Abstract
Substrate ingress and product egress from the active site of urease is tightly controlled by an active site flap. Molecular dynamics simulations of urease reveal a previously unobserved, wide-open flap state that, unlike the well-characterized closed and open states, allows ready access to the metal cluster in the active site. This state is easily reached, via low free energy barriers, from the closed and open states. Additionally, we find that even when the flap is closed, a region of the binding pocket is solvent exposed leading to the hypothesis that it may act as a substrate/product reservoir. The newly identified wide-open state offers further opportunities for small molecule drug discovery by defining a more extensive active site pocket than has been previously described.
Helicobacter pylori (H. pylori), a bacteria found in the stomach, causes stomach ulcers, adenocarcinoma of the distal stomach, and lymphoma of the gastric mucosa1–3 and has a 2–4% mortality rate among infected humans.4 According to the Center for Disease Control, two-thirds of people worldwide are infected with H. pylori, with the highest percentage from regions of Western Africa, South America, and Northern China.5 Currently, H. pylori infection cannot be cured with a single drug. Instead, a combination of medications are often used that cause side effects in approximately half of patients.6 This highlights the need for novel antimicrobials that target H. pylori and urease is one such target.7 However, the urease active site region in the available crystal structures is highly constrained around the metal ion,3 thereby only allowing small molecule inhibitors like acetohydroxamic acid,3 boric acid8 or phosphate9 that both bind the metal cluster while satisfying the active site constraints. Herein we identify a novel wide-open state of the urease active site that offers new opportunities for small-molecule drug discovery by defining a more extensive binding pocket that may or may not require a metal binding warhead.
The ureases are a group of closely related enzymes found in certain plants, bacteria and fungi.10,11 Notably associated with H. pylori and other pathogenic species, ureases offer attractive targets for drug design because of their role in protecting the pathogen from the highly acidic pH of the gut.12 Catalyzing the breakdown of urea into ammonia and carbamate,13 they are extremely efficient enzymes, speeding up this reaction by at least fourteen orders of magnitude and turning over several thousand substrate molecules per second.14–16
The ureases are multimeric, with each active site containing a dinickel cluster in its active site.17 The precise mechanism of the enzyme-catalyzed reaction is not yet fully understood,13,15,18–22 but in addition to breaking down urea, the catalytic cycle appears to facilitate large-scale protein motion such as diffusion of urease enzymes.23 Each active site is capped by a 33-residue flap, which governs access to and egress from the dinickel cluster.15 In this study, we used classical molecular dynamics (MD) simulations to study the motion of these flaps.
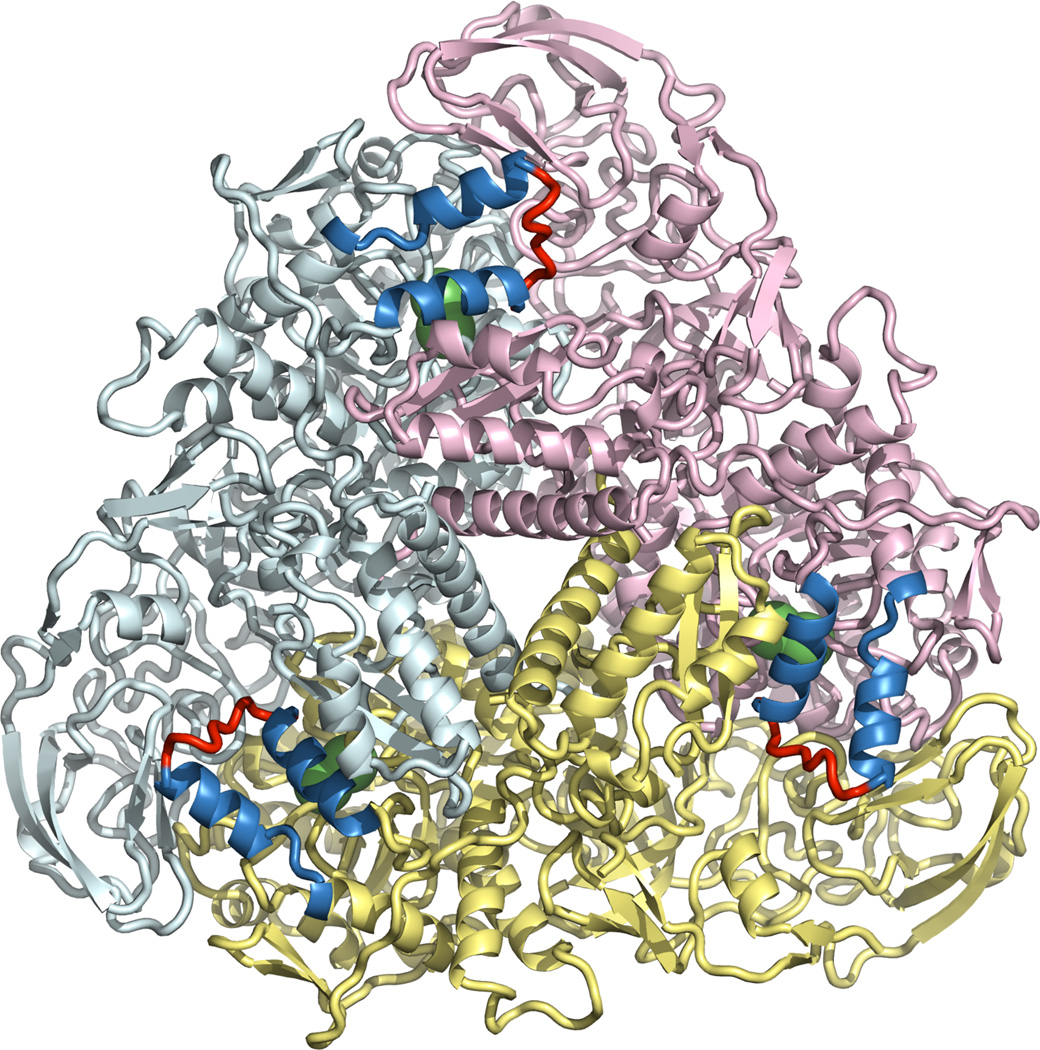
We chose the urease from the bacterium Klebsiella aerogenes (KA) for study (see Figure 1) rather than H. pylori urease because of the immense size (150,000 atoms for H. pylori, 35,000 atoms for KA) of the latter relative to the former. Moreover, the flap and active site regions are well conserved (vida infra) between the two systems allowing us to realistically extrapolate from KA to H. pylori with regards to dynamics in these regions. Nonetheless, simulations of H. pylori urease using modern GPU technology are underway and will be reported on in due course. KA urease is a homotrimer of heterotrimers, and contains three active sites.24 We ran two separate simulations, one starting from a structure in which all three flaps were closed (PDB ID 1FWJ), and the other from a structure in which all three flaps were open (PDB ID 1EJX). We performed a symmetry expansion to generate the (αβγ)3 form. Running 180 ns of simulation on the closed-flap structure and 100 ns of simulation on the open-flap structure, we generated 840 ns (280ns X 3 active sites) of flap dynamics.
Figure 1.
The initial structure of K. aerogenes urease in the open state, as used in our simulations. The trimeric subunits are shown in yellow, pink and cyan. The flaps are shown as α-helices (blue) and loops (red). Nickel ions are shown as green spheres.
The flap itself comprises three regions: two short α-helices and between them a flexible loop. The channel into the active site, protected by the flap, lies on the border between two trimeric subunits, so that a different subunit lies on the other side of the channel from the flap. Each of the helices is able to tilt away from this other subunit, bringing the flap into one of two partially open states; when both α-helices do so at the same time, the flap enters the open state. Inspection of the crystal structures from the Protein Data Bank and our simulation trajectories reveals that neither the partially open state, nor the open state, allows ready access to the active site region of urease, posing the fundamental question of how the substrate enters the dinickel active site.
Pertinent to this question, our simulations have revealed a new wide-open state. We propose that this state is important for substrate entry and product exit. It is distinguished from the open state by a loss of helical character in the α-helices with consequent extension of the loop into neighboring residues. Indeed, the loop itself appears to have characteristics of an intrinsically unstructured protein25 that has multiple states it can access in the resting state, but becomes ordered (forming the closed state) upon substrate or inhibitor binding. In this wide-open state, the extended loop moves away from the protein, opening up a wide pathway into the active site.
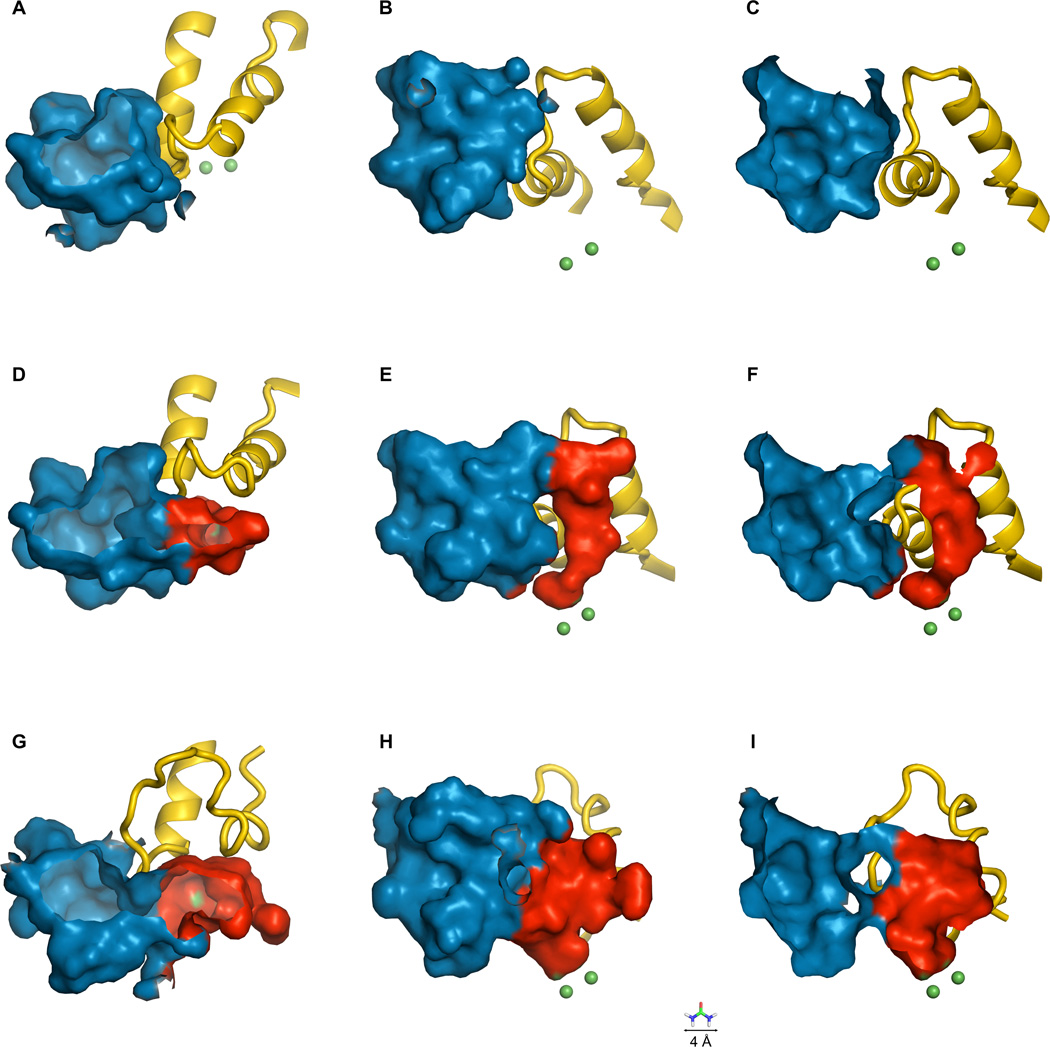
The closed, open and wide-open states are shown in Figure 2. In the closed state (Figure 2, A – C), the flap acts to seal the di-nickel cluster off from the bulk solvent entirely. In the open state (Figure 2, D – F), the flap moves slightly, enough to open only a very narrow pathway into the dinickel cluster. This pathway is too narrow to even admit a molecule of urea. Much of the obstruction of the active site channel is due to a single residue, His 320 in the α subunit. This residue is thrust into the binding pocket, dividing it into two parts and severely restricting access to the dinickel cluster. As part of the transition from open to wide-open, His 320 moves out of the way, opening up a wide pathway and allowing substrate molecules ready access to the nickel cluster (Figure 2, G – I; see also Supporting Information). In all three states, however, an interesting aspect of the binding pocket is revealed. An ancillary pocket remains open even while the flap is closed, although the function of this pocket is unknown. One hypothesis is that it may act as a substrate/product reservoir that is perfectly placed to take advantage of the increased access to the active site once the protein enters the wide-open state. However, it also may be a site for regulator binding or even simply the vestige of evolution.
Figure 2.
The flap, active site and ancillary binding pocket of K. aerogenes urease. The flap is shown in yellow, the active site in red and the ancillary binding pocket in blue. The nickel ions are shown as green spheres, and exposed nickel surface is also shown in green (in D – I). From left to right: Closed flap (A – C), open flap (D – F) and wide-open flap (G – I). Three views are shown: From the top, looking down into the binding pockets (A, D, G); from the side (B, E, H); and from the side in cutaway view (C, F, I). For scale, a urea molecule (4 Å across) is depicted at the bottom of the figure.
We investigated the possibility that the ancillary pocket acts as a substrate-specific binding site by comparing the PDB structure of K. aerogenes urease with the structures of three other ureases, namely those from H. pylori, B. pasteurii and Jack Bean. We found that in all four ureases the ancillary pocket exists and potentially fulfills the role of a reservoir; however, the amino acid residues that make a significant contribution to its surface are less conserved, on average, than in the protein at large. This may be contrasted with the flap itself, which is more highly conserved. Based on the structural and sequence analysis work we hypothesize that it is unlikely that the ancillary pocket acts as a ligand specific tight binding site.
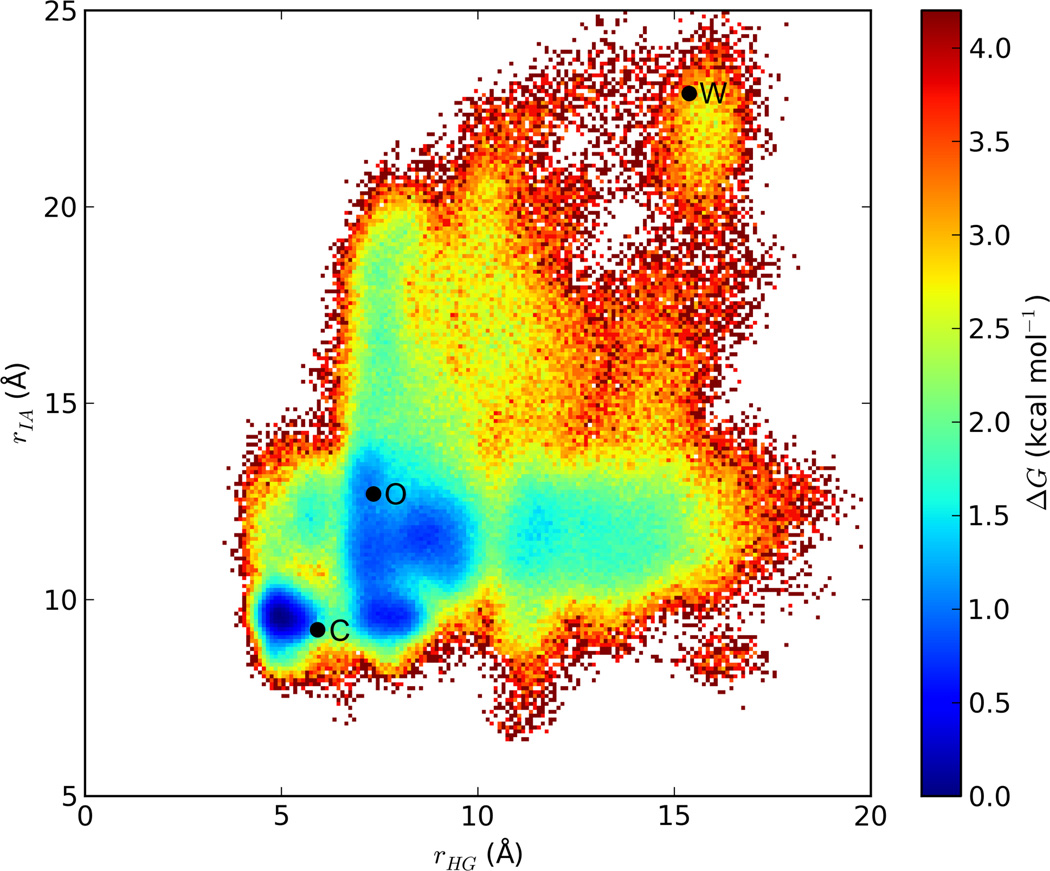
From our simulation data, we have computed a relative free energy map, based on two separate distances between α-carbon atoms in residues across the flap opening from each other. One of these distances, rHG, spans the gap between His 320 in the α subunit of trimer 1 and Gly 46 in the α subunit of trimer 3; the other, rIA, spans the gap between Ile 326 in the α subunit of trimer 1 and Ala 68 in the β subunit of trimer 3. Values for rHG and rIA were also obtained from the other two flaps, at the trimer 3—trimer 2 and trimer 2—trimer 1 interfaces. A figure depicting rHG and rIA is included in supporting information. This map (Figure 3) shows that transition between the closed and open states is facile, with an activation barrier of approximately 1.5 kcal mol−1 and a ΔG of less than 1 kcal mol−1. The wide-open state is less populated than either the closed state or the open state (ΔG ≈ 2.5 kcal mol−1). Nevertheless, it is readily accessible from the open state via two pathways, each with an activation barrier of no more than 4 kcal mol−1.
Figure 3.
Relative free energy map of the urease flap opening. The points marked by “C,” “O,” and “W” represent respectively the closed, open and wide-open structures shown in Figure 2. Points C and O represent the initial X-ray structures of the closed- and open-flap models, being PDB IDs 1FWJ and 1EJX respectively; for the latter structure, the flap was added by homology modeling. Point W is a representative wide-open conformation selected from the simulation trajectory of 1FWJ.
The present simulations reveal for the first time the entire ensemble of flap states available to urease and provide insight into how substrate gains access to the active site. We note that the flap itself appears to have the characteristics of an intrinsically unstructured protein,25 which becomes more ordered upon substrate binding to urease. Moreover, the computed free energy estimates are in accord with efficient catalysis, as is the secondary pocket, which we hypothesize may accelerate substrate/product ingress/egress in an assembly-line-like manner.
In conclusion, although the open structure is not open enough to allow substrate access to the active site, simulation data points to the existence and ready accessibility of the wide-open state, in which diffusion of substrate into the active site and departure of products should be rapid. This wide-open state reveals a more expansive active site pocket, which is more suitable for exploitation by small molecule inhibitors. Protein dynamics have been important for characterizing conformational states for key drug discovery efforts, such as for the classical case of HIV-1 Protease.26
Supplementary Material
Acknowledgement
B. P. R. and K. M. M. acknowledge financial support from the NIH (R01 GM066859). Supercomputer time was granted by the Large Allocations Resource Committee (TGMCA05T010). We would also like to think Dr. Robert Hausinger for helpful advice regarding the present manuscript.
Footnotes
Supporting Information Available: Details of computational methods and force field parameters, additional figures relating to our analysis of the simulations, and PDB text of K. aerogenes urease with a flap in the wide-open conformation. This information is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Covacci A, Telford JL, Del Giudice G, Parsonnet J, Rappuoli R. Science. 1999;284:1328. doi: 10.1126/science.284.5418.1328. [DOI] [PubMed] [Google Scholar]
- 2.Dunn BE, Cohen H, Blaser MJ. Clin Microbiol Rev. 1997;10:720. doi: 10.1128/cmr.10.4.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ha NC, Oh ST, Sung JY, Cha KA, Lee MH, Oh BH. Nat Struct Biol. 2001;8:505. doi: 10.1038/88563. [DOI] [PubMed] [Google Scholar]
- 4.Suerbaum S, Michetti P. N Engl J Med. 2002;347:1175. doi: 10.1056/NEJMra020542. [DOI] [PubMed] [Google Scholar]
- 5.Pounder RE, Ng D. Aliment Pharmacol Ther. 995;9(Suppl 2):33. [PubMed] [Google Scholar]
- 6.Chey WD, Wong BC. Am J Gastroenterol. 2007;102:1808. doi: 10.1111/j.1572-0241.2007.01393.x. [DOI] [PubMed] [Google Scholar]
- 7.Follmer CJ. Clin Pathol. 2010;63:424. doi: 10.1136/jcp.2009.072595. [DOI] [PubMed] [Google Scholar]
- 8.Benini S, Rypniewski WR, Wilson KS, Mangani S, Ciurli S. J Am Chem Soc. 2004;126:3714. doi: 10.1021/ja049618p. [DOI] [PubMed] [Google Scholar]
- 9.Benini S, Rypniewski WR, Wilson KS, Ciurli S, Mangani S. J Biol Inorg Chem. 2001;6:778. doi: 10.1007/s007750100254. [DOI] [PubMed] [Google Scholar]
- 10.Mobley HLT, Hausinger RP. Microbiol. Rev. 1989;53:85. doi: 10.1128/mr.53.1.85-108.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krajewska B. J. Mol. Catal. B: Enzym. 2009;59:9. [Google Scholar]
- 12.Follmer C. J. Clin. Pathol. 2010;63:424. doi: 10.1136/jcp.2009.072595. [DOI] [PubMed] [Google Scholar]
- 13.Dixon NE, Riddles PW, Gazzola C, Blakeley RL, Zerner B. Can. J. Biochem. 1980;58:1335. doi: 10.1139/o80-181. [DOI] [PubMed] [Google Scholar]
- 14.Sumner JB. J. Biol. Chem. 1926;69:435. [Google Scholar]
- 15.Pearson MA, Park I-S, Schaller RA, Michel LO, Karplus PA, Hausinger RP. Biochemistry. 2000;39:8575. doi: 10.1021/bi000613o. [DOI] [PubMed] [Google Scholar]
- 16.Callahan BP, Yuan Y, Wolfenden R. J. Am. Chem. Soc. 2005;127:10828. doi: 10.1021/ja0525399. [DOI] [PubMed] [Google Scholar]
- 17.Zambelli B, Musiani F, .; Benini S, Ciurli S. Acc. Chem. Res. 2011;44:520. doi: 10.1021/ar200041k. [DOI] [PubMed] [Google Scholar]
- 18.Carlsson H, Nordlander E. Bioinorg. Chem. Appl. 2010;2010:364891. doi: 10.1155/2010/364891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benini S, Rypniewski WR, ; Wilson KS, ; Ciurli S, Mangani S. J. Biol. Inorg. Chem. 2001;6:778. doi: 10.1007/s007750100254. [DOI] [PubMed] [Google Scholar]
- 20.Musiani F, Arnofi E, Casadio R, Ciurli S. J. Biol. Inorg. Chem. 2001;6:300. doi: 10.1007/s007750000204. [DOI] [PubMed] [Google Scholar]
- 21.Estiu G, Merz KM. J. Phys. Chem. B. 2007;111:10263. doi: 10.1021/jp072323o. [DOI] [PubMed] [Google Scholar]
- 22.Estiu G, Merz KM. J. Am. Chem. Soc. 2004;126:11832. doi: 10.1021/ja047934y. [DOI] [PubMed] [Google Scholar]
- 23.Muddana HS, Sengupta S, Mallouk TE, Sen A, Butler PJ. J. Am. Chem. Soc. 2010;132:2110. doi: 10.1021/ja908773a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jabri E, Carr MB, Hausinger RP, Karplus PA. Science. 1995;268:998. [PubMed] [Google Scholar]
- 25.Wright PE, Dyson HJ. J Mol Biol. 1999;293:321. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 26.Hornak V, Simmerling C. Drug Discov Today. 2007;12:132. doi: 10.1016/j.drudis.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.