Abstract
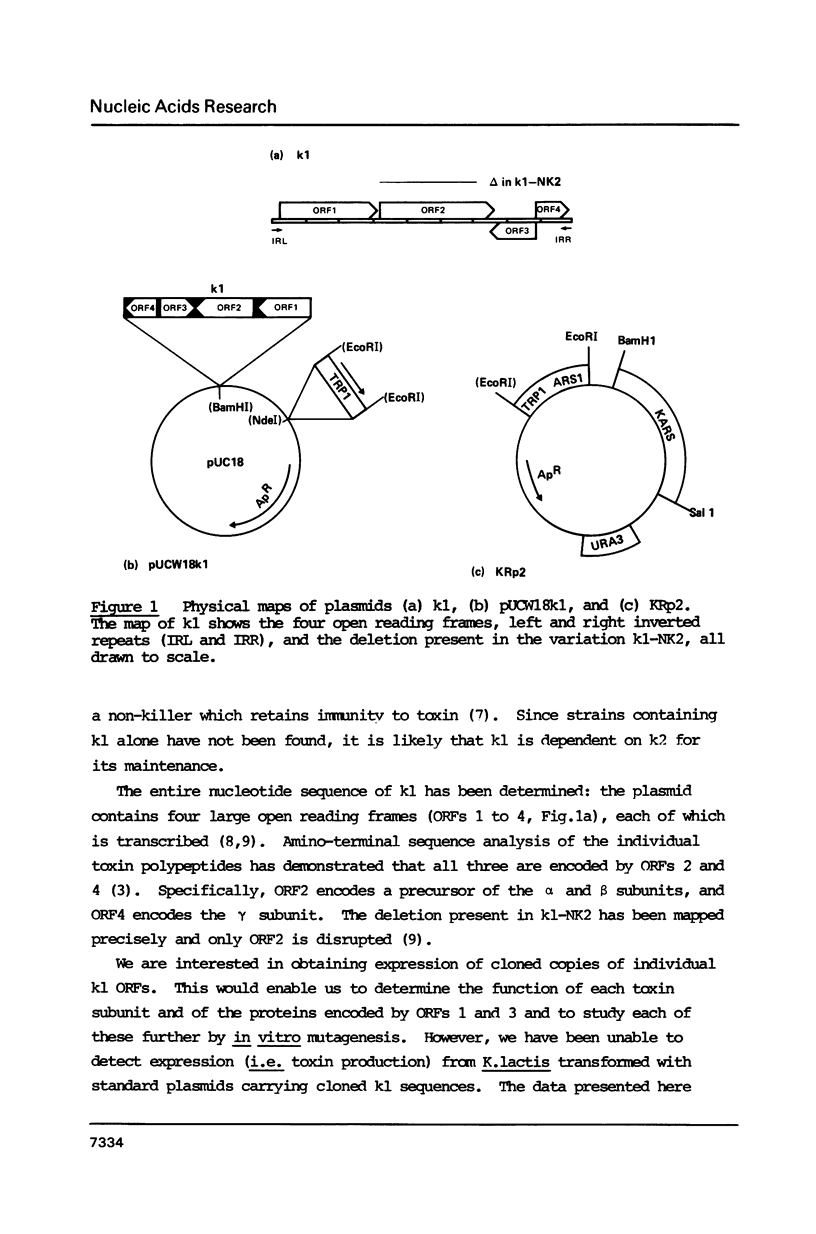
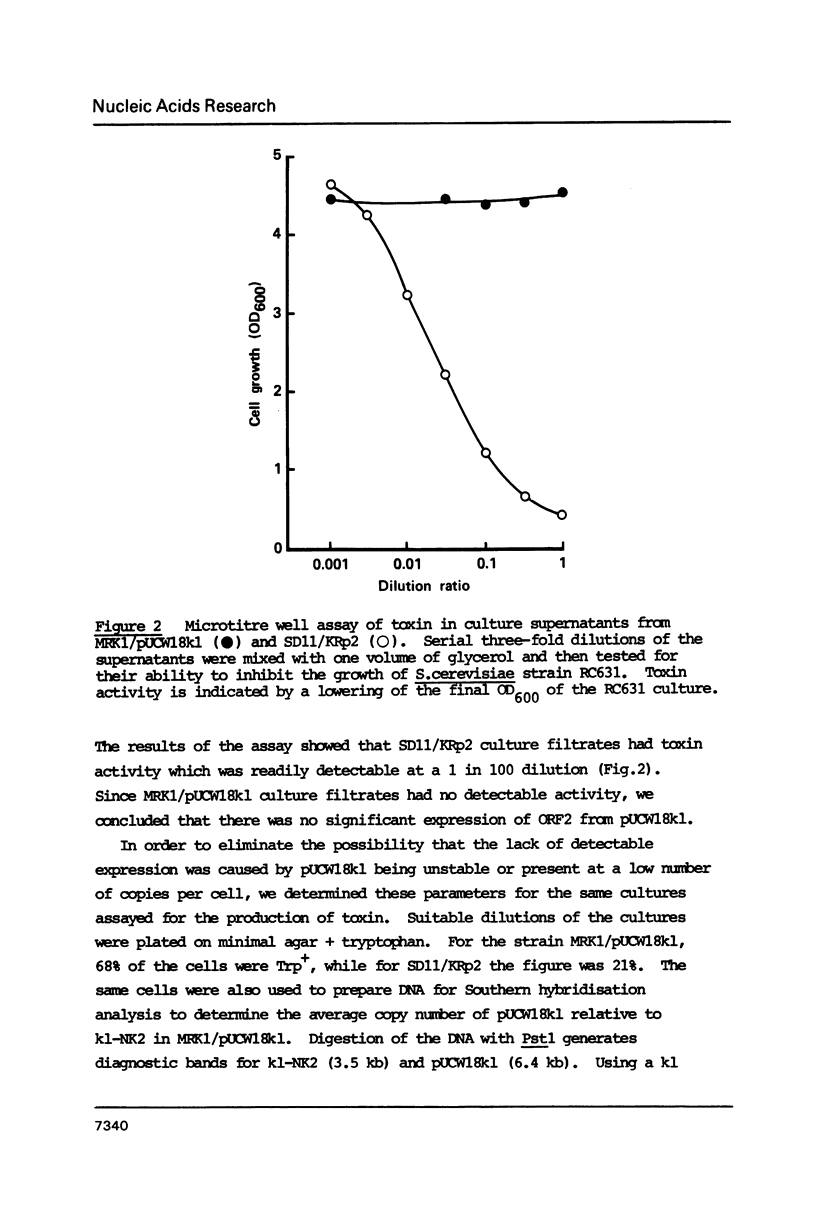
The killer toxin of Kluyveromyces lactis consists of three polypeptides encoded by the linear plasmid k1. We re-introduced the entire k1 sequence, cloned on a circular replicating plasmid, into K. lactis strains lacking k1, and found that the resulting transformants did not produce toxin. The barrier to expression was found to be transcriptional: the four transcripts of native k1 were absent, and instead shorter, aberrant k1 transcripts were made. We determined the precise initiation sites of the four transcripts of native k1: these had very short untranslated leaders and mapped about 14bp downstream of an "upstream conserved sequence" (UCS). It appears that k1 has novel promoters which are inactive on circular plasmids which replicate in the nucleus. This is consistent with the suggestion that native k1 resides in the cytoplasm.
Full text
PDF








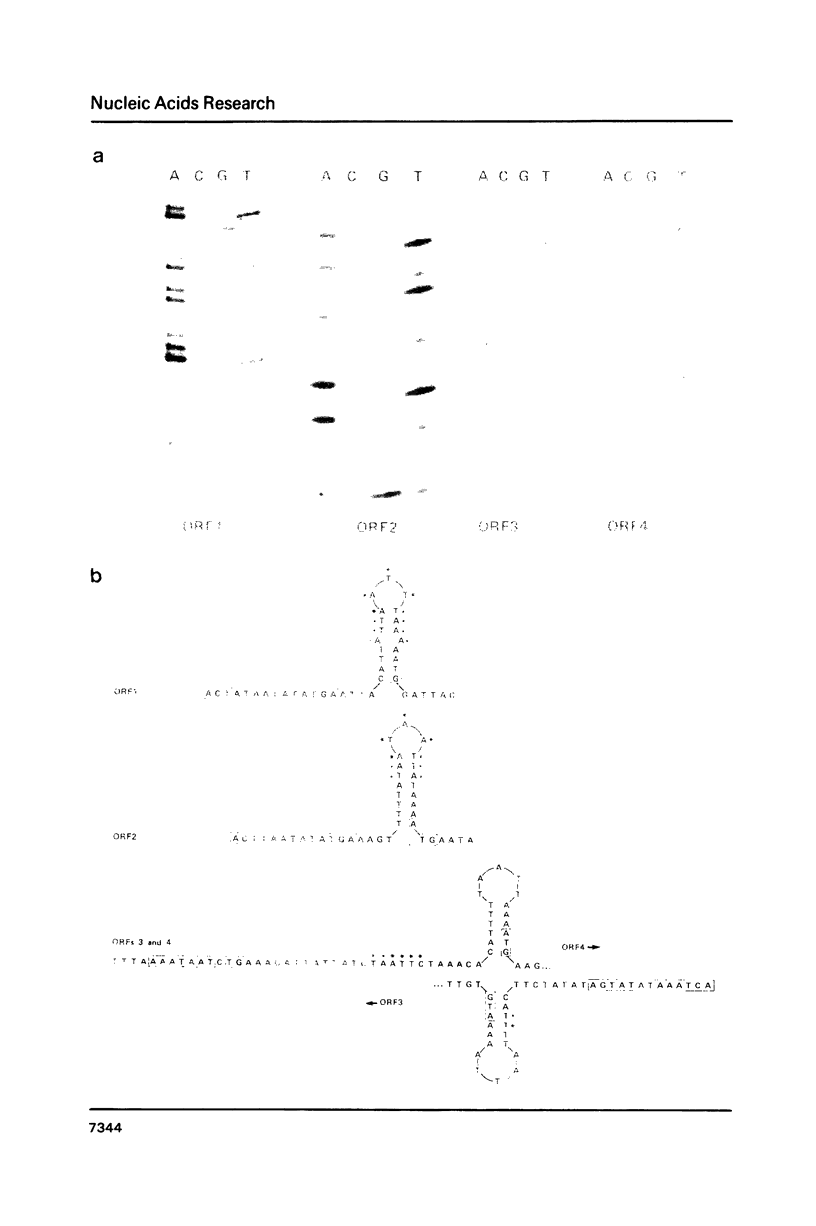






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bernad A., Zaballos A., Salas M., Blanco L. Structural and functional relationships between prokaryotic and eukaryotic DNA polymerases. EMBO J. 1987 Dec 20;6(13):4219–4225. doi: 10.1002/j.1460-2075.1987.tb02770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostian K. A., Lemire J. M., Halvorson H. O. Physiological control of repressible acid phosphatase gene transcripts in Saccharomyces cerevisiae. Mol Cell Biol. 1983 May;3(5):839–853. doi: 10.1128/mcb.3.5.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner D. G., Shaw W. V. The use of synthetic oligonucleotides with universal templates for rapid DNA sequencing: results with staphylococcal replicon pC221. EMBO J. 1985 Feb;4(2):561–568. doi: 10.1002/j.1460-2075.1985.tb03665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breunig K. D., Dahlems U., Das S., Hollenberg C. P. Analysis of a eukaryotic beta-galactosidase gene: the N-terminal end of the yeast Kluyveromyces lactis protein shows homology to the Escherichia coli lacZ gene product. Nucleic Acids Res. 1984 Mar 12;12(5):2327–2341. doi: 10.1093/nar/12.5.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Chen W., Struhl K. Yeast mRNA initiation sites are determined primarily by specific sequences, not by the distance from the TATA element. EMBO J. 1985 Dec 1;4(12):3273–3280. doi: 10.1002/j.1460-2075.1985.tb04077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Fleig U. N., Pridmore R. D., Philippsen P. Construction of LYS2 cartridges for use in genetic manipulations of Saccharomyces cerevisiae. Gene. 1986;46(2-3):237–245. doi: 10.1016/0378-1119(86)90408-7. [DOI] [PubMed] [Google Scholar]
- Gunge N., Murata K., Sakaguchi K. Transformation of Saccharomyces cerevisiae with linear DNA killer plasmids from Kluyveromyces lactis. J Bacteriol. 1982 Jul;151(1):462–464. doi: 10.1128/jb.151.1.462-464.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunge N., Sakaguchi K. Intergeneric transfer of deoxyribonucleic acid killer plasmids, pGKl1 and pGKl2, from Kluyveromyces lactis into Saccharomyces cerevisiae by cell fusion. J Bacteriol. 1981 Jul;147(1):155–160. doi: 10.1128/jb.147.1.155-160.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunge N., Tamaru A., Ozawa F., Sakaguchi K. Isolation and characterization of linear deoxyribonucleic acid plasmids from Kluyveromyces lactis and the plasmid-associated killer character. J Bacteriol. 1981 Jan;145(1):382–390. doi: 10.1128/jb.145.1.382-390.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunge N., Yamane C. Incompatibility of linear DNA killer plasmids pGKL1 and pGKL2 from Kluyveromyces lactis with mitochondrial DNA from Saccharomyces cerevisiae. J Bacteriol. 1984 Aug;159(2):533–539. doi: 10.1128/jb.159.2.533-539.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentschel C., Irminger J. C., Bucher P., Birnstiel M. L. Sea urchin histone mRNA termini are located in gene regions downstream from putative regulatory sequences. Nature. 1980 May 15;285(5761):147–151. doi: 10.1038/285147a0. [DOI] [PubMed] [Google Scholar]
- Hishinuma F., Nakamura K., Hirai K., Nishizawa R., Gunge N., Maeda T. Cloning and nucleotide sequences of the linear DNA killer plasmids from yeast. Nucleic Acids Res. 1984 Oct 11;12(19):7581–7597. doi: 10.1093/nar/12.19.7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G. H., Leavitt M. C., Ito J. Yeast killer plasmid pGKL1 encodes a DNA polymerase belonging to the family B DNA polymerases. Nucleic Acids Res. 1987 Nov 11;15(21):9088–9088. doi: 10.1093/nar/15.21.9088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y., Hirai K., Hishinuma F. The yeast linear DNA killer plasmids, pGKL1 and pGKL2, possess terminally attached proteins. Nucleic Acids Res. 1984 Jul 25;12(14):5685–5692. doi: 10.1093/nar/12.14.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipman D. J., Maizel J. Comparative analysis of nucleic acid sequences by their general constraints. Nucleic Acids Res. 1982 Apr 24;10(8):2723–2739. doi: 10.1093/nar/10.8.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marczynski G. T., Jaehning J. A. A transcription map of a yeast centromere plasmid: unexpected transcripts and altered gene expression. Nucleic Acids Res. 1985 Dec 9;13(23):8487–8506. doi: 10.1093/nar/13.23.8487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil J. B., Smith M. Saccharomyces cerevisiae CYC1 mRNA 5'-end positioning: analysis by in vitro mutagenesis, using synthetic duplexes with random mismatch base pairs. Mol Cell Biol. 1985 Dec;5(12):3545–3551. doi: 10.1128/mcb.5.12.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa O., Sakaguchi K., Gunge N. Curing of the killer deoxyribonucleic acid plasmids of Kluyveromyces lactis. J Bacteriol. 1981 Dec;148(3):988–990. doi: 10.1128/jb.148.3.988-990.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sor F., Wésolowski M., Fukuhara H. Inverted terminal repetitions of the two linear DNA associated with the killer character of the yeast Kluyveromyces lactis. Nucleic Acids Res. 1983 Aug 11;11(15):5037–5044. doi: 10.1093/nar/11.15.5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreekrishna K., Webster T. D., Dickson R. C. Transformation of Kluyveromyces lactis with the kanamycin (G418) resistance gene of Tn903. Gene. 1984 Apr;28(1):73–81. doi: 10.1016/0378-1119(84)90089-1. [DOI] [PubMed] [Google Scholar]
- Stam J. C., Kwakman J., Meijer M., Stuitje A. R. Efficient isolation of the linear DNA killer plasmid of Kluyveromyces lactis: evidence for location and expression in the cytoplasm and characterization of their terminally bound proteins. Nucleic Acids Res. 1986 Sep 11;14(17):6871–6884. doi: 10.1093/nar/14.17.6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark M. J., Boyd A. The killer toxin of Kluyveromyces lactis: characterization of the toxin subunits and identification of the genes which encode them. EMBO J. 1986 Aug;5(8):1995–2002. doi: 10.1002/j.1460-2075.1986.tb04455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark M. J., Mileham A. J., Romanos M. A., Boyd A. Nucleotide sequence and transcription analysis of a linear DNA plasmid associated with the killer character of the yeast Kluyveromyces lactis. Nucleic Acids Res. 1984 Aug 10;12(15):6011–6030. doi: 10.1093/nar/12.15.6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark M. J. Resolution of sequence discrepancies in the ORF1 region of the Kluyveromyces lactis plasmid k1. Nucleic Acids Res. 1988 Jan 25;16(2):771–771. doi: 10.1093/nar/16.2.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B. W. The replication of adenovirus DNA with purified proteins. Cell. 1983 Nov;35(1):7–9. doi: 10.1016/0092-8674(83)90201-5. [DOI] [PubMed] [Google Scholar]
- Sugisaki Y., Gunge N., Sakaguchi K., Yamasaki M., Tamura G. Kluyveromyces lactis killer toxin inhibits adenylate cyclase of sensitive yeast cells. Nature. 1983 Aug 4;304(5925):464–466. doi: 10.1038/304464a0. [DOI] [PubMed] [Google Scholar]
- Thompson A., Oliver S. G. Physical separation and functional interaction of Kluyveromyces lactis and Saccharomyces cerevisiae ARS elements derived from killer plasmid DNA. Yeast. 1986 Sep;2(3):179–191. doi: 10.1002/yea.320020306. [DOI] [PubMed] [Google Scholar]
- Tokunaga M., Wada N., Hishinuma F. Expression and identification of immunity determinants on linear DNA killer plasmids pGKL1 and pGKL2 in Kluyveromyces lactis. Nucleic Acids Res. 1987 Feb 11;15(3):1031–1046. doi: 10.1093/nar/15.3.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson D. H. The yeast ARS element, six years on: a progress report. Yeast. 1985 Sep;1(1):1–14. doi: 10.1002/yea.320010102. [DOI] [PubMed] [Google Scholar]