Abstract
Protein folding is often hampered by protein aggregation, which can be prevented by a variety of chaperones in the cell. A dataset that evaluates which chaperones are effective for aggregation-prone proteins would provide an invaluable resource not only for understanding the roles of chaperones, but also for broader applications in protein science and engineering. Therefore, we comprehensively evaluated the effects of the major Escherichia coli chaperones, trigger factor, DnaK/DnaJ/GrpE, and GroEL/GroES, on ∼800 aggregation-prone cytosolic E. coli proteins, using a reconstituted chaperone-free translation system. Statistical analyses revealed the robustness and the intriguing properties of chaperones. The DnaK and GroEL systems drastically increased the solubilities of hundreds of proteins with weak biases, whereas trigger factor had only a marginal effect on solubility. The combined addition of the chaperones was effective for a subset of proteins that were not rescued by any single chaperone system, supporting the synergistic effect of these chaperones. The resource, which is accessible via a public database, can be used to investigate the properties of proteins of interest in terms of their solubilities and chaperone effects.
Keywords: chaperonin, Hsp60, Hsp70, aggregates, proteome
Newly synthesized proteins emerging from the ribosome must fold into their native structures to acquire their functions (1). Although protein folding is a spontaneous process, in which the amino acid sequence dictates the native structure (2), nonproductive intermolecular interactions result in aggregate formation (1, 3, 4). Because protein stability is marginal in general, proteins always have the inherent risk of aggregation (3, 4). Perturbations of cellular proteostasis (4), such as cellular stresses or heterologous recombinant protein expression, often cause protein aggregation in the cell and the formation of inclusion bodies, which are one of the bottlenecks in various types of biological research, from traditional molecular biology to modern synthetic biology (3, 4).
To counteract the inevitable tendency toward protein aggregation, cells have evolved a variety of chaperones (5). Chaperones prevent irreversible aggregate formation by binding nonnative proteins and then assisting with productive folding (3, 4). Indeed, when more than 3,000 Escherichia coli proteins were synthesized by reconstituted cell-free translation under chaperone-free conditions, a substantial fraction of the proteome, a quarter of the proteins quantified, was aggregation-prone (6), implying that chaperones are required to rescue the aggregation-prone proteins.
The best-characterized chaperones are those in E. coli (4). In E. coli, three major chaperone systems are known to be involved in the folding of newly synthesized proteins in the cytoplasm (4). The first is trigger factor (TF), which directly associates with the ribosome and interacts with nascent chains cotranslationally (7). The second is DnaK, a member of the Hsp70 family that is widely conserved in all kingdoms of life and is considered to act on a broad spectrum of proteins in cooperation with the cochaperones, DnaJ and GrpE (4, 8, 9). The third is GroEL, which belongs to a well-conserved chaperonin family (4, 10–12). In the presence of ATP, GroEL forms a large cylindrical complex with the cochaperonin GroES, which encapsulates substrate proteins within its cavity to assist with folding (4, 10–12). These three chaperone systems are known to act cooperatively: TF and DnaK exhibit overlapping cotranslational roles in vivo (13–15). Overexpression of DnaK/DnaJ and GroEL/GroES in E. coli rpoH mutant cells, which are deficient in heat-shock proteins, prevents aggregation of newly translated proteins (16). GroEL is believed to be involved in folding after the polypeptides are released from the ribosome, although the possible cotranslational involvement of GroEL has also been reported (17–20).
Over the past two decades many efforts have been focused on elucidating the mechanism of each chaperone as a molecular machine (4, 9). The next question to be addressed is which chaperone is effective for certain proteins. A dataset on the substrate biases for each chaperone would provide an invaluable resource, not only for understanding the role of chaperones in protein folding but also for applications to protein science and engineering. However, the mechanisms by which chaperones recognize their substrates are not fully understood. Although global analyses of chaperone–protein interactions in cells have been conducted (4, 7, 8, 21–24), no systematic evaluation of chaperone effects under uniform conditions has been performed.
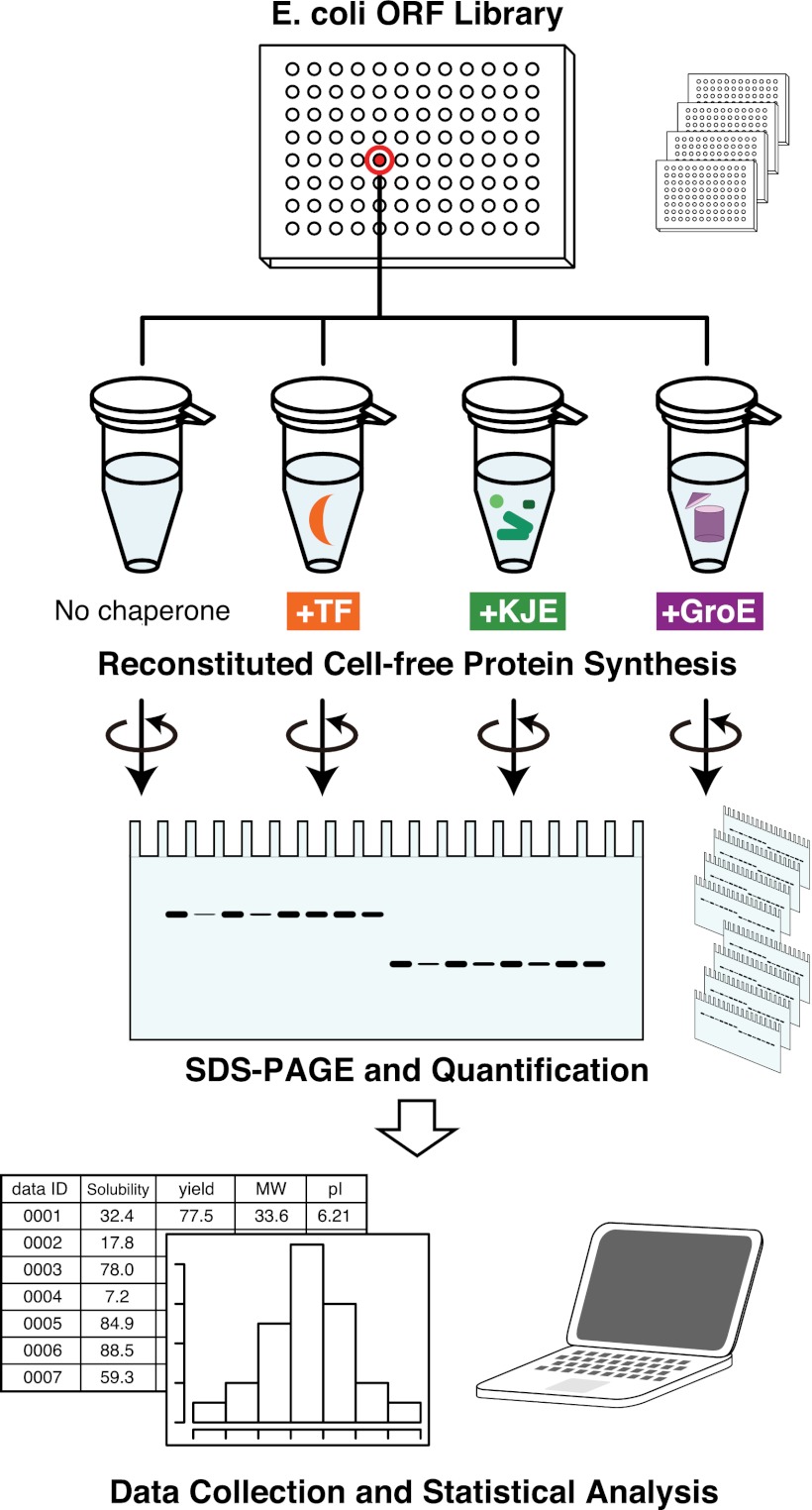
In this context, a reconstituted cell-free translation system that only contains the essential factors for protein synthesis is ideal to evaluate the role of chaperones, because the cell-free system is chaperone-free. Therefore, we chose an E. coli reconstituted cell-free system, the PURE system (25, 26). Using the PURE system, we previously analyzed the aggregation propensities of all E. coli proteins under a chaperone-free condition (6). In addition, the PURE system has been used to investigate the role of GroE in the folding of some newly synthesized proteins (19, 20, 27). We have extended those previous studies to a comprehensive evaluation of the major E. coli chaperones, TF, DnaK/DnaJ/GrpE (DnaKJE), and GroEL/GroES (GroE), on ∼800 aggregation-prone, cytosolic E. coli proteins. The scheme of the global analysis is shown in Fig. 1: the one-by-one synthesis of individual aggregation-prone proteins in the presence of each chaperone, the quantification of solubility by a centrifugation-based assay, and the statistical analyses of the collected data. This is an “in vitro (reconstituted) proteome” approach, in which the properties of thousands of proteins, including proteins with extremely low abundance in cells, are investigated individually after cell-free translation. This large dataset is an invaluable resource for investigations of the properties of proteins of interest. In addition, the statistical analysis of the data revealed many intriguing properties of chaperones in terms of substrate recognition.
Fig. 1.
An in vitro expressed proteome approach for global aggregation analysis. Schematic illustration of the experiment. Seven hundred and ninety-two aggregation-prone proteins were separately expressed with a reconstituted cell-free translation system, the PURE system, in the absence and the presence of the major E. coli chaperones (trigger factor, TF; DnaK/DnaJ/GrpE, KJE, GroEL/GroES, GroE). Each translation product was labeled with [35S]methionine. After translation, the uncentrifuged total fraction (Total) and the supernatant fraction after centrifugation (Sup) were electrophoresed and quantified by autoradiography. The ratio of the translation products in the Total and Sup fractions was defined as the solubility, which represented the aggregation propensity of the protein. The dataset (∼800 × 4) obtained from this experiment was subjected to statistical analyses to investigate the relationship between the effects of chaperones and the various properties of the proteins.
Results
Global Aggregation Analysis in the Presence of E. coli Chaperones.
In our previous global aggregation analysis, our aggregation-prone group was defined as the proteins with less than 30% solubility (6). The aggregation-prone group (1,234 proteins) includes proteins that are not located in the cytosol, such as integral membrane and periplasmic proteins. We limited the present analysis to the aggregation-prone E. coli proteins that are predicted to reside in the cytosol (792 proteins), because we used the major cytosolic E. coli chaperones, TF, DnaKJE and GroE, in the following analysis.
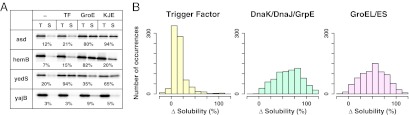
We synthesized all of the cytosolic aggregation-prone proteins by the PURE system at 37 °C for 60 min, in the absence or presence of each chaperone. Each chaperone was added at the approximate physiological concentration, based on previous assessments of chaperone activities under cell-free conditions (19, 28, 29). The in vitro activities of each chaperone were confirmed (Fig. S1 and Materials and Methods). The [35S]methionine-labeled proteins were electrophoresed on SDS gels and quantified (Fig. 2A). The aggregation propensity was examined by a centrifugation assay (6). Briefly, an aliquot of the translation mixture was centrifuged, and then the supernatant fractions were electrophoresed and quantified (Fig. 1). The solubility was defined as the proportion of the protein in the supernatant fraction to that in the uncentrifuged total protein sample. Typical results are shown in Fig. 2A. Almost all of the proteins (788 of 792) were quantified for their solubilities under each condition. Experimental error (defined as a SD) in the assay has been previously estimated to be 10% (6, 27). Indeed, the analysis in the absence of chaperones was reproducible, because the SD of the solubilities between the current and previous data (6) was less than 10% on average (Fig. S2A), and the solubilities of more than 90% of the translated proteins in the absence of a chaperone (718 of 788) were less than 30% (Fig. S2B).
Fig. 2.
Global analysis of chaperone effects on the prevention of aggregate formation. (A) Typical examples. SDS-gels of four aggregation-prone E. coli cytosolic proteins (asd, hemB, yedS, and yajB) in the absence and the presence of the chaperones are shown. The numbers below the electrophoretic pattern indicate the solubility values, calculated by the ratio of the amount of translation products in the Sup (S) and Total (T) fractions. (B) Histograms of solubilities in the presence of three E. coli chaperones. The aggregation-prevention effect is represented as Δsolubility, defined by subtracting the solubilities in the absence of chaperones from those in the presence of each chaperone (see Fig. S2B for raw data).
Because all of the translated proteins belong to the aggregation-prone group, which might occlude the exit tunnel in the ribosome, one might ask whether chaperones could facilitate protein synthesis by preventing aggregation. However, the presence of the chaperones had little influence on the yields of translated proteins (Fig. S3), suggesting that the overall translation efficiencies were not accelerated by any of the chaperones.
Overview of the Dataset.
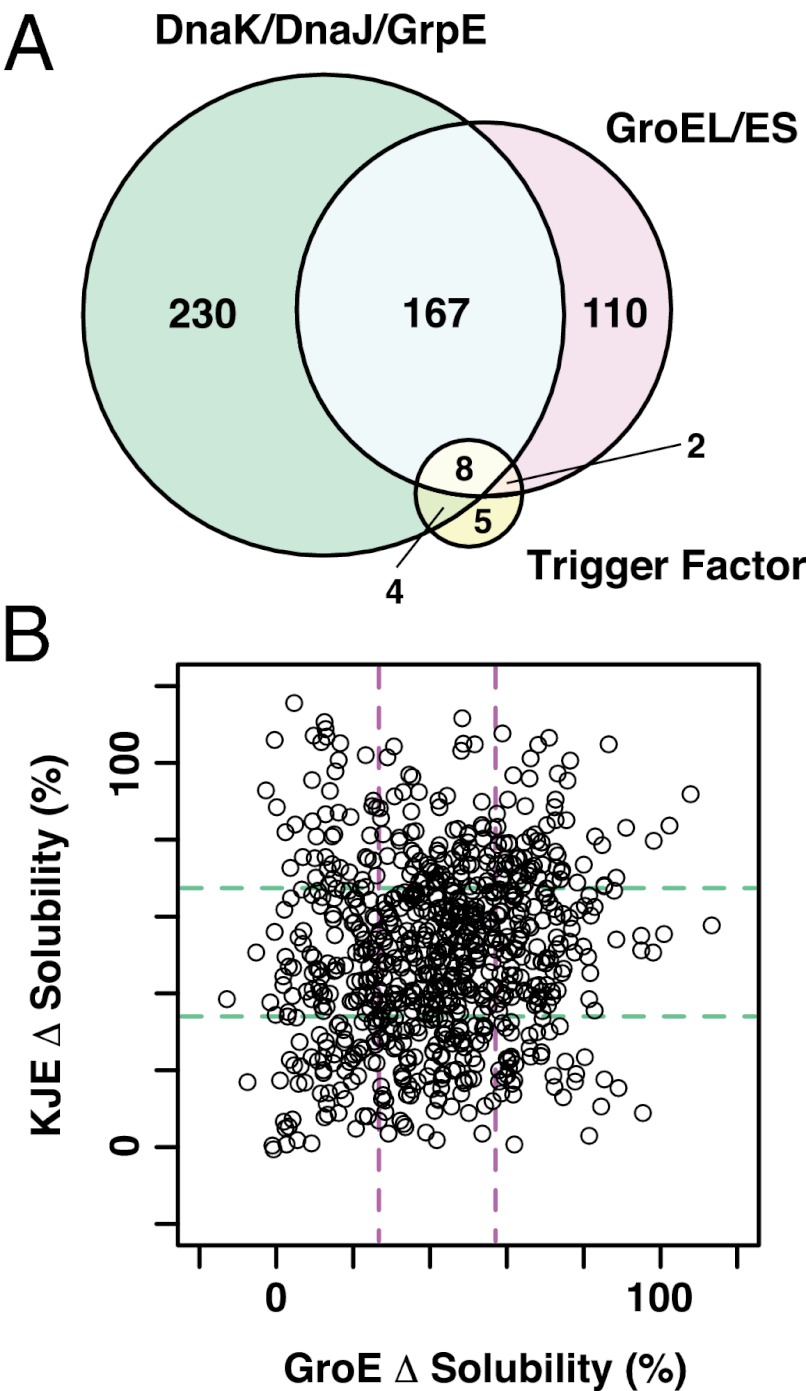
In total, more than 3,000 assays (788 proteins × 4 conditions = 3,152) were conducted (Dataset S1). The arranged data, combined with data obtained from our previous aggregation analysis, are freely accessible at our online database (eSol database: http://tp-esol.genes.nig.ac.jp). Overall, the chaperones tested here effectively increased the solubility of the aggregation-prone proteins (Fig. S2B). To manage the raw data (Fig. S2B), the chaperone effects were expressed as the difference (Δ) in solubility, calculated by subtracting the solubilities in the absence of each chaperone from those in its presence (Fig. 2B). We note that the analyses using the raw data (Fig. S2B) did not change the conclusions described below. Overall, the solubilities of two-thirds of the proteins (526 of 788) were drastically increased, defined as more than a 50% increase, in the presence of any one of the chaperones (Fig. 3A and Table S1). The proteins that were not rescued by any one of the chaperones, defined as less than a 20% increase in the solubility, represented only 3% of the total (24 of 788) (Table S1). Taken together, this comprehensive analysis explicitly confirms the global role of chaperones in preventing the aggregation of hundreds of proteins.
Fig. 3.
Overlaps and differences in chaperone effects. (A), A Venn diagram showing the overlap in the effects of chaperones. The numbers of proteins with solubilities that were drastically increased by at least one chaperone (defined as > +50% Δsolubility) are shown. See also Table S1. (B) Two-dimensional distribution plot of Δsolubilities for DnaKJE and GroE. Dashed lines represent the boundaries of the lower and upper quartiles [34, 67% and 26, 58% solubility values for DnaKJE (green) and GroE (purple), respectively].
Effects of Each Chaperone.
Next, we compared the effects of each chaperone in detail. It is noteworthy that TF had only a marginal effect (Fig. 2B and Fig. S2B). The number of proteins with >50% increase in the Δsolubility by TF was only 19 (Fig. 3A and Table S1). On average, TF generated only a 13% increase in the Δsolubility. On the other hand, DnaKJE and GroE increased the solubilities of many proteins (Fig. 2B and Table S1). The solubilities of 409 and 287 proteins with DnaKJE and GroE, respectively, were drastically (>50%) increased (Fig. 3A and Table S1). Approximately 30% of the proteins with >50% increase in solubility (175 proteins) were common between DnaKJE and GroE (Fig. 3A and Table S1), indicating that these overlapping proteins were rescued well by either DnaKJE or GroE. Taken together, the data clearly show the global effects of DnaKJE and GroE in preventing aggregation.
Relationship Between Chaperone Effects and Physicochemical Properties.
To further investigate the effects of DnaKJE and GroE, we plotted the data on DnaKJE and GroE in two dimensions (Fig. 3B). As already noted in the Venn diagram (Fig. 3A), a large number of proteins was plotted on the diagonal line, showing again that DnaKJE and GroE each rescued substantial amounts of proteins to similar extents (Fig. 3B). In addition to the overlapping effects, we also found that a fraction of the proteins was biased toward DnaKJE or GroE (Fig. 3B). The lower right area in Fig. 3B contains the proteins that were rescued by GroE but not by DnaKJE, termed the tentative GroE-specific proteins; the upper left area contains the tentative DnaK-specific proteins. These biases suggest that DnaKJE and GroE could have different recognition modes for substrates. To extract the possible preferences of DnaKJE and GroE, we analyzed the physicochemical properties of the proteins: the molecular weights, the deduced isoelectric points (pI), the structural classifications, and the oligomeric states.
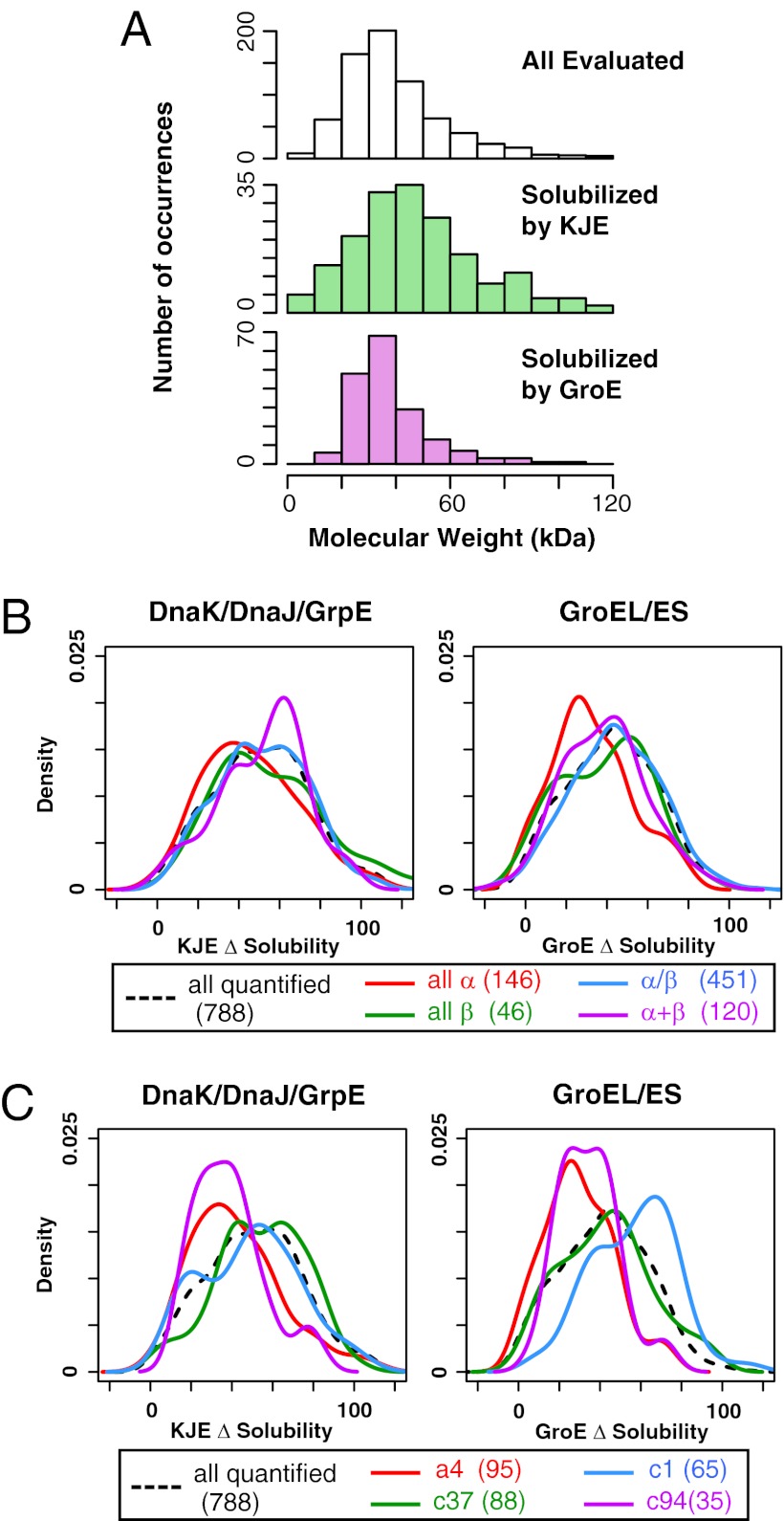
Regarding the molecular weights, the overall correlation between the solubility and the molecular weights was low (Fig. S4A). However, we found some biases when we compared the proteins that were well-solubilized by either DnaK or GroE, defined as the upper quartile (≥75th percentile) in the distribution (Fig. S4A). The histogram showed that GroE is biased toward lower molecular weight proteins (20∼50 kDa), whereas DnaKJE is effective for larger ones (> 60 kDa) (Fig. 4A). As for the pI, both DnaKJE and GroE were not effective for higher pI proteins, but these tendencies were weak (Fig. S4 B and C).
Fig. 4.
Correlation between chaperone effects and physicochemical properties. (A) Histograms of molecular weight for all evaluated proteins and the proteins that were rescued by DnaKJE or GroE. Well-solubilized proteins were defined as those in the upper quartile (≥75th percentile) in Δsolubility for DnaKJE or GroE. (B) Comparison between Δsolubility and SCOP classes (all α, all β, α/β, and α+β). The distributions of Δsolubility for DnaKJE and GroE are shown by Kernel-type density maps. The numbers in parentheses indicate the number of proteins categorized in each class. (C) Comparison between Δsolubility and SCOP folds. The four most abundant SCOP folds in the quantified proteins are shown by Kernel-type density maps. The numbers in parentheses indicate the number of proteins categorized in each fold. a4, DNA/RNA-binding 3-helical bundle; c1, TIM β/α-barrel; c37, P-loop containing nucleoside triphosphate hydrolases; c94, Periplasmic binding protein-like II.
To explore the contribution of the amino acid contents to Δsolubility by DnaKJE or GroE, we conducted the partial least-squares (PLS) regression analysis by using the ratio of 20 amino acid contents, molecular weight, and pI as the predictor variables (Fig. S5). However, the contribution of these parameters was fairly low. This fact suggests that other factors (e.g., a bias in the local amino acid composition and structural parameters) might contribute to the substrate recognition by chaperones.
Earlier works revealed that some of structural motifs were correlated with the aggregation propensity (6) and were enriched in GroE substrates (30, 31). Then, to address the correlation between the chaperone effects and the tertiary or quaternary structures, the Structural Classification of Proteins (SCOP) database (class and fold) (32) and the oligomeric states of proteins were compared, although only a small number of proteins was analyzed, because of the limited database size. When classified by the SCOP classes (all-α, all-β, α/β, and α+β), DnaKJE was effective for the α+β class, whereas GroE was not effective for the all-α class (Fig. 4B). Furthermore, we found some biases for DnaKJE and GroE in several SCOP folds (Fig. 4C). GroE was biased toward the c1 (TIM barrel) -fold, which is plausible because the most abundant fold in the in vivo obligate GroE substrates is the TIM barrel-fold (30, 31). Neither DnaKJE nor GroE was effective for the a4 (DNA/RNA-binding 3-helical bundle-fold) and c94 (periplasmic binding protein-like II) -folds (Fig. 4C).
The database on the oligomeric states of proteins is still insufficient, but it is partially available as the SUBUNIT annotation in the UniProt database (33). The distribution of DnaKJE tended to be effective for the heterooligomer group, and GroE was positively biased toward the monomer group (Fig. S6), although we should note that we individually translated the proteins one by one and did not translate the heterooligomeric pairs together.
Correlation Between Chaperone Effects and Known in Vivo Chaperone Substrates.
We compared our data with those for the previously identified in vivo chaperone substrates. Regarding DnaK and GroE, the mapping of the in vivo substrates (15, 31) showed a slight enrichment of both the DnaK and GroE substrates in the proteins with the DnaK- and GroE-biased proteins, respectively (Fig. S7A). In terms of the in vivo chaperone substrates, we applied a predictor for DnaK binding motifs (34) to our data and found that the overall correlation was poor (Fig. S7B).
Cooperative Effects of Chaperones on “Recalcitrant” Proteins.
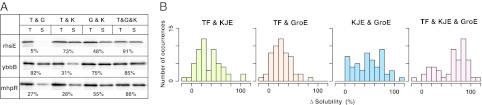
We found that neither DnaKJE nor GroE could rescue a subset of proteins mapped around the lower left area in the plot (Fig. 3B). As expected, none of these proteins were rescued by TF (Dataset S1), and thus they were named “recalcitrant” proteins. We then examined various combinations of chaperones to investigate whether these recalcitrant proteins could be solubilized. All of the recalcitrant proteins, which were defined as the proteins categorized in the lower quartiles in DnaKJE and GroE (53 proteins), were translated in the presence of chaperone combinations: TF+DnaKJE, TF+GroE, DnaKJE+GroE, and TF+DnaKJE+GroE. Typical results are shown in Fig. 5A, and all of the results are listed in Dataset S2. The solubilities under the TF+DnaKJE and TF+GroE conditions were slightly increased, whereas the combination of DnaKJE and GroE was more effective for some of these recalcitrant proteins (Fig. 5B), reflecting the consensus that GroE and DnaKJE synergistically assist with the folding of nascent polypeptides (28, 35). Strikingly, the addition of all three chaperones to the recalcitrant proteins drastically changed the solubility distribution: More than 70% of the recalcitrant proteins (38 of 53) showed significantly improved solubilities in the presence of all chaperones (Fig. 5B). These results suggest that TF also has the potential to act cooperatively with DnaKJE and GroE, although TF itself was not very effective in preventing aggregation.
Fig. 5.
Combined effects of chaperones on recalcitrant proteins. Fifty-three proteins, with Δsolubility in the presence of DnaKJE or GroEL that was lower than the boundaries of the lower quartiles of both DnaKJE and GroE (lower left area in Fig. 3B), were chosen as recalcitrant proteins. (A) Typical examples of the combination effect of chaperones on several recalcitrant proteins (nhsE, ybbB, and mhpR). The numbers below the electrophoretic pattern indicate the solubility values. T&G, TF and GroEL/ES; T&K, TF and DnaKJE; G&K, GroEL/ES, and DnaKJE; T&G&K, TF, GroEL/ES, and DnaKJE. (B) Histograms of Δsolubility obtained from the combination of chaperones.
Discussion
We performed a global analysis of the effects of the major E. coli chaperones on ∼800 aggregation-prone proteins, coupled with a reconstituted cell-free translation system [the PURE system (25, 26)]. This system is a significant extension of our previous global aggregation analysis under chaperone-free conditions (6). Thus far, only a handful of proteins have been individually examined in terms of chaperone function in vitro. We have now presented more than 3,000 raw datapoints to investigate the effect of chaperones. The global data themselves are unique and thus represent an invaluable resource for protein science and engineering (Dataset S1). The resource, which is also accessible via a public database (eSol database: http://tp-esol.genes.nig.ac.jp), can be used to investigate the properties of proteins of interest, in terms of their solubilities and chaperone effects, before a detailed analysis. In addition, the resources can be potentially extended to in silico analyses of proteins, such as prediction tools for protein aggregation and chaperone substrate recognition, as our previous aggregation analysis has already been used in many bioinformatics studies (e.g., refs. 31, 36, 37).
Chaperones were originally defined as proteins that assist with the correct folding of proteins by preventing intermolecular aggregation (4, 5). After the emergence of the chaperone concept, many in vitro studies have shown that chaperones prevent protein aggregation (4, 9). Because most of those studies were conducted on only a few proteins of interest, the data presented here are unique in being an explicit in vitro experimental demonstration of the global role of chaperones in preventing aggregation.
Our data clearly revealed that chaperones cope with a wide spectrum of aggregation-prone proteins. However, the data should be interpreted with caution because we collected the global data from a centrifugation-based assay (6). The data only indicated whether the target proteins were soluble or not. Soluble proteins do not always fold properly, as discussed previously (19). Alternatively, the soluble states of target proteins might only be achieved while binding to DnaK (or DnaJ) or GroEL. It is feasible that chaperones are associated with the nonnative forms of proteins even in the presence of ATP, which is constantly present in the cell-free translation system. Indeed, it is well known that the heterologous expression of a recombinant protein sometimes results in the formation of a binary complex between DnaK and the recombinant protein (3, 4). GroEL also associated with proteins that folded extremely slowly (38). Finally, as we discussed previously (6), we cannot exclude the possibility that the soluble fractions might include oligomeric species that were not precipitated under the present conditions.
Our analysis revealed the importance of the evolutionally conserved chaperonin (GroE) and Hsp70 (DnaK) families in the global prevention of aggregation. The solubilities of more than ∼66% of the aggregation-prone proteins were increased in the presence of either the DnaK or GroE system (Figs. 2B and 3, and Table S1). Importantly, DnaKJE and GroE each rescued substantial amounts of proteins to similar extents (Fig. 3B). These overlapping effects can be attributed to their abilities to bind the substrate proteins with fuzzy recognition, ensuring the robustness of the chaperone network in cells. As a result, these conserved chaperones play a global role in maintaining the proteostasis in cells (13, 16, 21).
Notably, some biases in the chaperone effects were found between DnaKJE and GroE (Fig. 3B). In particular, the molecular weight was weakly related to the chaperone effects; DnaKJE is effective for larger proteins and GroE is biased toward 20∼50 kDa proteins (Fig. 4A). The DnaK effect for larger proteins was previously reported for in vivo DnaK substrates (14, 39). As for GroE, the tendency is consistent with the observation that GroE acts mainly on < 60 kDa proteins, because of the steric limitation of its cylindrical cavity (30, 31, 38, 40), suggesting that GroEL would be most effective when the substrates are sequestered in the cavity of the GroEL-GroES complex. The sequestration of substrates could also explain why GroE is slightly biased toward monomeric proteins (Fig. S6), reflecting the GroE mechanism by which the substrate in the chaperonin cavity can fold into the native state, without interacting with other outside proteins. Other physicochemical properties, such as the pI and amino acid content, did not show any notable correlations with the substrate biases (Figs. S4 and S5). Therefore, besides the molecular weights, the other key properties for the chaperone preferences are still unknown. A bioinformatics approach, which integrates amino acid sequence and secondary and tertiary structure information, may be required to reveal the nature of the substrate recognition mechanisms of chaperones.
One of the striking results of this analysis was that TF by itself had only a modest effect on reducing aggregation (Fig. 2B). Similar marginal effects of TF were previously observed in in vitro translation experiments using S30 lysates from E. coli (28). In addition, a previous large-scale E. coli interactome analysis revealed that TF interacts with only 40 proteins, in contrast to the 310 DnaK and 776 GroEL interactors (21). One possible explanation for the marginal effect is that TF alone might not be sufficient to complete folding and minimize the aggregation. Unlike GroEL or DnaK, TF has no energy-consuming mechanism, and thus needs other chaperones to assist with correct folding to prevent aggregation (7). Indeed, this idea is compatible with the proposal that TF acts to delay the folding with translating ribosomes (28). This consideration is supported by our data that TF cooperated with DnaKJE and GroE to prevent the aggregation of recalcitrant proteins that were not rescued by either DnaKJE or GroE (Fig. 5B).
Finally, our approach has provided invaluable resources for a wide spectrum of protein research. In particular, cell-free synthesis-based proteomics could pave the way for investigations of low abundance proteins, which may not be detected by mass spectroscopy-based proteomics.
Materials and Methods
E. coli ORF Library.
For the expression of 792 agregation-prone cytosolic proteins by the in vitro translation system (PURE system), an E. coli ORF library (ASKA library) was used (6, 41–43). All ORFs were individually amplified by PCR for the cell-free expression by the PURE system, as described previously (6).
Preparation of Chaperones.
All chaperones were expressed in E. coli and purified by the following procedures. Hexahistidine-tagged trigger factor, DnaK, GrpE, and GroES were purified by metal-chelating chromatography and ion-exchange chromatography according to the previous reports, with slight modifications (27). For DnaK and GrpE, a 1 mM ATP wash step was performed before the elution in the metal-chelating chromatography. For GroES, the ion-exchange chromatography step was omitted because the metal-chelating chromatography process was sufficient for purification. Hexahistidine-tagged DnaJ was purified by metal-chelating chromatography in the presence of 0.05% (vol/vol) Brij58 detergent, according to the purification procedure for Mdj1p (a yeast Hsp40 homolog) reported previously (44). GroEL was prepared by hydrophobic-interaction chromatography and size-exclusion chromatography, according to the previous report (45).
The in vitro activities of each chaperone were confirmed by the following methods. For TF, the association of TF with ribosomes was assessed by sucrose cushion experiment (20), which revealed that at least half of the ribosome was associated with TF under the condition used (Fig. S1); for the DnaK system, the DnaKJE-assisted folding of firefly luciferase was monitored; and for GroEL and GroES, the ATPase activity and the GroE-assisted folding of rhodanese (46) were confirmed.
Cell-Free Protein Synthesis and Centrifugation-Based Aggregation Assay.
The procedure for the evaluation of the protein aggregation propensity was based on the method reported previously (6). Each ORF was individually translated with the PURE system under the four conditions (the absence of chaperone, +TF, +DnaKJE, +GroE). The components for the transcription-translation-coupled reaction were reported previously (including [35S]methionine for detection) (6), and the concentration of each chaperone was as follows: TF: 5.0 μM (monomer); DnaK, DnaJ, and GrpE: 5.0 (monomer), 2.0 (monomer), and 2.0 (monomer) μM, respectively; GroEL and GroES: 0.5 (tetradecamer) and 1.0 (heptamer) μM, respectively. Protein synthesis was performed at 37 °C for 60 min. After the reaction, an aliquot was withdrawn as the total fraction. The remainder was centrifuged at 21,600 × g for 30 min, and the supernatant fraction was collected. Both the total and supernatant fractions were separated by SDS/PAGE, and the band intensities were quantified by autoradiography. The ratio of the supernatant to the total protein was defined as the solubility, the index of protein aggregation propensity (6).
Data Analyses.
The molecular weight, pI, amino acid content, and SCOP classification were determined as described previously (6). Briefly, the molecular weight, pI, and amino acid content were calculated from the amino acid sequence obtained from GenoBase (http://ecoli.naist.jp/GB8/). The SCOP classification was determined by the dataset obtained from GenoBase, the annotation of which was based on the SUPERFAMILY database (42, 47). The oligomeric state of the proteins was determined from the SUBUNIT annotation in the UniProt database (http://www.uniprot.org) (33). The proteins with a SUBUNIT annotation containing the word “monomer,” “heterooligomer,” or “homooligomer” were included in each category. The PLS regression analysis was conducted by using the statistical software R (http://www.R-project.org.) with the PLS package. In this analysis, Δsolubilities were used as the objective variable and molecular weight, pI, and the content of 20 amino acids as the predictor variables.
Prediction of DnaK Binding Sites.
The prediction of DnaK binding sites was conducted with the LIMBO algorithm (34). The parameter values were obtained from the previous report (34) and the LIMBO Web site. The window value was set to 7, the threshold value was set to 8.26, which was used for the “high sensitivity prediction” condition, and the position specific scoring matrix was obtained from a previous report (34). The peak numbers were counted with a script developed in-house.
Supplementary Material
Acknowledgments
We thank Hirotada Mori and Tomoaki Matsuura for the gift of the ASKA library plasmid set; Bei-Wen Ying and Yoshihiro Shimizu for their technical advice and useful suggestions; and Millicent Masters for Escherichia coli MGM100 strain. This work was supported in part by Grant-in-Aid for Research Activity Start-Up Grant 22870010 (to T.N.); Grant-in-Aid for Scientific Research (A) Grant 18201040 (to T.U.); and Grant-in-Aid for Scientific Research on Priority Area Grants 19037007 and 19058002 (to H.T.) from The Ministry of Education, Culture, Sports, Science and Technology, Japan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1201380109/-/DCSupplemental.
References
- 1.Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 2.Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 3.Tyedmers J, Mogk A, Bukau B. Cellular strategies for controlling protein aggregation. Nat Rev Mol Cell Biol. 2010;11:777–788. doi: 10.1038/nrm2993. [DOI] [PubMed] [Google Scholar]
- 4.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 5.Ellis J. Proteins as molecular chaperones. Nature. 1987;328:378–379. doi: 10.1038/328378a0. [DOI] [PubMed] [Google Scholar]
- 6.Niwa T, et al. Bimodal protein solubility distribution revealed by an aggregation analysis of the entire ensemble of Escherichia coli proteins. Proc Natl Acad Sci USA. 2009;106:4201–4206. doi: 10.1073/pnas.0811922106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoffmann A, Bukau B, Kramer G. Structure and function of the molecular chaperone Trigger Factor. Biochim Biophys Acta. 2010;1803:650–661. doi: 10.1016/j.bbamcr.2010.01.017. [DOI] [PubMed] [Google Scholar]
- 8.Mayer MP, Bukau B. Hsp70 chaperones: Cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richter K, Haslbeck M, Buchner J. The heat shock response: Life on the verge of death. Mol Cell. 2010;40:253–266. doi: 10.1016/j.molcel.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 10.Thirumalai D, Lorimer GH. Chaperonin-mediated protein folding. Annu Rev Biophys Biomol Struct. 2001;30:245–269. doi: 10.1146/annurev.biophys.30.1.245. [DOI] [PubMed] [Google Scholar]
- 11.Taguchi H. Chaperonin GroEL meets the substrate protein as a “load” of the rings. J Biochem. 2005;137:543–549. doi: 10.1093/jb/mvi069. [DOI] [PubMed] [Google Scholar]
- 12.Horwich AL, Fenton WA, Chapman E, Farr GW. Two families of chaperonin: Physiology and mechanism. Annu Rev Cell Dev Biol. 2007;23:115–145. doi: 10.1146/annurev.cellbio.23.090506.123555. [DOI] [PubMed] [Google Scholar]
- 13.Deuerling E, Schulze-Specking A, Tomoyasu T, Mogk A, Bukau B. Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature. 1999;400:693–696. doi: 10.1038/23301. [DOI] [PubMed] [Google Scholar]
- 14.Teter SA, et al. Polypeptide flux through bacterial Hsp70: DnaK cooperates with trigger factor in chaperoning nascent chains. Cell. 1999;97:755–765. doi: 10.1016/s0092-8674(00)80787-4. [DOI] [PubMed] [Google Scholar]
- 15.Deuerling E, et al. Trigger Factor and DnaK possess overlapping substrate pools and binding specificities. Mol Microbiol. 2003;47:1317–1328. doi: 10.1046/j.1365-2958.2003.03370.x. [DOI] [PubMed] [Google Scholar]
- 16.Gragerov A, et al. Cooperation of GroEL/GroES and DnaK/DnaJ heat shock proteins in preventing protein misfolding in Escherichia coli. Proc Natl Acad Sci USA. 1992;89:10341–10344. doi: 10.1073/pnas.89.21.10341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vorderwülbecke S, et al. Low temperature or GroEL/ES overproduction permits growth of Escherichia coli cells lacking trigger factor and DnaK. FEBS Lett. 2004;559:181–187. doi: 10.1016/S0014-5793(04)00052-3. [DOI] [PubMed] [Google Scholar]
- 18.Genevaux P, et al. In vivo analysis of the overlapping functions of DnaK and trigger factor. EMBO Rep. 2004;5:195–200. doi: 10.1038/sj.embor.7400067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ying BW, Taguchi H, Kondo M, Ueda T. Co-translational involvement of the chaperonin GroEL in the folding of newly translated polypeptides. J Biol Chem. 2005;280:12035–12040. doi: 10.1074/jbc.M500364200. [DOI] [PubMed] [Google Scholar]
- 20.Ying BW, Taguchi H, Ueda T. Co-translational binding of GroEL to nascent polypeptides is followed by post-translational encapsulation by GroES to mediate protein folding. J Biol Chem. 2006;281:21813–21819. doi: 10.1074/jbc.M603091200. [DOI] [PubMed] [Google Scholar]
- 21.Arifuzzaman M, et al. Large-scale identification of protein-protein interaction of Escherichia coli K-12. Genome Res. 2006;16:686–691. doi: 10.1101/gr.4527806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gong Y, et al. An atlas of chaperone-protein interactions in Saccharomyces cerevisiae: Implications to protein folding pathways in the cell. Mol Syst Biol. 2009;5:275. doi: 10.1038/msb.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masters M, et al. Protein folding in Escherichia coli: The chaperonin GroE and its substrates. Res Microbiol. 2009;160:267–277. doi: 10.1016/j.resmic.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 24.Azia A, Unger R, Horovitz A. What distinguishes GroEL substrates from other Escherichia coli proteins? FEBS J. 2012;279:543–550. doi: 10.1111/j.1742-4658.2011.08458.x. [DOI] [PubMed] [Google Scholar]
- 25.Shimizu Y, et al. Cell-free translation reconstituted with purified components. Nat Biotechnol. 2001;19:751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- 26.Shimizu Y, Kanamori T, Ueda T. Protein synthesis by pure translation systems. Methods. 2005;36:299–304. doi: 10.1016/j.ymeth.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 27.Ying BW, Taguchi H, Ueda H, Ueda T. Chaperone-assisted folding of a single-chain antibody in a reconstituted translation system. Biochem Biophys Res Commun. 2004;320:1359–1364. doi: 10.1016/j.bbrc.2004.06.095. [DOI] [PubMed] [Google Scholar]
- 28.Agashe VR, et al. Function of trigger factor and DnaK in multidomain protein folding: Increase in yield at the expense of folding speed. Cell. 2004;117:199–209. doi: 10.1016/s0092-8674(04)00299-5. [DOI] [PubMed] [Google Scholar]
- 29.Hoffmann A, et al. Trigger factor forms a protective shield for nascent polypeptides at the ribosome. J Biol Chem. 2006;281:6539–6545. doi: 10.1074/jbc.M512345200. [DOI] [PubMed] [Google Scholar]
- 30.Kerner MJ, et al. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell. 2005;122:209–220. doi: 10.1016/j.cell.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 31.Fujiwara K, Ishihama Y, Nakahigashi K, Soga T, Taguchi H. A systematic survey of in vivo obligate chaperonin-dependent substrates. EMBO J. 2010;29:1552–1564. doi: 10.1038/emboj.2010.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: A structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 33.Jain E, et al. Infrastructure for the life sciences: Design and implementation of the UniProt website. BMC Bioinformatics. 2009;10:136. doi: 10.1186/1471-2105-10-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Durme J, et al. Accurate prediction of DnaK-peptide binding via homology modelling and experimental data. PLOS Comput Biol. 2009;5:e1000475. doi: 10.1371/journal.pcbi.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langer T, et al. Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature. 1992;356:683–689. doi: 10.1038/356683a0. [DOI] [PubMed] [Google Scholar]
- 36.Tartaglia GG, Dobson CM, Hartl FU, Vendruscolo M. Physicochemical determinants of chaperone requirements. J Mol Biol. 2010;400:579–588. doi: 10.1016/j.jmb.2010.03.066. [DOI] [PubMed] [Google Scholar]
- 37.Castillo V, Graña-Montes R, Ventura S. The aggregation properties of Escherichia coli proteins associated with their cellular abundance. Biotechnol J. 2011;6:752–760. doi: 10.1002/biot.201100014. [DOI] [PubMed] [Google Scholar]
- 38.Ewalt KL, Hendrick JP, Houry WA, Hartl FU. In vivo observation of polypeptide flux through the bacterial chaperonin system. Cell. 1997;90:491–500. doi: 10.1016/s0092-8674(00)80509-7. [DOI] [PubMed] [Google Scholar]
- 39.Mogk A, et al. Identification of thermolabile Escherichia coli proteins: Prevention and reversion of aggregation by DnaK and ClpB. EMBO J. 1999;18:6934–6949. doi: 10.1093/emboj/18.24.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sakikawa C, Taguchi H, Makino Y, Yoshida M. On the maximum size of proteins to stay and fold in the cavity of GroEL underneath GroES. J Biol Chem. 1999;274:21251–21256. doi: 10.1074/jbc.274.30.21251. [DOI] [PubMed] [Google Scholar]
- 41.Kitagawa M, et al. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): Unique resources for biological research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- 42.Riley M, et al. Escherichia coli K-12: A cooperatively developed annotation snapshot—2005. Nucleic Acids Res. 2006;34:1–9. doi: 10.1093/nar/gkj405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kazuta Y, et al. Comprehensive analysis of the effects of Escherichia coli ORFs on protein translation reaction. Mol Cell Proteomics. 2008;7:1530–1540. doi: 10.1074/mcp.M800051-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kubo Y, et al. Two distinct mechanisms operate in the reactivation of heat-denatured proteins by the mitochondrial Hsp70/Mdj1p/Yge1p chaperone system. J Mol Biol. 1999;286:447–464. doi: 10.1006/jmbi.1998.2465. [DOI] [PubMed] [Google Scholar]
- 45.Motojima F, et al. Hydrophilic residues at the apical domain of GroEL contribute to GroES binding but attenuate polypeptide binding. Biochem Biophys Res Commun. 2000;267:842–849. doi: 10.1006/bbrc.1999.2020. [DOI] [PubMed] [Google Scholar]
- 46.Koike-Takeshita A, Yoshida M, Taguchi H. Revisiting the GroEL-GroES reaction cycle via the symmetric intermediate implied by novel aspects of the GroEL(D398A) mutant. J Biol Chem. 2008;283:23774–23781. doi: 10.1074/jbc.M802542200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Madera M, Vogel C, Kummerfeld SK, Chothia C, Gough J. The SUPERFAMILY database in 2004: Additions and improvements. Nucleic Acids Res. 2004;32(Database issue):D235–D239. doi: 10.1093/nar/gkh117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.