Abstract
Stem cell activity fluctuates throughout an organism’s lifetime to maintain homeostatic conditions in all tissues. As animals develop and age, their organs must remodel and regenerate themselves in response to environmental and physiological demands. Recently, the highly conserved Hippo signaling pathway, discovered in Drosophila melanogaster, has been implicated as a key regulator of organ size control across species. Deregulation is associated with substantial overgrowth phenotypes and eventual onset of cancer in various tissues. Importantly, emerging evidence suggests that the Hippo pathway can modulate its effects on tissue size by the direct regulation of stem cell proliferation and maintenance. These findings provide an attractive model for how this pathway might communicate physiological needs for growth to tissue-specific stem cell pools. In this review, we summarize the current and emerging data linking Hippo signaling to stem cell function
Keywords: stem cells, Hippo signaling, Yap, size regulation, regeneration, tumorigenesis
Background: Stem cells and organ size
Many mammalian organs contain a subpopulation of undifferentiated stem cells (SC) involved in tissue replenishment and repair. Exquisite molecular mechanisms exist to balance SC proliferation, death, and fate decisions. Particularly important during development and regeneration, SC numbers and activity need to be tightly monitored to produce organs of a predetermined size. There seems to be a precedent for this in the case of the brain. In mice, a decrease in the number of neuronal progenitor cells leads to a reduced cortical size while increased numbers of progenitor cells leads to exencephalic forebrain overgrowth [1]. Similarly, pancreas size is also dependent on the number of progenitor cells during development [2]. Thus, it seems reasonable to hypothesize that the pathways that control mammalian organ size communicate with SC compartments because tissue expansion increases the need for SC numbers and/or activity. Our current insight into such communication, however, is scant.
Recently, Drosophila genetics has led to the emergence of a new signaling cascade, the Hippo pathway, which may constitute an intrinsic size regulator that stops growth when an organ reaches its normal size [3–11]. Mutations in components of this pathway display hugely overgrown organs, which are the result of an increase in mitosis and a decreased susceptibility to undergo cell death [reviewed in 12–14]. Importantly, emerging evidence suggests that the Hippo pathway can modulate its effects on tissue size by the direct regulation of SC proliferation and maintenance. Current work in flies and mammals has also implicated a role for cellular crowding and cell-cell contacts in regulating Hippo signaling, providing a particularly attractive model for how this pathway might communicate the physiological needs of organ growth to their tissue-specific SC pools [15–19]. In this review, we summarize the recent findings that tie Hippo signaling to the regulation and maintenance of SCs in the mammal, and highlight questions that remain unanswered in this promising new field.
Hippo signaling in mammals
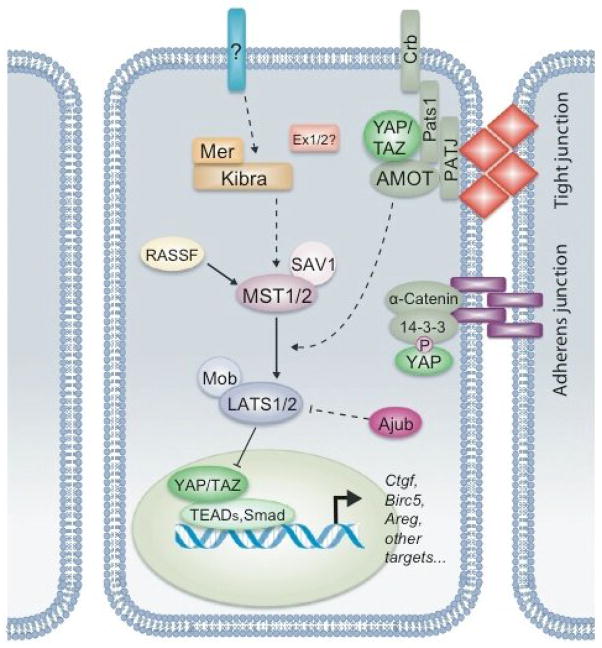
The Drosophila Hippo signaling pathway is highly conserved throughout evolution, with all core components having direct orthologs in mammals (Figure 1). Accordingly, several loss-of-function mutant phenotypes in flies can be rescued by the expression of their respective human homologs [20–23]. Signal transduction between the mammalian Hippo components is also analogous to that in flies. At the core of the signaling cascade are the Sterile 20-like kinases MST1 and MST2 and their regulatory protein, WW45 (also known as SAV1), which interact to form an activated complex. MST1/2 can also be activated by binding to the RASSF family proteins, which recruit this kinase to the cell membrane and promote its activity [24,25]. Activated MST1/2 can then directly phosphorylate the large tumor suppressor homolog kinases LATS1 and LATS2 [26–28]. LATS1/2 are regulated by MOBKL1A/B (collectively referred to as MOB1), which is also phosphorylated by MST1/2 to enhance binding in the LATS1/2-MOB1 complex [29]. In response to high cell densities, activated LATS1/2 phosphorylates the WW-domain containing transcriptional co-activators YAP at Ser127 and TAZ at Ser89, promoting 14-3-3 binding and thereby inhibiting their translocation into the nucleus [26,30–35]. Uninhibited YAP/TAZ localize to the nucleus where they serve as co-activators for the TEA-domain family member (TEAD) group of DNA-binding transcription factors [36,37]. Together, the YAP/TAZ-TEAD complex promotes proliferative and survival programs by inducing the expression of a yet unclear transcriptional program. Although our understanding of signal transduction within the core kinase cascade is well defined, the mechanisms and proteins involved in upstream regulation of the Hippo pathway are not as well established. Among many proteins postulated to be important in the initial steps of Hippo signal transduction, the only one functionally validated in vivo is the Neurofibromatosis2 gene product, NF2 (also known as Merlin) [38,39]. However, how the membrane-associated NF2 protein signals to MST or other downstream components is still a matter of major investigation. Recently, studies into the mammalian pathway have also highlighted important points of divergence, and Hippo signaling appears to be much more complicated and even context-specific in mammals [12–14].
Figure 1.
A schematic model of the Hippo signaling cascade in mammals. Cells, in blue with a dark blue lipid bilayer and a green nucleus, are shown with their respective cellular junctions. Blunted and arrowed lines indicate either inhibition or activation, respectively. Solid lines represent known interactions while dashed lines indicate unknown mechanisms. Crumbs (Crb), Expanded homologues (Ex1/2), Kibra, and Ajuba (Ajub) represent other potential regulators of Hippo signaling in mammals not discussed in the text.
YAP: A ‘stemness’ gene
During normal homeostatic conditions, adult SCs reside in defined, organ-specific progenitor cell compartments. For instance, the epithelium of the small intestine arises from actively cycling Lgr5+ SCs in the base of the crypts, and ‘mini-guts’ can be generated in vitro from a single Lgr5+ SC [40]. Similarly, skin SCs present at the hair follicle and interfollicular basal stem/progenitor compartments are responsible for organ homeostasis and regeneration of the tissue. One of the first pieces of evidence linking Hippo pathway activity to SC function came from the observations that YAP and/or TEAD expression was enriched in anatomical compartments containing stem/progenitor cells. In organs such as the small intestine and the developing brain, YAP expression is highly restricted to progenitor compartments, whereas other tissues such as the skin and skeletal muscle, show graded YAP levels based on differentiation status: nuclear (active) YAP expression in stem/progenitor cells, and cytoplasmic (inactive) YAP in mature cells [41–44]. This spatial organization linking YAP expression/activity to progenitor compartments in various organs indicates that the transcriptional activity of the YAP/TEAD complex could be important in the maintenance of SC traits in normal tissues. This conclusion is in agreement with other studies that have described YAP and TEAD as ‘stemness’ genes based on expression analyses of adult hematopoietic, neural and embryonic SCs [45].
Although the staining pattern of YAP in various tissues is generally well characterized, other data and tools for assaying the in vivo activity of this pathway remain elusive. Specifically, the precise expression pattern of other Hippo signaling components in tissues is mostly unclear. The generation of novel detection reagents, such as improved phosphospecific antibodies, to monitor cellular compartments where the pathway is active or inactive will be critical to understand the full mechanisms by which Hippo signaling controls SC biology. Additionally, an in vivo transcriptional reporter for YAP/TEAD transcriptional activity, akin to the TOPflash reporter for WNT activity is, as of yet, lacking in the Hippo field [46]. The generation of such a tool could prove important for marking and/or defining SCs in vivo, while simultaneously facilitating their isolation from various tissues. Finally, a major challenge in the Hippo field has been to determine the cell-specific effects that this pathway has in different tissues. Much of what is known about Hippo is based on conditional knockouts at the whole organ level. As such, it remains unclear whether this pathway would affect SCs and progenitors differently. Similarly, whether Hippo plays a cell- or non-cell-autonomous role in SC biology will have to be investigated, because the majority of experiments performed in mammals could affect the stem cell niche as much as the stem cells themselves. Therefore, direct manipulation of SCs and other organ-specific cells would be beneficial in revealing precisely which cell populations contribute to Hippo mutant phenotypes. Regardless of these issues, much progress has been made in exploring the cellular and molecular underpinnings of Hippo signaling in various types of SCs. We outline these findings in the next section (Table 1).
Table 1.
Known mechanisms/interactions with other major pathways that impinge on Hippo signaling in somatic and embryonic stem cells
| Stem cell type | Phenotype | Mechanistic insight | Refs |
|---|---|---|---|
| Skin | α-catenin cKO or Yap O.E. causes epidermal SC expansion; leads to SCC | α-catenin recruits and indirectly binds YAP through 14-3-3 at adherens junctions (AJs) | 19, 55 |
| Liver | MST1/2 cKO or WW45/MER cKO expands hepatocytes and/or oval cells leading to mixed HCC/CC tumors. | Canonical Hippo signaling, with MST and WW45/MER controlling YAP localization in hepatocytes and oval cells and in oval cells only, respectively. | 26, 39, 41, 48–51 |
| Intestine | O.E. of active YAP or MST/SAV1 cKO expands progenitor-like cells and blocks differentiation | Active YAP promotes WNT signaling by enhancing β-catenin transcriptional activity and induces expression of Notch targets. | 41, 59, 60 |
| Cardiac muscle | WW45/LATS/MST cKO or YAP O.E. promotes cardiomyocyte proliferation. YAP cKO leads to myocardial hypoplasia. | Nuclear YAP binds β-catenin while indirectly stimulating WNT signaling through the IGF pathway. | 61, 62 |
| CNS | MST/LATS cKO or YAP activation expands neural progenitor cells in neural tube. YAP O.E. expands CGNPs in the cerebellum and leads to medulloblastoma. |
Canonical Hippo signaling in the neural tube. Shh induces expression and nuclear localization of YAP in cerebellar granule neural precursors (CGNPs). Notch induces YAP expression in the cortex. |
64–67 |
| ESCs | Loss of TAZ in hESCs and loss of YAP or TEAD in mESCs results in a loss of self-renewal. YAP O.E. prevents differentiation in mESCs |
In hESCs, TAZ promotes self-renewal by mediating TGF-β signals and controlling the localization of SMAD2/3-4. In mESCs, YAP binds SMAD1 in response to BMP signaling for ESC maintenance. |
73–77 |
Abbreviations: cKO: conditional knockout; SCC: squamous cell carcinoma; AJs: adherens junctions; HCC: hepatocellular carcinoma; CC: cholangiocarcinoma; O.E.: overexpression; IGF: insulin-like growth factor; Shh: sonic hedgehog; CGNP: cerebellar granule neural precursor; CSC: cancer stem cell; EMT: epithelial-mesenchymal transition; hESCs: human embryonic stem cells; mESCs: mouse embryonic stem cells; TGF-β: transforming growth factor β; BMP: bone morphogenic protein
Hippo signaling and somatic stem cells
Hippo in the liver
Compared with other organs, growth in the liver has a number of unusual features. In adults, the hepatocytes that make up the majority of the liver are largely quiescent, dividing approximately once every year. These mature cell types are immensely important in this organ, as tissue replenishment is accomplished by differentiated hepatocytes rather than multipotent stem cells. If, however, hepatocyte proliferation is suppressed (i.e. in response to hepatotoxins), a putative, yet ill-defined stem cell population referred to as ‘oval cells’, found in periportal regions, expands and differentiates into both hepatocytes and cholangiocytes to regenerate lost liver tissue [reviewed in 47].
Landmark studies that initially supported the physiological relevance of the Hippo pathway in mammals were done in the liver, utilizing mouse models that conditionally overexpress YAP in hepatocytes [26,41]. YAP activation in the postnatal liver resulted in dramatic but reversible liver hyperplasia, with up to a fourfold increase in the total mass of the organ. At the cellular level, exacerbated proliferation of mature hepatocytes was shown to be the main component of the hyperplasia. These studies provided the initial demonstration that an ortholog of the Drosophila Hippo pathway could impact tissue size in mammals and laid the groundwork for further exploration of this pathway. More recently, other components of the Hippo pathway were postulated to repress proliferation in the liver [48–51]. Two separate studies showed that, following Mst1/2 deletion, livers overgrew and mice developed tumors with mixed hepatocellular carcinoma (HCC) and cholangiocarcinoma (CC) phenotypes, indicating that these malignancies originated from bipotential liver progenitor cells [48,49]. Accordingly, histological and biochemical examination showed an expansion of both hepatocytes and oval-like cells, a decrease in the level of phosphorylated YAP and LATS1/2 proteins, and increased nuclear YAP localization [48,49]. In cell lines derived from MST1/2 null livers, depletion of YAP caused growth inhibition and extensive apoptosis, findings that support the premise that YAP activation is the major mechanism underlying liver overgrowth seen with MST1/2 depletion.
Similar results were found in hepatocyte-specific WW45 and NF2 conditional knockout (cKO) mice, whose livers also overgrew and developed HCC/CC mixed tumors, but only showed increased numbers of oval cells without concomitant hepatocyte expansion [49–51]. In NF2 cKO livers, the downstream role of canonical Hippo pathway components was less clear, because opposing data regarding a connection to YAP have been published [39,51]. Overall, because the type of cell(s) expanded varied depending on the component deleted/overexpressed, and because these genetic alterations were manipulated at the whole organ level, key experiments using cell-specific Hippo alterations would clearly elucidate the need for this pathway in controlling growth of the various cell types that make up the liver. Notwithstanding, the aforementioned results clearly indicate that Hippo signaling is required, at least in a cell-autonomous way, to prevent the hyperactivation of YAP in mature and/or progenitor cells, thereby preventing aberrant hepatocyte and/or oval cell expansion, and malignant transformation.
Hippo and skin stem cells
Skin, the largest organ in mammals, protects the body from environmental hazards and prevents dehydration. In order to continuously regenerate and maintain its structural and functional integrity, the skin relies on the self-renewing abilities of epidermal SCs (eSCs) residing in the basal layer. Asymmetric divisions in this SC compartment produce short-lived progenitor cells that stratify, leaving the basal layer and moving up through the suprabasal layers to the surface of the organ as they terminally differentiate [52].
Recent studies have highlighted the importance of YAP in epidermal development and SC homeostasis [19,43]. Using a mouse model with skin-inducible expression of YAP, two independent groups demonstrated that the activation of YAP results in a severe thickening of the epidermal layer. Remarkably, this hyperplasia is driven by the expansion of undifferentiated interfollicular stem and progenitor cells [19]. The expanded cells displayed enhanced clonogenic activity and extended self-renewal as demonstrated by the use of colony-formation assays. In contrast, skin-specific deletion of YAP or genetic ablation of the YAP-TEAD interaction during epidermal development resulted in epidermal hypoplasia and failure of skin expansion [19]. This phenotype was attributed to the gradual loss of epidermal stem/progenitor cells and their limited capacity to self-renew.
Surprisingly, genetic analysis showed that in the skin YAP is not regulated by the canonical Hippo kinases. Instead, it was shown that α-catenin, a component of adherens junctions (AJs) and a known tumor suppressor in epithelial tissues, is an upstream negative regulator of YAP. Based on the massive overgrowth phenotypes obtained by deletion of α-catenin in the skin and developing brain, it was postulated that AJs could act as molecular biosensors of cell density and positioning [53–55]. The genetic and functional data linking YAP and α-catenin support and extend this idea and suggest that YAP is a critical mediator of a “crowd contro” molecular circuitry in the epidermis. In this model, increased cellular density (sensed by an increased number of AJs) limits SC expansion by inactivating YAP. Low basal cell density, as in a growing embryo or after wounding, would translate into nuclear YAP and proliferation. When this molecular network is defective (e.g. by deletion of α-catenin, inactivation of 14-3-3, or activation of YAP) hyper-proliferation and tumors can arise.
Hippo in the intestine
The intestinal epithelium is one of the most rapidly regenerating tissues in the body, turning over completely every 4 to 5 days through the continual proliferation of intestinal SCs (ISCs) located at both the +4 position and in the base of the crypt (Lgr5+) [reviewed in 56]. In Lgr5+ ISCs, Notch signaling functions synergistically with the Wnt pathway, the primary proliferation driver in the ISC compartment, to control the balance required for proper growth [57,58]. While endogenous YAP expression is typically restricted to the crypt compartment, expression of an inducible YAP-S127A protein in the intestine led to a reversible expansion of undifferentiated cells from the crypt, a phenotype very similar to the one observed after YAP activation in the skin. It was also shown that aberrant Notch activation was potentially responsible for the hyperplastic phenotype [41].
Recent studies have also begun to dissect the function of upstream Hippo regulators in this tissue. Conditional deletion of MST1/2 resulted in a similar intestinal phenotype as the YAP overexpressing model, with an expansion of progenitor cells, a disappearance of all secretory lineages, and the onset of colonic polyps, while SAV1 cKO mice showed a milder phenotype [59,60]. Accordingly, the authors noted a decrease in YAP phosphorylation and thus, prominent nuclear localization of YAP in both cKO guts. It was further suggested that YAP overexpression mediates the activation of Notch and Wnt signaling by enhancing β-catenin transcriptional activity and inducing the expression of Notch targets [59]. To this end, the authors showed that the ablation of one YAP allele sufficiently suppressed the excessive proliferation seen in MST1/2 cKO animals, a finding that placed YAP genetically downstream of these kinases. This, along with the finding that complete loss of YAP does not alter colonic development, highlights this protein as a promising drug target in gut malignancies. Together, these results are consistent with a model in which the canonical Hippo components, SAV1 and MST1/2, actively restrict YAP transcriptional activity in the ISC compartment to a level that is insufficient to promote proliferation, and that aberrant proliferation induced by YAP in ISCs is in part or wholly due to the activation of Wnt and Notch signaling.
Hippo signaling in the heart
Unlike other tissues, the role of Hippo signaling in muscles is not well characterized. Recent work has shown that cardiac-specific deletion of the upstream kinases (WW45, MST1/2, and LATS) or overexpression of constitutively active YAP resulted in embryos with dramatic cardiomegaly due to elevated cardiomyocyte number and proliferation. Conversely, YAP deletion caused the opposite result, ultimately leading to myocardial hypoplasia. Genetic studies revealed that YAP interacts with β-catenin to promote Wnt signaling, a promoter of stemness and proliferation in the heart [61,62]. Loss of β-catenin in SAV1 cKO hearts suppressed this overgrowth phenotype, confirming the aforementioned interaction data [61]. A second, independent group extended these results in their own study and suggested a model in which YAP activates the insulin-like growth factor (IGF) pathway, resulting in the inactivation of glycogen synthase kinase 3β (GSK-3β) and, therefore, inactivation of the Wnt degradation complex [62]. These results are in line with other studies in which BIO, a GSK-3β inhibitor, and PI3K-Akt signaling promote cardiomyocyte proliferation, although this study provides the first evidence linking all three pathways biochemically [62]. Therefore, in the heart, YAP promotes embryonic and neonatal cardiomyocyte proliferation by directly binding to β-catenin in the nucleus to promote an SC gene profile, while indirectly promoting Wnt signaling through the IGF pathway.
Hippo and nervous tissues
Neural progenitor cells reside along the ventricular zone in the developing vertebrate neural tube and are responsible for generating myriad cell types composing the mature central nervous system (CNS) [reviewed in 63]. YAP protein is expressed in this progenitor zone in mouse, frog, and chick neural tubes, and colocalizes with Sox2, a neural progenitor marker [64,65]. Here, loss of Mst1/2 or Lats1/2, or activation of YAP-TEAD lead to a marked expansion of neural progenitors, partially due to an upregulation of cell cycle re-entry and stemness genes, and a block to differentiate by suppressing key genes. Conversely, YAP loss-of-function results in increased cell death and precocious neural differentiation [64].
In the cerebellum, endogenous YAP is highly expressed in cerebellar granule neural precursors (CGNPs) and in tumor-repopulating cancer SCs in the perivascular niche [66]. An increase in cells showing an undifferentiated CGNP phenotype in this region of the brain, such as medulloblastomas common in children, also express high levels of YAP [66,67]. Given that CGNPs rely on Sonic hedgehog (Shh) signaling to expand, and that activation of the Shh pathway is implicated in human medulloblastomas, the connection between the Shh and Hippo pathways was investigated [66]. It was found that Shh signaling induces the expression and nuclear localization of YAP in CGNPs, and that YAP then drives the proliferation of these cells. Together, these studies suggest a new model for the brain, in which YAP promotes NSC proliferation by serving as a possible nexus between NSC proliferative pathways, such as Notch and Shh (and possibly others), which were traditionally thought to act in parallel to control brain development.
Hippo and embryonic stem cells
Embryonic SCs (ESCs), isolated from the inner cell mass of blastocysts, are the source of all tissues composing the developing embryo, fetus and ultimately adult organism. In vitro, human and mouse ESC (hESC and mESC respectively) depend on different signals for self-renewal: mESCs rely on the cytokine leukemia inhibitory factor (LIF) and signals from bone morphogenic proteins (BMPs), while hESC rely on fibroblast growth factor (FGF) signaling and a balance between transforming growth factor β (TGF-β)/Activin and BMP signaling [68–72]. Transcriptional regulation has also proven to be key for ESC self-renewal, plasticity and differentiation, as forced expression of various transcription factors can reprogram differentiated tissues into pluripotent SCs (induced pluripotent stem cells, iPSCs) capable of self-renewing and generating adult mice [68,69]. Recently, studies investigating YAP and TAZ have uncovered a role for these transcriptional co-activators in regulating ESC self-renewal and differentiation.
One study that links Hippo to ESC biology found that TAZ dominantly controls the localization of SMAD2/3-4 proteins, which are transcriptional regulators that mediate TGF-β signaling. Upon stimulation with TGF-β, TAZ binds SMAD2/3-4 proteins to facilitate their nuclear accumulation and couples them to the Mediator complex, thereby promoting their transcriptional activity [73]. Importantly, knocking down TAZ, but not YAP in hESCs resulted in a loss of self-renewal and differentiation into neuroectoderm, the same phenotype seen with TGF-β receptor inhibition. Conversely, knocking down LATS2 enhanced the generation of human iPS cells by preventing this kinase from inactivating TAZ [74]. In mouse ESCs (mESCs), YAP associates with SMAD1 to control Id gene transcription for ESC maintenance in response to BMP stimulation [75]. These studies indicated a link among YAP/TAZ-dependent BMP/TGF-β transcriptional output, ESC maintenance and fate decisions.
More recently, two studies found that during mESC differentiation, YAP is inactivated and that knockdown of this or TEAD proteins results in a loss of pluripotency [76,77]. Conversely, YAP is activated in iPSCs, increases reprogramming efficiency, and prevents differentiation in mESCs when it is ectopically overexpressed [76]. These studies also found that YAP-TEAD bind to and promote the transcription of known stemness genes -- such as Oct3/4, Sox2, PcG targets, LIF targets, Nanog and BMP signaling targets -- in mESCs but not in mature cells. Together, these data point to a model in which YAP/TAZ maintain ESC pluripotency in vitro by mediating BMP/TGF-β transcriptional activity and directly promoting the expression of important stemness genes.
Concluding remarks
Since its discovery in the past decade, much progress has been made in the Hippo field and it is now clear that this pathway and its effectors, YAP and TAZ, play critical roles in cell fate decisions, SC proliferation and regeneration. However, key questions regarding the identity and biological relevance of upstream Hippo modulators, and the mechanisms and contexts by which Hippo cross-talks with other SC regulatory pathways remain to be answered. A particular challenge in the field relates to discovering how Hippo signaling might sense and respond to physiological needs for growth and repair in particular organs. Interestingly, recent data from the mouse and the fly suggest that YAP/Yorkie activation might be crucial for injury-induced intestinal stem cell proliferation and regeneration in response to tissue damage [60,78–80]. Still, conclusive answers to these questions could bring important insight to the poorly understood problem of organ size control. To this end, it is important to realize that in addition to cell-autonomous signals, microenvironmental cues from the SC compartment, or the niche, are known to play a key role in enabling adult SCs to perceive and respond to environmental changes and needs [81].
Cell shape and polarity also have a profound effect on the outcome of cell divisions, and thus differentiation decisions, with cleavage-plane orientation determining whether divisions will be symmetric (producing identical daughter cells), or asymmetric (producing daughters with different fates) [reviewed in 82]. It is not surprising then, that the significance of cell junctions and polarity complexes in modulating Hippo signaling has become increasingly apparent [reviewed in 12–14]. In addition to its binding to α-catenin and adherens junctions, YAP can directly interact with members of the Crumbs polarity complex at tight junctions [83–87]. These observations suggest that YAP can physically localize to both adherens and tight junctions. Whether one particular adhesion complex is the most important regulator of YAP activity and localization will probably depend on the architecture of each particular tissue. Interestingly, Hippo pathway proteins Lats1, Mst1, and Drosophila Mats (Mob1 homolog) are reported to be activated by membrane targeting [88–90]. Therefore, these membrane adhesion complexes might serve as a platform for Hippo pathway phosphorylation events to occur. The challenge is now to validate these observations in vivo and place them in a cellular and physiological framework that could provide new insights into SC biology and organ growth.
It is now fair to speculate that proper tissue homeostasis, including the number of stem and mature cells, is achieved through a combination of cell- and non-cell-autonomous signaling, spatial control of YAP/TAZ localization by cell-cell contact, and mechanical cues dictated by tissue architecture. Further elucidation of these processes and how they ultimately converge on Hippo signaling will likely provide insight into molecular mechanisms that regulate development, SC maintenance, and tumorigenesis. Additional studies probing this exceptionally important stem cell pathway will thus be critical in the search for new, regenerative approaches to human medicine and disease.
Figure 2.
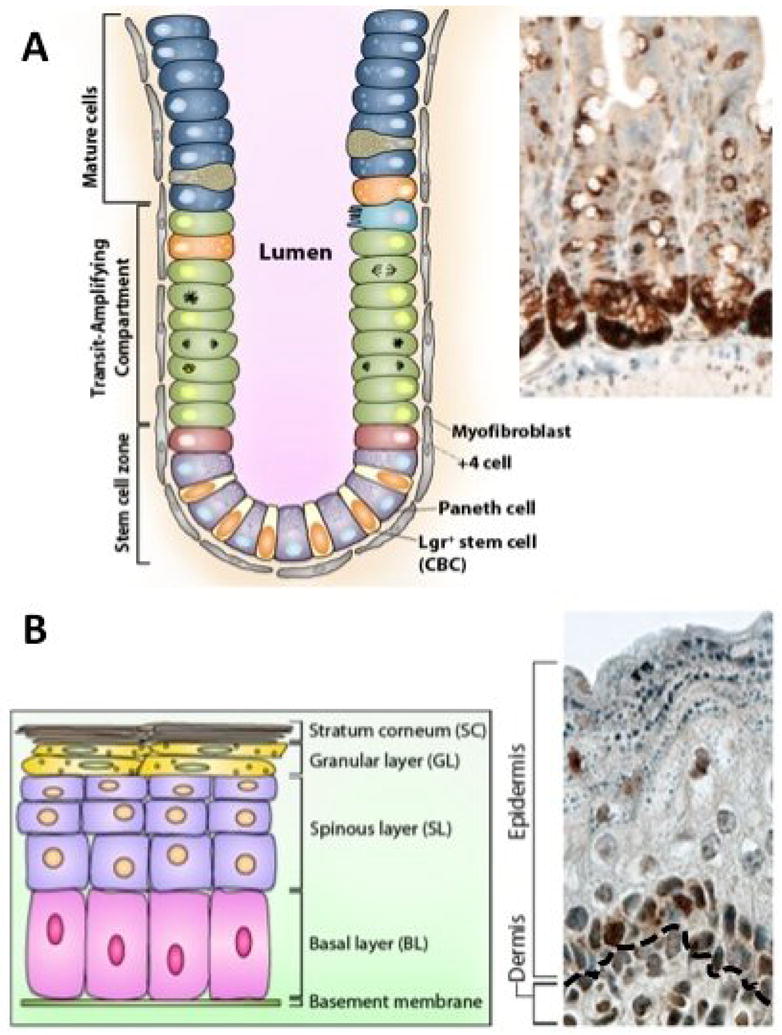
YAP expression in stem/progenitor cell compartments. (A) Intestinal crypt architecture with quiescent (+4) and active crypt base columnar (CBC, Lgr5+) stem cells shown. Also shown but not discussed in the text are mature cell types, the transit-amplifying compartment, and components of the intestinal stroma (myofibroblasts). Inset depicts YAP localization in crypts, in wildtype intestine. (B) Epidermal architecture with progenitor cells residing in the basal layer (BL). Asymmetric divisions in this compartment produces short-lived progenitor cells that stratify as they differentiate, leaving the basal layer and moving up into the spinous layer (SL), granular layer (GL), and stratum corneum (SC). Inset depicts significant YAP localization in the basal layer of wildtype skin. Black dotted line represents the border between the dermis and epidermis.
Acknowledgments
We apologize to those whose work we could not cite due to space constraints. We thank K. Schlegelmilch and E. Barry for their image contributions and general feedback. FDC is a Pew Scholar and is supported by grants from the NIH, Stand Up to Cancer Foundation and the Department of Defense
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Depaepe V, et al. Ephrin signalling controls brain size by regulating apoptosis of neural progenitors. Nature. 2005;435:1244–1250. doi: 10.1038/nature03651. [DOI] [PubMed] [Google Scholar]
- 2.Stanger BZ, et al. Organ size is limited by the number of embryonic progenitor cells in the pancreas but not the liver. Nature. 2007;445:886–891. doi: 10.1038/nature05537. [DOI] [PubMed] [Google Scholar]
- 3.Justice RW, et al. The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev. 1995;9:534–546. doi: 10.1101/gad.9.5.534. [DOI] [PubMed] [Google Scholar]
- 4.Xu TA, et al. Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development. 1995;121:1053–1063. doi: 10.1242/dev.121.4.1053. [DOI] [PubMed] [Google Scholar]
- 5.Dong J, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harvey KF, et al. The Drosophila Mst ortholog, hippo, restricts growth and cell proliferation and promotes apoptosis. Cell. 2003;114:457–467. doi: 10.1016/s0092-8674(03)00557-9. [DOI] [PubMed] [Google Scholar]
- 7.Pantalacci S, et al. The Salvador partner Hippo promotes apoptosis and cell-cycle exit in Drosophila. Nat Cell Biol. 2003;5:921–927. doi: 10.1038/ncb1051. [DOI] [PubMed] [Google Scholar]
- 8.Jia J, et al. The Drosophila Ste20 family kinase dMST functions as a tumor suppressor by restricting cell proliferation and promoting apoptosis. Genes Dev. 2003;17:2514–2519. doi: 10.1101/gad.1134003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Udan RS, et al. Hippo promotes proliferation arrest and apoptosis in the Salvador/Warts pathway. Nat Cell Biol. 2003;5:914–920. doi: 10.1038/ncb1050. [DOI] [PubMed] [Google Scholar]
- 10.Wu S, et al. Hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- 11.Tapon N, et al. salvador promotes both cell cycle exit and apoptosis in Drosophila and is mutated in human cancer cell lines. Cell. 2002;110:467–478. doi: 10.1016/s0092-8674(02)00824-3. [DOI] [PubMed] [Google Scholar]
- 12.Zhao B, et al. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat Cell Biol. 2011;13:877–883. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan D. The Hippo signaling pathway in development and cancer. Dev Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen CL, et al. The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc Natl Acad Sci USA. 2010;107:15810–15815. doi: 10.1073/pnas.1004060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grzeschik NA, et al. Lgl, aPKC, and Crumbs regulate the Salvador/Warts/Hippo pathway through two distinct mechanisms. Curr Biol. 2010;20:573–581. doi: 10.1016/j.cub.2010.01.055. [DOI] [PubMed] [Google Scholar]
- 17.Ling C, et al. The apical transmembrane protein Crumbs functions as a tumor suppressor that regulates Hippo signaling by binding to Expanded. Proc Natl Acad Sci USA. 2010;107:10532–10537. doi: 10.1073/pnas.1004279107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson BS, et al. Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila via the FERM-domain protein expanded. Curr Biol. 2010;20:582–590. doi: 10.1016/j.cub.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schlegelmilch K, et al. Yap1 acts downstream of α-catenin to control epidermal proliferation. Cell. 2011;144:782–795. doi: 10.1016/j.cell.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu S, et al. Hippo encodes a Ste-20 family protein kinase that restricts cell proliferation and promotes apoptosis in conjunction with salvador and warts. Cell. 2003;114:445–456. doi: 10.1016/s0092-8674(03)00549-x. [DOI] [PubMed] [Google Scholar]
- 21.Lai ZC, et al. Control of cell proliferation and apoptosis by mob as tumor suppressor, mats. Cell. 2005;120:675–685. doi: 10.1016/j.cell.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 22.Huang J, et al. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 23.Tao W, et al. Human homologue of the Drosophila melanogaster lats tumour suppressor modulates CDC2 activity. Nat Genet. 1999;21:177–181. doi: 10.1038/5960. [DOI] [PubMed] [Google Scholar]
- 24.Khokhlatchev A, et al. Identification of a novel Ras-regulated proapoptotic pathway. Curr Biol. 2002;12:253–265. doi: 10.1016/s0960-9822(02)00683-8. [DOI] [PubMed] [Google Scholar]
- 25.Oh HJ, et al. Role of the tumor suppressor RASSF1A in Mst1-mediated apoptosis. Cancer Res. 2006;66:2562–2569. doi: 10.1158/0008-5472.CAN-05-2951. [DOI] [PubMed] [Google Scholar]
- 26.Dong J, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan EH, et al. The Ste20-like kinase Mst2 activates the human large tumor suppressor kinase Lats1. Oncogene. 2005;24:2076–2086. doi: 10.1038/sj.onc.1208445. [DOI] [PubMed] [Google Scholar]
- 28.Hirabayashi S, et al. Threonine 74 of MOB1 is a putative key phosphorylation site by MST2 to form the scaffold to activate nuclear Dbf2-related kinase 1. Oncogene. 2008;27:4281–4292. doi: 10.1038/onc.2008.66. [DOI] [PubMed] [Google Scholar]
- 29.Praskova M, et al. MOBKL1A/MOBKL1B phosphorylation by MST1 and MST2 inhibits cell proliferation. Curr Biol. 2008;18:311–321. doi: 10.1016/j.cub.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oh H, Irvine KD. In vivo regulation of Yorkie phosphorylation and localization. Development. 2008;135:1081–1088. doi: 10.1242/dev.015255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hao Y, et al. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J Biol Chem. 2008;283:5496–5509. doi: 10.1074/jbc.M709037200. [DOI] [PubMed] [Google Scholar]
- 32.Zhao B, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J, et al. Negative regulation of YAP by LATS1 underscores evolutionary conservation of the Drosophila Hippo pathway. Cancer Res. 2008;68:2789–2794. doi: 10.1158/0008-5472.CAN-07-6205. [DOI] [PubMed] [Google Scholar]
- 34.Lei QY, et al. TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol Cell Biol. 2008;28:2426–2436. doi: 10.1128/MCB.01874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oka T, et al. Mst2 and Lats kinases regulate apoptotic function of Yes kinase-associated protein (YAP) J Biol Chem. 2008;283:27534–27546. doi: 10.1074/jbc.M804380200. [DOI] [PubMed] [Google Scholar]
- 36.Zhao B, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang H, et al. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial–mesenchymal transition. J Biol Chem. 2009;284:13355–13362. doi: 10.1074/jbc.M900843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hamaratoglu F, et al. The tumour-suppressor genes NF2/Merlin and Expanded act through Hippo signalling to regulate cell proliferation and apoptosis. Nat Cell Biol. 2006;8:27–36. doi: 10.1038/ncb1339. [DOI] [PubMed] [Google Scholar]
- 39.Zhang N, et al. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev Cell. 2010;19:27–38. doi: 10.1016/j.devcel.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;470:353–358. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 41.Camargo FD, et al. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 42.Li Y, et al. Genome-wide analysis of N1ICD/RBPJ targets in vivo reveals direct transcriptional regulation of Wnt, SHH, and Hippo Pathway effectors by Notch1. Stem Cells. 2012 doi: 10.1002/stem.1030. Advanced online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang H, et al. Yes-associated protein (YAP) transcriptional coactivator functions in balancing growth and differentiation in skin. Proc Natl Acad Sci USA. 2011;108:2270–2275. doi: 10.1073/pnas.1019603108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watt KI, et al. Yap is a novel regulator of C2C12 myogenesis. Biochem Biophys Res Commun. 2010;393:619–624. doi: 10.1016/j.bbrc.2010.02.034. [DOI] [PubMed] [Google Scholar]
- 45.Ramalho-Santos M, et al. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- 46.Molenaar M, et al. XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 47.Avruch J, et al. Mst1/2 signaling to Yap: gatekeeper for liver size and tumour development. Br J Cancer. 2011;104:24–32. doi: 10.1038/sj.bjc.6606011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song H, et al. Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc Natl Acad Sci USA. 2010;107:1431–1436. doi: 10.1073/pnas.0911409107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu L, et al. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci USA. 2010;107:1437–1442. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee KP, et al. The Hippo-Salvador pathway restrains hepatic oval cell proliferation, liver size and liver tumorigenesis. Proc Natl Acad Sci USA. 2010;107:8248–8253. doi: 10.1073/pnas.0912203107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benhamouche S, et al. Nf2/Merlin controls progenitor homeostasis and tumorigenesis in the liver. Genes Dev. 2010;24:1718–1730. doi: 10.1101/gad.1938710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fuchs E. Scratching the surface of skin development. Nature. 2007;445:834–842. doi: 10.1038/nature05659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lien WH, et al. alphaE-catenin controls cerebral cortical size by regulating the hedgehog signaling pathway. Science. 2006;311:1609–1612. doi: 10.1126/science.1121449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lien WH, et al. Cadherin-catenin proteins in vertebrate development. Curr Opin Cell Biol. 2006;18:499–506. doi: 10.1016/j.ceb.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 55.Silvis MR, et al. α-catenin is a tumor suppressor that controls cell accumulation by regulating the localization and activity of the transcriptional coactivator Yap1. Sci Signal. 2011;4:ra33. doi: 10.1126/scisignal.2001823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 57.Fre S, et al. Notch and Wnt signals cooperatively control cell proliferation and tumorigenesis in the intestine. Proc Natl Acad Sci USA. 2009;106:6309–6314. doi: 10.1073/pnas.0900427106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barker N, et al. The intestinal stem cell. Genes Dev. 2008;22:1856–1864. doi: 10.1101/gad.1674008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou D, et al. Mst1 and Mst2 protein kinases restrain intestinal stem cell proliferation and colonic tumorigenesis by inhibition of Yes-associated protein (Yap) overabundance. Proc Natl Acad Sci USA. 2011;108:E1312–20. doi: 10.1073/pnas.1110428108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cai J, et al. The Hippo signaling pathway restricts the oncogenic potential of an intestinal regeneration program. Genes Dev. 2010;24:2383–2388. doi: 10.1101/gad.1978810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heallen T, et al. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332:458–461. doi: 10.1126/science.1199010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xin M, et al. Regulation of insulin-like growth factor signaling by Yap governs cardiomyocyte proliferation and embryonic heart size. Sci Signal. 2011;4:ra70. doi: 10.1126/scisignal.2002278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ma DK, et al. Adult neural stem cells in the mammalian central nervous system. Cell Res. 2009;19:672–682. doi: 10.1038/cr.2009.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cao X, et al. YAP regulates neural progenitor cell number via the TEA domain transcription factor. Genes Dev. 22:3320–3334. doi: 10.1101/gad.1726608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gee ST, et al. Yes-associated protein 65 (YAP) expands neural progenitors and regulates Pax3 expression in the neural plate border zone. PLoS One. 2011;6:e20309. doi: 10.1371/journal.pone.0020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fernandez-L A, et al. YAP1 is amplified and up-regulated in hedgehog-associated medulloblastomas and mediates Sonic hedgehog-driven neural precursor proliferation. Genes Dev. 2009;23:2729–2741. doi: 10.1101/gad.1824509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Orr BA, et al. Yes-Associated Protein 1 is widely expressed in human brain tumors and promotes glioblastoma growth. J Neuropathol Exp Neuro. 2011;70:568–577. doi: 10.1097/NEN.0b013e31821ff8d8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Evans M. Discovering pluripotency: 30 years of mouse embryonic stem cells. Nat Rev Mol Cell Biol. 2011;12:680–686. doi: 10.1038/nrm3190. [DOI] [PubMed] [Google Scholar]
- 69.Chambers I, Smith A. Self-renewal of teratocarcinoma and embryonic stem cells. Oncogene. 2004;23:7150–7160. doi: 10.1038/sj.onc.1207930. [DOI] [PubMed] [Google Scholar]
- 70.Biswas A, Hutchins R. Embryonic stem cells. Stem Cells Dev. 2007;2:213–222. doi: 10.1089/scd.2006.0081. [DOI] [PubMed] [Google Scholar]
- 71.Darr H, Benvenisty N. Human embryonic stem cells: the battle between self-renewal and differentiation. Regen Med. 2006;3:317–325. doi: 10.2217/17460751.1.3.317. [DOI] [PubMed] [Google Scholar]
- 72.Xiao L, et al. Activin A maintains self-renewal and regulates fibroblast growth factor, Wnt, and bone morphogenic protein pathways in human embryonic stem cells. Stem Cells. 2006;6:1476–1486. doi: 10.1634/stemcells.2005-0299. [DOI] [PubMed] [Google Scholar]
- 73.Varelas X, et al. TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat Cell Biol. 2008;10:837–848. doi: 10.1038/ncb1748. [DOI] [PubMed] [Google Scholar]
- 74.Qin H, et al. Transcriptional analysis of pluripotency reveals the Hippo pathway as a barrier to reprogramming. Hum Mol Genet. 2012 Jan 27; doi: 10.1093/hmg/dds023. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alarcón C, et al. Nuclear CDKs drive Smad transcriptional activity and turnover in BMP and TGF-β pathways. Cell. 2009;139:757–769. doi: 10.1016/j.cell.2009.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lian I, et al. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev. 2010;24:1106–1118. doi: 10.1101/gad.1903310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tamm C, et al. Regulation of mouse embryonic stem cell self-renewal by a Yes-YAP-TEAD22 signaling pathway downstream of LIF. J Cell Sci. 2010;124:1136–1144. doi: 10.1242/jcs.075796. [DOI] [PubMed] [Google Scholar]
- 78.Ren F, et al. Hippo signaling regulates Drosophila intestine stem cell proliferation through multiple pathways. Proc Natl Acad Sci USA. 2010;107:21064–21069. doi: 10.1073/pnas.1012759107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shaw RL, et al. The Hippo pathway regulates intestinal stem cell proliferation during Drosophila adult midgut regeneration. Development. 2010;24:4147–4158. doi: 10.1242/dev.052506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Karpowicz P, et al. The Hippo tumor suppressor pathway regulates intestinal stem cell regeneration. Development. 2010;24:4135–4145. doi: 10.1242/dev.060483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Morrison SJ, Spradling AC. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132:598–611. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Panbianco C, Gotta M. Coordinating cell polarity with cell division in space and time. Trends Cell Biol. 2011;21:672–680. doi: 10.1016/j.tcb.2011.08.006. [DOI] [PubMed] [Google Scholar]
- 83.Varelas X, et al. The Crumbs complex couples cell density sensing to Hippo-dependent control of the TGF-β-SMAD pathway. Dev Cell. 2010;19:831–844. doi: 10.1016/j.devcel.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 84.Grzeschik NA, et al. Lgl, aPKC, and Crumbs regulate the Salvador/Warts/Hippo pathway through two distinct mechanisms. Curr Biol. 2010;20:573–581. doi: 10.1016/j.cub.2010.01.055. [DOI] [PubMed] [Google Scholar]
- 85.Robinson BS, et al. Crumbs regulates Salvador/Warts/Hippo signaling in Drosophila via the FERM-domain protein Expanded. Curr Biol. 2010;20:582–590. doi: 10.1016/j.cub.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen CL, et al. The apical-basal cell polarity determinant Crumbs regulates Hippo signaling in Drosophila. Proc Natl Acad Sci USA. 2010;107:15810–15815. doi: 10.1073/pnas.1004060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ling C, et al. The apical transmembrane protein Crumbs functions as a tumor suppressor that regulates Hippo signaling by binding to Expanded. Proc Natl Acad Sci USA. 2010;107:10532–10537. doi: 10.1073/pnas.1004279107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Avruch J, et al. Nore1 and RASSF1 regulation of cell proliferation and of the MST1/2 kinases. Methods Enzymol. 2006;407:290–310. doi: 10.1016/S0076-6879(05)07025-4. [DOI] [PubMed] [Google Scholar]
- 89.Hergovich A, et al. The human tumour suppressor LATS1 is activated by human MOB1 at the membrane. Biochem Biophys Res Commun. 2006;345:50–58. doi: 10.1016/j.bbrc.2006.03.244. [DOI] [PubMed] [Google Scholar]
- 90.Ho LL, et al. Mob as tumor suppressor is activated at the cell membrane to control tissue growth and organ size in Drosophila. Dev Biol. 2009;337:274–283. doi: 10.1016/j.ydbio.2009.10.042. [DOI] [PubMed] [Google Scholar]