Abstract
The RecBCD enzyme is important for both restriction of foreign DNA and recombinational DNA repair. Switching enzyme function from the destructive antiviral state to the productive recombinational state is regulated by the recombination hotspot, χ (5′-GCTGGTGG-3′). Recognition of χ is unique in that it is recognized as a specific sequence within single-stranded DNA (ssDNA) during DNA translocation and unwinding by RecBCD. The molecular determinants of χ recognition and the subsequent alteration in function are unknown. Consequently, we mutated residues within the RecC subunit that comprise a channel where ssDNA is thought to be scanned for a χ sequence. These mutants were characterized in vivo with regard to χ recognition, UV-sensitivity, phage degradation, and recombination proficiency. Of 38 residues mutated, 11 were previously undescribed mutations that altered χ recognition. The mutants fell into two classes: five that failed to respond to χ, and six that suggested a relaxed specificity for χ recognition. The location of the first set of mutations defines a recognition structure responsible for sequence-specific binding of ssDNA. The second set defines a highly conserved structure, linked to the recognition structure, which we hypothesize regulates conversion of RecBCD from a molecular machine that destroys DNA to one that repairs it. These findings offer insight into the evolution of enzymes with alternate χ recognition specificities.
Keywords: helicase, nuclease, protein–DNA interactions
Homologous recombination in Escherichia coli is initiated by the heterotrimeric helicase/nuclease, RecBCD (1, 2). RecBCD comprises two motor subunits (RecB and RecD) with opposite translocation polarities that travel along the complementary DNA strands (2–4). The enzyme unwinds and degrades double-stranded DNA (dsDNA) from an end until it encounters an 8-nt sequence, called Chi [crossover hotspot instigator (χ): 5′-GCTGGTGG-3′] (5, 6). When the enzyme recognizes this single-stranded DNA (ssDNA) sequence, its nucleolytic activities are altered: the vigorous 3′→5′ nuclease is down-regulated and the 5′→3′ nuclease is up-regulated (7, 8). In addition to these changes, χ-activated RecBCD displays the capacity to load RecA protein, an essential factor for DNA strand exchange, onto the χ-containing ssDNA (9–11). Thus, by switching in response to χ recognition, RecBCD changes from an enzyme that provides an antiviral function to cells by degrading foreign DNA and bacteriophages, to one that provides a recombinational dsDNA-break repair function to maintain chromosome integrity (2, 12). As a reflection of this important function, the χ sequence is overrepresented in the E. coli genome by ∼sixfold and is found on average every 4.5 kb (13).
To date, a few RecBCD mutants with a deficiency in χ recognition have been reported (14–20). Because recognition of χ results from a number of interdependent steps, defects in χ recognition can result from a number of molecular events (2). Lack of recognition can arise from: (i) a defective RecB motor, which is needed to pump the χ-containing ssDNA through a channel in RecBCD to a χ-scanning site in RecC (15–17); (ii) loss of the RecD subunit, which results in an enzyme, RecBC, that is constitutive for recombination functions (21–23); (iii) the inability to recognize χ (18–20); or (iv) the inability to couple the χ recognition to the requisite conformational change.
The crystal structure of the RecBCD enzyme showed that the dsDNA is separated into two strands at a pin structure located in RecC, such that the 5′- and 3′-DNA strands are positioned to bind the motor domains of the RecD and RecB subunits, respectively (4). The structure also reveals the presence of two channels: one each for the 5′- and 3′-terminated DNA strands. Moreover, the exit for the 3′-ssDNA channel is facing the nuclease domain of RecBCD, which is located at the C terminus of the RecB subunit; consequently, unless the conformation of RecBCD is changed, the 3′-terminated DNA strand has a high probability of being degraded. The RecC subunit was proposed to scan the ssDNA for the χ sequence as the unwound DNA is pumped through the internal channel (4). Pseudorevertants (recC* mutants) of a recC-null strain were found that did not respond to the canonical χ but were inferred to recognize an altered sequence (14). All of these mutants arose from insertions or deletion downstream of the original frame-shift mutation to restore the reading frame and to change 7–9 amino acid residues of RecC (19). One mutant (recC1004) did not respond to normal χ but recognized an altered χ sequence, χ*, 5′-GCTGGTGCTCG-3′ (19, 20, 24). By virtue of displaying altered specificity, rather than ambiguous loss of recognition, this is the only mutant that directly implicates a subunit, RecC, in the χ recognition process.
Because the χ sequence is essentially used as a bar-code for DNA self-recognition, its sequence varies in divergent bacteria (25). Bacillus subtilis uses 5′-AGCGG-3′ as its cognate χ sequence to regulate its RecBCD homolog, AddAB (2, 26), and Lactococcus lactis uses 5′-GCGCGTG-3′ as χ to regulate its RecBCD homolog, RexAB enzyme, which is an AddAB ortholog (27). The χ sequences of the Gram-negative bacterium, Haemophilus influenzae, are the χ variants 5′-GNTGGTGG-3′ and 5′-G[C/G]TGGAGG-3′ (28). Finally the χ sequence of E. coli was found to attenuate the endogenous ATP-dependent dsDNA exonuclease in a variety of related enteric bacteria, including Shigella Sonnei, Citrobacter freundii, Klebsiella pneumoniae, Serratia marcescens, Salmonella typhimurium, and Proteus mirabilis, suggesting a common or degenerate variant in these closely related species (29–31). Soley from statistical analysis of genome sequences, it has become possible to identify overrepresented sequences with the genomic characteristics of χ sequences (25). This strategy led to the identification of 5′-GAAGCGG-3′ in Staphylococcus aureus and 5′-GCGCGTG-3′ in several other Streptococci as χ-like sequences. Thus, there is great variety in the sequences of χ-like elements, and the strategy of using a DNA sequence-regulated helicase/nuclease is broadly used within the Bacteria.
Although the structure of RecBCD provided considerable insight into RecBCD function, it did not identify the χ recognition motifs because the ssDNA had not penetrated into the RecC subunit. Predicting the domain responsible for recognition is not possible because, despite the multitude of structures that reveal the nature of sequence-specific recognition of dsDNA, there is relatively little known regarding the molecular determinants of sequence-specific recognition of ssDNA. Consequently, we identified and mutated candidate residues on the surface of the channel within the RecC subunit that could be involved in χ recognition. We found 11 different mutations that result in an altered response to χ. The mutations fell into two categories: one category behaved in vivo as though it has lost the capacity to recognize χ; the second behaved as although the sequence determinant for χ recognition had become relaxed. These mutations define the χ interaction region and offer insights into an allosteric relay system that transmits recognition of a specific regulatory sequence within ssDNA: recognition that results in a switch in enzymatic and functional personality of RecBCD.
Results
Identification of Amino Acids Within the RecC Channel That Potentially Interact with χ.
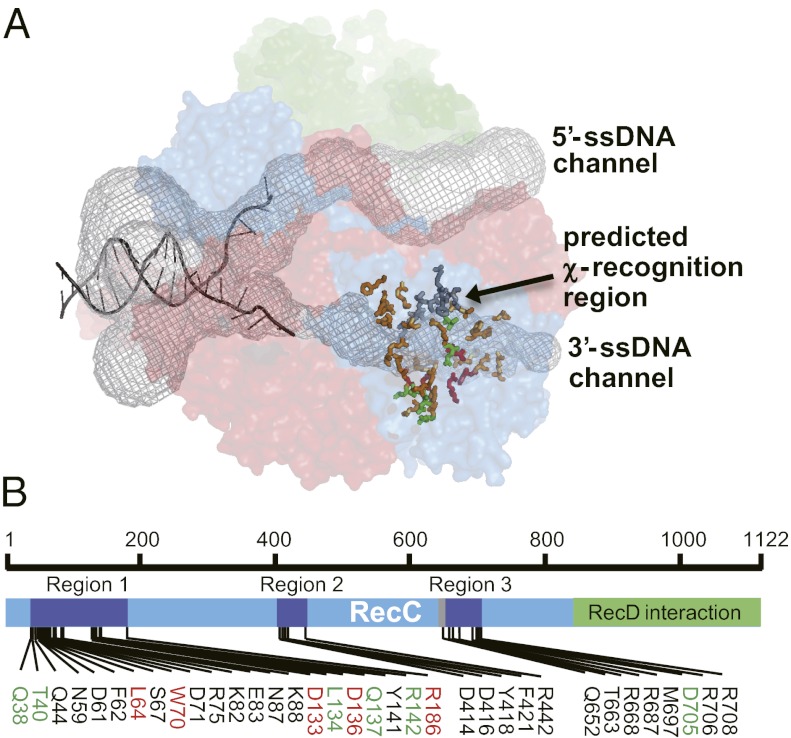
The crystal structure was used to identify candidate residues that are surface-accessible along the putative χ recognition channel (Fig. 1A) (4). An initial group of 22 residues was selected and, based on the subsequent identification of positive candidates, another group of more focused residues was mutagenized. In the end, 35 amino acids were individually changed to alanine (Fig. 1B). An additional three mutations (E399G, V450G, and A766V) were generated as a consequence of PCR mutagenesis errors and did not alter phenotype. The selected residues are widely spread on the RecC polypeptide but cluster in three regions of primary structure (Fig. 1B): Q38 to R186, D414 to F442, and Q652 to R708. The last region is close to the position of the recC*1004 alteration (20).
Fig. 1.
The RecC subunit of the RecBCD complex. (A) Structure of RecBCD showing the location of DNA and channels (4). RecB, RecC, and RecD are red, blue, and green, respectively. The two internal channels (black mesh) were calculated using MOLE (47). Type 1 or type 2 residues are red or green, respectively; other residues mutated in this study are bronze-orange sticks; and those altered in recC1004 (19) are gray sticks. (B) Primary structure of RecC showing residues mutated here. Type 1 or type 2 mutations are in red or green, respectively. The region altered in recC* mutants is gray (19).
RecC-Channel Mutants Retain the Capacity to Nucleolytically Destroy T4 Phage in Vivo.
The individual recC mutations were inserted into a plasmid, transformed into a recC-null strain, and then assessed for functions in vivo. All of the mutants at least partially complemented the null strain with regard to UV survival, showing that at least partially functional proteins were being produced (Table S1). Of the mutants examined, all displayed RecBCD-dependent nuclease activity, as judged by the inability of mutant T4 gene 2− bacteriophage to plate on these cells (Table S1; some mutants were not fully characterized because, based on initial screening, their phenotypes did not pass the criteria defined below). The gene 2 product of bacteriophage T4 is a DNA end-binding protein that normally protects the linear dsDNA of the phage from degradation by RecBCD enzyme; T4 phage lacking gene 2 function are restricted for growth because of RecBCD nuclease function (15, 32). Consequently, each of these RecBCD mutants is proficient for dsDNA exonuclease activity. Note that T4 gene 2-defective phage grow well on cells lacking recD (22); this is because the RecBC enzyme is a helicase but not a nuclease, and the nuclease activity of RecBCD is needed to block survival of T4 gene 2− phage (21). Thus, the failure of this phage to grow on our recC mutant strains shows that none of the mutations resulted in the loss of the RecD subunit from RecBCD to produce a recD−-like phenotype. This finding is significant because the RecBC enzyme does not recognize χ (23): had any of our mutations resulted in the loss of RecD (15), then they may have appeared as false-negatives with respect to loss of χ recognition.
RecC-Channel Mutants Display Two Distinctive Classes of Defects with Regard to Their Response to χ.
The recognition of χ was examined by plating bacteriophage λ, without or with a χ sequence, on bacteria expressing the mutant RecBCD (20, 33). Phage λ propagates by two routes: one is recombination and the other is rolling circle replication to make a tandemly repeated genome for packaging into phage particles. Both routes are stimulated by the interaction between RecBCD and χ, when the phage’s recombination functions (red and gam genes) are inactive: χ protects the λ genome from degradation by RecBCD, and χ also activates RecBCD to enhance recombination frequency. Therefore, a λ phage with a χ sequence (χ+) produces more progeny than wild-type λ (which does not have a χ sequence, χ0) in wild-type cells (6). As a result, χ+ phage make larger plaques compared with isogenic χ0 phage. Large plaques can also appear if the nuclease activity of the RecBCD enzyme is reduced; however, because all of our mutant RecBCD enzymes (except one) display nuclease activity as measured by the T4 gene 2− plating assay, then such a possibility can be excluded.
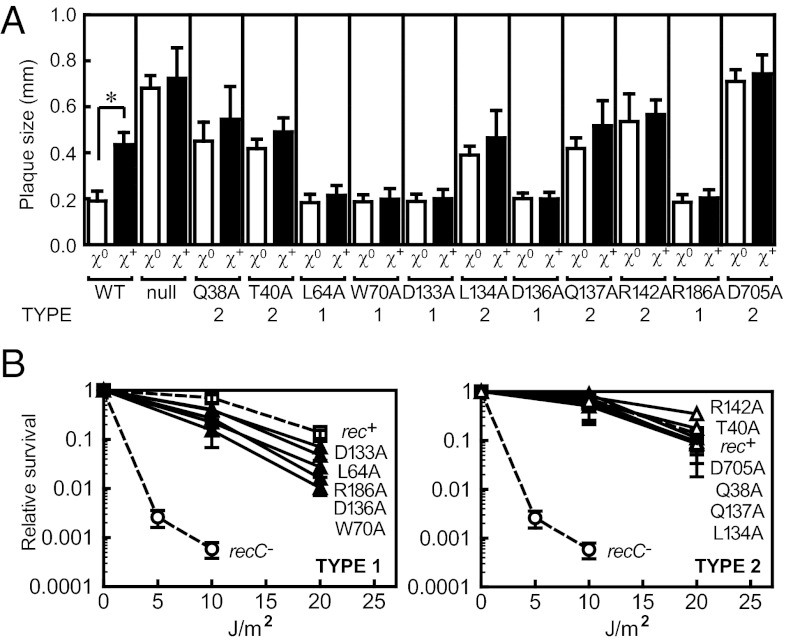
The results of plating phage λ on all 38 mutant recBCD cells are summarized in Table S1 (columns four and five). Wild-type bacteria show the typical response that results in plaques for χ+ phage that are about two- to threefold larger than for χ0 phage. Because the RecBCD nuclease restricts λ phage growth, phage without a χ sequence forms large plaques on a recC− strain. Therefore, to identify RecC-channel mutants that potentially lacked χ recognition, we applied the following phenotypic criteria: (i) the mutant displayed nuclease activity [i.e., blocked plating of T4 gene 2− mutant phage (Table S1, column three)]; (ii) the mutant cells produced plaques for χ0 phage λ that are the same size as wild-type cells (Table S1, column four); and (iii) the mutant cells produced plaques for χ+ phage λ that are the same size as for the χ0 phage (Table S1, column five). There were five mutants that met these criteria: L64A, W70A, D133A, D136A, and R186A (Fig. 1A); they are summarized in Table S2 and classified as “type 1.” For each mutant, the growth of either χ0 or χ+ phage λ results in the same plaque size, and the size is the same as for χ0 phage λ grown on wild-type cells (Fig. 2A).
Fig. 2.
Phenotypic assays reveal defects in recognition of χ by the RecC-channel mutants. (A) The sizes of phage λ plaques distinguish χ recognition for the recC mutants. Each bar represents average with SD of 10 individual plaques; *P < 0.005 in t test. (B) UV resistance of recC mutants. (Left) Type 1 mutants, ▲; (Right) type 2 mutants, △; recC+, □; and recC− (without plasmid), ○. Averages with SD from at least two independent experiments.
A second possible type of χ recognition mutant is an enzyme that either recognized a new χ sequence that was present on both χ0 and χ+ phages, or that displayed a relaxed recognition of sequences that were a subset of the canonical χ sequence. To identify such candidates, the following phenotypic criteria were applied: (i) the mutant displayed nuclease activity [blocked plating of T4 gene 2− mutant phage (Table S1, column three)]; (ii) the mutant cells produced a large plaque size for χ0 phage λ that was at least as large as the plaque for χ+ phage λ on wild-type cells (Table S1, column four); and (iii) the mutant cells produced plaque sizes for χ+ phage λ that are the same as for χ0 phage (Table S1, column five). There were six mutants that met these criteria: Q38A, T40A, L134A, Q137A, R142A, and D705A (Fig. 1A); they are also summarized in Table S2 and classified as “type 2.” For each mutant, the growth of χ0 or χ+ phage λ results in a large plaque; in fact, the plaque size is as large, or larger, than that for χ+ phage growing on wild-type cells (Fig. 2A). Interestingly, for the D705A mutation, the plaque sizes approach the size observed for the recBCD-null strain, but clearly neither the D705A mutation nor the other type 1 and 2 mutations result in a null phenotype for UV sensitivity (Fig. 2B) or for T4 gene 2− growth, showing that they are not simply inactive. Finally, when we combined some of the type 2 mutations into a single RecC polypeptide for reasons that will become apparent below, the phenotype of Q137A-D705A, R142A-D705A, and Q137A-R142A-D705A was the same as for the single mutations (Table S1), showing that they were not synergistic.
Both Type 1 and Type 2 Mutants Reveal Alterations in χ-Mediated Recombination.
To verify that these newly isolated RecBCD mutants are altered for χ recognition, recombination between red− gam− λ phages (which are defective for phage-encoded recombination function) with or without a χ sequence was measured in vivo (20). The cross is designed to determine whether a sequence inserted into the int gene region (between J and cI) behaves as a recombination hotspot by measuring the ratio of clear and turbid plaques formed by the recombinant progeny (Fig. 3A) (20). Recombinants possessing the S+ and Jh markers are assayed to determine whether their phenotype with regard to the intervening immunity region is turbid (cI+) or clear (cI). If spontaneous recombination occurred evenly along the DNA, then the recombination frequency on both the left and right sides of the immunity region should be the same (1:1); if a recombination hotspot is inserted at the int locus, then turbid plaques will be prominent (20). All of the type 1 and 2 mutants were proficient for basal χ-independent recombination (Table S1) that occurs because of a low frequency of spontaneous activation of RecBCD (18).
Fig. 3.
In vivo λ recombination assays. (A) Schematic of λ recombination assay. (B) Recombination frequency (%). (C) Hotspot activity (ratio of turbid/clear plaques). Averages with SDs from at least three assays. Recombination frequency of χ0 and χ+ λ for wild-type strain (chromosomal recC+ gene) was 0.63 ± 0.16 and 2.8 ± 0.5, and hotspot activity was 1.8 ± 0.2 and 7.2 ± 0.6, respectively. *P < 0.005 in t test.
For the wild-type protein, the response to χ is evident in both the recombination frequency and the hotspot activity: both measurements are increased [2.6 (± 0.5)-fold and 3.8 (± 1.2)-fold, respectively] by χ (Fig. 3 B and C and Table S2). We note that in our assay, the mutant recC genes are expressed from a multicopy plasmid under the control of the Ptac promoter. It was previously reported that overexpression of the RecC subunit reduces χ hotspot activity (34). In agreement, even though the expression in our work was in the absence of isopropyl-β-d-thiogalactopyranoside (IPTG), we also observed that the increased gene dosage of RecC diminished the measured χ response (Fig. S1). The control with wild-type RecC expressed from the chromosome showed a 1.8-fold higher χ hotspot activity when compared to the same assay with RecC expressed from the plasmid, which is the same fold-difference previously noted (34). All of the type 1 mutants (L64A, W70A, D133A, D136A, and R186A) displayed recombination frequencies similar to wild-type for the χ0 phage and, significantly, none showed an increased frequency in response to χ (Fig. 3B). In addition, none of the type 1 mutants showed higher hotspot activity in the χ+ cross (Fig. 3C). We also produced a histidine variant at position R186 (R186H) because this mutation is present in the previously characterized recC2145 allele, which does not recognize χ in vivo (15). The recC2145 allele actually has two substitutions in RecC (R186H and G304S) (2). Given that R186H and R186A have the same χ recognition deficiencies, we conclude the R186 position is responsible for the phenotype of the recC2145 allele. Thus, all of the type 1 mutants fail to recognize χ, and they define residues that are likely to be directly involved in binding the recombination hotspot.
The type 2 mutants, however, revealed a complex recombination behavior. The L134A, R142A, and D705A mutations showed higher recombination frequency than wild-type for χ0 and χ+ phage (Fig. 3B) but apparently no hotspot activity (Fig. 3C). On the other hand, the Q38A mutant showed approximately wild-type levels of recombination frequency for χ0 phage (Fig. 3B) and an increased hotspot activity (Fig. 3C). However, if these mutations caused an altered or relaxed recognition specificity, then interpretation of the phage recombination measurements can be complicated if the alternative χ sequence precedes the recombination interval being studied. If these mutant RecBCD enzymes recognize novel (perhaps shorter) χ-like sequences (see Discussion), then they could respond to the putative χ-like sequence first, which would preclude recognition of the distal wild-type χ sequence (35). Phage λ does indeed possess at least two χ-like sequences (cCTGGTGG at 541 bp and GtTGGTGG at 2,763 bp from the right end) upstream of the selected S+ marker. If any of the type 2 mutants are activated by an upstream χ variant, then recombination of both χ0 and χ+ phage will be increased (Fig. 3B, D705A, for example), resulting in a ratio of hotspot activity of only 1.0 (Fig. 3C, D705A, for example).
To determine whether the two classes of mutants represent a gain or loss of function, we measured their genetic behavior in cells containing wild-type RecBCD. One allele (R186A or D705A) from each class was investigated (Fig. S2). The D705A allele (type 2) is completely dominant over the wild-type, resulting in large plaques for both χ0 and χ+ phage in the wild-type background. In contrast, the R186A allele (type 1) has no effect on χ0 phage but reduced the plaque size for χ+ phage; therefore, the R186A mutant is codominant to the wild-type RecBCD.
Residues Identified by Mutagenesis Are Highly Conserved in all RecC Homologs.
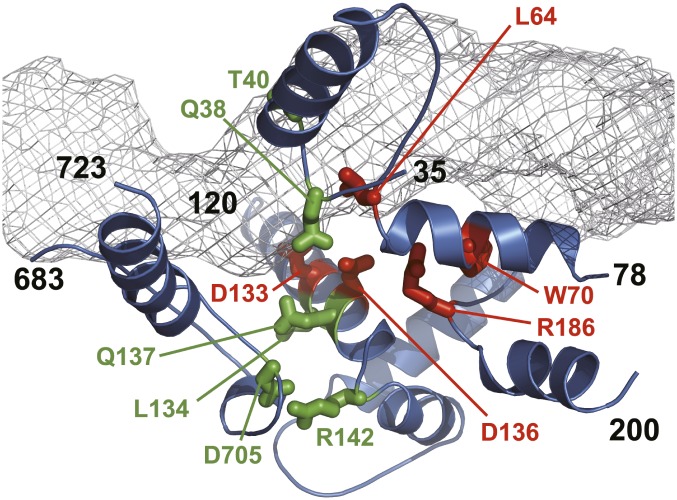
To determine whether the residues identified by mutational analysis in this study are generally important to RecBCD function in other bacteria, we examined their conservation in recC orthologs. The sequences of 112 unique bacterial recC genes were aligned, showing clear islands of conservation (Fig. S3). The residues identified here fall into five regions of conservation (Fig. 4). Many of these χ recognition residues are highly conserved, with several showing extraordinary preservation. For example, at position 142, arginine is found in 97% of the 112 orthologs, and at position 705, aspartate is nearly always (90%) found and the residue is always acidic (aspartic or glutamic acid). Interestingly, although some amino acids (e.g., R706) are highly conserved, they have no effect on χ recognition, at least when mutated to an alanine (Table S1). In the RecBCD structure (Fig. 5), R142 and D705 form a salt bridge that, based on the conservation, must be a crucial and universal structural element of the χ response. The significance of the locations of the other residues involved in χ recognition, and their conservation, will be made apparent in the Discussion.
Fig. 4.
Conservation of the amino acid residues involved in χ recognition among RecC orthologs. Five regions of highly conserved sequence implicated in χ recognition are shown in WebLogo format (48, 49). Type 1 residues are in red, and type 2 in green. Residues in gray were mutated but did not affect χ recognition. Gap in line 2 reflects a very small number of RecC sequences (not including E. coli) having a single amino acid insert there (Fig. S3).
Fig. 5.
Locations of residues in RecC structure implicated in χ recognition. Channel for 3′-ssDNA is gray mesh calculated using MOLE (47). Residues important for χ recognition are in stick format, and red for type 1 or green for type 2. Secondary structural elements are from regions of conservation in Fig. 4; black residue numbers indicate the starts and ends. Note that the first two elements, as well as the third and fourth, are continuous; the fifth is separate and comprises a loop joined by the ionic interaction between the highly conserved R142 and D705 residues.
Discussion
The crystal structure of RecBCD revealed that each DNA strand passes through a different interior channel and engages with a motor domain in either the RecB or RecD subunit (4). The 3′-ssDNA tail is translocated by the RecB motor into a proposed χ-scanning channel in RecC (17), eventually exiting the complex at the nuclease domain, which is at the C terminus of RecB. Several crystal structures of RecBCD have revealed the details of the interaction between the 3′-tail and the RecB motor, but this tail was always too short to cross the RecB/RecC interface (36). However, the channel through RecC in the protein complex is a clear structural feature and, together with several previous studies, suggested that χ sequence recognition occurs in the RecC subunit (14, 19, 20). Indeed, here we established that this region serves as the χ-scanning site of RecBCD, and we identified residues that are crucial to the recognition and regulation process.
Among the 35 individual mutations to alanine that we intentionally constructed, 11 mutants showed a phenotype characteristic of altered χ recognition (Fig. 5). Because of PCR mutagenesis errors, sequencing uncovered several other substitutions. The accidental channel mutations, R186C and D705H, and the intentional substitution, R186H, have the same phenotypes as their respective alanine substitution, showing that at least for these positions, the phenotype is not unique to alanine substitution. Therefore, in all, we analyzed 41 different mutations at 38 amino acid positions, of which 11 unique loci resulted in altered χ recognition (Fig. 1, Table S1, and Table S2). Based on their in vivo characteristics, these mutations fell into two groups: type 1 mutations, which resulted in a loss of χ recognition or response, and type 2 mutations, which resulted in, we infer, an altered or relaxed recognition of χ.
The type 1 residues (L64, W70, D133, D136, and R186) seem to define regions that are directly responsible for either recognition of, or response to, χ. These RecBCD mutant proteins either fail to recognize the canonical χ sequence or they fail to switch to the χ-activated form of the enzyme. These mutants could have completely lost the capacity to recognize any χ sequence or they might require a sequence that is not present in bacteriophage λ. Interestingly, cells expressing the type 1 mutations were not drastically UV sensitive (Fig. 2B). A likely explanation is that these mutant enzymes show a low but meaningful basal level of repair mediated by χ-like or nonspecific sequences, or a low level of spontaneous activation (18), which is augmented by the RecF-pathway components to effect repair via a “hybrid” pathway (37). Biochemical analysis of the mutant RecBCD enzymes, which is described in the companion article by Yang et al. (38), shows that they are unable to recognize χ.
The RecC structure itself resembles a Superfamily 1 (SF1) helicase and may interact with ssDNA using an equivalent region of the structure (4). The most striking structural feature of the type 1 residues is that they form an almost continuous patch of surface on subdomain 1B near the rear exit of the RecC tunnel (Fig. 5 and Fig. S4), defining a part of the χ-scanning site. Although not contiguous within the primary structure, D133-D136-R186-W70 form an approximately linear array that comprises the ssDNA-recognition motif that could read the sequence of the incoming ssDNA in the 3′ to 5′ direction, offering a possible mechanism for χ recognition. Because alteration of any one of these residues prevents response of RecBCD to χ, we imagine that both the acidic aspartate and the basic arginine are forming specific hydrogen bonds with the nucleotides of the χ sequence, and that both the aromatic tryptophan and hydrophobic leucine (L64) are making hydrophobic interactions with the bases in the χ sequence. Interestingly, arginine interacts predominantly with G and secondarily with T, and aspartate interacts with C (39): these are the nucleotides that constitute the χ sequence.
Although much is known about sequence-specific binding to dsDNA (see, for example, ref. 39), there are relatively few examples of sequence-specific recognition of ssDNA. The RNA polymerase of N4 bacteriophage interacts with ssDNA as a template, but requires a DNA stem-loop structure (40); the structure of this complex shows that an arginine and lysine hydrogen-bond with guanine bases, and a tryptophan stacks on the guanine in the ssDNA loop; in addition, a specificity loop makes base-specific contacts with the stem via aspartate and arginine residues and salt bridges with lysine residues (41). The σ70-ssDNA complex is another example: it uses an amphipathic helix with exposed residues to directly contact bases in the ssDNA formed upon melting (42). The hnRNP K protein binds ssDNA by a network of hydrogen bonds between isoleucine residues and two specific bases (43). Finally, the telomeric end-binding protein of Oxytricha binds (T4G4)2 specifically via stacking interactions of tyrosine on bases, and H-bonding of tyrosine, glutamine, and arginine residues with the bases (44). Characterization of the χ recognition locus of RecBCD adds to this limited set of sequence-specific ssDNA interactions.
The type 2 mutations define a particularly interesting and previously undescribed class of altered RecBCD enzymes. These mutants show high levels of recombination even in the absence of χ, yet they possess nuclease activity, suggesting that they are either constitutively activated to promote recombination or they recognize a DNA sequence that is much more frequent than the canonical 8-nt χ sequence. If these mutant enzymes were to be activated for recombination by a 6- or 7-nt sequence, then they would initiate recombination in phage λ crosses in an interval before the one being measured (downstream of the canonical χ). Thus, their recombination activity would be high but they would appear to be unresponsive to the canonical χ because the mutant RecBCD would have been activated to promote a crossover before the wild-type χ sequence; in addition, this gain of function—that is, the capacity to recognize many more χ-like sequences—would be consistent with the dominant phenotype (Fig. S2). The type 2 residues (Q38, T40, L134, Q137, R142, and D705) are found in the subdomains 1A, 1B, and 2A of RecC and, based on the crystal structure of RecBCD, it is evident that all of these residues cannot interact directly with χ (Fig. 5). However, we identified a polypeptide loop lining the channel that may have a role in this process (4, 36). The structure of RecC is related to SF1 helicases but it also has an additional loop inserted in the 2A domain. One of the χ-recognition mutants (D705) is located in this loop and it makes an ion pair with another χ-recognition mutant (R142) situated in the 1B domain. Although these residues are remote from the channel, R142 is at the far end of an α-helix—the χ-recognition helix in the 1B domain—that contains a cluster of other χ-recognition mutations that do line the channel (D133, L134, D136, and Q137), suggesting a network of interactions that constitute a structure important for transmission of an allosteric signal from the χ-recognition helix to the ion-pair structure. This signal could mediate the conformational changes needed to alter the functionality of RecBCD (Fig. S5).
Interestingly, some of the amino acid residues identified here as being involved in χ recognition show a remarkable degree of conservation with 112 other bacterial RecC proteins (Fig. 4). In particular, the nearly absolute maintenance of the two residues that comprise the ion pair (R142 and D/E705) uncovers the universal importance of this structural element to RecBCD function; in agreement, this structure, although different in detail, is conserved in AddAB enzyme (45). In addition, most of the other residues important to χ recognition are also very well conserved and are found in conserved regions.
The χ sequence is used as a bar code to permit bacteria to identify DNA that has a high probability of being its own chromosomal DNA, which is in need of recombinational DNA repair (1, 2): if RecBCD enzyme does not encounter a cognate χ sequence while unwinding DNA, then it will nucleolytically destroy it. For this reason, the cognate χ sequences are the most abundant sequences in their respective organisms (25, 46); in E. coli they are found every ∼4.5 kb in the genome [1,009 sites for the 4,639,675-bp genome (13)]. Our identification of the molecular determinants for χ recognition provides insight into the specificity of χ sequence recognition by RecBCD homologs. It is known that many enteric bacteria use the same sequence, 5′-GCTGGTGG, as χ (29, 30). In fact, even though Salmonella’s RecC is somewhat diverged, all of the χ recognition residues identified here are conserved. Interestingly, Haemophilus RecC has T40P and L64M substitutions (Table S3), and here we showed that mutations at these positions in E. coli RecC change its χ recognition. In fact, the Haemophilus χ sequence is 5′-GNTGGTGG, or 5′-G[C/G]TGGAGG (28). Given that T40 is also mutated to a proline in both P. mirabilis and Serratia proteamaculans, which recognize the same χ as E. coli or a degenerate version, this fact is consistent with our interpretation herein that T40 confers a possible relaxed specificity of χ recognition. Thus, T40 and L64 may well contribute to the recognition of the second or sixth letter of the χ sequence, and it is easy to imagine how the recognition of divergent χ sequences could evolve via slight modification of these or the other residues identified herein. Detailed in vitro analysis (38) and a crystal structure of RecBCD in a χ recognition complex will provide further insight into the mechanism of ssDNA sequence-specific χ recognition and its effects on the biochemical properties of RecBCD.
Materials and Methods
In Vivo Assays: UV Sensitivity, Bacteriophage λ Plaque Size, T4 Gene 2− Bacteriophage Plating, and Bacteriophage λ Recombination.
E. coli, bacteriophage, plasmid, construction of RecC-channel mutants and other details are provided in SI Materials and Methods and Tables S4 and S5. For the phenotypic analyses, cells expressing recC from a relatively low-copy plasmid were grown in the absence of IPTG induction. UV sensitivity was measured using exponentially growing cultures diluted and spread on L agar plates. Plates were irradiated with UV light (254 nm). Bacteriophage λ plaque sizes were measured as described previously (33). RecBCD nuclease activity in vivo was examined using the T4 gene 2− bacteriophage plating assay (32). Bacteriophage λ recombination crosses were performed as described previously (20, 24), except that IPTG induction was not necessary. The adsorption frequency of parental phages was 96.0 ± 3.1% (n = 42).
Sequence Alignments.
The sequences of RecC polypeptide were manually selected at KEGG GENES database (http://www.genome.jp/kegg/genes.html) to ensure that there were no redundant entries (e.g., fragments or mutant strains). These sequences were aligned to the E. coli protein using MAFFT (http://www.genome.jp/tools/mafft/). For the alignment, the 112 unique bacterial genes listed in the SI Materials and Methods were used.
Supplementary Material
Acknowledgments
This work was supported by the following institutions: grants from the Japan Society for the Promotion of Science Postdoctoral Fellowship for Research Abroad, The Naito Foundation, the Kato Memorial Bioscience Foundation, the Takeda Foundation, and the Sumitomo Foundation, and Grants-in-Aid for Scientific Research from both the Japan Society for the Promotion of Science and the Ministry of Education, Culture, Sports, Science and Technology (to N.H.); a Royal Society and Wellcome Trust grant (to M.S.D.); Global COE Genome Information Big Bang, Grants-in-Aid for Scientific Research, and Grant-in-Aid for Scientific Research on Innovative Areas (to I.K.); and National Institutes of Health Grant GM41347 (to S.C.K.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1206076109/-/DCSupplemental.
References
- 1.Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dillingham MS, Kowalczykowski SC. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol Mol Biol Rev. 2008;72:642–671. doi: 10.1128/MMBR.00020-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dillingham MS, Spies M, Kowalczykowski SC. RecBCD enzyme is a bipolar DNA helicase. Nature. 2003;423:893–897. doi: 10.1038/nature01673. [DOI] [PubMed] [Google Scholar]
- 4.Singleton MR, Dillingham MS, Gaudier M, Kowalczykowski SC, Wigley DB. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature. 2004;432:187–193. doi: 10.1038/nature02988. [DOI] [PubMed] [Google Scholar]
- 5.Ponticelli AS, Schultz DW, Taylor AF, Smith GR. Chi-dependent DNA strand cleavage by RecBC enzyme. Cell. 1985;41:145–151. doi: 10.1016/0092-8674(85)90069-8. [DOI] [PubMed] [Google Scholar]
- 6.Lam ST, Stahl MM, McMilin KD, Stahl FW. Rec-mediated recombinational hot spot activity in bacteriophage lambda. II. A mutation which causes hot spot activity. Genetics. 1974;77:425–433. doi: 10.1093/genetics/77.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dixon DA, Kowalczykowski SC. The recombination hotspot χ is a regulatory sequence that acts by attenuating the nuclease activity of the E. coli RecBCD enzyme. Cell. 1993;73:87–96. doi: 10.1016/0092-8674(93)90162-j. [DOI] [PubMed] [Google Scholar]
- 8.Anderson DG, Kowalczykowski SC. The recombination hot spot χ is a regulatory element that switches the polarity of DNA degradation by the RecBCD enzyme. Genes Dev. 1997;11:571–581. doi: 10.1101/gad.11.5.571. [DOI] [PubMed] [Google Scholar]
- 9.Spies M, Kowalczykowski SC. The RecA binding locus of RecBCD is a general domain for recruitment of DNA strand exchange proteins. Mol Cell. 2006;21:573–580. doi: 10.1016/j.molcel.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 10.Churchill JJ, Kowalczykowski SC. Identification of the RecA protein-loading domain of RecBCD enzyme. J Mol Biol. 2000;297:537–542. doi: 10.1006/jmbi.2000.3590. [DOI] [PubMed] [Google Scholar]
- 11.Anderson DG, Kowalczykowski SC. The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a χ-regulated manner. Cell. 1997;90:77–86. doi: 10.1016/s0092-8674(00)80315-3. [DOI] [PubMed] [Google Scholar]
- 12.Handa N, Ichige A, Kusano K, Kobayashi I. Cellular responses to postsegregational killing by restriction-modification genes. J Bacteriol. 2000;182:2218–2229. doi: 10.1128/jb.182.8.2218-2229.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blattner FR, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 14.Schultz DW, Taylor AF, Smith GR. Escherichia coli RecBC pseudorevertants lacking chi recombinational hotspot activity. J Bacteriol. 1983;155:664–680. doi: 10.1128/jb.155.2.664-680.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amundsen SK, Neiman AM, Thibodeaux SM, Smith GR. Genetic dissection of the biochemical activities of RecBCD enzyme. Genetics. 1990;126:25–40. doi: 10.1093/genetics/126.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holbeck SL, Smith GR. Chi enhances heteroduplex DNA levels during recombination. Genetics. 1992;132:879–891. doi: 10.1093/genetics/132.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spies M, Dillingham MS, Kowalczykowski SC. Translocation by the RecB motor is an absolute requirement for χ-recognition and RecA protein loading by RecBCD enzyme. J Biol Chem. 2005;280:37078–37087. doi: 10.1074/jbc.M505521200. [DOI] [PubMed] [Google Scholar]
- 18.Arnold DA, Bianco PR, Kowalczykowski SC. The reduced levels of χ recognition exhibited by the RecBC1004D enzyme reflect its recombination defect in vivo. J Biol Chem. 1998;273:16476–16486. doi: 10.1074/jbc.273.26.16476. [DOI] [PubMed] [Google Scholar]
- 19.Arnold DA, Handa N, Kobayashi I, Kowalczykowski SC. A novel, 11 nucleotide variant of χ, χ*: One of a class of sequences defining the Escherichia coli recombination hotspot χ. J Mol Biol. 2000;300:469–479. doi: 10.1006/jmbi.2000.3861. [DOI] [PubMed] [Google Scholar]
- 20.Handa N, Ohashi S, Kusano K, Kobayashi I. Chi-star, a chi-related 11-mer sequence partially active in an E. coli recC1004 strain. Genes Cells. 1997;2:525–536. doi: 10.1046/j.1365-2443.1997.1410339.x. [DOI] [PubMed] [Google Scholar]
- 21.Chaudhury AM, Smith GR. A new class of Escherichia coli recBC mutants: Implications for the role of RecBC enzyme in homologous recombination. Proc Natl Acad Sci USA. 1984;81:7850–7854. doi: 10.1073/pnas.81.24.7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Amundsen SK, Taylor AF, Chaudhury AM, Smith GR. recD: The gene for an essential third subunit of exonuclease V. Proc Natl Acad Sci USA. 1986;83:5558–5562. doi: 10.1073/pnas.83.15.5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Churchill JJ, Anderson DG, Kowalczykowski SC. The RecBC enzyme loads RecA protein onto ssDNA asymmetrically and independently of chi, resulting in constitutive recombination activation. Genes Dev. 1999;13:901–911. doi: 10.1101/gad.13.7.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Handa N, Kowalczykowski SC. A RecA mutant, RecA(730), suppresses the recombination deficiency of the RecBC(1004)D-chi* interaction in vitro and in vivo. J Mol Biol. 2007;365:1314–1325. doi: 10.1016/j.jmb.2006.10.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halpern D, et al. Identification of DNA motifs implicated in maintenance of bacterial core genomes by predictive modeling. PLoS Genet. 2007;3:1614–1621. doi: 10.1371/journal.pgen.0030153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chédin F, Noirot P, Biaudet V, Ehrlich SD. A five-nucleotide sequence protects DNA from exonucleolytic degradation by AddAB, the RecBCD analogue of Bacillus subtilis. Mol Microbiol. 1998;29:1369–1377. doi: 10.1046/j.1365-2958.1998.01018.x. [DOI] [PubMed] [Google Scholar]
- 27.el Karoui M, Ehrlich D, Gruss A. Identification of the lactococcal exonuclease/recombinase and its modulation by the putative Chi sequence. Proc Natl Acad Sci USA. 1998;95:626–631. doi: 10.1073/pnas.95.2.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sourice S, Biaudet V, El Karoui M, Ehrlich SD, Gruss A. Identification of the Chi site of Haemophilus influenzae as several sequences related to the Escherichia coli Chi site. Mol Microbiol. 1998;27:1021–1029. doi: 10.1046/j.1365-2958.1998.00749.x. [DOI] [PubMed] [Google Scholar]
- 29.McKittrick NH, Smith GR. Activation of Chi recombinational hotspots by RecBCD-like enzymes from enteric bacteria. J Mol Biol. 1989;210:485–495. doi: 10.1016/0022-2836(89)90125-3. [DOI] [PubMed] [Google Scholar]
- 30.Rinken R, de Vries J, Weichenhan D, Wackernagel W. The recA-recBCD dependent recombination pathways of Serratia marcescens and Proteus mirabilis in Escherichia coli: Functions of hybrid enzymes and hybrid pathways. Biochimie. 1991;73:375–384. doi: 10.1016/0300-9084(91)90104-9. [DOI] [PubMed] [Google Scholar]
- 31.Smith GR, Roberts CM, Schultz DW. Activity of Chi recombinational hotspots in Salmonella typhimurium. Genetics. 1986;112:429–439. doi: 10.1093/genetics/112.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oliver DB, Goldberg EB. Protection of parental T4 DNA from a restriction exonuclease by the product of gene 2. J Mol Biol. 1977;116:877–881. doi: 10.1016/0022-2836(77)90276-5. [DOI] [PubMed] [Google Scholar]
- 33.Handa N, Ichige A, Kobayashi I. Contribution of RecFOR machinery of homologous recombination to cell survival after loss of a restriction-modification gene complex. Microbiology. 2009;155:2320–2332. doi: 10.1099/mic.0.026401-0. [DOI] [PubMed] [Google Scholar]
- 34.Rinken R, Wackernagel W. Inhibition of the recBCD-dependent activation of Chi recombinational hot spots in SOS-induced cells of Escherichia coli. J Bacteriol. 1992;174:1172–1178. doi: 10.1128/jb.174.4.1172-1178.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stahl FW, Thomason LC, Siddiqi I, Stahl MM. Further tests of a recombination model in which chi removes the RecD subunit from the RecBCD enzyme of Escherichia coli. Genetics. 1990;126:519–533. doi: 10.1093/genetics/126.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saikrishnan K, Griffiths SP, Cook N, Court R, Wigley DB. DNA binding to RecD: Role of the 1B domain in SF1B helicase activity. EMBO J. 2008;27:2222–2229. doi: 10.1038/emboj.2008.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ivancić-Baće I, et al. RecFOR function is required for DNA repair and recombination in a RecA loading-deficient recB mutant of Escherichia coli. Genetics. 2003;163:485–494. doi: 10.1093/genetics/163.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang L, et al. Alteration of χ recognition by RecBCD reveals a regulated molecular latch and suggests a channel-bypass mechanism for biological control. Proc Natl Acad Sci USA. 2012;109:8907–8912. doi: 10.1073/pnas.1206081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luscombe NM, Laskowski RA, Thornton JM. Amino acid-base interactions: A three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. 2001;29:2860–2874. doi: 10.1093/nar/29.13.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davydova EK, Santangelo TJ, Rothman-Denes LB. Bacteriophage N4 virion RNA polymerase interaction with its promoter DNA hairpin. Proc Natl Acad Sci USA. 2007;104:7033–7038. doi: 10.1073/pnas.0610627104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gleghorn ML, Davydova EK, Rothman-Denes LB, Murakami KS. Structural basis for DNA-hairpin promoter recognition by the bacteriophage N4 virion RNA polymerase. Mol Cell. 2008;32:707–717. doi: 10.1016/j.molcel.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malhotra A, Severinova E, Darst SA. Crystal structure of a sigma 70 subunit fragment from E. coli RNA polymerase. Cell. 1996;87:127–136. doi: 10.1016/s0092-8674(00)81329-x. [DOI] [PubMed] [Google Scholar]
- 43.Braddock DT, Baber JL, Levens D, Clore GM. Molecular basis of sequence-specific single-stranded DNA recognition by KH domains: Solution structure of a complex between hnRNP K KH3 and single-stranded DNA. EMBO J. 2002;21:3476–3485. doi: 10.1093/emboj/cdf352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peersen OB, Ruggles JA, Schultz SC. Dimeric structure of the Oxytricha nova telomere end-binding protein alpha-subunit bound to ssDNA. Nat Struct Biol. 2002;9:182–187. doi: 10.1038/nsb761. [DOI] [PubMed] [Google Scholar]
- 45.Saikrishnan K, et al. Insights into Chi recognition from the structure of an AddAB-type helicase-nuclease complex. EMBO J. 2012;31:1568–1578. doi: 10.1038/emboj.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Touzain F, Petit MA, Schbath S, El Karoui M. DNA motifs that sculpt the bacterial chromosome. Nat Rev Microbiol. 2011;9:15–26. doi: 10.1038/nrmicro2477. [DOI] [PubMed] [Google Scholar]
- 47.Petrek M, Kosinová P, Koca J, Otyepka M. MOLE: A Voronoi diagram-based explorer of molecular channels, pores, and tunnels. Structure. 2007;15:1357–1363. doi: 10.1016/j.str.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 48.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schneider TD, Stephens RM. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.