Abstract
Objective
To research computational methods for coreference resolution in the clinical narrative and build a system implementing the best methods.
Methods
The Ontology Development and Information Extraction corpus annotated for coreference relations consists of 7214 coreferential markables, forming 5992 pairs and 1304 chains. We trained classifiers with semantic, syntactic, and surface features pruned by feature selection. For the three system components—for the resolution of relative pronouns, personal pronouns, and noun phrases—we experimented with support vector machines with linear and radial basis function (RBF) kernels, decision trees, and perceptrons. Evaluation of algorithms and varied feature sets was performed using standard metrics.
Results
The best performing combination is support vector machines with an RBF kernel and all features (MUC score=0.352, B3=0.690, CEAF=0.486, BLANC=0.596) outperforming a traditional decision tree baseline.
Discussion
The application showed good performance similar to performance on general English text. The main error source was sentence distances exceeding a window of 10 sentences between markables. A possible solution to this problem is hinted at by the fact that coreferent markables sometimes occurred in predictable (although distant) note sections. Another system limitation is failure to fully utilize synonymy and ontological knowledge. Future work will investigate additional ways to incorporate syntactic features into the coreference problem.
Conclusion
We investigated computational methods for coreference resolution in the clinical narrative. The best methods are released as modules of the open source Clinical Text Analysis and Knowledge Extraction System and Ontology Development and Information Extraction platforms.
Keywords: Coreference resolution, natural language processing, biomedical informatics, information extraction, nlp, machine learning, human-computer interaction and human-centered computing, Intelligent tutoring and tailored information representation, Improving the education and skills training of health professionals, providing just-in-time access to the biomedical literature and other health information, applications that link biomedical knowledge from diverse primary sources (includes automated indexing), linking the genotype and phenotype, discovery
Background and significance
The field of natural language processing (NLP) has been steadily moving toward semantic parsing. Central to this are the tasks of relation detection and classification. Anaphora is a relation between linguistic expressions where the interpretation of one linguistic expression (the anaphor) is dependent on the interpretation of another (the antecedent). When the anaphor and the antecedent point to the same referent in the real world, they are termed coreferential.1
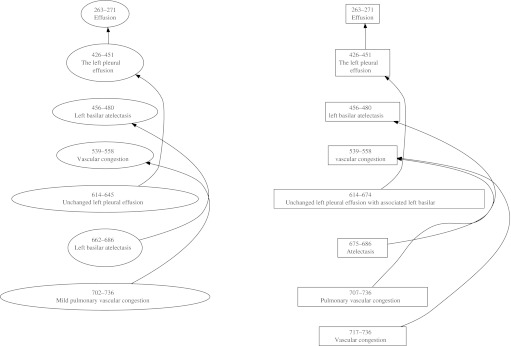
The identification of entity mentions (or named entities, NEs) referring to the same world object (‘coreference resolution’) is critical for comprehensive information extraction (IE). The set of such entity mentions forms a chain. The left side of figure 1 presents the coreferential entities that one would find in this particular radiology report. There are three chains—one linking the mentions ‘effusion,’ ‘the left pleural effusion,’ and ‘unchanged left pleural effusion’; another linking ‘vascular congestion’ and ‘mild pulmonary vascular congestion’; and the third linking ‘left basilar atelectasis’ and ‘left basilar atelectasis.’ Each chain consists of pairs, for example, ‘effusion’ (antecedent)—'the left pleural effusion' (anaphor), and ‘the left pleural effusion’ (antecedent)—‘the unchanged left pleural effusion’ (anaphor). Without asserting coreferential relations between the mentions, the seven mentions in these three coreference chain examples would be stored as separate, completely independent instances, leading to fragmented clinical information and potentially multiple clinical events where only three events actually occurred.
Figure 1.
Example of coreferential chains from one report. Ovals are gold standard chains, rectangles are system output.
A detailed review of coreference resolution systems in the general, bio-, and clinical domains has been presented elsewhere,1–3 concluding that there are only a handful of efforts4–6 addressing coreference in the clinical domain. Coreference resolution in the clinical domain is the shared task for the upcoming 2011 i2b2/VA NLP challenge (https://www.i2b2.org/NLP/Coreference/Call.php), which is expected to significantly advance the field. A significant barrier to progress in coreference resolution in the clinical domain has been the lack of a shared annotated corpus to serve as a training and test bed for both rule-based and machine learning methods, with the latter requiring much more data. In our earlier work, we built one such corpus (the Ontology Development and Information Extraction corpus, ODIE).7 In this manuscript, our objective is to describe our research on computational methods for coreference resolution in the clinical narrative and to evaluate the system we developed using the ODIE corpus. We present what to our knowledge is the first end-to-end coreference system for the clinical narrative (1) which is built off and evaluated on a shared annotated clinical corpus, (2) in which entity mentions and features are automatically generated without any human intervention (we utilize the clinical Text Analysis and Knowledge Extraction System (cTAKES)8 9), (3) which is integrated within an open-source comprehensive clinical IE system, cTAKES. We distribute our coreference system open source as part of cTAKES and ODIE.10
Materials and methods
Materials
The ODIE project adopts the Message Understanding Conference 711 (MUC-7) terminology and extends it to the clinical domain, defining markable as a linguistic expression signifying a clinical concept and participating in coreferential relations within the given document. The ODIE coreference corpus consists of 105 082 tokens of clinical notes from two institutions. For full details consult Savova et al.7 In summary, the Mayo Clinic set comprises 100 notes equally distributed between clinical and pathology notes. The University of Pittsburgh Medical Center (UPMC) set comprises 80 notes equally distributed among four types of narrative: emergency department notes, discharge summaries, surgical pathology notes, and radiology notes. Three domain experts created the gold standard annotations for coreference pairs and chains, resulting in 7214 markables, 5992 pairs, and 1304 chains. The overall inter-annotator agreement on the Mayo Clinic dataset is 0.6607 and on the UPMC dataset 0.4172. Each report averaged 40 markables, 33 pairs, and seven chains. The markables belong to one of eight semantic categories: (1) Person; (2) the UMLS Anatomy semantic group; (3) the UMLS Disorders semantic group (excluding the semantic types of Sign or symptom (see (5)) and Finding); (4) the UMLS Procedures semantic group; (5) the UMLS Sign or symptom semantic type; (6) the UMLS Laboratory or test result semantic type; (7) the UMLS Indicator, reagent, or diagnostic aid semantic type; and (8) the UMLS Organ or tissue function semantic type (cf Bodenreider and McCray12).
This work also makes use of syntactic phrase structure information, and thus requires gold standard parsed corpora for training a constituency parser. We use both general domain and clinical domain corpora for training. The widely used Wall Street Journal section of the Penn Treebank13 with an additional nominal modifier structure14 is our general domain data (about 50 000 sentences). Clinical domain data contain text from Mayo Clinic clinical notes, clinical questions,15 randomly selected Medpedia16 paragraphs, and a sample of queries describing exchanges between clinical investigators and retrievalists (about 15 000 sentences). The clinical domain corpus was annotated with phrase structure information as part of the Multi-source integrated Platform for Answering Clinical Questions (MiPACQ) project.17 A publication describing that corpus and detailed agreement statistics is forthcoming, but for now we will just mention that the annotations of clinical data following the Penn Treebank phrase structure guidelines were very reliable, with inter-annotator agreements above 90%.
System description
General architecture
The coreference system is a modular extension to the existing cTAKES system. Built within the Unstructured Information Management Architecture (UIMA) engineering framework,18 cTAKES is an open source IE platform for processing clinical notes; its current release identifies clinical NEs, including diseases/disorders, signs/symptoms, anatomical sites, procedures, and medications, and their ontological mapping code, negation status, and context. cTAKES components include modules for preprocessing, sentence detection, tokenization, lemmatization, part-of-speech tagging, shallow parsing, dependency parsing, and named entity recognition (NER). The coreference module follows the NER component, consumes the clinical NEs and pronouns, and links them into coreferential chains. The OpenNLP19 maximum entropy-based constituency parser with a new model trained on clinical data is also wrapped into cTAKES as an additional module to provide syntactic information.
The coreference module is split into three submodules—markable detection, markable consolidation, and chain creation, executed in that order. Since the goal of our coreference module is to be end-to-end, we do not assume gold standard markable mentions, thus the markable detection submodule first generates candidate mentions for subsequent resolution. All clinical NEs discovered by the NER component except medications and all pronouns not referring to persons (including relative pronouns) are considered markable candidates. The decision to exclude person references (‘I,’ ‘we,’ ‘you,’ etc) is based on the observation that people described in the subject of the clinical narratives include usually, if not always, the patient, and occasionally the patient's family members, and the healthcare practitioner. Resolution of pronouns in this constrained setting has been resolved through existing methods.20
The purpose of the markable consolidation submodule is to align the markable candidates created in the previous step to proper noun phrase boundaries identified by the constituency parser. The alignment enables subsequent components to obtain syntactic information (see ‘Features’ section below) from the parse trees. Markables were associated with constituents in the parse tree by finding the constituent that spanned the same word sequence as the markable. In the case where there was not an exact match, the smallest subtree covering the entire markable string was used. For example, if the markable covers the string ‘left shoulder,’ the associated constituent may cover the span ‘his left shoulder’ if the parser returns a structure with no constituent exactly spanning the words ‘left shoulder’ (this is in fact how noun phrase structure is represented in the Penn Treebank13).
The chain creation submodule iterates through each candidate markable and determines its antecedent, if any. The pairwise decisions are then clustered into chains of markables that refer to the same entity. The three types of markables—common noun phrase (NP) markables (eg, ‘pain’), relative pronoun markables (eg, ‘which’), and selected personal pronoun markables (those not referring to people, eg, ‘it’)—are processed differently (demonstrative pronouns were not resolved in this work because of a lack of training data). NP markables undergo a two-stage process. The first stage uses an anaphoricity classifier to determine the probability of the markable itself being coreferential, using a support vector machine (SVM)21 learner trained on the same data (see table 1 for features used). This probability then becomes a feature in the second stage, which estimates the probability of it being coreferential with a candidate antecedent. Candidate antecedents for NP anaphors are drawn from the previous 10 sentences, a window derived from the corpus itself where about 90% of the coreferential markables within a pair occur within a distance of 10 sentences. An alternative to having a preset window is to consider all markables as candidates. Preliminary investigation found that beyond 10 sentences the ratio of false antecedents to true antecedents was too high. These candidates are then further filtered to have matching semantic categories with the proposed anaphor.
Table 1.
Features used by support vector machine
| Feature | Description | |
| Baseline | TokenDistance | Number of tokens between markables |
| SentenceDistance | Number of sentences between markables | |
| ExactMatch | Markables are exact string matches | |
| StartMatch | Markables match at the start | |
| EndMatch | Markables match at the end | |
| SoonStr | Markables match besides determiners | |
| Pronoun1 | Proposed antecedent is pronoun | |
| Pronoun2 | Proposed anaphor is pronoun | |
| Definite1 (A) | Proposed antecedent is definite | |
| Definite2 | Proposed anaphor is definite | |
| Demonstrative2 | Proposed anaphor is demonstrative | |
| NumberMatch (A) | Markables have same number | |
| WnClass (A) | Markables have same named entity semantic category | |
| Alias | Markables have UMLS Concept Unique Identifier (CUI) overlap | |
| ProStr (A) | Markables are same and are pronouns | |
| SoonStrNonpro | Markables are same and are not pronouns | |
| WordOverlap | Markables share at least one word | |
| WordSubstr | With stopwords removed, one is substring of the other | |
| BothDefinites | Both markables are definite | |
| BothPronouns | Both markables are pronouns | |
| Indefinite (A) | Antecedent is indefinite | |
| Pronoun | Antecedent is pronoun and anaphor is not | |
| ClosestComp | Antecedent is closest semantically compatible markable | |
| NPHead (A) | Antecedent span ends noun phrase (NP) span | |
| Anaph | Probability output by anaphoricity classifier (NP markables only) | |
| PermStrDist | String similarity under various permutations | |
| Manually selected syntactic features | PathLength | Length of path between markables in syntax tree |
| NPunderVP1 | Antecedent node is child of VP (verb phrase) node | |
| NPunderVP2 | Anaphor node is child of VP node | |
| NPunderS1 | Antecedent node is child of S (sentence) node | |
| NPunderS2 | Anaphor node is child of S node | |
| NPunderPP1 | Antecedent node is child of PP (prepositional phrase) node | |
| NPunderPP2 | Anaphor node is child of PP node | |
| NPSubj1 | Antecedent node has SBJ (subject) function tag | |
| NPSubj2 | Anaphor node has SBJ function tag | |
| NPSubjBoth | Both nodes have SBJ function tag | |
| Automatically selected syntactic features | Path n-grams | See text in the ‘Features’ section |
Italicized features indicate those directly taken from Ng and Cardie.22 ‘(A)’ indicates the feature is used in the anaphoricity classifier.
Anaphor and antecedent pairs from this filtered set are processed individually and the most probable one according to a probabilistic classifier is proposed as the true antecedent. If the highest probability is below 0.5i, this is interpreted to mean that it is more likely than not that the proposed anaphor is not coreferential with the best antecedent, and thus that the proposed anaphor is in fact not coreferential. This anaphoricity feature method of anaphoricity detection stands in contrast with an anaphoricity filtering approach, first classifying for anaphoricity and only attempting to find an antecedent for those markables surviving the filter. The filtering approach was considered, but it is often only possible to conclusively detect anaphoricity in the context of other referents, in which case the filtering and classification phases become redundant. We prefer an approach in which standalone anaphoricity information is included as a feature in resolution, and a final decision of non-anaphoricity is simply an inability to resolve an entity mention with any previous entity mentions.
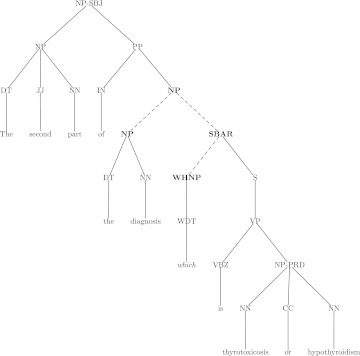
Relative pronouns are resolved using a strict syntactic tree-matching heuristic (see figure 2). Personal pronouns use an SVM classifier as in NP markables except with several modifications—there is no anaphoricity classifier input, the sentence distance is limited to the empirically derived window of three, the linking is greedy (the first link with probability over 0.5 is accepted), and the traversal order follows Hobbs' algorithm.23
Figure 2.
Syntactic tree demonstrating tree matching heuristic for relative pronouns. Constituents in bold represent the markables. Dashed lines represent the section of the tree that must match. Categories follow Penn Treebank definitions: CC, coordinating conjunction; DT, determiner; IN, preposition; JJ, adjective; NN, common noun; NP, noun phrase; PP, prepositional phrase; PRD, predicate function tag; S, sentence; SBAR, subordinate clause; SBJ, subject function tag; WHNP, Wh-noun phrase; WDT, Wh-determiner; VP, verb phrase; VBZ, 3rd person singular present verb.
Features
For each markable pair considered by the pairwise coreference classifiers, a set of features is extracted using many information sources. The features are considered in two categories: baseline features and syntactic features. The syntactic features are further divided into manually selected features and automatically selected features.
The baseline feature set overlaps significantly with the work of Ng and Cardie22 on coreference in the general domain (see table 1). These features are mainly concerned with string similarity and shallow syntactic information with semantic features taking advantage of knowledge resources like UMLS.
Phrase-structure information (syntactic features) can be a valuable additional source of information for coreference resolution, as indicated by its use in 19 of the 21 systems participating in the 2011 CoNLL3 shared task on coreference resolution. As described above, we added a constituency parser module to cTAKES, that is, a wrapper around the OpenNLP parser implementing Ratnaparkhi's maximum entropy parser.24 While this parser is no longer the state of the art, it is still very accurate, relatively fast, and has license compatibility.
We trained the parser model on concatenated general domain and clinical domain data described above. Since the focus of this work is coreference and not parsing, we did not attempt to optimize the parser training regimen nor evaluate the parser. Preliminary results showed that the parser achieves a labeled F score of 0.81, which is lower than the general domain state of the art but difficult to contextualize here due to the lack of other reported work in the clinical domain.
The bottom section of table 1 shows the set of manually selected syntax features. These features are easily extracted from the automatic parses of the clinical notes and are intended to represent linguistically important information very concisely. For example, the NPunder* features indicate whether the markable is a child of a sentence (S) node, verb phrase (VP) node, or prepositional phrase (PP) node. This may be useful for determining the salience of an antecedent based on syntactic position as posited by linguistic theory.23 25
The automatically selected feature set is a first step in the direction of simplifying the task of applying syntactic information to NLP tasks in the clinical domain. It has long been assumed that syntax can improve performance on downstream NLP tasks, but if linguistic expertise is required to select syntactic features on a task-by-task basis (as in the manually selected features), the pace of progress may be considerably slower. We investigated the utility of large numbers of simple features by extracting paths from parser output, using automatic feature selection to reduce sparsity and overfitting.
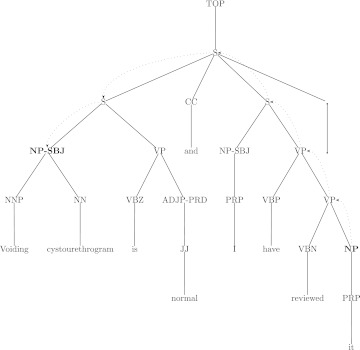
The automatically derived features used here are n-gram segments of the path in the syntax tree between the anaphor and antecedent nodes. Figure 3 illustrates a syntax tree containing an anaphor and its correct antecedent. The dashed arcs in the tree represent the path between the anaphor and antecedent. This path can be represented as the string ‘S<S>S>VP>VP’ with ‘<’ indicating an upward traversal in the tree and ‘>’ indicating a downward traversal toward the anaphor. In the case that the antecedent is not in the same sentence as the anaphor, the tree is extended one level higher, with a node labeled TOP, taken to be the root of all sentence trees in a document. Subpaths of n nodes are extracted from each path, for all n from 3 to 5 in this work. The choice of path fragment lengths is based on the observation that path lengths of <2 are frequent enough and thus may not be very discriminative, and path lengths >5 are quite sparse and may lead to overfitting. The direction of the path is maintained, so that it is clear in any n-gram whether that fragment of the path is going up or down. The set of path 3-grams in figure 3, for example, includes ‘S<S>S,’ ‘S>S>VP,’ and ‘S>VP>VP.’
Figure 3.
Syntactic tree for ‘Voiding cystourethrogram is normal and I have reviewed it.’ Bold indicates markable nodes, while dashed arcs indicate the syntactic path from anaphor to antecedent. Categories follow Penn Treebank definitions: ADJP, adjective phrase; CC, coordinating conjunction; JJ, adjective; NN, common noun; NNP, proper noun; NP, noun phrase; PRD, predicate function tag; PRP, personal pronoun; S, sentence; SBJ, subject function tag; VBN, past participle verb; VBP, non-3rd person singular present verb; VBZ, 3rd person singular present verb; VP, verb phrase.
Feature selection
The Weka machine learning software (version 3.6)26 was used for selecting a set of relevant features from several thousand automatically generated syntactic features. The feature selection (FS) algorithms are grouped into three types. The first type is ranking algorithms, which rank each feature individually according to the χ2 statistic, information gain, gain ratio, or symmetrical uncertainty. For our data, the results of ranking algorithms were very similar. The second FS type is subset selection, which evaluates a subset of features by considering its predictive ability along with its degree of redundancy. Subsets of features that have high correlation with the class label and less redundancy are preferred. Heuristic methods such as Forward or Backward search, Greedy Hill Climbing, Best First Search, and Genetic Algorithm (GA) were employed for searching the best subset. GA performed better than other heuristic searches, since GA is less likely to be trapped at a particular local optimum through crossover and mutation. The last FS type is the wrapper method, which uses a classifier and re-sampling techniques to choose a feature subset.
To get the advantages of both ranking and subset selection algorithms, we used the intersection of these two sets as our FS output. As a result, the selected features are highly relevant as a group and are informative individually. In addition, the total number of features was reduced greatly to about 100, so that the downstream learning could operate faster and more effectively.
Data sampling
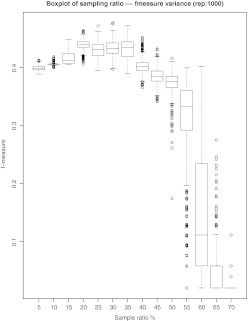
In the pairwise comparison paradigm for coreference resolution used here, each prospective anaphor is compared to all markables before it. In many cases there may be 10 or 20 markables between an anaphor and its closest antecedent. As a result, the training regimen will generate 10 or 20 negative instances for the one ‘true’ (coreferent) pair. In the training corpus as a whole, there are thus many more negative instances than positive ones. This can cause the classifier to be overwhelmed by the dominant negative class and output all negative predictions. One solution to this issue is to use sampling approaches, either down-sampling the dominant class or up-sampling the minority class. Maloof27 showed empirically that down-sampling provides a similar effect as up-sampling, which was confirmed by our sampling experiments. For down-sampling, we sampled without replacement at varied ratios from the negative instances, and repeated 1000 times for variance calculation. For up-sampling, we duplicated the positive examples multiple times. An SVM with a radial basis function (RBF) kernel was trained on each up-sampled or down-sampled set. The model performance was measured by the f-score of predictions on a separate validation set. The f-score is the harmonic mean of recall (R) and precision (P): (F=(2*P*R)/(P+R)), where recall is (R=TP/(TP+FN)) and precision is (P=TP/(TP+FP)).
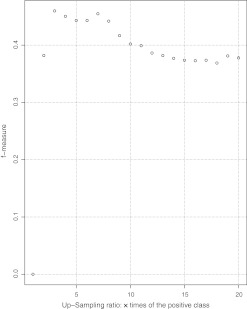
Figures 4 and 5 display the f-score variance of both sampling methods. The best down-sampling ratio is around 20% and its f-score is comparable with that of duplicating the positive instances three times. We used down-sampling at a 20% ratio because it reduced the computational load of training. The variance of f-score at 20% down-sampling is low, which ensures robustness.
Figure 4.
Down-sampling box plot.
Figure 5.
Up-sampling positive examples.
Study design
Experimental setup
The tests are intended to be an end-to-end evaluation of a coreference system, so that no gold standard information about the test set is given to the system. This differs from most work on coreference in the general domain, in which gold standard markables, entity types, parse trees, or other information sources are sometimes supplied. In our system, all features used at test time by the coreference resolution component were automatically extracted from cTAKES modules, including sentence segmentation, word tokenization, word lemmatization, part of speech tagging, NER, constituency parses, and markable identification.
There are some variations between the way markables are annotated and the way the system detects them that are relevant for scoring. For example, the gold standard may only have the head of an NP annotated as the markable, while the system identifies the entire NP. Thus, for scoring it was necessary to correctly align overlapping markables. We computed a minimum edit distance using a dynamic programming algorithm,28 assigning negative points to alignment of non-overlapping spans, zero points to inserting a gap in the alignment (non-alignment of a pair), and positive points to an alignment proportional to the amount of overlap (normalized by the combined span length).
The dataset was divided into separate training and testing data. The testing data contained 36 notes that were not used for anything other than the experiments reported here. The training set is comprised of the remaining 144 notes. Feature selection and preliminary experiments were performed on a subset of the training data.
Configurations
For this first work on the problem of coreference resolution in the clinical domain, we designed experiments to examine three variables.
First, we compared four classifiers on the pairwise classification—SVMs with RBF and linear kernels (LIBSVM29), decision trees, and multilayer perceptrons (Weka26). In all classifier experiments, we used all selected features and the same anaphoricity classifier for the anaphoricity feature, which used an SVM with an RBF kernel with outputs mapped to probabilities.
The second variable we examined was the impact of the three types of features we used: (1) baseline features; (2) manually selected syntactic features; and (3) automatically selected syntactic path n-gram features. We tested variations of the system using baseline only, baseline plus manual syntax, and baseline plus automatic syntax. In all of these experiments we used an SVM with an RBF kernel, as this was the best performing method in development.
The final variable measures differences across institutions. Mayo Clinic and UPMC data differ partially due to conventions at different institutions and partially due to different genres (discharge summaries, pathology reports, etc). In general, one expects improved performance for a given machine learning task when the amount of training data is increased. However, that assumption is based on the data being produced by the same distribution. In the third experiment, we examine the validity of that assumption by training and testing separately on Mayo Clinic and UPMC data only.
Evaluation metrics
We evaluate our system on the commonly used metrics, MUC, B3, CEAF, and BLANC (for details, consult Zheng et al2). MUC compares the equivalence classes formed by the individual coreference pairs. Following the standard information retrieval paradigm, MUC recall errors are calculated by the number of missing pair links in the system output, and MUC precision errors are calculated by reversing the role of system output and gold standard. This metric does not give credit for recognizing singletons, markables that do not refer back to another markable. B3 is designed to address this shortcoming. It also follows the information retrieval approach of calculating precision and recall. CEAF has been developed to address an issue with B3 that an entity can be used more than once during score calculation. CEAF aligns the gold standard chains and the system-generated chains to solve the problem of reusing entities in B3. BLANC has been recently proposed to address the shortcomings of MUC, B3, and CEAF, by implementing a Rand index style metric.
Results
Table 2 shows the results of all three experiments in three sections. The top rows indicate the results of the classifier experiments showing a small but consistent advantage for the SVM with an RBF kernel.
Table 2.
Results
| Classifier | Dataset | Features | BLANC | B3 | MUC | CEAF |
| RBF | All | All | 0.596 | 0.690 | 0.352 | 0.486 |
| Linear | All | All | 0.592 | 0.695 | 0.329 | 0.474 |
| Decision tree | All | All | 0.582 | 0.680 | 0.310 | 0.464 |
| Perceptron | All | All | 0.593 | 0.677 | 0.332 | 0.477 |
| RBF | All | Baseline | 0.577 | 0.657 | 0.301 | 0.461 |
| RBF | All | Baseline+manual | 0.589 | 0.680 | 0.336 | 0.476 |
| RBF | All | Baseline+auto | 0.575 | 0.666 | 0.302 | 0.458 |
| RBF | Mayo | All | 0.642 | 0.712 | 0.472 | 0.551 |
| RBF | UPMC | All | 0.579 | 0.673 | 0.304 | 0.462 |
Mayo, Mayo Clinic; RBF, radial basis function; UPMC, University of Pittsburgh Medical Center.
The middle rows of the table show the results of the feature set experiments. Baseline results were 2–5 points lower than the result using all the features (from the first experiment). The manually selected syntax features (referred to as ‘manual’ in table 2) showed a consistent improvement over the baseline features, while the automatically selected n-gram path features (referred to as ‘auto’ in table 2) did not seem to be a clear improvement on their own. However, the manual and automatic features together (as in the first experiment) performed better than any partial feature set.
The bottom section of the table shows the results of the experiment broken down by data source. The system trained and tested on only Mayo Clinic records scored markedly better than the system trained and tested on all the data. The system trained and tested on UPMC data, on the other hand, showed much lower performance than the system trained on all the data.
We randomly selected 12 test documents (33%) from the total of 36 test documents and manually reviewed the disagreements (N=109) (table 3). The most frequent error source is sentence distance between the two coreferential markables exceeding the window of 10 sentences (37.61%). This is observed in documents where the clinical mentions in the Final Diagnosis and History of Present Illness sections point to the same entity, yet the sections are far apart. The next most frequent error (19.27%) is due to NER, that is, the system failed to discover the markables. For example, two mentions of ‘peritubular capillaries’ are not discovered as NEs and therefore not linked in a coreferential relation. The inability to recognize some synonyms and use ontology knowledge to relate more general terms to more specific terms is another source of errors (10.09%), for example, ‘disease’ is not linked to ‘unresectable non-small-cell lung cancer’ although both mentions refer to the same entity and are ontologically related.
Table 3.
Distribution of manually reviewed disagreements (a sample of 109 randomly selected disagreements from 12 test documents)
| Type of error | Raw count | % |
| Sentence distance | 41 | 37.61 |
| Named entity recognition (NER) errors | 21 | 19.27 |
| Classifier error | 9 | 8.26 |
| Annotation error (missing in gold standard) | 9 | 8.26 |
| Synonyms | 6 | 5.50 |
| Demonstratives | 6 | 5.50 |
| Ontology knowledge | 5 | 4.59 |
| Miscellaneous errors (eg, word sense disambiguation, linguistic errors, etc) | 12 | 11.01 |
| Total | 109 | 100 |
Discussion
This work describes one of the first systems to perform end-to-end coreference in the clinical domain, and is the first to be trained and tested on the ODIE dataset. This can serve as a baseline result which can be a comparison point for many of the systems in the 2011 i2b2/VA NLP challenge (which uses the ODIE corpus as part of its challenge data). The module and models were released in December 2011 as part of the open source cTAKES project under an Apache license. The ODIE corpus will be available under a data use agreement as well. In addition, we are preparing to release the evaluation code used here into the public domain to make it easier for different groups to compare results and to encourage the use of multiple metrics for the notoriously difficult task of scoring coreference resolution.
The main source of error in this system was sentence distances exceeding the empirically derived limit of 10 sentences between markables. Of course, since sentence distance is an early filter, many of those links may have been missed for other reasons, but it is still an important problem to solve because we cannot get these links right if we do not get a chance to score them. Simply increasing the limit will greatly increase the number of negative examples, making the training data even more imbalanced and potentially increasing false positives. A possible solution to this problem is hinted at by the fact that coreferent markables sometimes occurred in predictable (although distant) sections of the note. If we first segment a clinical note into sections and model the sections as discourse units, it may be easier to represent the way that coreference occurs between sections that are sometimes far away from each other.
Future work will explore the parser performance on clinical data and methods for optimizing the in-domain performance using both domain specific and open domain data. Future work will also evaluate other means of applying syntactic information to the coreference problem, for example, by using tree kernel approaches30 in combination with feature-based kernels.31 We will also investigate the use of dependency structure in addition to phrase structure information.
Finally, this work is difficult to compare to general domain work for many reasons, most importantly because of data differences and task setup differences. The CoNLL 2011 challenge track allowed outside resources analogous to UMLS and also did not provide gold standard mentions. In those trials, MUC scores ranged from 19.98 to 59.95, B3 scores from 50.46 to 68.79, CEAF scores from 31.68 to 56.37, and BLANC scores from 51.12 to 73.71.
Conclusion
This paper described one of the first coreference systems for the clinical domain, which can be used as a baseline for future work. The system, released open source in December 2011, was trained and tested on the ODIE corpus and produced results which are better than might be expected given the difficulty of the task and lack of work in this domain.
Footnotes
Funding: This work was funded by NIH grant R01 CA127979 (PI Crowley), RC1LM010608 (PI Savova), SHARP award 90TR0002 (PI Chute), and U54LM008748 (PI Kohane).
Competing interests: None.
Contributors: All authors of this manuscript contributed to (1) conception and design, acquisition of data, or analysis and interpretation of data, (2) drafting the article or revising it critically for important intellectual content, and (3) final approval of the version published.
Provenance and peer review: Not commissioned; externally peer reviewed.
Of course one could treat this value as a threshold parameter to the algorithm and experiment with values providing the best results; in this case values >0.5 were interpreted in the literal probabilistic sense of being more likely than not.
References
- 1.Mitkov R. Anaphora resolution. Pearson Education. 1st edn Edinburgh, UK: Longman, 2002 [Google Scholar]
- 2.Zheng J, Chapman W, Crowley R, et al. Coreference resolution: a review of general methodologies and applications in the clinical domain. J Biomed Inform 2011;44:1113–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pradhan S, Ramshaw L, Marcus M, et al. CoNLL-2011 shared task: modeling unrestricted coreference in OntoNotes. Proceedings of the 15th Conference on Computational Natural Language Learning Shared Task. Portland, Oregon: Association of computational Linguistics, 2011:1–27 [Google Scholar]
- 4.Coden A, Savova G, Sominsky I, et al. Automatically extracting cancer disease characteristics from pathology reports into a cancer disease knowledge model. J Biomed Inform 2009;42:937–49 [DOI] [PubMed] [Google Scholar]
- 5.Roberts A, Gaizauskas R, Hepple M, et al. Building a semantically annotated corpus of clinical text. J Biomed Inform 2009;42:950–66 [DOI] [PubMed] [Google Scholar]
- 6.Hahn U, Romacker M, Schulz S. Medsyndikate-a natural language system for the extraction of medical information from findings report. Int J Med Inform 2002;67:63–74 [DOI] [PubMed] [Google Scholar]
- 7.Savova G, Chapman W, Zheng J, et al. Anaphoric relations in the clinical narrative: corpus creation. J Am Med Inform Assoc 2011;18:459–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Savova G, Masanz J, Ogren P, et al. Mayo clinical text analysis and knowledge extraction system (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010;17:507–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clinical Text Analysis and Knowledge Extraction System (cTAKES). http://ohnlp.svn.sourceforge.net/viewvc/ohnlp/trunk/cTAKES/ (accessed 6 Dec 2011). [DOI] [PMC free article] [PubMed]
- 10.ODIE Ontology Development and Information Extraction (ODIE) toolset. http://www.bioontology.org/ODIE (accessed 6 Dec 2011).
- 11.MUC-7 MUC-7 Coreference task definition. http://www-nlpir.nist.gov/related_projects/muc/proceedings/co_task.html (accessed 6 Dec 2011).
- 12.Bodenreider O, McCray A. Exploring semantic groups through visual approaches. J Biomed Inform 2003;36:414–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marcus M, Santorini B, Marcinkiewicz M. Building a large annotated corpus of English: the Penn treebank. Comput Ling 1994;19:313–30 [Google Scholar]
- 14.Vadas D, Curran J. Adding noun phrase structure to the Penn Treebank. Proceedings of the Association of Computational Linguistics. Prague, Czech Republic, 2007:240–7 [Google Scholar]
- 15.Ely J, Osheroff J, Ebel M, et al. Analysis of questions asked by family doctors regarding patient care. BMJ 1999;319:358–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Medpedia http://www.medpedia.com/ pp 10–18 (accessed 6 Dec 2011).
- 17.Cairns B, Nielsen R, Masanz J, et al. The MiPACQ clinical question answering system. Proceedings of the American Medical Informatics Association Annual Symposium. Washington, DC, 2011:170–80 [PMC free article] [PubMed] [Google Scholar]
- 18.UIMA http://uima.apache.org (accessed 6 Dec 2011).
- 19.OpenNLP http://opennlp.sourceforge.net/ (accessed 6 Dec 2011).
- 20.Harkema H, Dowling J, Thornblade T, et al. ConText: an algorithm for determining negation, experiencer, and temporal status from clinical reports. J Biomed Inform 2009;42:839–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cortes C, Vapnik V. Support-vector networks. Mach Learn 1995;10:273–97 [Google Scholar]
- 22.Ng V, Cardie C. Improving machine learning approaches to coreference resolution. Proceedings of the 40th annual meeting Association for Computational Linguistics. Philadelphia, PA, 2002:104–11 [Google Scholar]
- 23.Hobbs J. Resolving pronoun references. Lingua 1978;44:311–38 [Google Scholar]
- 24.Ratnaparkhi A. A linear observed time statistical parser based on maximum entropy models. Proceedings of the Second Conference on Empirical Methods in Natural Language Processing (EMNLP). Providence, Rhode Island: Brown University, 1997:1–10 [Google Scholar]
- 25.Gundel J, Hedberg N, Zacharski R. Cognitive status and the form of referring expressions in discourse. Language 1993;69:274–307 [Google Scholar]
- 26.Hall M, Frank E, Holmes G, et al. The WEKA data mining software: an update. SIGKDD Explorations 2009;11 [Google Scholar]
- 27.Maloof M. Learning when data sets are imbalanced and when costs are unequal and unknown. Proceedings of the International Conference of Machine Learning Workshop on Learning from Imbalanced Data Sets. Washington, DC, 2003 [Google Scholar]
- 28.Cormen T, Leiserson C, Rivest R, et al. Introduction to Algorithms. Cambridge, MA: Massachusetts Instutute of Technology, 2009 [Google Scholar]
- 29.LIBSVM http://www.csie.ntu.edu.tw/∼cjlin/libsvm/ (accessed 6 Dec 2011).
- 30.Collins M, Duffy M. Convolution Kernels for Natural Language. Advances in Neural Information Processing Systems Conference (NIPS). London, UK: MIT Press, 2001:625–32 [Google Scholar]
- 31.Nguyen T, Moschitti A, Riccardi G. Convolution kernels on constituent, dependency and sequential structures for relation extraction. Proceedings of the 2009 Conference on Empirical Methods in Natural Language Processing; 6–7 August 2009, Singapore. 2009:1378–87 [Google Scholar]