Abstract
The complex, partially redundant gene regulatory architecture underlying vertebrate heart formation has been difficult to characterize. Here, we dissect the primary cardiac gene regulatory network in the invertebrate chordate, Ciona intestinalis. The Ciona heart progenitor lineage is first specified by Fibroblast Growth Factor/Map Kinase (FGF/MapK) activation of the transcription factor Ets1/2 (Ets). Through microarray analysis of sorted heart progenitor cells, we identified the complete set of primary genes upregulated by FGF/Ets shortly after heart progenitor emergence. Combinatorial sequence analysis of these co-regulated genes generated a hypothetical regulatory code consisting of Ets binding sites associated with a specific co-motif, ATTA. Through extensive reporter analysis, we confirmed the functional importance of the ATTA co-motif in primary heart progenitor gene regulation. We then used the Ets/ATTA combination motif to successfully predict a number of additional heart progenitor gene regulatory elements, including an intronic element driving expression of the core conserved cardiac transcription factor, GATAa. This work significantly advances our understanding of the Ciona heart gene network. Furthermore, this work has begun to elucidate the precise regulatory architecture underlying the conserved, primary role of FGF/Ets in chordate heart lineage specification.
Keywords: Ciona intestinalis, heart development, gene regulatory network, FGF, Ets, Map Kinase, chordate
Vertebrate heart formation is regulated by a complex, multi-layered gene regulatory network (GRN) (Srivastava, 2006). Previous research has identified extensive sets of transcription and signaling factors that contribute to vertebrate heart formation (Niu et al., 2007; Wolf and Basson, 2010; Xu and Baldini, 2007). However, vertebrate heart morphogenesis is a dynamic, multi-step process (Evans et al., 2010; Vincent and Buckingham, 2010) regulated by extensive, partially redundant GRN components (Dunwoodie, 2007; Singh et al., 2009; Targoff et al., 2008). Thus, comprehensive documentation of the vertebrate heart GRN will be challenging (Sperling, 2011). Studies in a range of organisms have highlighted the importance of GRN topology in developmental patterning. In particular, comprehensive, validated invertebrate networks illustrate how inter linked regulatory sub circuits mediate robust gene expression outputs (Bolouri and Davidson; Davidson, 2010; Davidson and Levine, 2008; Maduro, 2009; Sandmann et al., 2007; Tsachaki and Sprecher, 2012). Within each sub circuit, linkages encoded by cis-regulatory elements (CREs) generate topologies tailored for specific biological functions (Peter and Davidson, 2009). It is therefore essential to decipher the topology of heart regulatory sub-circuits in order to understand how complex spatio-temporal processes are coordinated during cardiogenesis.
The genomic simplicity and experimental tractability of Ciona intestinalis has made this invertebrate chordate a potent, emerging model for comprehensive, validated GRN studies (Christiaen et al., 2008; Imai et al., 2006; Imai et al., 2009; Lemaire, 2011). Ciona is a member of the tunicates, a group of basal chordates that diverged prior to vertebrate genome duplications (Dehal et al., 2002). Accordingly, the C. intestinalis genome contains a single ortholog of most vertebrate transcription and signaling factors. The Ciona genome is also highly condensed, containing greatly reduced amounts of non-coding DNA in comparison with most vertebrate genomes (Di Gregorio and Levine, 2002). Additionally, sequencing of two Cionid genomes (C. intestinalis and C. savignyi) permits efficient identification of conserved, candidate cis-regulatory elements through phylogenetic foot printing (Johnson et al., 2004; Satoh et al., 2003). Lastly, transfection through electroporation allows rapid testing of candidate elements through transgenic reporter gene analysis (Corbo et al., 1997; Shi et al., 2005). However, the derived nature of Ciona embryogenesis limits its potential as a model for vertebrate development (Lemaire, 2011). One important exception is Ciona heart development (Davidson, 2007).
Recent studies indicate that initial aspects of Ciona and vertebrate cardiogenesis are surprisingly well conserved (Christiaen et al., 2008; Davidson, 2007; Davidson et al., 2006; Ragkousi et al., 2011; Satou et al., 2004; Stolfi et al., 2010). The Ciona heart is derived from two founder cells, the B7.5 blastomeres. In vertebrate and Ciona embryos, the helix-loop-helix transcription factor Mesp plays an important, potentially chordate specific, role in establishing cardiac founder cell identity (Bondue et al., 2008; Davidson et al., 2005; Saga et al., 2000; Satou et al., 2004). After one round of division, the four B7.5 lineage cells express the Ets family transcription factor Ets1/2 (Davidson et al., 2006). A recent study has shown that the mouse ortholog, Ets1, is expressed early in the emerging myocardium (Schachterle et al., 2012). The Mesp, Ets1/2 expressing B7.5 blastomeres then divide asymmetrically, creating two distinct cell lineages. Fibroblast growth factor (FGF)/Map Kinase (MapK) signaling differentially activates Ets1/2 in the smaller daughters, termed the trunk ventral cells (TVCs, Fig. 1A). Ets1/2 activity establishes TVC transcriptional identity, mediating the expression of the conserved cardiac transcription factors GATAa and Nkx (Davidson et al., 2006). Ets1/2 also regulates TVC migration by directly upregulating the Forkhead family transcription factor FoxF (Beh et al., 2007). As observed with vertebrate heart progenitors, the TVCs associate with the endoderm and migrate as bilateral precursors towards the ventral midline (Christiaen et al., 2008; Ragkousi et al., 2011). The TVCs subsequently give rise to definitive heart precursors as well as islet expressing pharyngeal muscle precursors (Stolfi et al., 2010). Parallel deployment of FGF signaling, cardiac transcription factors and migratory cell behavior strongly support homology between the TVCs and vertebrate heart progenitor field. Studies of the relatively simple TVC gene regulatory network are therefore likely to provide critical insights into related but much more complex vertebrate heart progenitor networks.
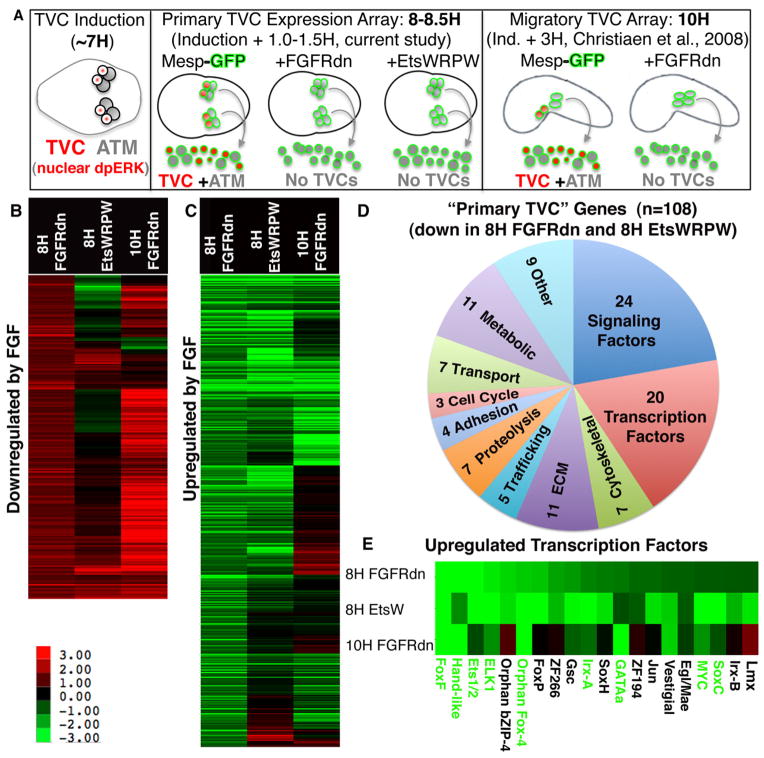
Figure 1. Primary FGF/Ets dependent gene expression in the TVC lineage.
(A) Schematic illustrating staged harvesting of dissociated founder lineage cells for microarray analysis relative to the time of initial TVC induction. H = hours post fertilization in this and all subsequent figures. Nuclear dpERK = nuclear localization of di-phosphorylated Extracellular-signal Regulated Kinase indicative of MapK signaling cascade activity (B, C) log2 fold changes (scale shown below panel B) for all probe sets showing a robust (fc > log20.8) and significant (p< 0.06) increase (B) or decrease (C) in the Mesp-FGFRdn samples (8H FGFRdn). These cut-off values were modeled on those employed for the 10H array (Christiaen et al., 2008). For each panel, average fold change values from the corresponding probe sets from the early Mesp-EtsWRPW samples (8H EtsWRPW) and previously published 10H Mesp-FGFRdn samples (Christiaen et al., 2008) are also shown. (D) Pie chart showing the predicted functions of all 108 annotated “primary TVC genes.” (E) Fold change data for the 20 primary TVC transcription factors, green lettering indicates confirmation of early TVC expression by in situ expression assay (see Fig. 2).
The simple, binary nature of FGF:MapK:Ets1/2 dependent TVC transcription represents a powerful system for deciphering the downstream cardiac GRN. Through targeted transgenic manipulations of transcription factor activity in the B7.5 founder lineage, the TVC gene network can be either potentiated or disrupted. Transgenically altered B7.5 lineage cells can then be isolated and used for whole-genome micro-arrays. Comparative array studies have led to the comprehensive identification of Ets1/2 and FoxF dependent gene expression associated with TVC migration (Christiaen et al., 2008). Additionally, cis regulatory analysis of the crucial TVC migratory gene, RhoDF, indicate that Ets1/2 and FoxF act through a coherent feed-forward network to activate migratory cell behavior (Christiaen et al., 2008). However, the primary role of Ets1/2 in establishing TVC identity remains poorly defined.
Here, we employ microarray analysis on sorted B7.5 lineage cells to identify primary TVC genes upregulated shortly after localized Ets1/2 activation. We utilized bioinformatics to identify Ets1/2 co-motifs enriched in predicted cis-regulatory domains of candidate primary Ets1/2 target genes. We then tested the most highly enriched co-motif, ATTA, through reporter analysis. Our results indicate that associated Ets1/2 and ATTA motifs demarcate cis-regulatory regions driving primary TVC gene expression. Thus it appears that differential co-binding sites encode a temporal cascade of Ets1/2 dependent gene expression. Ets and ATTA binding sites encode primary TVC gene expression while Ets and FoxF binding sites encode subsequent, migratory TVC gene expression. The structure of this “dual input” feed-forward sub-circuit may reflect a fundamental mechanism employed to layer the expression of Map Kinase target genes during multiple developmental and physiological processes.
Methods
Embryological techniques
Ciona intestinalis adults were purchased from M-REP (San Diego). Fertilization, dechorionation and electroporation were performed as previously described (Corbo et al., 1997). Embryos were staged according to the developmental timeline established in Hotta et al., 2007.
Histochemistry and in situ hybridization
X-gal staining, anti-body staining and in situ hybridization were performed as previously described (Cooley et al., 2011). Antisense RNA probes were created from Gene Collection library clones (Satou et al., 2002) as listed in Supplemental Fig. 1.
Molecular Cloning
Genomic regions used for enhancer analysis were amplified using sequence specific primers carrying appropriate restriction sites according to established protocols. Deletions, insertions and point mutations were introduced through PCR according to established protocols (Davidson et al., 2005). The FoxF minimal enhancer region (−1135/840:Fkh:lacZ) is as described in Beh et al, 2007 except that the BamH1 site immediately proceeding the Fkh basal promoter was converted into a Sph1 site to eliminate an Ets binding site motif encoded by the BamHI restriction site. Complete sequence information for most amplified regions and specific mutational alterations are displayed in Figs. 3–5 and Supplemental Figs. 4 and 5. For regions where the precise sequence is not shown we listed relevant primers in Supplemental Table 4.
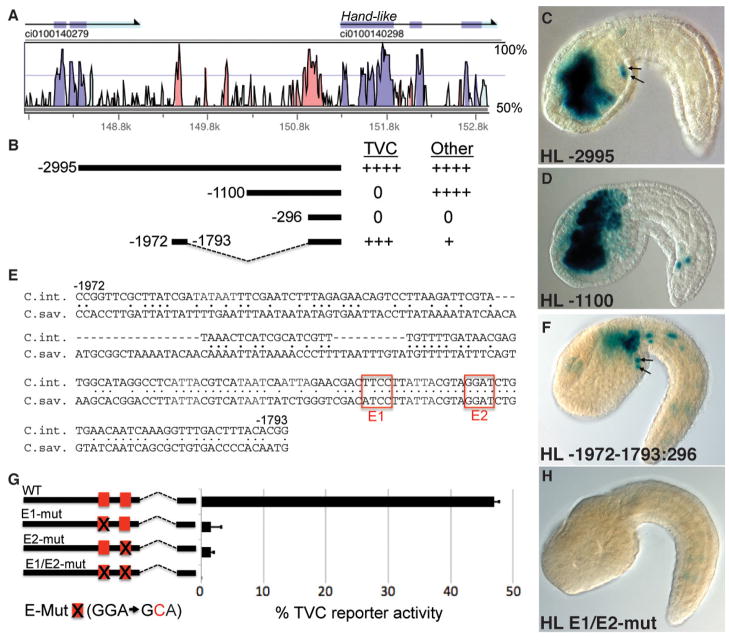
Figure 3. Cis-regulatory analysis of Hand-like.
(A) Vista alignment depicts sequence conservation between C. intestinalis and C. savignyi in the 5′ intergenic region (Couronne et al., 2003). (B) Diagram of Hand like reporter constructs aligned with (A) and numbered by their distance from the translation start codon. For each construct, relative reporter expression in the TVCs and other lineages is indicated to the right (++++ = nearly all transgenic embryos display denoted reporter staining; +++ = >50% of stained embryos; + = <5% of stained embryos; 0 = no expression). (C–D) Representative Stage 22-Hand-like 2995:+1:lacZ and Hand-like 1100:+1:lacZ embryos, arrows indicate reporter expression in TVCs. (E) Alignment of the highly conserved 179bp distal element, C. int. = Ciona intestinalis, C. sav. = Ciona savignyi, red boxes highlight two conserved Ets1/2 binding sites. (F) Representative embryo showing TVC reporter expression (arrows) driven by the 179bp distal element fused to the Hand-like 296bp basal promoter. (G) Diagram of point mutations, indicated by an X (sequence provided below), introduced into putative Ets1/2 binding sites (red boxes) in the Hand-like -1972-1793:296:lacZ reporter construct (error bars indicate standard deviation, three trials, N>600 for each sample). (H) Representative embryo displaying loss of reporter activity when the two Ets binding motifs are mutated (E1/E2-mut, as diagrammed in G).
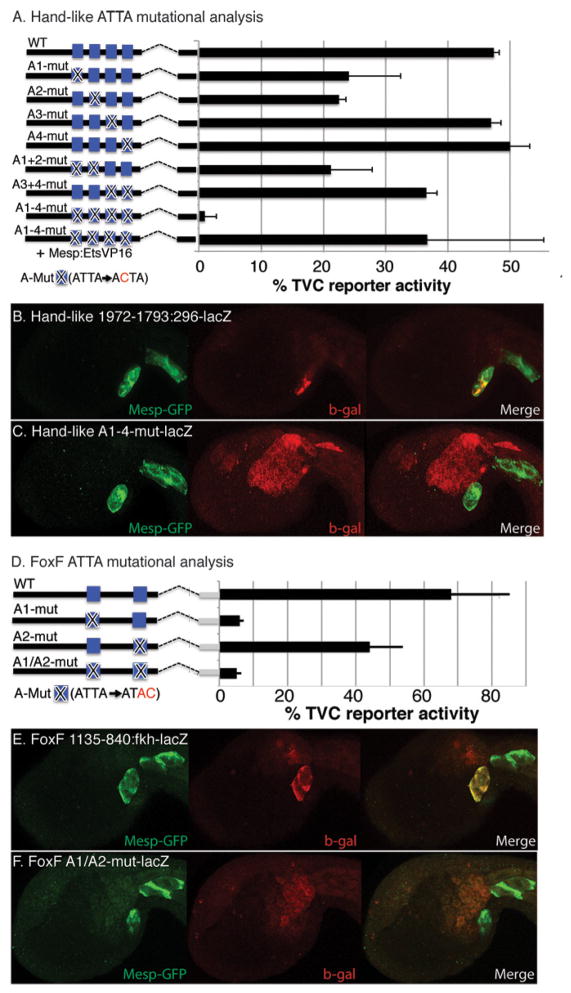
Figure 5. The ATTA co-motif contributes to TVC reporter expression in both Hand-like and FoxF regulatory elements.
(A, D) Diagrams illustrating mutational analysis of ATTA co-motifs in (A) the Hand-like TVC distal CRE (−1972-1793:296:lacZ) and (D) the FoxF TVC CRE (−1135-840:Fkh:lacZ). Point mutations, indicated by an X, were introduced into ATTA co-motif sequences (blue boxes) and the sequence of these mutations is shown below each diagram. Barplots depict percent of embryos showing TVC reporter expression (error bars indicate standard deviation, three trials, N>47 for each sample). (B, E) These panels display representative embryos co-electroporated with a Mesp-GFP construct to visualize the B7.5 lineage, and either (B) the Hand-like CRE reporter construct or (E) the FoxF CRE reporter. (C, F) Same as (B, E) except that the reporter constructs contain mutations in the ATTA motifs, as diagrammed in (A, D), three trials, N>161 for each sample.
Cell Sorting and Microarrays
Fluorescent activated sorting of Mesp GFP labeled B7.5 founder lineage cells was performed using the FACSAria (BD Bioscienes, San Jose, CA) sorter as previously described (Christiaen et al., 2008) with modifications appropriate for stringent exclusion of YFP labeled cells and cell clusters as appropriate for this instrument (performed at AZCC/ARL-Division of Biotechnology Cytometry Core Facility funded by the Cancer Center Support Grant, CCSG-CA 023074). RNA extraction and probe synthesis were conducted as described in Christiaen et al., 2008 and hybridization to the Ciona intestinalis custom-design GeneChipTM was conducted using standard Affymetrix procedures at the University of Arizona Genomics Core (http://www.azcc.arizona.edu/research/shared-services/gss).
Microarray data analysis
Raw expression data sets (CEL files) were read and analyzed using R Programming tools (www.r-project.org) and the Affy and LIMMA libraries provided by the Bioconductor community (www.bioconductor.org). Custom probe sets for Ciona were background subtracted and normalized using the Affy RMA function. The 3 or 4 biological replicas for each experimental condition were then fitted to a linear statistical model. Lists of probes that varied significantly between comparisons of interest including fold change and probability of fold change corrected for false discovery rate were obtained using LIMMA functions. The resulting lists were then annotated using community-provided spreadsheets (Smyth, 2005). Cluster 3.0 and TreeView 1.1.1 were used to cluster and display fold-changes as described in (Christiaen et al., 2008). Raw CEL files, RMA processed data sets and appropriate descriptions of the experimental conditions will be deposited in the ArrayExpress database upon publication of this manuscript.
Results
Microarray analysis identifies primary Ets1/2 target genes
We employed whole-genome microarray analysis of sorted B7.5 lineage cells to identify primary FGF:MapK:Ets1/2 target genes (Fig. 1, Supplemental Table 1). For this analysis, B7.5 lineage cells were labeled with the Mesp-GFP reporter. Adjacent lineages were labeled with a MyoD-YFP reporter permitting stringent fluorescent activated cell sorting of the B7.5 lineage cells. This approach was previously utilized to identify 539 genes upregulated by FGF:MapK in migrating TVCs at 10 hours post-fertilization (HPF) (Christiaen et al., 2008). However, this 10H target gene set is likely to contain numerous genes upregulated indirectly following initial MapK mediated Ets1/2 activation at Stage 15 (7HFP). To identify primary Ets1/2 target genes we examined gene expression from B7.5 lineage cells isolated between 8–8.5HPF, 0.5–1 hour after FGF:MapK:Ets1/2 target genes are first detected at St. 16, 7.5HPF (Ragkousi et al., 2011). We will subsequently refer to this array as the 8H or primary TVC dataset. We have provided a schematic diagram to clarify the relationship between the current and previous array studies (Fig. 1A). Identification of candidate primary TVC genes involved a comparison between wild-type controls and two different transgenic embryonic samples. In the first experimental sample, we targeted a dominant negative form of the sole Ciona FGF receptor (FGFRdn) to the B7.5 lineage using the Mesp enhancer (Mesp-FGFRdn). This construct has previously been shown to completely abrogate FGF:MapK:Ets1/2 mediated induction of the TVC lineage (Davidson et al., 2006). Therefore, we anticipated that comparative gene expression analysis between control and transgenic Mesp-FGFRdn B7.5 lineage cells would provide a comprehensive overview of primary FGF dependent gene expression. In the second experimental sample, we targeted a dominant repressor form of Ets1/2 to the B7.5 lineage using the Mesp enhancer (Mesp-EtsWRPW). This construct is designed to repress Ets1/2 target gene transcription and has previously been shown to abolish TVC induction (Davidson et al., 2006). We anticipated that comparative gene expression analysis between control, Mesp-FGFRdn and Mesp-EtsWRPW B7.5 lineage cells would help to further distinguish direct Ets target genes within the FGF dependent gene set. To ensure rigorous statistical analysis we obtained three or four distinct biological replicates for each transgenic manipulation. LIMMA analysis of the resulting dataset identified 2329 probe sets displaying significant changes (p<0.06) in expression when comparing wild-type to sorted FGFRdn B7.5 lineage cells. Of these, 241 probe sets displayed a robust (FC>20.8) increase in the FGFRdn background (Fig. 1B), while 389 were robustly decreased (Fig. 1C). In comparing these results to the previous 10HPF array data, we found that 188/241 probe sets (78%) were robustly increased in both FGFRdn samples while 156/389 probe sets (41%) were robustly decreased in both FGFRdn samples. Thus it appears that genes repressed during primary specification (Fig. 1B) are more likely to be maintained in a stable off state (as previously noted in Davidson, 2011) while FGF activated genes (Fig. 1C) tend to undergo subsequent dynamic alterations, including a small subset (29/389) that appeared to be down-regulated by FGF in the 10H array.
We next employed the Mesp EtsWRPW array data to predict candidate direct Ets target genes within the primary gene set. We anticipated that genes directly upregulated by the FGF:MapK:Ets1/2 pathway (Fig. 1C) would display reduced expression in both the FGFRdn and EtsWRPW samples. This pattern was observed for the majority of FGFRdn down-regulated probe sets (218/389 displayed a robust, FC>20.8 and significant, p<.06 decrease in EtsWRPW samples, Fig. 1C). Of the remaining FGFRdn down regulated probe sets, many showed no significant response to EtsWRPW (135/171) or a mild but significant down regulation (24/171) leaving 12 probe sets displaying significant upregulation in the EtsWRPW background. These results suggest that a subset of FGF target genes is either modulated by an Ets-independent transcriptional pathway or by an indirect Ets pathway (repression of a repressor). Alternatively, the disparity in the response to EtsWRPW and FGFRdn may reflect the premature action of Mesp-EtsWRPW. Based on the detection of Mesp driven reporters, Ets-WRPW protein is likely to be produced at significant levels by St. 12, ~5.5HPF (Cooley et al., 2011). In comparison, Mesp-FGFRdn will not impact gene expression until FGF mediated expression changes first occur at St. 16, ~7.5HPF (Cooley et al., 2011; Ragkousi et al., 2011). Thus, our Mesp-EtsWRPW 8–8.5H probe set expression levels may reflect secondary, compensatory regulation.
We next examined the impact of Mesp EtsWRPW within the FGFRdn upregulated probe sets (Fig. 1B). We anticipated that direct repression by FGF:MapK:Ets1/2 would be reflected by a substantial subset of probes displaying increased expression in the FGFRdn samples and decreased expression in the EtsWRPW samples. Surprisingly, only 17 of the 241 FGFRdn upregulated probe sets (7%) displayed robust and significant decreases in expression within the Ets-WRPW samples. These results may reflect Ets independent down-regulation. Additionally, 55/241 FGFRdn upregulated probe sets (23%) were also upregulated in the Ets-WRPW samples. These cases most likely reflect indirect repression (upregulation of a downstream repressor). Thus, our EtsWRPW array data appears to include some secondary Ets1/2 mediated expression changes (due to premature Mesp-EtsWRPW repression as discussed above).
We next examined the composition of the 218 probe sets predicted to represent the primary TVC genes directly upregulated by Ets. These probe sets match 161 unique genes, including 108 with discernable orthology to genes with characterized domains or functions. A functional classification of these presumed direct target genes is shown in Fig. 1D. Approximately 40% of these genes (44/108) represent either signaling components or transcription factors. These include the previously characterized primary TVC transcription factors FoxF, Hand-like (also annotated as Notrlc) and GATAa (Beh et al., 2007; Davidson, 2007; Imai et al., 2003). We also detected FGF/Ets mediated upregulation of two Irx family transcription factors (Irx-A and Irx-B) which may represent functional orthologs to the early vertebrate heart field genes Irx1,2,4 and 5 (Christoffels et al., 2000). This set also includes 7 orthologs to genes involved in FGFR, MapK or Ets regulatory feedback loops that are likely to modulate signaling dynamics following initial TVC specification (Cbl, sprouty, Map KIK1, Dusp6.9, Odin, Ets1/2 and Edl). We also detected groups of genes associated with the cytoskeleton, ECM, vesicle trafficking, proteolysis and adhesion (33/108). These genes may serve to prime the emergent TVCs for migration. In particular, these genes may serve to alter the adhesion properties of the TVCs allowing them to detach from their sister cells (the anterior tail muscle precursors) and attach to the adjacent endoderm precursors. In this vein, it is interesting to note upregulation of orthologs to three genes associated with Drosophila myoblast fusion (Sticks and Stones, Roughest and Rolling Stone) which function to mediate adhesive interactions between muscle founder cells and fusion competent myoblasts (Krauss, 2010).
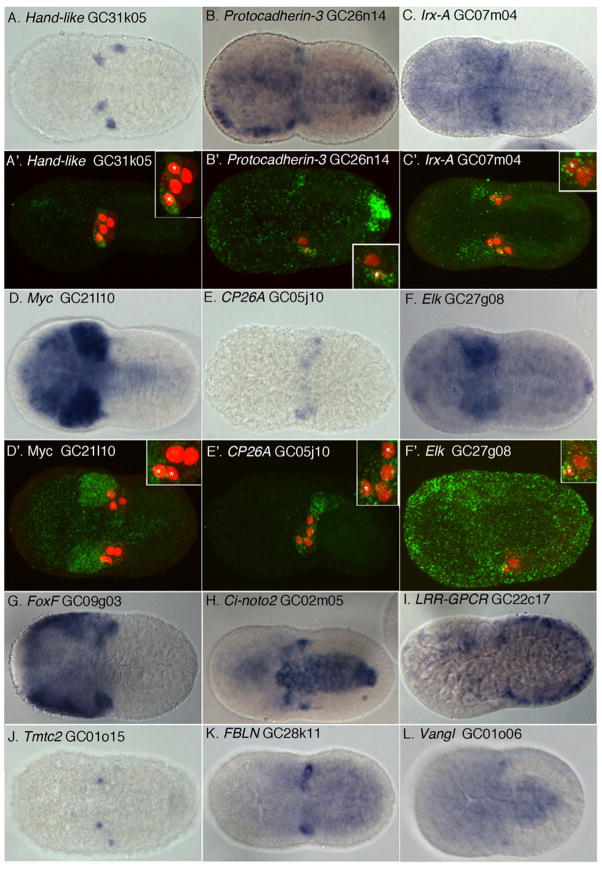
To validate our microarray results, we performed in situ expression assays for the subset of our 160 candidate FGF/MapK /Ets target genes predicted to have detectable expression levels (130 with expression >6, Fig. 2, Supplemental Fig. 1). To mirror the array, this assay was performed on 8.5HPF embryos. At this stage, expression in the newly born TVCs (clearly demarcated by Hand-like expression, Fig. 2A) can be somewhat difficult to identify if a gene is also expressed in the adjacent mesenchyme and tail muscle lineages (see for example Fig. 2F). To compensate for this issue, we employed confocal microscopy of fluorescently labeled transcripts for a select set of candidate genes (Fig. 2A’–F’). Overall, we observed staining for 47 candidate gene antisense probes, 80% of which displayed apparent expression in the TVCs (Supplemental Fig. 1). These results indicate that the 8–8.5H microarray dataset provides an accurate representation of initial FGF/Ets targets in the newly born TVC lineage.
Figure 2. Expression of candidate primary TVC genes in the newly emergent TVC lineage.
In-situ hybridizations (A–L) including fluorescent tyramide staining (A′–F′) for a select set of candidate RNA probes (green), founder cell lineage labeled by Mesp-lacZ (red). TVC nuclei indicated by asterisks in inset, magnified panels. Ventral views are shown except for panels A′, E′ and F′ which are lateral confocal sections.
Characterization of cis regulatory elements for Hand-like expression
To initiate our analysis of primary TVC gene regulation we characterized the minimal regulatory element required for TVC expression of Hand-like (Fig. 3). Previous research had identified a ~3000bp stretch of 5′ intergenic DNA capable of driving lacZ reporter expression in the endogenous Hand-like expression domain (TVCs, endoderm, trunk lateral cells and anterior neural tube) when fused to the Fkh basal promoter (Davidson and Levine, 2003). To simplify further analysis we examined reporter activity mediated through the native Hand-like promoter. We characterized a 296bp Hand-like basal promoter region that displayed no independent reporter activity. Through reporter testing of upstream regions fused to this basal promoter, we identified two distinct regulatory elements within the 5′ intergenic region (Fig. 3A–F). The proximal element (beginning at 1100bp upstream of the start codon) drove reporter expression in all non-TVC Hand-like expression domains (Fig. 3B,D). A short, well-conserved distal element (from 1972bp to 1793bp upstream of the start codon) drove robust reporter expression in the TVCs along with weaker staining in neural, mesenchyme and tail muscle lineages (Fig. 3B,E,F).
To evaluate the presumed role of Ets1/2 in direct upregulation of Hand-like, we mutated the two Ets consensus binding site motifs in the minimal, distal Hand-like TVC element (red boxes in Fig. 3E,G). Single base pair mutation (GGAW → GCAW) of either one of these two binding sites was sufficient to abrogate nearly all reporter activity (Fig. 3G,H).
Identification of candidate Ets co-motifs by sequence analysis
Ets family factors acts in partnership with a wide array of co-factors to regulate target genes (Sharrocks, 2001). Indeed, recruitment of a specific partner is a key determinant in target gene selection (Verger and Duterque-Coquillaud, 2002). We therefore predicted that primary heart genes would be co-regulated by Ets1/2 in association with a specific co factor. Furthermore, we anticipated that co-factor binding sites would be associated with Ets binding sites within primary TVC cis-regulatory elements (CREs). Based on these predictions, we employed comparative sequence analysis and whole-genome motif ranking to identify candidate co factor binding sites would be associated with Ets binding sites within primary TVC cis-regulatory elements (CREs). Based on these predictions, we employed comparative sequence analysis and whole-genome motif ranking to identify candidate co-factor binding sites.
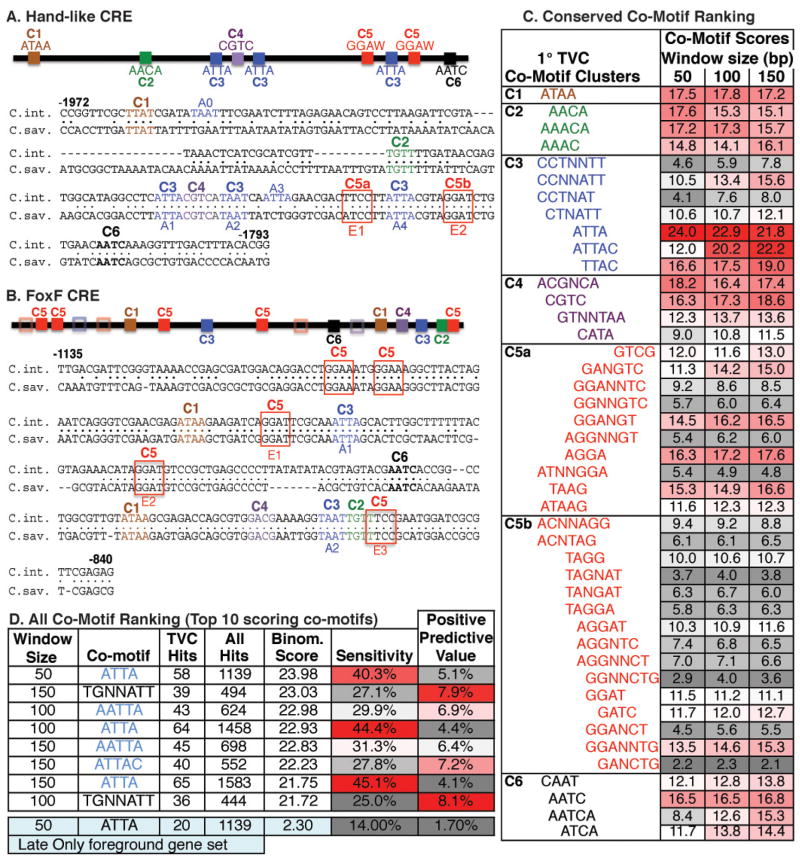
We first compared the Hand-like and FoxF primary TVC cis-regulatory elements. The Hand-like (Fig. 3) and FoxF CREs (Beh et al., 2007) both direct rapid onset reporter expression in newly arisen TVCs. These two elements also contain functionally critical Ets binding motifs (Fig. 3G, H; Beh et al., 2007). We therefore predicted that the Hand-like and FoxF CREs are bound by Ets1/2 in association with a shared co-factor. We searched for candidate co-factor binding motifs (co-motifs) by identifying all consensus sequences conserved in both the Hand-like and FoxF CREs (Fig. 4A–C). We employed consensus sequences rather than weight matrices as these are easier to handle computationally. We generated a set of all possible 4- and 5- nucleotide motifs with N and NN spacers added after the 2nd nucleotide (e.g. AAAA, AANNAA, AANAA, AANNAAA and all combinations of nucleotides). We then matched these motifs against C. intestinalis vs. C. savignyi alignments of the Hand-like and FoxF regulatory elements (Fig. 4A, B). Of the 2704 motifs examined, only 44 motifs were present and phylogenetically conserved in both primary TVC elements. These motifs can be roughly grouped into 6 clusters based on their alignments to the Hand-like CRE. (Fig. 4C).
Figure 4. Bio-informatic prediction of Ets Co-motifs in primary TVC regulatory elements.
(A,B) Schematic and sequence alignments of the Hand-like or FoxF CREs showing the distribution of 6 clusters (C1–C6) composed of 47 shared co-motifs. Clusters are grouped by the first appearance of a co-motif in the Hand-like CRE. Hollow boxes indicate partial matches to motif clusters (indicated by color) in the FoxF CRE. ATTA (A0-4) and Ets binding motifs (E1–E3) are also annotated. (C) Co-motif scores for all shared co-motifs grouped by clustering in the Hand-like CRE. Note that some sequences have been reverse-complemented to clarify their alignment within a cluster. Cluster 2 may represent a FoxF binding site involved in maintenance of TVC gene expression. Cluster 4 may represent a CREB binding site (CGTCA, Matsumoto et al., 2007) potentially involved in co-recruitment of a CBP co-activator (Foulds et al., 2004). (D) Data on the top 10 scoring co-motifs from the primary TVC CRE sequence set, along with the ATTA co-motif from the “late only” TVC CRE sequence set.
We next ranked the 44 shared co-motifs by quantifying their enrichment in predicted primary TVC CREs. The analysis was modelled on a previous effort used to generate “motif-tissue scores”, as described in detail in Auger et al., 2009 and Haeussler et al., 2010. To generate a ‘foreground’ set of predicted primary TVC CREs, we collected all conserved non-coding sequence (CNS) adjacent to the 144 annotated Ets1/2primary target genes. We then filtered these CNS regions by nucleosome binding scores, retaining regions predicted to have low nucleosome occupancy. This step was based on a recent finding demonstrating a strong correlation between low-nucleosome binding scores and regulatory function in Ciona non-coding sequences (Khoueiry et al., 2010). We used the same process to generate a “background” set of predicted CREs for all 15,569 Ciona gene models. Foreground and background CRE sequences were then scanned for conserved co-motifs adjacent to conserved ETS binding motifs (GGAW). For this analysis, we employed a series of window sizes (50bp, 100bp or 150bp) based on the spacing range of the Ets and co-motif binding sites in the defined Hand-like and FoxF CREs. Each Ets-motif+co-motif CRE match was assigned to the nearest adjacent gene. This analysis generated a list of “hits” within the foreground “primary TVC” gene set and the background “all gene” set. We could then calculate the sensitivity (TVC gene hits/144 TVC genes) and positive predictive value (TVC gene hits/All gene Hits) of each Ets-motif+co-motif combination (Supplemental Table 1; Markstein and Levine, 2002). We also generated logarithmic “co-motif scores” based on the probability of obtaining a similar sensitivity and predictive value with foreground genes drawn randomly from the genome (Haeussler et al., 2010). A high “co-motif” score represents a high level of selective concentration around primary TVC candidate genes. Logarithmic scores were calculated for all 44 Ets motif+ co-motif pairs (color-coded in Fig. 4C, listed in Supplemental Table 1). Among the 44 co-motifs, ATTA displayed a notably higher score in all three window sizes. The extended ATTAC motif showed a similarly high score in the larger 100bp and 150bp windows. We also conducted a less biased analysis using all 2704 possible tetramer and pentamer co-motifs (Supplemental Table 2, Fig. 4D). Even in this much larger data set, ATTA containing motifs occupied the top scoring positions in all three window sizes. Furthermore, this analysis indicates that the enriched co-motif consensus may extend to AATTAR. To determine whether these co-motif predictions are specific to the primary TVC gene set, we conducted the same analysis using the set of genes that appear to be robustly upregulated by FGF in migrating TVCs (10H array, Christiaen et al., 2008) but not in the primary TVC data set (8H array). To ensure that this “late only” gene set was the same size as our primary TVC gene list, we only included the 144 most severely down-regulated genes (cut off was an ~ 4 fold decrease) in the FGFRdn 10H array. In the predicted “late only” CREs, the ATTA motif was no longer among the top scoring motifs (Supplemental Table 3). Interestingly, the top scoring “late only” motif (TGNTAT) overlaps with the one alternative top-scoring motif in the primary TVC CREs (TGNNATT) suggesting enrichment of “TGNNAT” reflects a broad abundance of this co-motif in heart gene CREs while ATTA enrichment is temporally specific. We also noted that the co-motif scores in the “late only” CRE set were substantially lower than scores in the primary TVC CRE set. This score difference results from a much higher prevalence of conserved regions adjacent to primary TVC genes (average of 2330 conserved base-pairs/gene, of which 962 bp constitute “nucleosome-free” sequence) in comparison to the “late only” TVC genes (average of 961 conserved base-pairs/gene, 256 bp of “nucleosome-free” sequence). The observed difference in conservation levels has interesting implications regarding variable evolutionary constraints on elements regulating these two gene sets but it does not impact the relative ranking of co-motif scores. Overall, these results strongly suggest that an ATTA binding transcription factor works with Ets1/2 to specifically co-regulate primary TVC genes. To test this model, we initiated functional analysis of primary TVC regulatory elements.
Primary target gene regulation requires the ATTA motif
We employed site-directed mutagenesis of the Hand-like and FoxF regulatory elements to test the predicted role of the ATTA co-motif in early heart gene regulation (Fig. 5). The proximal, more conserved 100bp region of the Hand-like TVC CRE contains four ATTA motifs (three of which are conserved). Site-directed mutation of the first or second conserved ATTA motifs (A1 or A2, ATTA→ACTA) greatly reduced TVC reporter activity (Fig. 5A). In contrast, mutation of the third conserved motif (A4) or the adjacent non conserved ATTA motif (A3) had no discernable impact (Fig. 5A). However, simultaneous mutation of both A3 and A4 cause a more notable disruption of reporter activity (Fig. 5A). Additionally, mutation of all four ATTA motifs abrogated nearly all reporter activity (Fig. 5A–C). The previously characterized 237bp FoxF TVC CRE contains two conserved ATTA motifs (Beh et al., 2007). Site-directed mutation of the first conserved ATTA severely reduced TVC reporter activity (Fig. 5D) while mutation of the second ATTA motif only slightly reduced TVC reporter expression (Fig. 5D). This analysis supports our bio-informatic prediction, indicating that expression of both Hand-like and FoxF in the TVCs requires cis-regulatory elements containing Ets binding sites and associated ATTA co-motifs. We also employed the Hand-like A1-4 mutant construct for cis-trans complementation assays (Beh et al., 2007). We found that targeted expression of activated Ets1/2 (Mesp-EtsVP16) is able to restore TVC expression of the Hand-like A1-4 mut–lacZ reporter (Fig. 5A). This result supports predicted in-vivo binding of the Ets1/2 DNA binding domain to Ets consensus motifs in the Hand-like TVC CRE.
Regulation of the secondary TVC gene ASB does not require the ATTA motif
We next began to test whether the ATTA co-motif is specifically required for primary heart gene expression by characterizing the cis-regulation of ASB (Ankyrin-Socs Box). Microarray assays indicate that ASB has a delayed response to FGF signaling, displaying significant FGF dependent upregulation only in the 10H array. To verify the timing of ASB expression we conducted staged in-situ expression assays (Supplemental Fig. 2). ASB expression is first apparent in the TVCs at St. 18 (~9HPF), one hour after robust expression of Hand-like, FoxF and other early heart genes (St. 16/17, 8H).
Initially we determined that a 2.98 kb region of intergenic DNA 5′ to ASB was sufficient to drive lacZ reporter expression in the TVCs. A series of truncated 5′ intergenic DNA constructs were generated in order to identify the ASB minimal TVC enhancer (Supplemental Fig. 3). Through this analysis we identified a 105 bp TVC regulatory element (832 – 727 bps upstream of the ASB transcriptional start site). When fused to a 500 bp basal promoter, this 105bp CRE drove reporter expression in the TVCs, mesenchyme and tail muscle (Supplemental Fig. 3). Our informatic analysis suggests that ATTA sites are involved in early target gene regulation but not in late target gene regulation. To test this prediction we mutated the two ATTA motifs in the ASB minimal enhancer. Disruption of the ATTA motifs had no discernible impact on TVC reporter gene expression (Supplemental Fig. 3). In contrast, mutation of FoxF consensus binding motifs (TRTTT) in the ASB regulatory element led to robust disruption of TVC reporter expression (Supplemental Fig. 3). These data suggest that the ATTA co-motif is specifically associated with primary heart gene regulation while later, migratory TVC gene regulation involves FoxF (as previously observed in the cis-regulatory analysis of the critical “migratory TVC gene” RhoD/F, Christiaen et al., 2008). However, analysis of ATTA motif function in a broader set of migratory TVC gene enhancers is required to definitively address the hypothesized temporal specificity of the ATTA motif.
Clustered Ets/ATTA motifs designate primary heart gene enhancers
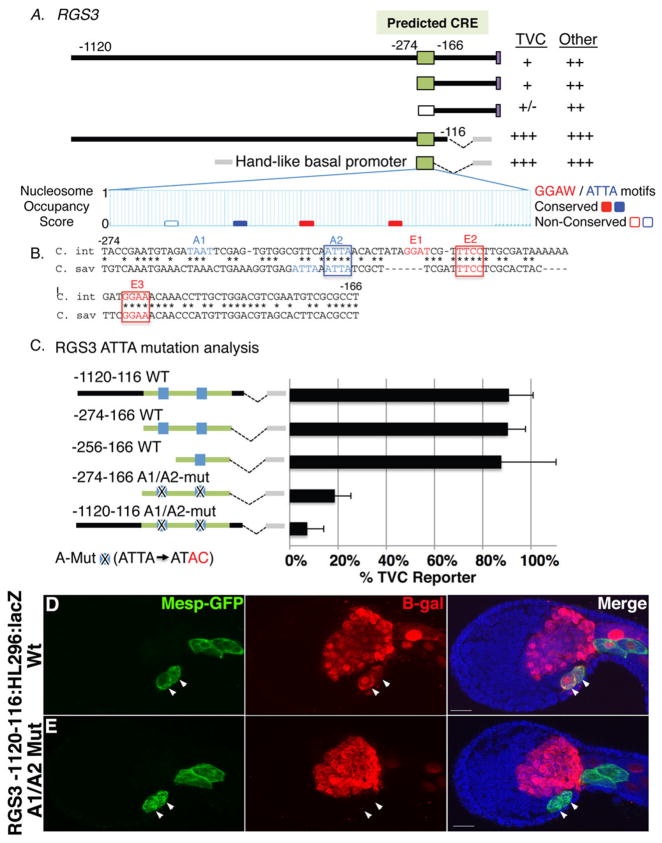
Our analysis indicates that primary TVC gene expression is encoded by Ets/ATTA motif combinations. To further test this hypothesis, we attempted to identify and test the role of the Ets/ATTA motif combination in a broader group of primary TVC regulatory elements (Fig. 6 and Supplemental Figs. 4–5). We employed two approaches to obtain and test further primary TVC elements. Initially, we employed serial deletion analysis of the 5′ non-coding DNA adjacent to one of the highly enriched primary heart genes from the array, LGR (Leucine-rich-repeat-rich G-protein coupled Receptor). We found that a 2879bp upstream region was sufficient to drive reporter expression in the endogenous LGR expression domain (TVCs, neural tube and tail muscle) and that deletion of the distal 84 bp region of this construct led to a complete loss of TVC reporter expression (Supplemental Fig. 4E). Although this 84 bp region is not phylogenetically conserved, it does contain four Ets motifs and three adjacent ATTA motifs in a region of partial nucleosome occupancy (Supplemental Fig. 4E, 5). Subsequently, we used informatic software (http://chordator.vze.com/; Haeussler et al., 2010) to search for conserved Ets/ATTA motif combinations adjacent to our primary heart gene set, isolated these predicted CREs, and tested their ability to drive TVC reporter expression. Based on the characterized Hand-like and FoxF enhancers, we set the search parameters for conserved clusters of two GGAW motifs and one ATTA motif in a 50 base pair window. We also required that these clusters fall within 10,000 base pairs of a candidate gene. This search identified 825 clusters adjacent to 615 different genes (~4% of all 15,569 annotated genes). Of these, 64 clusters flanked 33 of the candidate primary TVC genes (~23% of 144 annotated genes). Thus the Ets/ATTA combination motif displays close to a six-fold enrichment in proximity to foreground, primary TVC genes over background. Candidate TVC CREs were selected for functional analysis based on three criteria: low nucleosome occupancy scores; ease of cloning the predicted CRE along with the endogenous promoter and confirmed TVC expression of the adjacent gene. Six candidate CREs were isolated and tested by reporter analysis (Fig. 6 and Supplemental Fig. 4). This set included four CREs contained in intergenic regions upstream of candidate primary TVC genes (RGS3-aniseedV3_8034, unc5-aniseedV3_6051, aniseed VS_11904 and aniseedV3 1839), one CREs located downstream of Hand-like and one CRE in the first intron of the conserved cardiac transcription factor GATAa (Ragkousi et al., 2011).
Figure 6. Reporter analysis of the predicted RGS3 primary TVC regulatory element.
(A) Diagram of RGS3 reporter constructs numbered by their distance from the translation start codon along with the resulting expression levels in the TVCs and other lineages (+++ and + as described in Fig. 3; ++ = ~10–20% of stained embryos; +/−= no expression in some trials but sporadic expression at very low levels in other trials). At the bottom of panel (A) is a graphical representation of nucleosome occupancy scores (Segal et al; 2006) for the predicted CRE taken from the Aniseed Genome Browser (http://www.aniseed.cnrs.fr/). (B) Alignment of the conserved 108bp CRE, labeling as described in Fig. 3. (C) Mutational analysis of ATTA co-motifs in RGS3 reporter constructs as indicated, labeling as described for previous figures (2–3 trials, N>80 for each sample). (D–E) Representative RGS3-1120-116:HL296:lacZ transgenic embryos co-electroporated with a Mesp-GFP construct. (D) Wildtype reporter construct drives robust reporter expression (red) in the TVCs (arrowheads). (E) ATTA mutations (A1/A2-mut, as diagrammed in C) abrogate TVC reporter expression (arrowheads). Scale bar = 20um.
Our analysis of the GATAa intronic element is summarized in Supplemental Fig. 4. We first amplified a fragment including 1937bp of upstream intergenic DNA along with the first exon and intron, fusing the lacZ reporter in frame with the first few codons of the second exon. This construct (GATAa −1937+2112:lacZ) drove robust reporter expression in the TVCs and endoderm, recapitulating endogenous GATAa expression (Ragkousi et al., 2011). In contrast, a large fragment of upstream intergenic DNA (GATAa −4977) drove no TVC reporter activity. To facilitate further dissection of the intronic TVC CRE, we fused the ~2000bp first intron upstream of a 589 basal promoter fragment. The resulting construct (GATAa +81/+2112:−589:lacZ) preserved strong TVC reporter expression. We also observed strong TVC reporter expression when we removed all intronic sequence upstream of the predicted CRE (GATAa+1155/+2112:589:lacZ, Supplemental Fig. 4B). As anticipated, removal of the predicted ~300bp CRE (GATAa+1428/+2112:589:lacZ, Supplemental Fig. 4C) led to a complete loss of TVC reporter expression.
Our analysis of the remaining candidate CREs is displayed in Fig. 6 and Supplemental Fig. 4. A large ~2000 bp region containing the predicted 3′ Hand-like CRE fused to the 296bp native Hand-like promoter failed to drive reporter expression in the TVCs (Supplemental Fig. 4F). For the four predicted upstream CREs, 1–3kb intergenic 5′ regions containing the predicted clusters along with the endogenous promoter region and predicted ATG start codons were amplified and fused in frame to a lacZ reporter (Supplemental Fig. 4D,G,H and Fig. 6A). Two of these constructs, #11904 (−907:lacZ) and unc5 (−2248:lacZ), displayed no TVC reporter activity. However, the other two predicted 5′ CREs, #1839 (−2005:lacZ) and RGS3 (−1120:lacZ), displayed weak reporter expression in the TVC lineage along with strong expression in adjacent lineages. To further examine the presumed role of the candidate RGS3 and #1839 CREs in TVC expression, we tested minimal intergenic fragments beginning just prior to the predicted CRE domain (RGS3-274:lacZ, #1839-1124:lacZ) and found that both retained weak TVC lineage reporter activity (Fig. 6A, Supplemental Fig. 5D). We then attempted to test whether removal of the predicted CRE domain would abrogate TVC lineage reporter expression (RGS3-166:lacZ, #1839-849:lacZ, Fig. 6A, Supplemental Fig. 5D). Although removal of the CRE region did appear to reduce TVC expression, the low penetrance of TVC reporter expression driven by the CRE containing constructs (< 5% of transgenic embryos) made it difficult to rigorously assess the impact of CRE removal. We therefore attempted to boost the expression levels of these enhancer elements by fusing them to the 296bp Hand-like basal promoter (RGS3-1120-116:HL296:lacZ, #1839-2005-441:HL296:lacZ). Transgenic #1839-2005-441:HL296:lacZ embryos displayed no TVC reporter expression. However, transgenic RGS3-1120-116:HL296:lacZ embryos showed robust and consistent TVC reporter expression (Fig. 6C,D). Using this stronger base construct, we were able to specifically test the predicted contribution of the ATTA co-motif through deletion and targeted mutagenesis (Fig. 6C,E). We first found that the minimal intergenic fragment containing only the predicted CRE domain (RGS3-274-166:HL296:lacZ) retained strong TVC lineage reporter activity. This minimal fragment contains two ATTA motifs (Fig. 6B). A 5′ deletion removing one of these ATTA motifs (RGS3-256-116:HL296:lacZ) had no discernable impact on reporter activity. However, targeted mutagenesis of both ATTA motifs in the context of the minimal predicted element (-274-166 A1/A2-mut) nearly eliminated TVC reporter expression. Most strikingly, targeted mutagenesis of both ATTA motifs in the context of the full length ~1000bp RGS3 intergenic enhancer fragment also greatly reduced TVC reporter activity (-1120-116 A1/A2-mut; Fig. 6C,E). In summary, our functional analysis strongly supports a central role for the Ets/ATTA motif combination in the primary TVC cis-regulatory code.
Discussion
This study provides significant insights regarding initial deployment of the Ciona cardiogenic regulatory network. First, we identified a discrete set of primary genes upregulated by FGF:MapK:Ets1/2 shortly after heart progenitor (TVC) emergence. We have also begun to decipher the regulatory logic underlying primary TVC gene expression. Through sequence analysis of predicted primary TVC regulatory elements, we determined that Ets binding sites were consistently associated with the ATTA co-motif. We then confirmed the functional importance of the ATTA co-motif through mutational analysis in two characterized primary TVC regulatory elements. Using this Ets/ATTA combination motif, we identified additional primary TVC CREs. These results clarify the composition and topology of the Ciona heart GRN. In the following sections, we discuss the implications of Ciona heart GRN architecture in regards to chordate cardiogenesis and Map Kinase mediated gene regulatory networks.
Our results represent a valuable resource for deciphering the composition of the chordate cardiogenetic regulatory network. Although studies in Drosophila embryos and cultured mammalian cell lines (Bryantsev and Cripps, 2009; Musunuru et al., 2010; Reim and Frasch, 2009; Schlesinger et al., 2011) have provided powerful insights into vertebrate cardiogenesis, each of these approaches has inherent limitations. Insights into cell-line cardiogenic GRNs are ideally suited for optimization of in-vitro cardiomyocyte differentiation, but may not accurately represent the embryonic network. Studies of the Drosophila heart gene network have revealed a core regulatory “kernel” and many downstream effectors that display similar, potentially conserved roles in the vertebrate heart GRN (Davidson, 2006; Olson, 2006). However, there are indications of fundamental differences between initial heart specification programs in Drosophila and chordate embryos. In particular, Mesp mediated establishment of the nascent heart field appears to be chordate specific (Bondue and Blanpain, 2010). Studies in Ciona therefore fill an important niche in delineating chordate specific aspects of the cardiac GRN. We anticipate that the primary TVC gene-set revealed in this study includes numerous genes with undocumented or poorly characterized roles in vertebrate heart specification. For example, two of the 20 primary TVC transcription factors identified in this study are Irx family genes. Vertebrates Irx orthologs (Irx1,2,4, and 6) play essential but incompletely characterized roles in early vertebrate cardiogenesis (Bruneau et al., 2000; Cheng et al., 2011; Christoffels et al., 2000; Costantini et al., 2005; He et al., 2009). To pursue this line, we will characterize the function of these primary TVC genes in Ciona, focusing on morpholino knockdown and targeted manipulations of the 20 primary heart TFs.
This study also further establishes Ciona as a potent model for systems level analysis of chordate cardiogenesis. The complexity of vertebrate heart development has made it difficult to conduct comprehensive, systems level analysis of cardiogenic regulatory networks. Notably, the roles of specific vertebrate Ets family members and co-factors during FGF:MapK mediated cardiac cell specification remain poorly characterized (Lunn et al., 2007; Schachterle et al., 2012). Future work will focus on determining whether the Ets/ATTA co-motif revealed by this study reflects a conserved regulatory signature for early, FGF regulated heart genes in vertebrate embryos. To pursue this question, we are first attempting to identify the factor that binds to the ATTA co-motif. Our initial efforts are focused on homeodomain genes expressed in the founder cells during TVC specification. More broadly, we are interested in exploring how Ciona heart GRN topology is structured to regulate discrete, temporal cascades of target gene expression. To pursue this, we are attempting to sub-divide early TVC genes by the precise timing of initial expression and dissect the regulatory elements for groups of temporally co-regulated targets. We are also developing a high-throughput in-vivo method for temporal analysis of reporter gene expression. These future studies will help decipher how early signaling events potentiate cascades of gene expression leading to activation of the conserved heart regulatory kernel. Additionally, this research will help to illuminate how GRN topology coordinates progressive changes in progenitor cell behavior required for heart formation and other morphogenetic processes.
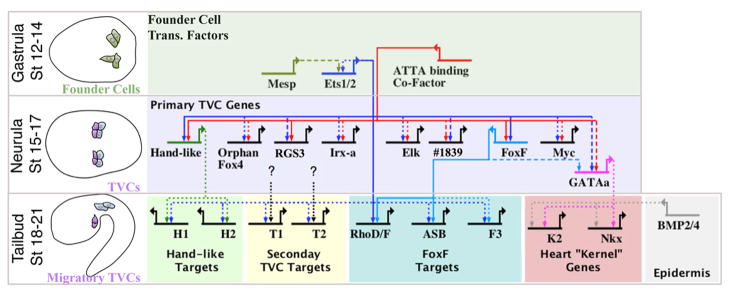
This study also has implications regarding GRN encoded MapK:Ets temporal specificity. Our results indicate that Ets1/2 co-regulates primary TVC genes in association with an “ATTA binding” co-factor. Previous research indicates that Ets1/2 continues to participate in TVC gene expression at later stages in partnership with one of these primary TVC genes, FoxF (Christiaen et al., 2008). Such coherent feed-forward circuits are a common element in developmental gene networks and are thought to generate a sign-sensitive delay (Alon, 2007), and provide robust temporal output despite noisy regulatory input (Mangan and Alon, 2003; Sandmann et al., 2007). Generally, feed-forward regulation is modeled as a single input circuit for early targets, followed by recruitment of a downstream gene to generate a dual-input circuit for later targets. During Ciona heart development, it appears that both early and late TVC gene batteries are regulated by dual inputs. Ets1/2 thereby mediates a temporal cascade of gene expression by swapping an early co-regulator for a late, feed forward co-regulator (Fig. 7).
Figure 7. Model of the TVC gene regulatory network.
BioTapestry (Longabaugh, 2011) was employed to construct a hypothetical TVC GRN. Schematics to the left indicate relevant stage. Top row displays initial network in the B7.5 founder cells. Middle row displays hypothesized Ets1/2 regulation of representative primary TVC genes as detailed in the current study. Bottom row displays presumed Ets1/2 feed-forward regulation of later, secondary TVC genes (Christiaen et al., 2008) and presumed conserved regulation of “kernel” genes (Davidson, 2006) by GATAa and Smad (downstream of epidermal BMP signaling, Christiaen et al. 2009). GATAa is positioned to reflect the apparent delay in the expression of this gene relative to other primary TVC genes such as Hand-like and FoxF (Ragkousi et al., 2011) and includes potential regulation through conserved FoxF binding motifs in the intronic TVC enhancer element (see Supplemental Fig. 4). We have also included hypothetical feed-forward regulation of sub-sets of secondary TVC genes by Ets1/2 and other primary TVC transcription factors (black dotted lines). We are interested in testing whether this hypothesized differential feed-forward architecture may serve to regulate discrete morphogenetic modules involved in TVC behavior. Line format represents the varying levels of evidence for hypothesized cis-regulatory connections: solid lines = validation through mutation of presumed binding sites; dashed lines = presence of binding motif in experimentally tested enhancer; dotted lines = no experimental evidence. H1, H2, T1, T2, F3 and K2 represent hypothetical sets of differential Ets1/2 target genes as labeled.
The structure of this network may reflect a general mechanism employed to layer the temporal expression of Map Kinase target genes during developmental and physiological processes. Testing this hypothesis will require further delineation of Ets1/2:FoxF feed forward regulation in a broader set of TVC migratory genes. In particular, it will be important to test the alternative possibility that primary and migratory TVC gene expression are regulated by temporally segregated Ets family factors (Ets1/2 vs. ELK respectively). We are also working to develop chromatin immunoprecipitation assays to examine the in-vivo, lineage specific binding of Ets1/2 and it co-factors to presumed regulatory regions at different embryonic stages. However, this approach will be difficult due to low trunk ventral cell numbers in developing embryos.
More generally, this work highlights the potential to elucidate MapK:Ets regulatory architecture through research on tunicate development. Tunicate embryos repeatedly deploy the FGF:MapK:Ets1/2 pathway to specify a variety of progenitor lineages (Bertrand et al., 2003; Lemaire, 2009; Miya and Nishida, 2003; Squarzoni et al., 2011). A combination of efficient experimental cis-regulatory analysis and newly developed in-silico methods are beginning to reveal how GRN topology delineates these varied specification events. In the notochord and neural lineages, extensive cis and trans-regulatory analysis has identified and confirmed discrete Ets1/2 co-transcription factors (Zic and FoxA in notochord progenitors (Kumano et al., 2006; Matsumoto et al., 2007) vs. GATAa in neural progenitors (Bertrand et al., 2003; Khoueiry et al. 2010). Starting with a characterized Ets + co-factor motif (GGAW+GATA) combination in the Otx neural lineage CRE, Khoueiry et al., (2010) generated accurate in-silico predictions of additional neural regulatory elements. Using a converse approach, Haeussler et al., 2010 employed lineage specific expression data to reveal a duplicated motif that encodes neuroectodermal gene regulation. We have employed a novel approach built on the methodologies pioneered in these and other recent studies (Bailey et al., 2006; Brown et al., 2007; Das and Dai, 2007; Erives, 2009; Hallikas et al., 2006; Jeziorska et al., 2009; Johnson et al., 2005; Kusakabe et al., 2004; Li et al., 2009; Rister and Desplan, 2010; Van Loo and Marynen, 2009). Our parameter-free computational detection method searches explicitly for motif pairs, starting with a known motif and searching for the unknown partner motif. Thus, our algorithm takes into account the combinatorial nature of regulation. The in-silico/in-vivo methodology refined in this study has the potential to generate a comprehensive GRN diagram of Ets mediated cell specification. Due to the pleiotropic use of MapK:Ets in tunicate embryogenesis, this diagram will cover a significant portion of primary patterning events. Furthermore, this comprehensive GRN model will provide profound insights into the regulatory topology underlying MapK/Ets specificity during a wide range of developmental and homeostatic processes.
Supplementary Material
Table 1. List of Ets binding site (GGAW) + co-motif combinations. These 44 co-motifs were conserved in both the FoxF and Hand-like regulatory elements and tested for enrichment near the 144 genes whose matching probe sets were robustly and significantly down-regulated in the Ets-WRPW and FGFRdn 8H array samples. Shown are the allowed window size, number of total matching genes (background of all Annotated Ciona genes), number of hits within the foreground gene set (true positive genes, TP), resulting sensitivity and binomial scores.
Table 2. Same format as Table 1, but showing results when all possible tetramer and pentamer co-motifs are included. Only the top 100 scoring co-motifs are displayed.
Table 3. Same format as Table 2, but co-motifs are ranked against a list of 144 genes with matching probe sets that were the most severely down-regulated genes in the FGFRdn 10H array samples (Christiaen et al., 2008) but not down-regulated in the FGFRdn 8H array samples.
Table 4. Primers used to generate reporter constructs. Reverse primers were used to generate full length fragments usually containing the native translation start site (bold) fused in frame to lacZ. The GATAa -589f primers was used to generate a native basal promoter fragment fused to the annotated intronic enhancer fragment (Fig. S4) using a shared BamH1 site.
Highlights.
Comprehensive I.D. of primary FGF and Ets1/2 target genes in the heart lineage.
Informatics used to predict a primary heart gene cis regulatory code.
Verification of regulatory code through mutation of predicted binding site motifs.
Successful prediction of new primary heart enhancers using motif combination.
Insights into the topology of Map Kinase/Ets regulated gene networks.
Acknowledgments
We would like to that thank Paul Kreig, Carol Gregorio and Parkin Antin for their advice and support. The work was supported by grants to B.D. from the AHA (0730345N) and NIH (R01HL091027) along with generous support from the Sarver Heart Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–61. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- Auger H, Lamy C, Haeussler M, Khoueiry P, Lemaire P, Joly JS. Similar regulatory logic in Ciona intestinalis for two Wnt pathway modulators, ROR and SFRP-1/5. Dev Biol. 2009;329:364–73. doi: 10.1016/j.ydbio.2009.02.018. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–73. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beh J, Shi W, Levine M, Davidson B, Christiaen L. FoxF is essential for FGF-induced migration of heart progenitor cells in the ascidian Ciona intestinalis. Development. 2007;134:3297–305. doi: 10.1242/dev.010140. [DOI] [PubMed] [Google Scholar]
- Bertrand V, Hudson C, Caillol D, Popovici C, Lemaire P. Neural tissue in ascidian embryos is induced by FGF9/16/20, acting via a combination of maternal GATA and Ets transcription factors. Cell. 2003;115:615–27. doi: 10.1016/s0092-8674(03)00928-0. [DOI] [PubMed] [Google Scholar]
- Bolouri H, Davidson EH. The gene regulatory network basis of the “community effect,” and analysis of a sea urchin embryo example. Dev Biol. 2009;340:170–8. doi: 10.1016/j.ydbio.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondue A, Blanpain C. Mesp1: a key regulator of cardiovascular lineage commitment. Circ Res. 2010;107:1414–27. doi: 10.1161/CIRCRESAHA.110.227058. [DOI] [PubMed] [Google Scholar]
- Bondue A, Lapouge G, Paulissen C, Semeraro C, Iacovino M, Kyba M, Blanpain C. Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell. 2008;3:69–84. doi: 10.1016/j.stem.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Brown CD, Johnson DS, Sidow A. Functional architecture and evolution of transcriptional elements that drive gene coexpression. Science. 2007;317:1557–60. doi: 10.1126/science.1145893. [DOI] [PubMed] [Google Scholar]
- Bruneau BG, Bao ZZ, Tanaka M, Schott JJ, Izumo S, Cepko CL, Seidman JG, Seidman CE. Cardiac expression of the ventricle-specific homeobox gene Irx4 is modulated by Nkx2-5 and dHand. Dev Biol. 2000;217:266–77. doi: 10.1006/dbio.1999.9548. [DOI] [PubMed] [Google Scholar]
- Bryantsev AL, Cripps RM. Cardiac gene regulatory networks in Drosophila. Biochim Biophys Acta. 2009;1789:343–53. doi: 10.1016/j.bbagrm.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Wang J, Su D, Pan H, Huang G, Li X, Li Z, Shen A, Xie X, Wang B, Ma X. Two novel mutations of the IRX4 gene in patients with congenital heart disease. Hum Genet. 2011;130:657–62. doi: 10.1007/s00439-011-0996-7. [DOI] [PubMed] [Google Scholar]
- Christiaen L, Davidson B, Kawashima T, Powell W, Nolla H, Vranizan K, Levine M. The transcription/migration interface in heart precursors of Ciona intestinalis. Science. 2008;320:1349–52. doi: 10.1126/science.1158170. [DOI] [PubMed] [Google Scholar]
- Christiaen L, Stolfi A, Levine M. BMP signaling coordinates gene expression and cell migration during precardiac mesoderm development. Dev Biol. 2009;340:179–87. doi: 10.1016/j.ydbio.2009.11.006. [DOI] [PubMed] [Google Scholar]
- Christoffels VM, Keijser AG, Houweling AC, Clout DE, Moorman AF. Patterning the embryonic heart: identification of five mouse Iroquois homeobox genes in the developing heart. Dev Biol. 2000;224:263–74. doi: 10.1006/dbio.2000.9801. [DOI] [PubMed] [Google Scholar]
- Cooley J, Whitaker S, Sweeney S, Fraser S, Davidson B. Cytoskeletal polarity mediates localized induction of the heart progenitor lineage. Nat Cell Biol. 2011;13:952–7. doi: 10.1038/ncb2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbo JC, Levine M, Zeller RW. Characterization of a notochord specific-enhancer from the Brachyury promoter region of the ascidian, Ciona intestinalis. Development. 1997;124:589–602. doi: 10.1242/dev.124.3.589. [DOI] [PubMed] [Google Scholar]
- Costantini DL, Arruda EP, Agarwal P, Kim KH, Zhu Y, Zhu W, Lebel M, Cheng CW, Park CY, Pierce SA, Guerchicoff A, Pollevick GD, Chan TY, Kabir MG, Cheng SH, Husain M, Antzelevitch C, Srivastava D, Gross GJ, Hui CC, Backx PH, Bruneau BG. The homeodomain transcription factor Irx5 establishes the mouse cardiac ventricular repolarization gradient. Cell. 2005;123:347–58. doi: 10.1016/j.cell.2005.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couronne O, Poliakov A, Bray N, Ishkhanov T, Ryaboy D, Rubin E, Pachter L, Dubchak I. Strategies and tools for whole-genome alignments. Genome Res. 2003;13:73–80. doi: 10.1101/gr.762503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das MK, Dai HK. A survey of DNA motif finding algorithms. BMC Bioinformatics. 2007;8(Suppl 7):S21. doi: 10.1186/1471-2105-8-S7-S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson B. Ciona intestinalis as a model for cardiac development. Semin Cell Dev Biol. 2007;18:16–26. doi: 10.1016/j.semcdb.2006.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson B, Levine M. Evolutionary origins of the vertebrate heart: Specification of the cardiac lineage in Ciona intestinalis. Proc Natl Acad Sci U S A. 2003;100:11469–73. doi: 10.1073/pnas.1634991100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson B, Shi W, Beh J, Christiaen L, Levine M. FGF signaling delineates the cardiac progenitor field in the simple chordate, Ciona intestinalis. Genes Dev. 2006;20:2728–38. doi: 10.1101/gad.1467706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson B, Shi W, Levine M. Uncoupling heart cell specification and migration in the simple chordate Ciona intestinalis. Development. 2005;132:4811–8. doi: 10.1242/dev.02051. [DOI] [PubMed] [Google Scholar]
- Davidson EH. Gene Regulatory Networks in Development and Evolution. Academic Press/Elsevier; San Diego: 2006. The Regulatory Genome. [Google Scholar]
- Davidson EH. Emerging properties of animal gene regulatory networks. Nature. 2010;468:911–20. doi: 10.1038/nature09645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH. Evolutionary bioscience as regulatory systems biology. Dev Biol. 2011;357:35–40. doi: 10.1016/j.ydbio.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH, Levine MS. Properties of developmental gene regulatory networks. Proc Natl Acad Sci U S A. 2008;105:20063–6. doi: 10.1073/pnas.0806007105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De Tomaso A, Davidson B, Di Gregorio A, Gelpke M, Goodstein DM, Harafuji N, Hastings KE, Ho I, Hotta K, Huang W, Kawashima T, Lemaire P, Martinez D, Meinertzhagen IA, Necula S, Nonaka M, Putnam N, Rash S, Saiga H, Satake M, Terry A, Yamada L, Wang HG, Awazu S, Azumi K, Boore J, Branno M, Chin-Bow S, DeSantis R, Doyle S, Francino P, Keys DN, Haga S, Hayashi H, Hino K, Imai KS, Inaba K, Kano S, Kobayashi K, Kobayashi M, Lee BI, Makabe KW, Manohar C, Matassi G, Medina M, Mochizuki Y, Mount S, Morishita T, Miura S, Nakayama A, Nishizaka S, Nomoto H, Ohta F, Oishi K, Rigoutsos I, Sano M, Sasaki A, Sasakura Y, Shoguchi E, Shin-i T, Spagnuolo A, Stainier D, Suzuki MM, Tassy O, Takatori N, Tokuoka M, Yagi K, Yoshizaki F, Wada S, Zhang C, Hyatt PD, Larimer F, Detter C, Doggett N, Glavina T, Hawkins T, Richardson P, Lucas S, Kohara Y, Levine M, Satoh N, Rokhsar DS. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–67. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- Di Gregorio A, Levine M. Analyzing gene regulation in ascidian embryos: new tools for new perspectives. Differentiation. 2002;70:132–9. doi: 10.1046/j.1432-0436.2002.700402.x. [DOI] [PubMed] [Google Scholar]
- Dunwoodie SL. Combinatorial signaling in the heart orchestrates cardiac induction, lineage specification and chamber formation. Semin Cell Dev Biol. 2007;18:54–66. doi: 10.1016/j.semcdb.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Erives A. Non-homologous structured CRMs from the Ciona genome. J Comput Biol. 2009;16:369–77. doi: 10.1089/cmb.2008.20TT. [DOI] [PubMed] [Google Scholar]
- Evans SM, Yelon D, Conlon FL, Kirby ML. Myocardial lineage development. Circ Res. 2010;107:1428–44. doi: 10.1161/CIRCRESAHA.110.227405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulds CE, Nelson ML, Blaszczak AG, Graves BJ. Ras/mitogen-activated protein kinase signaling activates Ets-1 and Ets-2 by CBP/p300 recruitment. Mol Cell Biol. 2004;24:10954–64. doi: 10.1128/MCB.24.24.10954-10964.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeussler M, Jaszczyszyn Y, Christiaen L, Joly JS. A cis-regulatory signature for chordate anterior neuroectodermal genes. PLoS Genet. 2010;6:e1000912. doi: 10.1371/journal.pgen.1000912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallikas O, Palin K, Sinjushina N, Rautiainen R, Partanen J, Ukkonen E, Taipale J. Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell. 2006;124:47–59. doi: 10.1016/j.cell.2005.10.042. [DOI] [PubMed] [Google Scholar]
- He W, Jia Y, Takimoto K. Interaction between transcription factors Iroquois proteins 4 and 5 controls cardiac potassium channel Kv4.2 gene transcription. Cardiovasc Res. 2009;81:64–71. doi: 10.1093/cvr/cvn259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta K, Mitsuhara K, Takahashi H, Inaba K, Oka K, Gojobori T, Ikeo K. A web-based interactive developmental table for the ascidian Ciona intestinalis, including 3D real-image embryo reconstructions: I. From fertilized egg to hatching larva. Dev Dyn. 2007;236:1790–805. doi: 10.1002/dvdy.21188. [DOI] [PubMed] [Google Scholar]
- Imai KS, Levine M, Satoh N, Satou Y. Regulatory blueprint for a chordate embryo. Science. 2006;312:1183–7. doi: 10.1126/science.1123404. [DOI] [PubMed] [Google Scholar]
- Imai KS, Satoh N, Satou Y. A Twist-like bHLH gene is a downstream factor of an endogenous FGF and determines mesenchymal fate in the ascidian embryos. Development. 2003;130:4461–72. doi: 10.1242/dev.00652. [DOI] [PubMed] [Google Scholar]
- Imai KS, Stolfi A, Levine M, Satou Y. Gene regulatory networks underlying the compartmentalization of the Ciona central nervous system. Development. 2009;136:285–93. doi: 10.1242/dev.026419. [DOI] [PubMed] [Google Scholar]
- Jeziorska DM, Jordan KW, Vance KW. A systems biology approach to understanding cis-regulatory module function. Semin Cell Dev Biol. 2009;20:856–62. doi: 10.1016/j.semcdb.2009.07.007. [DOI] [PubMed] [Google Scholar]
- Johnson DS, Davidson B, Brown CD, Smith WC, Sidow A. Noncoding regulatory sequences of Ciona exhibit strong correspondence between evolutionary constraint and functional importance. Genome Res. 2004;14:2448–56. doi: 10.1101/gr.2964504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Zhou Q, Yagi K, Satoh N, Wong W, Sidow A. De novo discovery of a tissue-specific gene regulatory module in a chordate. Genome Res. 2005;15:1315–24. doi: 10.1101/gr.4062605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoueiry P, Rothbacher U, Ohtsuka Y, Daian F, Frangulian E, Roure A, Dubchak I, Lemaire P. A cis-regulatory signature in ascidians and flies, independent of transcription factor binding sites. Curr Biol. 2010;20:792–802. doi: 10.1016/j.cub.2010.03.063. [DOI] [PubMed] [Google Scholar]
- Krauss RS. Regulation of promyogenic signal transduction by cell-cell contact and adhesion. Exp Cell Res. 2010;316:3042–9. doi: 10.1016/j.yexcr.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumano G, Yamaguchi S, Nishida H. Overlapping expression of FoxA and Zic confers responsiveness to FGF signaling to specify notochord in ascidian embryos. Dev Biol. 2006;300:770–84. doi: 10.1016/j.ydbio.2006.07.033. [DOI] [PubMed] [Google Scholar]
- Kusakabe T, Yoshida R, Ikeda Y, Tsuda M. Computational discovery of DNA motifs associated with cell type-specific gene expression in Ciona. Dev Biol. 2004;276:563–80. doi: 10.1016/j.ydbio.2004.09.037. [DOI] [PubMed] [Google Scholar]
- Lemaire P. Unfolding a chordate developmental program, one cell at a time: invariant cell lineages, short-range inductions and evolutionary plasticity in ascidians. Dev Biol. 2009;332:48–60. doi: 10.1016/j.ydbio.2009.05.540. [DOI] [PubMed] [Google Scholar]
- Lemaire P. Evolutionary crossroads in developmental biology: the tunicates. Development. 2011;138:2143–52. doi: 10.1242/dev.048975. [DOI] [PubMed] [Google Scholar]
- Li Q, Ritter D, Yang N, Dong Z, Li H, Chuang JH, Guo S. A systematic approach to identify functional motifs within vertebrate developmental enhancers. Dev Biol. 2009;337:484–95. doi: 10.1016/j.ydbio.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longabaugh WJ. BioTapestry: a tool to visualize the dynamic properties of gene regulatory networks. Methods Mol Biol. 2011;786:359–94. doi: 10.1007/978-1-61779-292-2_21. [DOI] [PubMed] [Google Scholar]
- Lunn JS, Fishwick KJ, Halley PA, Storey KG. A spatial and temporal map of FGF/Erk1/2 activity and response repertoires in the early chick embryo. Dev Biol. 2007;302:536–52. doi: 10.1016/j.ydbio.2006.10.014. [DOI] [PubMed] [Google Scholar]
- Maduro MF. Structure and evolution of the C. elegans embryonic endomesoderm network. Biochim Biophys Acta. 2009;1789:250–60. doi: 10.1016/j.bbagrm.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci U S A. 2003;100:11980–5. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markstein M, Levine M. Decoding cis-regulatory DNAs in the Drosophila genome. Curr Opin Genet Dev. 2002;12:601–6. doi: 10.1016/s0959-437x(02)00345-3. [DOI] [PubMed] [Google Scholar]
- Matsumoto J, Kumano G, Nishida H. Direct activation by Ets and Zic is required for initial expression of the Brachyury gene in the ascidian notochord. Dev Biol. 2007;306:870–82. doi: 10.1016/j.ydbio.2007.03.034. [DOI] [PubMed] [Google Scholar]
- Miya T, Nishida H. An Ets transcription factor, HrEts, is target of FGF signaling and involved in induction of notochord, mesenchyme, and brain in ascidian embryos. Dev Biol. 2003;261:25–38. doi: 10.1016/s0012-1606(03)00246-x. [DOI] [PubMed] [Google Scholar]
- Musunuru K, Domian IJ, Chien KR. Stem cell models of cardiac development and disease. Annu Rev Cell Dev Biol. 2010;26:667–87. doi: 10.1146/annurev-cellbio-100109-103948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Z, Li A, Zhang SX, Schwartz RJ. Serum response factor micromanaging cardiogenesis. Curr Opin Cell Biol. 2007;19:618–27. doi: 10.1016/j.ceb.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–7. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter IS, Davidson EH. Modularity and design principles in the sea urchin embryo gene regulatory network. FEBS Lett. 2009;583:3948–58. doi: 10.1016/j.febslet.2009.11.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragkousi K, Beh J, Sweeney S, Starobinska E, Davidson B. A single GATA factor plays discrete, lineage specific roles in ascidian heart development. Dev Biol. 2011;352:154–63. doi: 10.1016/j.ydbio.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reim I, Frasch M. Genetic and genomic dissection of cardiogenesis in the Drosophila model. Pediatr Cardiol. 2009;31:325–34. doi: 10.1007/s00246-009-9612-1. [DOI] [PubMed] [Google Scholar]
- Rister J, Desplan C. Deciphering the genome’s regulatory code: the many languages of DNA. Bioessays. 2010;32:381–4. doi: 10.1002/bies.200900197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saga Y, Kitajima S, Miyagawa-Tomita S. Mesp1 expression is the earliest sign of cardiovascular development. Trends Cardiovasc Med. 2000;10:345–52. doi: 10.1016/s1050-1738(01)00069-x. [DOI] [PubMed] [Google Scholar]
- Sandmann T, Girardot C, Brehme M, Tongprasit W, Stolc V, Furlong EE. A core transcriptional network for early mesoderm development in Drosophila melanogaster. Genes Dev. 2007;21:436–49. doi: 10.1101/gad.1509007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh N, Satou Y, Davidson B, Levine M. Ciona intestinalis: an emerging model for whole-genome analyses. Trends Genet. 2003;19:376–81. doi: 10.1016/S0168-9525(03)00144-6. [DOI] [PubMed] [Google Scholar]
- Satou Y, Imai KS, Satoh N. The ascidian Mesp gene specifies heart precursor cells. Development. 2004;131:2533–41. doi: 10.1242/dev.01145. [DOI] [PubMed] [Google Scholar]
- Satou Y, Yamada L, Mochizuki Y, Takatori N, Kawashima T, Sasaki A, Hamaguchi M, Awazu S, Yagi K, Sasakura Y, Nakayama A, Ishikawa H, Inaba K, Satoh N. A cDNA resource from the basal chordate Ciona intestinalis. Genesis. 2002;33:153–4. doi: 10.1002/gene.10119. [DOI] [PubMed] [Google Scholar]
- Schachterle W, Rojas A, Xu SM, Black BL. ETS-dependent regulation of a distal Gata4 cardiac enhancer. Dev Biol. 2012;361:439–49. doi: 10.1016/j.ydbio.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger J, Schueler M, Grunert M, Fischer JJ, Zhang Q, Krueger T, Lange M, Tonjes M, Dunkel I, Sperling SR. The cardiac transcription network modulated by Gata4, Mef2a, Nkx2.5, Srf, histone modifications, and microRNAs. PLoS Genet. 2011;7(2):e1001313. doi: 10.1371/journal.pgen.1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–8. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol. 2001;2:827–37. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- Shi W, Levine M, Davidson B. Unraveling genomic regulatory networks in the simple chordate, Ciona intestinalis. Genome Res. 2005;15:1668–74. doi: 10.1101/gr.3768905. [DOI] [PubMed] [Google Scholar]
- Singh MK, Li Y, Li S, Cobb RM, Zhou D, Lu MM, Epstein JA, Morrisey EE, Gruber PJ. Gata4 and Gata5 cooperatively regulate cardiac myocyte proliferation in mice. J Biol Chem. 2009;285:1765–72. doi: 10.1074/jbc.M109.038539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Limma: linear models for microarray data. In: Gentleman VCR, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. Springer; New York: 2005. pp. 397–420. [Google Scholar]
- Sperling SR. Systems biology approaches to heart development and congenital heart disease. Cardiovasc Res. 2011;91:269–78. doi: 10.1093/cvr/cvr126. [DOI] [PubMed] [Google Scholar]
- Squarzoni P, Parveen F, Zanetti L, Ristoratore F, Spagnuolo A. FGF/MAPK/Ets signaling renders pigment cell precursors competent to respond to Wnt signal by directly controlling Ci-Tcf transcription. Development. 2011;138:1421–32. doi: 10.1242/dev.057323. [DOI] [PubMed] [Google Scholar]
- Srivastava D. Making or breaking the heart: from lineage determination to morphogenesis. Cell. 2006;126:1037–48. doi: 10.1016/j.cell.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Stolfi A, Gainous TB, Young JJ, Mori A, Levine M, Christiaen L. Early chordate origins of the vertebrate second heart field. Science. 2010;329:565–8. doi: 10.1126/science.1190181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Targoff KL, Schell T, Yelon D. Nkx genes regulate heart tube extension and exert differential effects on ventricular and atrial cell number. Dev Biol. 2008;322:314–21. doi: 10.1016/j.ydbio.2008.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsachaki M, Sprecher SG. Genetic and developmental mechanisms underlying the formation of the Drosophila compound eye. Dev Dyn. 2012;241:40–56. doi: 10.1002/dvdy.22738. [DOI] [PubMed] [Google Scholar]
- Van Loo P, Marynen P. Computational methods for the detection of cis-regulatory modules. Brief Bioinform. 2009;10:509–24. doi: 10.1093/bib/bbp025. [DOI] [PubMed] [Google Scholar]
- Verger A, Duterque-Coquillaud M. When Ets transcription factors meet their partners. Bioessays. 2002;24:362–70. doi: 10.1002/bies.10068. [DOI] [PubMed] [Google Scholar]
- Vincent SD, Buckingham ME. How to make a heart: the origin and regulation of cardiac progenitor cells. Curr Top Dev Biol. 2010;90:1–41. doi: 10.1016/S0070-2153(10)90001-X. [DOI] [PubMed] [Google Scholar]
- Wolf M, Basson CT. The molecular genetics of congenital heart disease: a review of recent developments. Curr Opin Cardiol. 2010;25:192–197. doi: 10.1097/HCO.0b013e328337b4ce. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Baldini A. Genetic pathways to mammalian heart development: Recent progress from manipulation of the mouse genome. Semin Cell Dev Biol. 2007;18:77–83. doi: 10.1016/j.semcdb.2006.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 1. List of Ets binding site (GGAW) + co-motif combinations. These 44 co-motifs were conserved in both the FoxF and Hand-like regulatory elements and tested for enrichment near the 144 genes whose matching probe sets were robustly and significantly down-regulated in the Ets-WRPW and FGFRdn 8H array samples. Shown are the allowed window size, number of total matching genes (background of all Annotated Ciona genes), number of hits within the foreground gene set (true positive genes, TP), resulting sensitivity and binomial scores.
Table 2. Same format as Table 1, but showing results when all possible tetramer and pentamer co-motifs are included. Only the top 100 scoring co-motifs are displayed.
Table 3. Same format as Table 2, but co-motifs are ranked against a list of 144 genes with matching probe sets that were the most severely down-regulated genes in the FGFRdn 10H array samples (Christiaen et al., 2008) but not down-regulated in the FGFRdn 8H array samples.
Table 4. Primers used to generate reporter constructs. Reverse primers were used to generate full length fragments usually containing the native translation start site (bold) fused in frame to lacZ. The GATAa -589f primers was used to generate a native basal promoter fragment fused to the annotated intronic enhancer fragment (Fig. S4) using a shared BamH1 site.