007;17:973–980. doi: 10.1016/j.cub.2007.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda S, Oceguera-Yanez F, Kato T, et al. Cdc42 and mDia3 regulate microtubule attachment to kinetochores. Nature. 2004;428:767–771. doi: 10.1038/nature02452. [DOI] [PubMed] [Google Scholar]
- Zinkowski RP, Meyne J, Brinkley BR. The centromere-kinetochore complex: a repeat subunit model. J Cell Biol. 1991;113:1091–1110. doi: 10.1083/jcb.113.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]


