Abstract
Background & Aims
Prostaglandin-endoperoxide synthase (Ptgs)2 is an enzyme involved in prostaglandin production during the response to mucosal damage. Its expression is regulated, in part, by mRNA-binding proteins that control the stability of Ptgs2 mRNA. We used a precise system of colonic injury and repair to identify Ptgs2 mRNA-binding proteins.
Methods
We used endoscopy-guided mucosal excision to create focal injury sites in colons of mice. Wound beds from wild-type, Ptgs2−/−, Ptgs2+/−, and Myd88−/− mice were analyzed at 2-day intervals following injury for aspects of repair and Ptgs2 expression. We used cultured colonic mesenchymal stem cells (cMSCs) that express Ptgs2 to identify and analyze molecules that regulate Ptgs2 expression.
Results
Ptgs2−/− mice had defects in wound repair, validating the biopsy technique as a system to study the regulation of Ptgs2. Ptgs2+/− mice had similar defects in wound healing, so full induction of Ptgs2 is required for wound repair. In wild-type mice, levels of Ptgs2 mRNA increased significantly in the wound bed 2 and 4 days after injury; the highest levels of Ptgs2 were observed in cMSCs. In a functional small hairpin RNA knockdown screen, we identified Igf2bp1, a VICKZ mRNA-binding protein, as a regulator of Ptgs2 expression in cMSCs. Igf2bp1 also interacted physically with Ptgs2 mRNA. Igf2bp1 expression was induced exclusively in wound-bed cMSCs, and full induction of Ptgs2 and Igf2bp1 during repair required Myd88.
Conclusions
We identified Igf2bp1 as a regulator of Ptgs2 mRNA in mice. Igf2bp1 is required for full induction of Ptgs2 mRNA in cMSCs.
Keywords: Cox-2, Imp-1, ZBP1, colorectal cancer, inflammatory bowel disease
Introduction
Ptgs2 (Prostaglandin-endoperoxide synthase 2) is an important enzyme for localized prostaglandin production during the response to mucosal damage and the pathogenesis of colorectal cancer.1 Ptgs2 and Ptgs1 are isoforms that are the two rate-limiting enzymes in the synthesis of all prostaglandins, which affect downstream responses such as angiogenesis, proliferation, and apoptosis.1 Ptgs1 is widely expressed and constitutive, whereas Ptgs2 is focally inducible with limited constitutive expression (primarily colon and testes).2 During inflammation, damage signals and cytokines signal through Myd88 to activate NF-κB and induce Ptgs2 transcription.3 Specialized mRNA binding/stabilizing proteins bind the AU-rich elements (ARE) in the 3'-UTR of Ptgs2 mRNA and prevent its degradation.4 Additional, unknown mRNA-binding proteins likely regulate Ptgs2 expression, since mutations in the 3'-UTR outside of the ARE regions also affect Ptgs2 stability.5
To study the regulation and effects of Ptgs2 in vivo, we and others have used various mouse models of intestinal injury including dextran sodium sulfate (DSS),6–9 2,4,6-trinitrobenzene sulfonic acid,10 and irradiation.11 Here, Ptgs2 affects epithelial proliferation and resolution of inflammation. Despite the requirement for Ptgs2 during repair, understanding the mechanisms that control Ptgs2 expression has been challenging due to the variable timing and location of injuries in these models. To discover novel Ptgs2 regulatory proteins expressed after mucosal damage, we created small, focal injury sites in the mouse colon using endoscopy-guided forceps.12–14 The precise timing and location of injuries in this system allowed us to investigate the cellular source, target, and timing of specific genes that regulate Ptgs2 expression.
We previously showed that repair following biopsy injury proceeds reproducibly in defined stages.12 In the first phase, neutrophils are recruited to the wound and an immature protective epithelial barrier (wound associated epithelium; WAE) forms over the nascent wound bed. This WAE layer consists of a single layer of post-mitotic cells that emerge from crypts that are adjacent to the wound and migrate towards that center of the wound bed.12 The second phase of healing involves increased epithelial proliferation in the adjacent crypts as well as expansion of the wound bed mesenchyme by the influx and/or proliferation of macrophages and additional stromal cells.12 In the final stage, the wound bed is replaced by new crypts.
In this study, we were able to utilize this precise injury system to identify a novel Ptgs2 mRNA-binding protein, Insulin-like Growth Factor 2 Binding Protein 1 (Igf2bp1). We found that maximal Ptgs2 expression was required for proper mucosal healing, and that the highest levels of Ptgs2 expression were localized to colonic mesenchymal stem cells (cMSCs). Using an shRNA knockdown screen in cultured cMSCs, we demonstrated that Igf2bp1, a VICKZ mRNA binding protein, was necessary for maximal expression of Ptgs2 mRNA. Igf2bp1 interacted with Ptgs2 mRNA and expression of both genes was induced in wound bed cMSCs in a Myd88-dependent manner. These data demonstrate that Igf2bp1 was necessary for the maximal Ptgs2 expression in cMSCs that was required for proper colonic injury repair.
Materials and Methods
Mice
Animal experiments were performed in accordance with approved protocols from the Washington University School of Medicine Animal Studies Committee. Ptgs2+/− mice15 were bred to generate littermate WT, Ptgs2+/− and Ptgs2−/− progeny for experiments. Myd88+/− mice16 from Jackson Lab were bred to generate Myd88−/− and Myd88+/− littermate controls. All mice were on a C57Bl/6 background.
Colonic biopsy
We used a high-resolution miniaturized colonoscope system to visualize the lumen of the colon and discretely injure the mucosal layer in 10–16 week old anesthetized mice. After inflating the colon with PBS, we inserted 3 French flexible biopsy forceps into the sheath adjacent to the camera. We removed 3–5 full-thickness areas of the entire mucosa and submucosa that were distributed along the dorsal side of the colon. All genetically modified mice were injured with the same technique. For this study, we evaluated wounds that averaged ~1 mm2, which is equivalent to removal of ~250–300 crypts.
Colonic tissue preparation
Wounded mice were sacrificed 2–6 days post-injury and each wound was individually frozen in OCT to make frozen sections. See Supplementary Materials and Methods for detailed descriptions.
In situ hybridization
A cDNA clone for Ptgs2 (clone ID: 30059181) was purchased from Thermo Fisher Scientific to create digoxigenin-labeled antisense RNA probes. See Supplementary Materials and Methods for detailed descriptions.
Immunofluorescence
Staining was performed as previously published17. See Supplementary Materials and Methods for detailed descriptions.
RNA isolation and quantitative RT-PCR
For wound RNA isolation, the wound bed mucosa or adjacent uninjured mucosa was dissected with forceps under the guidance of a whole mount microscope, leaving the underlying muscularis propria intact. RNA was isolated using a NucleoSpin RNA II kit (Machery-Nagel, Duren, Germany). cDNAs were synthesized using Superscript III reverse transcriptase (Invitrogen). Quantitative RT-PCR reactions using SYBR-green master mix (Clontech) were analyzed on an Eppendorf realplex Mastercycler. RNA from wounds and uninjured mucosa were normalized to Gapdh. See Supplementary Materials and Methods for primer sequences.
Immunoprecipitation and RNA isolation in NIH3T3 cells
The open reading frame of Igf2bp1 was cloned into p3XFLAG-CMV-14 (Sigma-Aldrich). See Supplementary Materials and Methods for detailed descriptions.
shRNA Transfection and immunoblotting
HEK293T cells were transfected with Mission shRNA constructs specific for Igf2bp1 or non-targeting controls. Lentivirus obtained from transfected cells was used to infect cMSCs for knockdown of specific genes. See Supplementary Materials and Methods for detailed descriptions.
Cell isolation and culture conditions
Colonic mesenchymal stem cell isolation and culture was performed as previously reported.17 See Supplementary Materials and Methods.
Statistics
All statistical analyses were performed using GraphPad Prism 3.0 (GraphPad Software). Significant differences were evaluated by Student's t test, Analysis of Variance (ANOVA) followed by Tukey's Multiple Comparison test, or Fisher's exact test, as noted.
Results
Two functional Ptgs2 alleles are required for colonic mucosal wound repair
To determine if Ptgs2 expression was required for colonic mucosal repair of biopsy injuries, we assessed the gross and microscopic features of wound beds from Ptgs2−/− and control WT littermate mice at two-day intervals post-injury. Starting at day 6 post-injury (second stage of repair), Ptgs2−/− mice displayed severe defects that included loss of muscularis propria and an incomplete WAE barrier (Figure 1A-E). Specifically, the muscularis propria showed complete loss of α-smooth muscle actin (α-SMA) underlying wound beds of Ptgs2−/− but not WT mice (Figure 1A-D). The damaged muscularis propria was replaced by granulation tissue comprised of endothelial cells and myeloid cells that extended through the serosa (Supplemental Figure 1 A–F). This phenotype resembled the histology of perforated human colonic ulcers that can occur after prolonged NSAID use.18 Importantly, the α-SMA staining pattern was intact in the muscularis propria at day 2 post-injury in Ptgs2−/− mice, indicating that this phenotype was a delayed biological response to injury and was not due to a more fragile mucosa in the Ptgs2−/− mice (Supplemental Figure 1G-I).
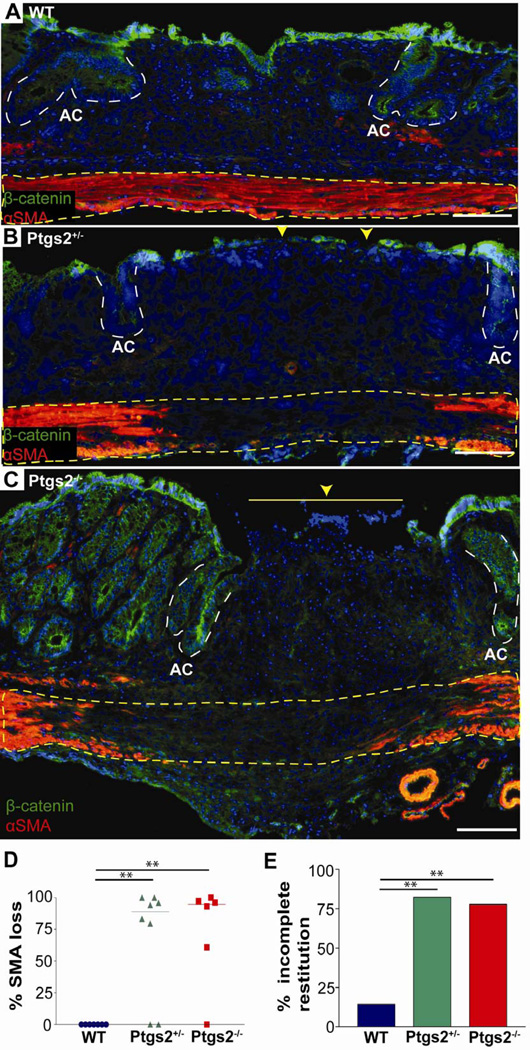
Figure 1. Two functional alleles of Ptgs2 are required for proper mucosal healing.
(A) Colonic sections from WT, (B) Ptgs2+/−, and (C) Ptgs2−/− mice 6 days post-injury were stained with antibodies against α-smooth muscle actin (α-SMA) (red), β-catenin (green), and bis-benzamide (nuclei, blue). Dotted yellow lines=muscularis propria, white dashed lines=adjacent crypts (AC), yellow arrowheads=epithelial barrier interruptions. Bars=100µm. (D) Graph of the percent loss of α-SMA underlying day 6 wound beds (gap length/wound bed length) that included data points (n≥6/genotype), median bar and significance by one-way ANOVA and post hoc Tukey's test: F2,18=10.33, **P<0.01. (E) Graph of the percentage of wound bed sections with incomplete epithelial coverage at day 6 post-injury. Wound bed sections were scored as either fully or incompletely covered by epithelial cells (β-catenin-positive). Fisher's exact test for the resulting data contingency table compared incomplete restitution in WT (2/14) to Ptgs2+/− (14/17) (P<0.001) and WT to Ptgs2−/− (7/9) (P<0.001).
The epithelial barrier at day 6 post-injury was also defective in Ptgs2−/− mice as WAE cells incompletely covered these wounds. WAE cells are claudin-4-positive, ZO-1-negative post-mitotic epithelial cells that emerge from adjacent crypts and completely migrate across the wound bed during the first stage of repair.12 In contrast to WT mice, most Ptgs2−/− wounds were only partially covered by an epithelial barrier at day 6 post-injury as shown by gross morphologic examination (Supplemental Figure 2) and by immunofluorescence microscopy (Figure 1A,C,E). In addition, the epithelial cells that partially covered Ptgs2−/− wounds were still immature WAE cells, whereas WT wounds were covered by a complete layer of mature surface epithelial cells (Supplemental Figure 3A-D). There were also defects in the final stage of healing in Ptgs2−/− wounds as they contained fewer regenerated crypts at 14 day post-injury (Supplemental Figure 4A-D). In summary, the complex repair defects in Ptgs2−/− mice were observed in the latter phases of repair.
Interestingly, by these parameters, littermate Ptgs2+/− mice also demonstrated defective wound repair (Figure 1B). At day 6 post-injury, Ptgs2+/− mice showed loss of the muscularis propria underlying the wound bed (Figure 1D) and an incomplete epithelial layer (Figure 1E). These data demonstrated that loss of just one functional Ptgs2 allele had severe effects on colonic mesenchymal and epithelial regeneration. Because this injury system was sensitive to a two-fold decrease of gene dosage, we proposed that it would be useful to discover novel proteins that regulate maximal Ptgs2 expression.
Ptgs2 expression is transiently and focally elevated during colonic wound repair
Given the profound wound repair defects in Ptgs2+/− mice, we next evaluated the timing and location of Ptgs2 expression in WT colonic wounds during two-day intervals post-injury. We first quantified Ptgs2 mRNA levels by qRT-PCR using mRNAs isolated from the mucosa of biopsy wounds. Compared to uninjured colonic mucosa, Ptgs2 levels peaked during the first stage of repair (an average of 1,640-fold higher at day 2 and 2,170-fold at day 4) and were subsequently down-regulated during the second stage of repair (only 93-fold elevation at day 6) (Figure 2A). In contrast, Ptgs1 mRNA levels did not differ between wounds and adjacent mucosa (Supplemental Figure 5). Interestingly, Ptgs2 mRNA expression was also significantly lower in the wound beds of Ptgs2+/− mice compared to WT mice at day 4 post-injury (Supplemental Figure 6). This intermediate expression of Ptgs2 in Ptgs2+/− mice was not sufficient for proper wound repair (Figure 1D,E), supporting the conclusion that maximal induction of Ptgs2 is a critical component of mucosal repair.
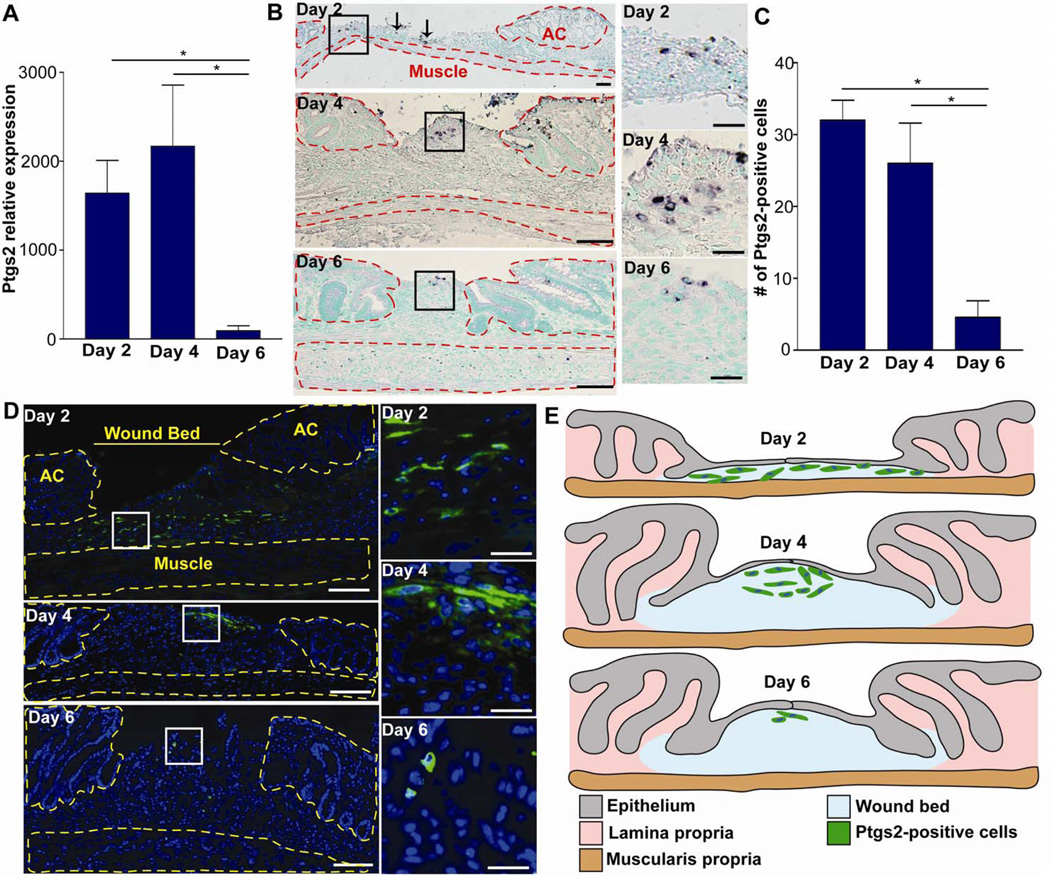
Figure 2. Ptgs2 is induced in colonic wounds in a spatial and temporal manner.
(A) Graph of Ptgs2 mRNA expression (qRT-PCR) wounds at days 2, 4 and 6 post-injury (baseline=uninjured mucosa; n≥5 wounds per day). Data displayed as means ±SEM and significance by one-way ANOVA and post-hoc Tukey's test: F2,18=5.33, *P<0.05. (B) Colonic sections from WT mice at days 2, 4 and 6 post-injury stained by in situ hybridization for Ptgs2. Solid black boxes=insets (right panels). Arrows indicated additional scattered Ptgs2 high-expressing cells outside of boxed area at day 2. The adjacent crypts (AC) and muscularis propria were outlined in red dashed lines. Bars=200µm; (50µm insets). (C) Graph of the number of Ptgs2 high-expressing cells per wound section. Data displayed as means ±SEM and significance by one-way ANOVA and post-hoc Tukey's test: F2,9=11.6, *P<0.05. (D) Colonic sections from WT mice at days 2, 4 and 6 post-injury stained with anti-Ptgs2 antisera (green) and bis-benzamide (blue). White boxes=insets (right panels). Yellow dashed lines marked adjacent crypts (AC) and muscularis propria. Bars=100 µm (25µm inset). (E) Cartoon depicting the typical positional and temporal relationship of Ptgs2 high-expressing cells in the wound bed post-injury.
We then performed in situ hybridization for Ptgs2 in WT mice at two-day intervals after injury. Because we wanted to identify cells that expressed the highest levels of Ptgs2, we limited the exposure time of the detection reagent. Throughout injury, Ptgs2 high-expressing cells were only detected within the mesenchyme of the wound bed and were not observed in either the underlying muscularis propria or crypts adjacent to the wound bed (Figure 2B). The Ptgs2 high-expressing cells were scattered in the wound bed at day 2 post-injury and were clustered beneath WAE cells at days 4 and 6 post-injury (Figure 2B). At day 6 post-injury, the number of Ptgs2 high-expressing cells was greatly diminished (Figure 2B). Thus, the cells with the highest levels of Ptgs2 mRNA in the upper wound during the first phase of repair were not detected during the second stage of repair.
We corroborated the in situ findings by immunofluorescence localization of Ptgs2 protein in wound bed sections. We used dilutions of primary antibody (1:5,000) that did not detect constitutive Ptgs2 expression in colonic mesenchymal stem cells (cMSCs) located outside of the wound bed or other cell types known to express lower levels of Ptgs2.7 In WT wound beds, we found a similar spatial and temporal pattern of Ptgs2 protein induction (Figure 2A). The number of Ptgs2 high-expressing cells in the wound bed was significantly higher at days 2 and 4 post-injury as compared to day 6 post-injury (Figure 2C). As with in situ hybridization, Ptgs2-high expressing cells detected by immunofluorescence were present only in the wound bed mesenchyme (Figure 2D). As additional controls, we performed in parallel in situ hybridization and immunofluorescence studies for localization of Ptgs2 mRNA and protein within wounds of Ptgs2−/− mice. We found no detectable signal in Ptgs2−/− mice for either assay (Supplemental Figure 7). Thus, Ptgs2 was maximally induced in upper wound bed mesenchymal cells during the first stage of repair (Figure 2E).
Ptgs2 high-expressing cells in the wound bed are colonic mesenchymal stem cells (cMSCs)
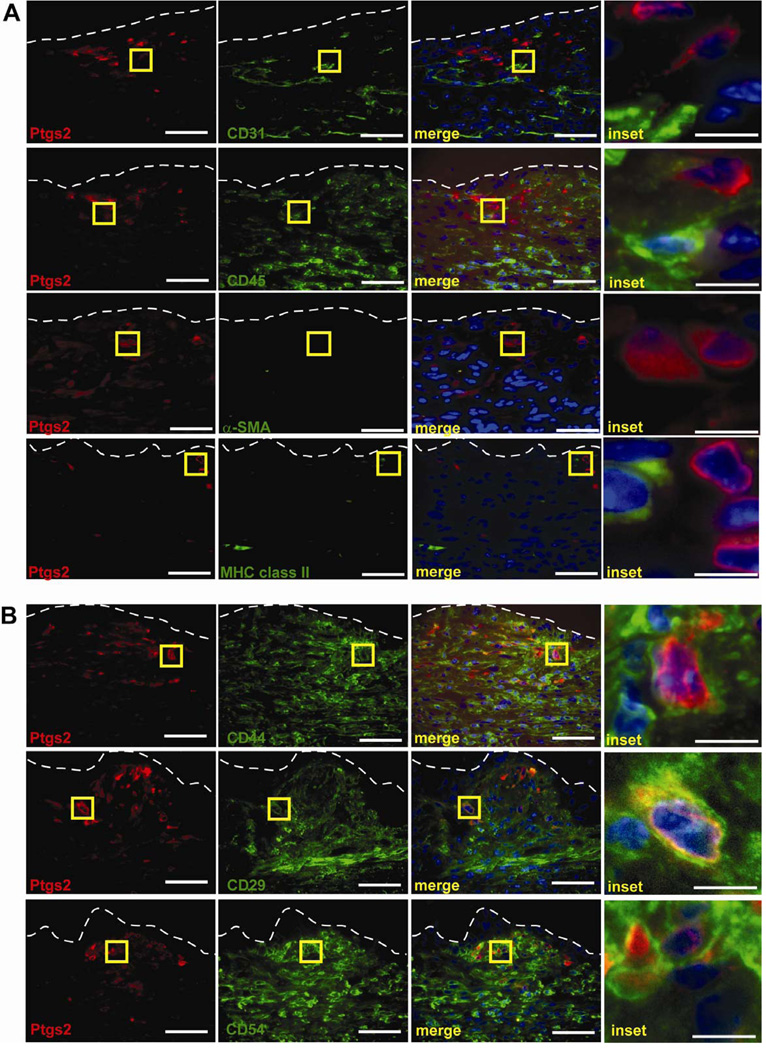
In order to identify novel Ptgs2 regulators during colonic repair, we first defined the mesenchymal cell lineages that produced the highest levels of Ptgs2. Candidates included, myeloid cells,19 endothelial cells,20 colonic mesenchymal stem cells,7 and myofibroblasts.21 Multi-label immunofluorescence imaging of Ptgs2 high-expressing cells and various lineage markers at day 4 post-injury (Figure 3) demonstrated that these cells were neither endothelial nor hematopoietic lineages as they did not co-label with either CD31 or CD45, respectively (Figure 3A). Instead, Ptgs2 high-expressing cells showed dual labeling for multiple cMSC markers 17 including CD44, CD29, and CD54 (Figure 3B). Additional studies at days 2 and 6 post-injury showed that the Ptgs2 high-expressing cells showed dual labeling with these same markers (data not shown). Since myofibroblasts and cMSCs share many of these markers, we evaluated definitive myofibroblast markers. We did not detect α-SMA cells in cells that expressed high levels of Ptgs2 (Figure 3A). Additionally, IFN-γ is expressed in wounds12 which can down-regulate α-SMA22 and increase MHC class II on myofibroblasts23. Therefore, we showed that Ptgs2 high-expressing cells were also MHC class II-negative in the wound bed to exclude resting or activated myofibroblasts as Ptgs2 high-expressing cells (Figure 3A).
Figure 3. Ptgs2 high-expressing cells in colonic wound beds are cMSCs.
(A) Sections of WT wounds at day 4 post-injury labeled with anti-Ptgs2 (red) lineage markers (green) for endothelial cells (anti-CD31), hematopoietic cells (anti-CD45), and mesenchymal cells (α-SMA and MHC class II). (B) WT day 4 wounds labeled with anti-Ptgs2 (red) and cMSC markers anti-CD44, anti-CD29, and anti-CD54. Yellow boxes=insets (right). White dashed lines=WAE basal surface. Bars=50 µm (10µm inset).
We hypothesized that cMSCs appeared in wound beds in part by migration from adjacent uninjured tissue. We injected GFP-expressing cultured cMSCs into the mucosa adjacent to wounds. We found that cMSCs migrated into wounds indicating these cells have the potential to migrate from the adjacent mucosa (Supplemental Figure 8). As a control for our isolation technique, we showed that Fstl1, a marker that distinguishes myofibroblasts from intestinal MSCs,24 was expressed in cultured cMSCs but not MIC 216 cells (Supplemental Figure 9).
To demonstrate that the defects seen in Ptgs2−/− mice were due to loss of prostaglandin synthesis, we injected stable analogues of PGE2 and PGI2 into Ptgs2−/− mice twice daily. This procedure rescued the muscle degradation phenotype at day 6 post-injury (Supplemental Figure 10). We chose these two prostaglandins as cMSCs express detectable levels of prostaglandin synthases for PGE2 and PGI2 but not for PGD2 or PGF2α.17 Since the Ptgs2 high-expressing cells in wounds were consistent with cMCSs, we further investigated cultured cMSCs for novel regulators of Ptgs2.
Igf2bp1 interacts with Ptgs2 mRNA
Our previous studies in cMSCs showed that mRNA-binding proteins including CUGbp2 play a major role in the post-transcriptional regulation of Ptgs2 expression through mRNA stabilization.17, 25 However, the ARE-binding protein CUGbp2 does not account for complete stabilization of Ptgs2 mRNA in cMSCs.17 Furthermore, previous reviews have proposed that additional, unknown mRNA-binding proteins will regulate Ptgs2 mRNA in a cooperative fashion.4
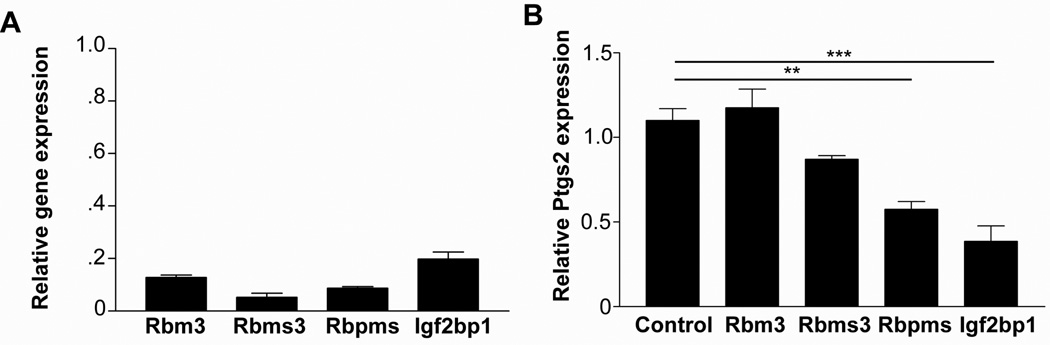
We hypothesized that additional transacting protein(s) outside of the ARE-binding family will interact with and regulate Ptgs2 mRNA in cMSCs. Because cMSCs express 10-fold higher levels of Ptgs2 than bone marrow MSCs, 17 we analyzed microarray data for these two cell types17 in order to identify candidate mRNA-binding proteins that were preferentially expressed in cMSCs. This screen identified known ARE-binding proteins (HuR, CUGbp1and CUGbp2) as well as candidate Ptgs2 mRNA-binding proteins (Rmb3, Rbms3, Rbpms and Igf2bp1). We functionally screened the non-ARE-binding protein candidates individually by shRNA-mediated knockdown in cMSCs. We generated multiple shRNAs for each gene, and the two shRNAs that had the greatest knockdown of the target gene (at least 80% knockdown efficiency) were used in further experiments (Figure 4A). Of all these candidates, we found that Igf2bp1-deficient cMSCs expressed the lowest levels of Ptgs2 mRNA (Figure 4B). Igf2bp1 was of further interest because it is an mRNA-binding protein26 that had not been previously shown to interact with Ptgs2 mRNA.
Figure 4. Igf2bp1 is required for maximal Ptgs2 expression in cMSCs.
(A) Graph depicting the relative mRNA expression (means ±SEM) of candidate Ptgs2 regulators after shRNA knockdown (two most efficient for each gene) versus control shRNA cells. (B) Graph depicting the relative expression of Ptgs2 mRNA in cMSCs that contain knockdown of candidate mRNA regulators (n=4). Data displayed as means ±SEM and significance by one-way ANOVA and post-hoc Tukey's test: F4,15=20.25, **P<0.01, ***P<0.001.
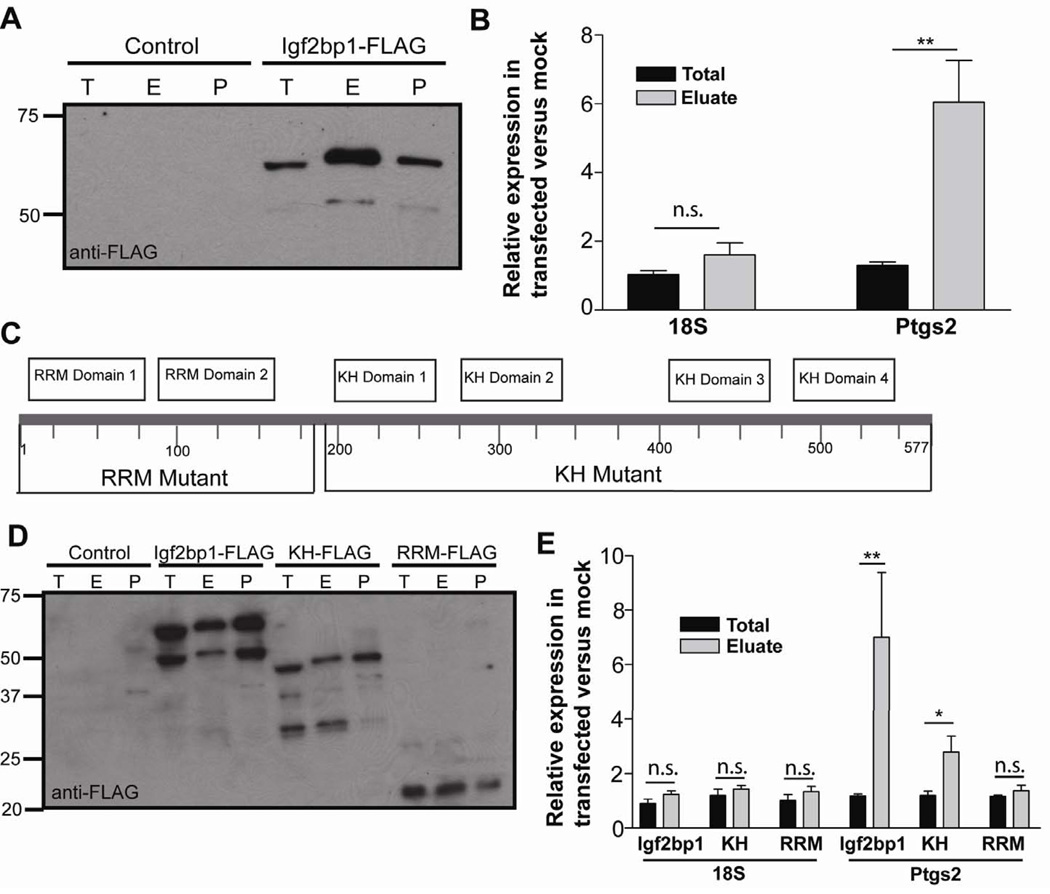
We next tested if Igf2bp1 interacted with Ptgs2 mRNA. We immunoprecipitated FLAG-tagged Igf2bp1 and eluted Igf2bp1 from the pellet fraction with 3X FLAG peptide. We then immunoblotted total protein, eluate, and pellet fractions to confirm immunoprecipitation of correct size proteins (Figure 5A). Protein was degraded in each fraction and the associated mRNAs were quantified by qRT-PCR (Figure 5B). Ptgs2 mRNA was enriched in the eluate as compared to the total fraction indicating immunoprecipitation of Igf2bp1 with Ptgs2 mRNA. (Figure 5B). As a control, 18S RNA was not enriched in the eluate. These data demonstrate the interaction of Igf2bp1 protein and Ptgs2 mRNA.
Figure 5. Igf2bp1 interacts with Ptgs2 mRNA.
(A) Immunoblot of mock-transfected (control) and Igf2bp1-FLAG transfected cells for anti-FLAG-HRP. T=total fraction, before immunoprecipitation, E=elution fraction, post-3X-FLAG incubation, and P=pellet fraction post-elution. (B) Graph of the relative RNA fractions from transfected versus mock-transfected cells. Data displayed as means ±SEM with significance by one-way ANOVA and post-hoc Tukey's test: F3,16=14.04, **P<0.01. (C) Graphical representation of the Igf2bp1 gene displaying domains and constructs. Amino acids were numbered at intervals. A C-terminal FLAG tag was included for RRM-FLAG (aa185) KH-FLAG (aa577) and Igf2bp1-FLAG. (D) Immunoblot with anti-FLAG-HRP for control, and cells transfected with Igf2bp1-FLAG and deletion mutants. (E) Graph of the relative expression of RNAs in transfected versus mock-transfected cells. Data displayed as means ±SEM with significance by one-way ANOVA and post-hoc Tukey's test: F11,28=6.14, *P<0.05, **P<0.01. Immunoblots represent>3 experiments. Graphs (N=3–4 experiments).
We then designed two FLAG-tagged truncation mutants of Igf2bp1, one containing two RRM RNA-binding motifs in the amino terminus (RRM-FLAG) and a second containing the four KH RNA-binding motifs in the carboxy terminus (KH-FLAG) (Figure 5C). We observed that KH-FLAG similarly enriched for Ptgs2 mRNA while RRM-FLAG did not (Figure 5D,E). These data suggested that the KH domains of Igf2bp1 were required for interaction with Ptgs2 mRNA.
Igf2bp1 is induced in Ptgs2 high-expressing cMSCs during the early phase of repair
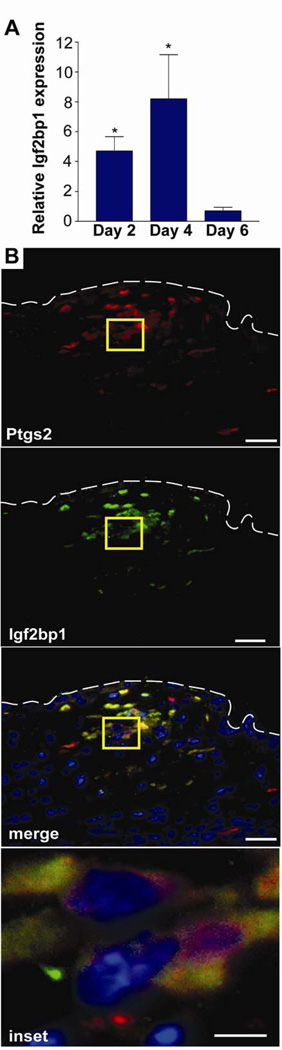
We next wanted to determine the spatial and temporal expression of Igf2bp1 in wound-associated cMSCs as well as the mechanism that controls its expression. We found that Igf2bp1 mRNA was enriched in the wound mucosa at days 2 and 4 post-injury (but not day 6 post-injury) as compared to uninjured mucosa (Figure 6A). This temporal pattern of Igf2bp1 induction/reduction was similar to Ptgs2 expression during repair. We also assessed whether Igf2bp1 co-localized with Ptgs2 high-expressing cMSCs by multi-label immunofluorescence and found that 100% of the Igf2bp1 signal co-labeled with Ptgs2 high-expressing cMSCs (Figure 6B). Thus, Igf2bp1 was expressed during wound repair at the appropriate time (first phase of healing) and location (cMSCs) to regulate maximal Ptgs2 expression.
Figure 6. Igf2bp1 is expressed in Ptgs2 high-expressing cMSCs during the first phase of wound repair.
(A) Graph of relative Igf2bp1mRNA expression (biopsy-injured compared to uninjured mucosa) at days 2, 4 and 6 post-injury (n≥5 wounds per day). Data displayed as means ±SEM with Student's t-test comparing injured to uninjured mucosa at each day; *P<0.05. (B) A section of WT colon 2 days post-biopsy was stained with anti-Ptgs2 (red) and goat anti-Igf2bp1 (green) antibodies. Individual and merged channels were shown. White dashed lines=WAE cells basal surface. Yellow box=inset. Bars=50 µm (10µm inset).
Myd88 signaling is required for induction of Igf2bp1 in cMSCs
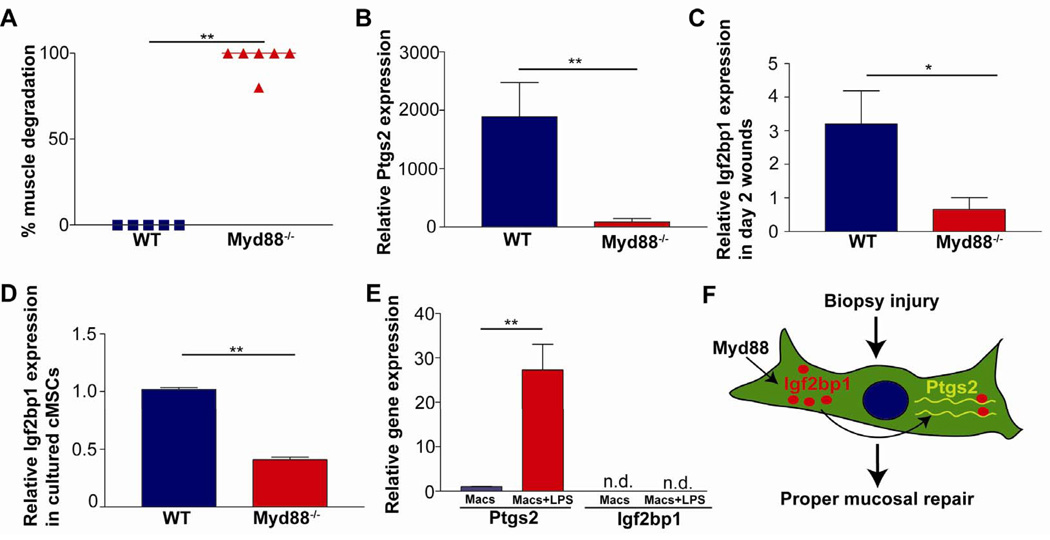
The temporal and cellular co-induction of Igf2bp1 and Ptgs2 suggested that these genes may be regulated by the same pathway. One candidate that regulates Ptgs2 expression in various cell types is Myd88 signaling3. Therefore, we hypothesized that Myd88 was required for induction of Igf2bp1 and maximal Ptgs2 expression in cMSCs. We first biopsy-injured Myd88−/− mice and found a significant repair defect as indicated by loss of α-SMA expression in the muscularis propria at day 6 post-injury (Figure 7A). This phenotype was similar to wounded Ptgs2+/− mice (Figure 1D), suggesting that Myd88 signaling was required for maximal Ptgs2 expression. Therefore, we next evaluated Ptgs2 expression in wounds 2 days post-injury, and found that the magnitude of Ptgs2 induction was 30-fold lower in Myd88−/− compared to WT mice (Figure 7B). We hypothesized that the dramatic difference in Ptgs2 expression in wounded Myd88−/− mice was due to defective Ptgs2 regulation in Ptgs2 high-expressing cMSCs. In support of this hypothesis, we found that in contrast to WT littermate controls, Igf2bp1 was not induced in Myd88−/− mice at day 2 post-injury (Figure 7C). To test whether cell-intrinsic Myd88 signaling was required in cMSCs, we isolated cMSCs and found significantly lower expression levels of Igf2bp1 in Myd88−/− compared to WT cMSCs (Figure 7D). From these data, we conclude that Myd88 signaling was required for induction of Igf2bp1 expression in cMSCs.
Figure 7. Myd88 is required for Igf2bp1 expression in cMSCs.
(A) Graph of loss of α-SMA in WT and Myd88−/− sections 6 days post-injury (n≥5 per genotype). (B–C) Graphs showing relative Ptgs2 (B) or Igf2bp1 (C) mRNA expression comparing day 2 wounds to uninjured mucosa (n≥ 4 per genotype) for WT and Myd88−/− mice. (D) Graph of Igf2bp1 mRNA relative expression comparing cultured Myd88−/− and WT cMSCs. (n=3 experiments with 3–4 replicates/sample). (E) Graph of relative Ptgs2 and Igf2bp1 mRNA expression comparing unstimulated bone marrow macrophages (Macs) to stimulated macrophages (Macs+LPS) (n=4/treatment). Data displayed as means ±SEM and Student's t-test; *P<0.05; **P<0.01. (F) Model: In WT mice, Myd88 signaling induced Igf2bp1 expression in cMSCs during the early phase of mucosal repair. Igf2bp1 interacted with Ptgs2 mRNA and was required for maximal Ptgs2 expression. Maximal expression of Ptgs2 was required for proper mucosal healing observed during the later phase of repair.
Since Ptgs2 can also be induced in macrophages by a Myd88-dependent, cell-intrinsic mechanism3, we tested whether Myd88 signaling was required for the expression of Igf2bp1 in this cell type. We treated WT bone marrow-derived macrophages with LPS, which dramatically induced Ptgs2 mRNA expression as expected (Figure 7E). We found that Igf2bp1 expression was not detectable in either LPS-treated or untreated macrophages. Thus, not all cell types capable of expressing Ptgs2 also express Igf2bp1 (Figure 7F). Therefore, the Myd88-dependent upregulation of Igf2bp1 may be a specific mechanism in MSCs.
Discussion
Igf2bp1 is a novel regulator of Ptgs2 in cMSCs
Igf2bp1 is a VICKZ mRNA-binding protein and its homologs found in other organisms have multiple names (Vg1RBP, Imp1, CRD-BP, KOC, and ZBP-1).26 ZBP-1 was identified as a zip-code binding factor that localizes β-actin mRNA to the leading edge of chicken embryo fibroblasts.27 Igf2bp1 regulates target mRNAs by several, complex mechanisms26 including: i) direct binding to target mRNAs that affect their stability or translation;4 ii) scaffolding with other mRNA-binding proteins;28 and iii) controlling mRNA intracellular localization through the formation of Igf2bp1-containing ribonucleoprotein granules (complex structures of mRNAs and proteins). Since Igf2bp1 has both shared and unique mechanisms of regulation compared to the known ARE-binding proteins, Igf2bp1 will be a key tool to elucidate principles that mediate the overall regulation of Ptgs2 in cMSCs.
The expression pattern of Igf2bp1 and the phenotype of Igf2bp1-deficient mice demonstrate that Igf2bp1 is important in development.29, 30 Igf2bp1 is an oncofetal protein that is expressed in embryos and various cancers but not adult tissues. Igf2bp1-deficient mice had dwarfism and impaired gut formation,30 thus, colonic biopsy of Igf2bp1-deficient mice was not feasible. An inducible Igf2bp1 deletion is required to test function in the adult.
Igf2bp1 is overexpressed in certain cancers including 81% of colorectal carcinomas.31 Since Ptgs2 is important for the progression of colon cancer and Igf2bp1 is frequently expressed in these cancers, the mechanism of the Ptgs2/Igf2bp1 interaction is highly relevant. Additionally, as Igf2bp1 is virtually undetectable in healthy tissue, its interaction with and stabilization of Ptgs2 makes it an intriguing candidate target for drug development. Drugs inhibiting the interaction of Igf2bp1 with Ptgs2 mRNA may potentially have fewer side effects than traditional NSAIDs or Ptgs2-inhibitors.
MSCs as mobilizable sources of Ptgs2 during repair
Although many cell types can express Ptgs2 in the colon, we showed that cMSCs expressed the highest detectable levels of Ptgs2 mRNA and protein after mucosal biopsy. Because Ptgs2+/− mice had defective healing similar to Ptgs2−/− mice, we concluded that maximal expression of Ptgs2 was necessary for proper mucosal repair. Previous studies identified numerous cell types capable of expressing Ptgs2,19–21 and lineage-specific Ptgs2 knockouts demonstrated that myeloid and endothelial expression of Ptgs2 was necessary for proper intestinal healing after DSS.8 We acknowledge that cMSCs are not the only cell type that expresses Ptgs2 during intestinal repair, and these previous studies highlight the importance of multiple Ptgs2-expressing cells, including myofibroblasts32, during healing. The relative contributions of different cell types are unknown. However, we show that cMSCs express the highest levels of Ptgs2 after severe mucosal injury; therefore the regulation of Ptgs2 in these cells is highly relevant to mucosal healing.
MSCs are an emerging cellular therapy for inflammatory diseases, such as inflammatory bowel disease.33 MSCs have at least two possible functions following intestinal injury; differentiation into stromal lineages to replace damaged tissue and expression of immunomodulatory molecules, including prostaglandins.33 The location of Ptgs2 high-expressing cMSCs directly beneath the epithelium may be functionally relevant since prostaglandins have a short half-life and act within a small area. Also, Ptgs2 is expressed during the first phase of repair when WAE cells begin migrating, and Ptgs2−/− mice have a defective epithelial barrier. We were able to rescue the muscle degradation phenotype in Ptgs2−/− mice, demonstrating that the main function of Ptgs2 is the production of prostaglandins. We were also able to inject cMSCs adjacent to wounds and show that they were capable of migrating towards sites of inflammation from adjacent uninjured tissue. We believe this may be one major method of homing for cMSCs, though we cannot rule out the migration of cMSCs from the bone marrow.
The accepted markers for MSCs are a constellation of positive markers including CD29, CD44 and CD54, as well as negative lineage markers.33, 34 A challenge in the field is that there is no single marker that only marks cMSCs. Because Igf2bp1 protein was only found in cMSCs in wound beds, we will further investigate whether Igf2bp1 may be such a marker.
In conclusion, we demonstrate the importance of maximal expression of Ptgs2 during mucosal repair, and identify Igf2bp1 as a novel mRNA-binding protein necessary for maximal Ptgs2 expression in cMSCs. Our study has implications for understanding complex mechanisms of Ptgs2 regulation by mRNA-binding proteins. Additional work will be needed to determine how Igf2bp1 regulates Ptgs2 expression and whether it interacts with ARE-binding proteins in cMSCs or affects Ptgs2 localization.
Supplementary Material
Acknowledgements
We thank Dr. William Stenson for critically reading the manuscript, and Dr. Deborah Rubin for providing the MIC 216 cells.
Grant Funding: R01 DK071619, P30-DK52574 (Washington University Digestive Disease Research Core), NIH/NHLBI T32 HL007317 Pre-doctoral Pulmonary and Critical Care Training Grant and Pew Scholars in the Biomedical Sciences.
Abbreviations
- Igf2bp1
Insulin-like growth factor 2 binding protein 1
- Ptgs2
Prostaglandin-endoperoxide synthase 2
- ARE
AU-rich elements
- cMSC
colonic mesenchymal stem cell
- WAE
wound associated epithelial cell
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Involvement: N.A.M and M.R.D. designed and performed the experiments; wrote the manuscript. H.M. performed experiments. T.S.S. oversaw the project/manuscript.
Disclosures: None
References
- 1.Wang D, Mann JR, DuBois RN. The role of prostaglandins and other eicosanoids in the gastrointestinal tract. Gastroenterology. 2005;128:1445–1461. doi: 10.1053/j.gastro.2004.09.080. [DOI] [PubMed] [Google Scholar]
- 2.Ishikawa T, Jain NK, Taketo MM, et al. Imaging cyclooxygenase-2 (Cox-2) gene expression in living animals with a luciferase knock-in reporter gene. Mol Imaging Biol. 2006;8:171–187. doi: 10.1007/s11307-006-0034-7. [DOI] [PubMed] [Google Scholar]
- 3.Wu KK. Control of cyclooxygenase-2 transcriptional activation by pro-inflammatory mediators. Prostaglandins Leukot Essent Fatty Acids. 2005;72:89–93. doi: 10.1016/j.plefa.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Barreau C, Paillard L, Osborne HB. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 2005;33:7138–7150. doi: 10.1093/nar/gki1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cok SJ, Morrison AR. The 3'-untranslated region of murine cyclooxygenase-2 contains multiple regulatory elements that alter message stability and translational efficiency. J Biol Chem. 2001;276:23179–23185. doi: 10.1074/jbc.M008461200. [DOI] [PubMed] [Google Scholar]
- 6.Morteau O, Morham SG, Sellon R, et al. Impaired mucosal defense to acute colonic injury in mice lacking cyclooxygenase-1 or cyclooxygenase-2. J Clin Invest. 2000;105:469–478. doi: 10.1172/JCI6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown SL, Riehl TE, Walker MR, et al. Myd88-dependent positioning of Ptgs2- expressing stromal cells maintains colonic epithelial proliferation during injury. J Clin Invest. 2007;117:258–269. doi: 10.1172/JCI29159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishikawa T, Oshima M, Herschman HR. Cox-2 deletion in myeloid and endothelial cells, but not in epithelial cells, exacerbates murine colitis. Carcinogenesis. 2011;32:417–426. doi: 10.1093/carcin/bgq268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng L, Riehl TE, Stenson WF. Regulation of colonic epithelial repair in mice by Toll-like receptors and hyaluronic acid. Gastroenterology. 2009;137:2041–2051. doi: 10.1053/j.gastro.2009.08.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang L, Lu YM, Dong XY. Effects and mechanism of the selective COX-2 inhibitor, celecoxib, on rat colitis induced by trinitrobenzene sulfonic acid. Chin J Dig Dis. 2004;5:110–114. doi: 10.1111/j.1443-9573.2004.00164.x. [DOI] [PubMed] [Google Scholar]
- 11.Riehl TE, Newberry RD, Lorenz RG, et al. TNFR1 mediates the radioprotective effects of lipopolysaccharide in the mouse intestine. Am J Physiol Gastrointest Liver Physiol. 2004;286:G166–G173. doi: 10.1152/ajpgi.00537.2002. [DOI] [PubMed] [Google Scholar]
- 12.Seno H, Miyoshi H, Brown SL, et al. Efficient colonic mucosal wound repair requires Trem2 signaling. Proc Natl Acad Sci U S A. 2009;106:256–261. doi: 10.1073/pnas.0803343106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pickert G, Neufert C, Leppkes M, et al. STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J Exp Med. 2009;206:1465–1472. doi: 10.1084/jem.20082683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Normand S, Delanoye-Crespin A, Bressenot A, et al. Nod-like receptor pyrin domain-containing protein 6 (NLRP6) controls epithelial self-renewal and colorectal carcinogenesis upon injury. Proc Natl Acad Sci U S A. 2011;108:9601–9606. doi: 10.1073/pnas.1100981108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morham SG, Langenbach R, Loftin CD, et al. Prostaglandin synthase 2 gene disruption causes severe renal pathology in the mouse. Cell. 1995;83:473–482. doi: 10.1016/0092-8674(95)90125-6. [DOI] [PubMed] [Google Scholar]
- 16.Adachi O, Kawai T, Takeda K, et al. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity. 1998;9:143–150. doi: 10.1016/s1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- 17.Walker MR, Brown SL, Riehl TE, et al. Growth factor regulation of prostaglandin-endoperoxide synthase 2 (Ptgs2) expression in colonic mesenchymal stem cells. J Biol Chem. 2010;285:5026–5039. doi: 10.1074/jbc.M109.032672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson MH, Wheatley T, Leach IH. Nonsteroidal antiinflammatory drug-induced colonic stricture. An unusual cause of large bowel obstruction and perforation. Dig Dis Sci. 1995;40:315–319. doi: 10.1007/BF02065416. [DOI] [PubMed] [Google Scholar]
- 19.Pouliot M, Baillargeon J, Lee JC, et al. Inhibition of prostaglandin endoperoxide synthase-2 expression in stimulated human monocytes by inhibitors of p38 mitogen-activated protein kinase. J Immunol. 1997;158:4930–4937. [PubMed] [Google Scholar]
- 20.Jones MK, Wang H, Peskar BM, et al. Inhibition of angiogenesis by nonsteroidal anti-inflammatory drugs: insight into mechanisms and implications for cancer growth and ulcer healing. Nat Med. 1999;5:1418–1423. doi: 10.1038/70995. [DOI] [PubMed] [Google Scholar]
- 21.Mifflin RC, Saada JI, Di Mari JF, et al. Regulation of COX-2 expression in human intestinal myofibroblasts: mechanisms of IL-1-mediated induction. Am J Physiol Cell Physiol. 2002;282:C824–C834. doi: 10.1152/ajpcell.00388.2001. [DOI] [PubMed] [Google Scholar]
- 22.Desmouliere A, Rubbia-Brandt L, Abdiu A, et al. Alpha-smooth muscle actin is expressed in a subpopulation of cultured and cloned fibroblasts and is modulated by gamma-interferon. Exp Cell Res. 1992;201:64–73. doi: 10.1016/0014-4827(92)90348-c. [DOI] [PubMed] [Google Scholar]
- 23.Saada JI, Pinchuk IV, Barrera CA, et al. Subepithelial myofibroblasts are novel nonprofessional APCs in the human colonic mucosa. J Immunol. 2006;177:5968–5979. doi: 10.4049/jimmunol.177.9.5968. [DOI] [PubMed] [Google Scholar]
- 24.Geske MJ, Zhang X, Patel KK, et al. Fgf9 signaling regulates small intestinal elongation and mesenchymal development. Development. 2008;135:2959–2968. doi: 10.1242/dev.020453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mukhopadhyay D, Houchen CW, Kennedy S, et al. Coupled mRNA stabilization and translational silencing of cyclooxygenase-2 by a novel RNA binding protein, CUGBP2. Mol Cell. 2003;11:113–126. doi: 10.1016/s1097-2765(03)00012-1. [DOI] [PubMed] [Google Scholar]
- 26.Yisraeli JK. VICKZ proteins: a multi-talented family of regulatory RNA-binding proteins. Biol Cell. 2005;97:87–96. doi: 10.1042/BC20040151. [DOI] [PubMed] [Google Scholar]
- 27.Ross AF, Oleynikov Y, Kislauskis EH, et al. Characterization of a beta-actin mRNA zipcode-binding protein. Mol Cell Biol. 1997;17:2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weidensdorfer D, Stohr N, Baude A, et al. Control of c-myc mRNA stability by IGF2BP1-associated cytoplasmic RNPs. RNA. 2009;15:104–115. doi: 10.1261/rna.1175909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yaniv K, Yisraeli JK. The involvement of a conserved family of RNA binding proteins in embryonic development and carcinogenesis. Gene. 2002;287:49–54. doi: 10.1016/s0378-1119(01)00866-6. [DOI] [PubMed] [Google Scholar]
- 30.Hansen TV, Hammer NA, Nielsen J, et al. Dwarfism and impaired gut development in insulin-like growth factor II mRNA-binding protein 1-deficient mice. Mol Cell Biol. 2004;24:4448–4464. doi: 10.1128/MCB.24.10.4448-4464.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ross J, Lemm I, Berberet B. Overexpression of an mRNA-binding protein in human colorectal cancer. Oncogene. 2001;20:6544–6550. doi: 10.1038/sj.onc.1204838. [DOI] [PubMed] [Google Scholar]
- 32.Mifflin RC, Pinchuk IV, Saada JI, et al. Intestinal myofibroblasts: targets for stem cell therapy. Am J Physiol Gastrointest Liver Physiol. 2011;300:G684–G696. doi: 10.1152/ajpgi.00474.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manieri NA, Stappenbeck TS. Mesenchymal stem cell therapy of intestinal disease: are their effects systemic or localized? Curr Opin Gastroenterol. 2011;27:119–124. doi: 10.1097/MOG.0b013e3283423f20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dominici M, Le Blanc K, Mueller I, et al. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.