Abstract
We have previously shown that broadly neutralizing antibodies reactive to the conserved stem region of the influenza virus hemagglutinin (HA) were generated in people infected with the 2009 pandemic H1N1 strain. Such antibodies are rarely seen in humans following infection or vaccination with seasonal influenza virus strains. However, the important question remained whether the inactivated 2009 pandemic H1N1 vaccine, like the infection, could also induce these broadly neutralizing antibodies. To address this question, we analyzed B-cell responses in 24 healthy adults immunized with the pandemic vaccine in 2009. In all cases, we found a rapid, predominantly IgG-producing vaccine-specific plasmablast response. Strikingly, the majority (25 of 28) of HA-specific monoclonal antibodies generated from the vaccine-specific plasmablasts neutralized more than one influenza strain and exhibited high levels of somatic hypermutation, suggesting they were derived from recall of B-cell memory. Indeed, memory B cells that recognized the 2009 pandemic H1N1 HA were detectable before vaccination not only in this cohort but also in samples obtained before the emergence of the pandemic strain. Three antibodies demonstrated extremely broad cross-reactivity and were found to bind the HA stem. Furthermore, one stem-reactive antibody recognized not only H1 and H5, but also H3 influenza viruses. This exceptional cross-reactivity indicates that antibodies capable of neutralizing most influenza subtypes might indeed be elicited by vaccination. The challenge now is to improve upon this result and design influenza vaccines that can elicit these broadly cross-reactive antibodies at sufficiently high levels to provide heterosubtypic protection.
Keywords: immunity, stalk, immunoglobulin, neutralization
Despite the availability of vaccines that can reduce the incidence and severity of disease, influenza remains the most common cause of morbidity and mortality by respiratory infection worldwide (1), with young children, immunocompromised patients, and the elderly at particular risk. Vaccine effectiveness is limited by antigenic variability, which occurs by antigenic drift or shift. The latter can cause devastating pandemics when lack of preexisting immunity is combined with mutations that increase viral pathogenicity. Current vaccines rely on trivalent inactivated (TIV) or live attenuated vaccines that contain components from prevailing strains of influenza A/H1N1, A/H3N2, and B. These vaccines elicit neutralizing antibodies directed against hemagglutinin and, to a lesser extent, neuraminidase that are key to their protective efficacy but are overcome by rapid antigenic evolution. Developing a vaccine effective in the face of antigenic variation is the central challenge for influenza research.
Although it was long believed that all effective neutralizing Abs were directed against the receptor-binding site in the globular head of hemagglutinin (HA), several recent reports have indicated that Abs against the stem region can occur in humans (2–7). These Abs were shown to have neutralizing activity against multiple influenza subtypes and were identified using a number of systems including phage display (2–4) and immortalization of memory B cells (5). We then demonstrated that these stem-reactive Abs were readily detected in patients infected with the pandemic H1N1 2009 influenza (referred to as pH1N1 2009 from here on) (7). By generating monoclonal Abs (mAbs) from plasmablasts isolated ex vivo, we showed that broadly cross-reactive stem-binding Abs dominated the B cell response in some patients. A vaccine that could elicit these Abs at a sufficiently high level to provide heterosubtypic protection would be an important achievement. Recent studies in mice have shown that sequential infection with influenza virus strains expressing different HAs (8) or vaccination with stem-only constructs can induce broadly cross-reactive Abs (9). However, rarely have these been found in humans following seasonal influenza vaccination (10, 11), and it was therefore uncertain whether such Abs could be induced in humans following immunization with the inactivated pH1N1 2009 vaccine.
To address this question, we examined the Ab responses in healthy adults vaccinated with the monovalent pH1N1 vaccine in 2009, the year the pandemic strain emerged. Our data demonstrate that it is indeed possible to induce broadly cross-reactive Abs by vaccination. The majority of HA-specific mAbs from healthy adult volunteers given the pH1N1 2009 vaccine displayed broad cross-reactivity to the HA head; in addition, three mAbs were shown to bind the HA stem. Although the frequency of stem-reactive Abs was low and not seen in all vaccinees, they were readily identified and demonstrated exceptionally broad cross-reactivity.
Results
Subunit Pandemic H1N1 2009 Vaccine Induces Rapid Expansion of Antigen-Specific Plasmablasts.
We examined humoral immune responses in 24 healthy adults (26–64 y old) immunized with the subunit pH1N1 2009 vaccine (Table S1). Subjects entered the study ∼6 mo after the first reports of pH1N1 2009 cases. The vaccine was given separately after the 2009 seasonal influenza vaccine, which contained a different H1N1 strain (A/Brisbane/59/07). Seventeen individuals (71%) receiving pH1N1 2009 vaccine demonstrated an increase in hemagglutination inhibition (HAI) titer at 28 d postvaccination (with at least a fourfold increase in HAI titer) (Fig. 1A).
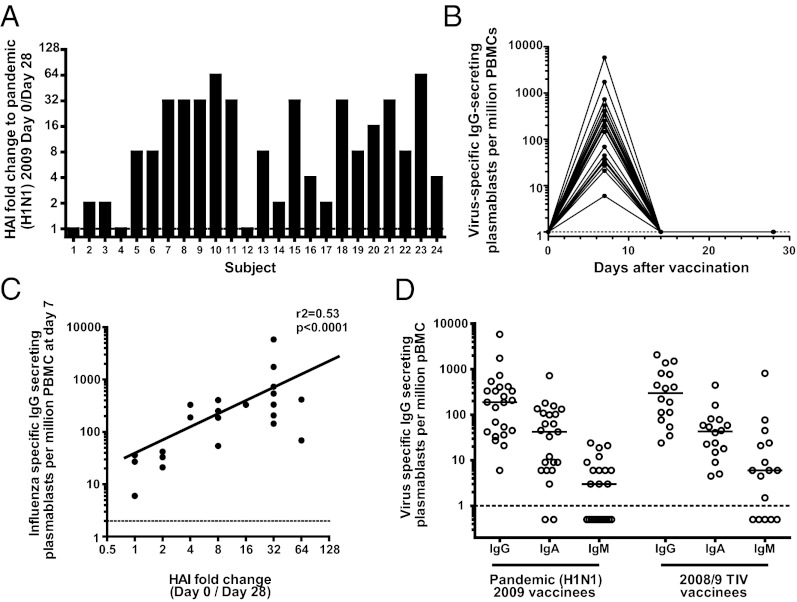
Fig. 1.
Rapid and potent plasmablast and serological responses after vaccination with the subunit pH1N1 2009 vaccine. Healthy adult volunteers were vaccinated with the subunit pH1N1 2009 monovalent vaccine. (A) Fold change in serum antibody titers between day 0 and 28 determined by HAI. (B) The number of vaccine-specific IgG-producing plasmablasts were determined by ELISPOT after vaccination. (C) The number of vaccine-specific plasmablasts correlates with increased serum antibody titers by HAI (Spearman’s correlation). (D) The numbers of influenza-specific IgG-, IgA-, and IgM-producing plasmablasts at day 7 as determined by ELISPOT after immunization with the monovalent pH1N1 2009 vaccine compared with the response elicited by the seasonal 2008/2009 TIV. These TIV data are from a cohort vaccinated in 2008. Dotted lines indicate limit of detection.
We previously demonstrated that seasonal TIV vaccination led to a large, transient expansion of Ab-secreting cells (plasmablasts) in the blood (11). Here, we show that the vaccine-specific plasmablast response to pH1N1 vaccination also peaked at day 7 before returning to background levels by day 14 (Fig. 1B). These kinetics were identical to vaccinees who were given the 2008/09 TIV [containing influenza A/Brisbane/59/07 H1N1, A/Brisbane 10/07 (H3N2), and B/Florida/4/06] (Fig. S1). Following immunization with the pH1N1 2009 vaccine, there was a positive correlation between the increases in HAI titer and peak plasmablast numbers (r2 = 0.53, P < 0.0001; Fig. 1C). The rapidity of the plasmablast response suggested a recall rather than primary response and, moreover, IgG-producing greatly outnumbered IgM-producing cells (Fig. 1D). This was also seen in the response to 2008/09 TIV.
Plasmablasts Induced by the Subunit Pandemic H1N1 2009 Vaccine Cross-React with the Most Recent Seasonal Influenza Strain.
The features of the plasmablast response to the pH1N1 2009 vaccine were suggestive of memory recall, so we wished to determine the extent to which plasmablasts could also be induced that were reactive against the seasonal influenza strain from the previous 2 y found in the 2009/10 TIV (A/Brisbane/59/07). The HA of the pH1N1 2009 strain diverged considerably from that of influenza A/Brisbane/59/07, with only 79% sequence homology (Fig. S2). Despite this, most individuals, after vaccination with the pH1N1 2009 vaccine, generated a large number of plasmablasts that reacted with the 2009/10 TIV (Fig. S3A). To enrich for plasmablasts, we sorted these cells by flow cytometry from 10 individuals at day 7 (Fig. S3B). A high proportion of sorted plasmablasts were antigen specific (Fig. S3C). In addition, we detected plasmablasts with specificity for A/Brisbane/59/07 HA as well as pH1N1 2009 HA in all sorted samples (Fig. S3D). These data show that the humoral response to vaccination had activity against both the homologous antigen and a heterologous HA from the seasonal influenza strain of the preceding 2 y.
Pandemic H1N1 2009 Vaccine Can Induce Antibodies That Bind the HA Stem.
To examine the specificities of the Ab response to pH1N1 2009 vaccine at the monoclonal level, we used single-cell RT-PCR of individual plasmablasts to produce mAbs as described (7, 11, 12). In total, we generated 78 mAbs from eight subjects. By ELISA, 58% (45 of 78) bound to purified pH1N1 2009 virus (Fig. 2A). Of these, 62% (28 of 45) bound to recombinant HA from the pandemic strain. Of the mAbs that demonstrated HA-specific binding, 89% (25 of 28) were shown to have functional activity against pH1N1 2009 virus by HAI and/or neutralization assay (Fig. 2B).
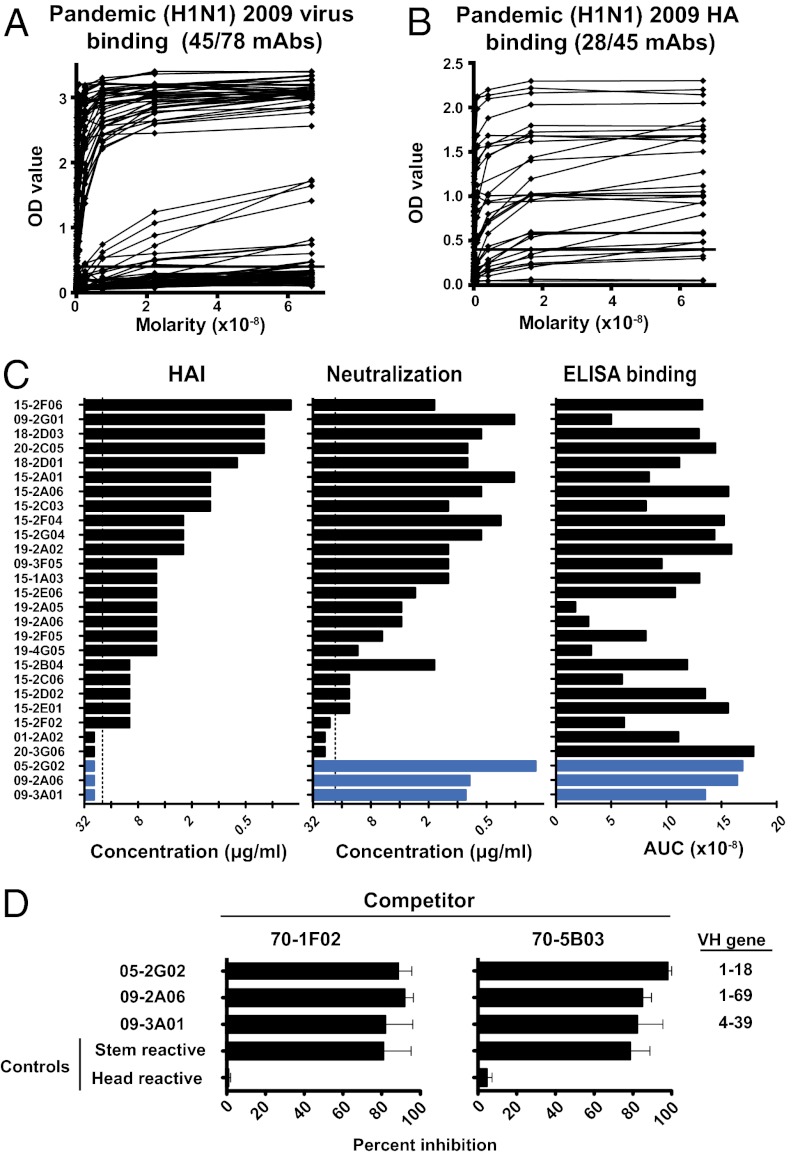
Fig. 2.
Stem-binding antibodies are induced following pH1N1 2009 vaccination. Human mAbs were generated from plasmablasts isolated from pH1N1 2009 vaccinees. (A) Binding of mAbs to the pH1N1 2009 virus by ELISA. (B) Binding to pH1N1 2009 HA by ELISA. (C) HA-binding mAbs were tested for HAI and neutralization activity. Three putative stem-binding mAbs are shown in blue. Dotted lines show the highest concentration of mAb tested. Data are representative of two to four repeat experiments. (D) The three putative stem-binding mAbs were tested by competition ELISA with two known stem-binding mAbs (70-1F02 and 70-5B03) (7). The reciprocal stem-binding mAb was used as a positive control, and a previously described HA-head-specific antibody (EM4C04) was used as a negative control. Bars represent means ± SEM for three repeats. The VH gene use of the individual stem-binding mAbs is shown on the right.
We previously showed that mAbs recognizing epitopes in the globular head of the influenza HA demonstrated binding by ELISA, positive HAI, and virus neutralization (7). In contrast, stem-binding mAbs showed binding by ELISA and neutralization, but negative HAI. In this set of mAbs, three mAbs (05-2G02, 09-2A06, and 09-3A01) were found to have no HAI activity despite binding by ELISA and neutralization, a pattern suggestive of stem-binding mAbs. To confirm this, we proceeded to compare their binding with known stem-reactive Abs (70-1F02 or 70-5B03) using competition ELISA (Fig. 2C). All three potential stem-binding mAbs were inhibited by greater than 80%, comparable to the stem-binding mAbs used as positive controls. Thus, by competition ELISA, we demonstrated that mAbs 05-2G02, 09-2A06, and 09-3A01 all compete for binding to epitope(s) in the same region of the HA stem.
The three stem-binding mAbs all used different VH gene segments (Fig. 2C and Table S2). This contrasted with the pH1N1 2009 infection, where the majority of mAbs used the VH1–69 gene segment, which was also shared by other reported stem-binding Abs (3, 4). Here, only one mAb used the VH1–69, although a second used the similar VH1–18. Taken together, our data suggest that stem-reactive Abs can indeed be elicited by the subunit pH1N1 2009 vaccine, but occur at a lower frequency than we observed after infection with the 2009 pandemic strain (7).
Monoclonal Antibodies Elicited by Pandemic H1N1 2009 Vaccine Cross-React with Antigenically Divergent Strains.
We tested all HA-specific mAbs for cross-reactivity against a panel of antigens and virus strains, including the pH1N1 1918 strain and more antigenically diverse H1N1, H5N1, and H3N2 strains. Strikingly, the majority of mAbs that bound the HA head also demonstrated broad cross-reactivity (Fig. 3A). Eighteen of 28 Abs were able to bind all three H1N1 HAs, while 6 of 28 bound both pH1N1 2009 and p1918 influenza HAs. The high degree of cross-reactivity suggested that many plasmablasts had arisen by secondary expansion of cross-reactive memory B cells targeting conserved epitopes. Comparing the binding of these mAbs to the most recent prepandemic seasonal H1N1 strain (A/Brisbane/59/07), the patterns of cross-reactivity generally conformed to three categories (Fig. S4). Half (14 of 28) of the Abs bound better to the pH1N1 2009 HA, suggesting ongoing adaptation through affinity maturation. Other Abs bound equally well to both HAs (9 of 28), whereas the last category (5 of 28) bound better to the Brisbane HA, consistent with original antigenic sin (OAS). Furthermore, the three stem-binding mAbs demonstrated the widest cross-reactivity by ELISA with detectable binding to all of the H1 HAs tested plus HA from the H5N1 strain (A/Indonesia/05/2005) (Fig. 3A). Finally, 05-2G02 displayed even greater cross-reactivity by also binding H3, albeit weakly.
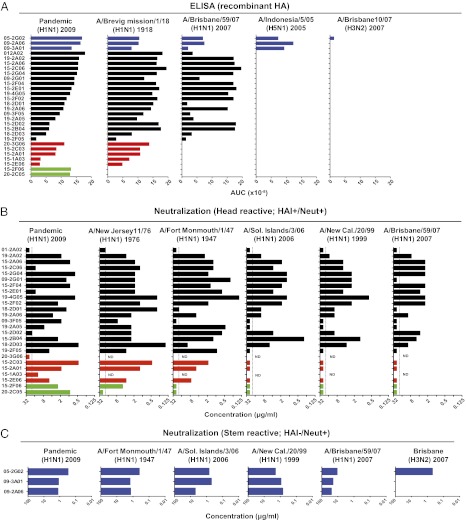
Fig. 3.
The pH1N1 2009 vaccine induces highly cross-reactive HA-specific antibodies. (A) pH1N1 2009 HA-binding mAbs were tested for binding to HAs from the indicated influenza strains by ELISA. Monoclonal antibodies are arranged according to degree of binding by ELISA to pH1N1 2009 HA and grouped according to cross-reactivity by ELISA (blue: stem-binders, bind all H1N1, H5N1, and H3N2; black: bind all H1N1; red: bind A/California/04/09 and A/Brevig Mission/1/18; green: bind A/California/04/09 only). (B) HA head-binding mAbs were tested for neutralizing activity against the indicated panel of H1N1 virus strains. Two mAbs (20-3G06 and 15-1A03) expressed poorly and were not tested for cross-reactivity (ND). (C) Three stem-binding mAbs were tested for neutralizing activity against various influenza virus strains. Influenza strains are arranged from left to right in order of sequence similarity to the pH1N1 2009. Dotted lines represent limits of detection. Data are representative of two to four repeats.
We next performed HAI and neutralization assays with the head-reactive antbodies using a more extensive panel of H1N1 virus strains including recent seasonal strains and historic outbreak strains (Fig. 3B and Fig. S5). These displayed a broad range of sequence diversity compared with the pH1N1 2009 virus. As expected from sequence homology (Fig. S2), the highest degree of cross-reactivity by neutralization assay was seen with A/New Jersey/76, with 68% of mAbs cross-neutralizing. Of the more recent seasonal strains, up to 43% of mAbs demonstrated cross-reactivity between the seasonal strains and pH1N1 2009 virus. In general, the fraction of cross-neutralizing mAbs paralleled sequence homology. Still, given the large antigenic differences measured by standard reference sera, the fraction of cross-neutralizing mAbs was much larger than expected.
We next analyzed neutralization by the stem-reactive Abs (Fig. 3C). All three stem-binding mAbs demonstrated broad cross-reactivity with the capacity to neutralize all H1N1 strains tested. The exceptional breadth of 05-2G02 was again shown, with neutralizing activity against H3N2 as well as the H1N1 strains. Abs that neutralize influenza strains from both phylogenetic group 1 and group 2 are exceedingly rare and have only been reported once in the literature (6). These data demonstrate the high degree of cross-reactivity of mAbs generated following pH1N1 2009 vaccination. This was true not only of the stem-reactive mAbs, one of which had unusually broad cross-reactivity against H1, H5, and H3, but also of the majority of non-stem-binding mAbs, which demonstrated substantial cross-reactivity within H1N1 strains in contrast to the more strain-specific mAbs generated following seasonal TIV (11).
Pandemic H1N1 2009 Vaccine Induces Monoclonal Antibodies with High Levels of Somatic Hypermutation.
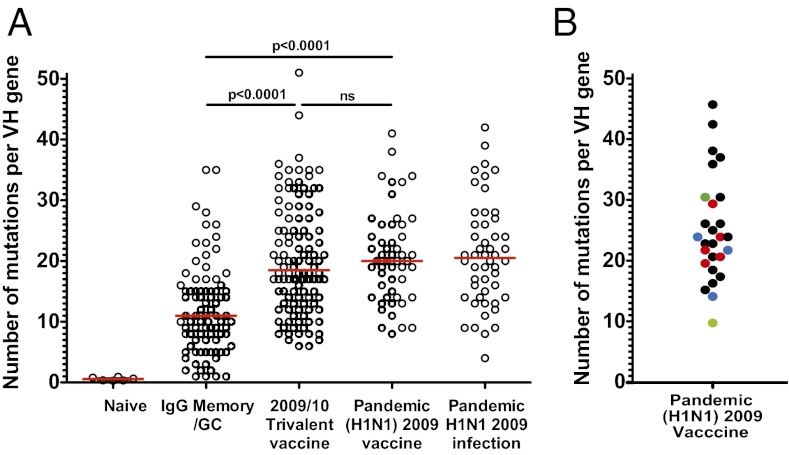
The kinetics of the response, the dominance of IgG-secreting cells and the remarkable cross-reactivity of individual mAbs point to a memory origin for most clones. This was supported by sequence analysis of virus-specific mAbs (Fig. 4A), which showed that most clones carried a very high number of VH gene mutations (median 21, range 8–41). This was significantly higher (P < 0.0001) than the average IgG-producing memory B cell or germinal center B cell (median 11, range 1–35) but similar to the number found in memory B cells responding to seasonal influenza vaccination (median 18.5, range 6–51) or results of our previous study of mAbs from patients infected with pH1N1 2009 virus. When the HA-specific mAbs were analyzed alone (Fig. 4B), they displayed similar levels of mutation compared with the virus-specific mAbs as a whole. There was no obvious correlation between the number of mutations and the degree of cross-reactivity of each individual mAb.
Fig. 4.
Monoclonal antibodies induced following the pH1N1 2009 vaccine display high levels of somatic hypermutation consistent with a recall response. Variable genes from plasmablasts induced following the pH1N1 2009 vaccine were amplified by single-cell RT-PCR and scored for numbers of somatic mutations. (A) The number of mutations per VH gene following pH1N1 2009 vaccination is compared with published data (11, 23–25). The red line shows the mean (P values by Student t tests). (B) The number of mutations per VH gene in HA-specific mAbs only. Colors represent the degree of cross-reactivity as in Fig 3.
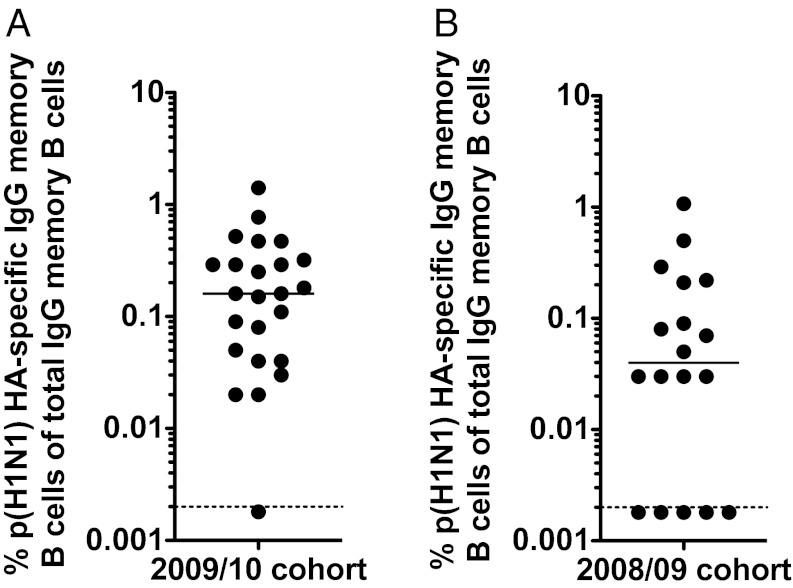
Memory B Cells Reactive to the Pandemic Strain Are Present Before Its Emergence.
We hypothesized that cross-reactive memory B cells capable of reacting to the pH1N1 2009 HA were already present before vaccination. We therefore analyzed prevaccination samples by memory B-cell assays (11) (Fig. 5A). All subjects had detectable memory B cells reactive against pH1N1 2009 HA before vaccination (median 0.4%, range 0.01–1.98%). However, subclinical infections with influenza that induce seroconversion without symptoms do occur and go unreported (13). It was therefore possible that some of the vaccinees had been exposed to pH1N1 2009 virus between its emergence (March 2009) and their vaccination (October 2009). To exclude this possibility, we tested baseline samples from a healthy cohort taken in 2008/09 before the emergence of the novel pandemic strain (Fig. 5B). Once again, memory B cells that reacted against the pH1N1 2009 HA could be detected in the majority of these specimens. In summary, these data show that memory B cells reactive to pH1N1 HA were indeed present in individuals, strongly suggesting that the pH1N1 2009 vaccine preferentially activates these cross-reactive memory B cells, thereby producing a humoral response with broad neutralizing activity.
Fig. 5.
Memory B cells reactive to the pH1N1 2009 influenza are detectable even before the emergence of the pandemic strain. Peripheral blood mononuclear cells (PBMCs) obtained before vaccination were tested for the presence of memory B cells reactive against the pH1N1 2009 HA as described (11). The pH1N1 HA-specific IgG memory B cell frequencies are shown in subjects from the year that the pH1N1 2009 emerged (2009/10) (A) and the previous year (2008/09) (B).
Discussion
Here, we examined the B-cell response in humans immunized with the inactivated 2009 pH1N1 influenza vaccine. Our results make the following points: (i) The inactivated 2009 pandemic influenza vaccine induces a broadly cross-reactive antibody response. (ii) Cross-reactive antibodies were generated against the conserved stem region of the HA molecule. (iii) In addition, antibodies generated against the variable head region of the HA also showed greater breadth than is typically seen after vaccination with the seasonal influenza vaccine. (iv) All of these antibodies exhibited high levels of somatic hypermutaion. (v) Memory B cells reactive to the HA of the pH1N1 2009 strain were detectable at low frequencies even before the emergence of the pandemic strain. Taken together, these results show that immunization with a dramatically altered HA induces a recall response in humans that favors broadly cross-reactive memory B cells. These findings have important implications for influenza immunization strategies and for the design of “universal” influenza vaccines.
In this study, we interrogated the B-cell response by generating human monoclonal antibodies from vaccine-induced plasmablasts. This approach has two major advantages. First, these were influenza-specific B cells proliferating in response to the specific vaccine and not resting memory B cells, which consist of a range of clones generated by a variety of previous exposures. Second, whereas other techniques use antigen to preferentially enrich for B cells with the specificities of interest, analysis of all plasmablasts allowed us to examine the repertoire with minimal bias toward a specific form of the antigen. Using this approach, we showed that the B-cell responses to the pH1N1 2009 and seasonal vaccines were comparable in many ways (11). Both vaccines induced large and rapid plasmablast responses. Both responses were predominantly made up of isotype-switched IgG-producing plasmablasts and mAbs generated from both showed extensive somatic hypermutation. These features imply that the response to the pH1N1 2009 vaccine arises from preexisting memory B cells. This was supported by the presence of memory B cells specific for pH1N1 2009 HA in individuals even before the emergence of the new virus, strongly implying they were induced by exposure to previous strains.
However, the Ab response to pH1N1 2009 vaccine was distinctive in one important respect: the high degree of cross-reactivity. Unlike previous studies of seasonal TIV, our data showed that cross-reactive Abs against both the head and stem of HA made up a large proportion of the response. Cross-reactive Abs induced by the seasonal TIV have been described in humans using a number of systems (2–5, 14), but their involvement in protective immunity has been difficult to elucidate. The extent to which this reflects their low concentration, poor function, or an underestimation of their importance is uncertain. Stem-binding Abs identified following vaccination were found using phage display libraries (2–4) or immortalization of memory B cells (5), and it has therefore not been possible to assess their overall contribution to the vaccine-induced response. Interestingly, a recent study (16) showed that the MF59 adjuvant can enhance the diversity and affinity of the antibody response to pandemic influenza vaccine, and another study showed that DNA priming can enhance antibody responses to the H5N1 vaccine (17).
Three stem-binding mAbs were identified in our study. One of them, 05-2G02, demonstrated an extraordinary breadth of neutralizing activity, with activity against all H1N1 strains tested as well as binding to H5N1 HA and neutralization of a H3N2 strain. This is reminiscent of a recently identified highly cross-reactive mAb that neutralized both group 1 and group 2 influenza viruses, with binding to all 16 HA subtypes (6). Interestingly, these two mAbs use different VH genes, suggesting that the capacity to recognize HAs from both phylogenetic groups may not be dependent on a unique antigen-binding structure. Both Abs provide important proof of concept that a universal vaccine is possible. However, it is clear that cross-reactive stem-binding Abs are very rare after seasonal vaccination. Studies describing stem-binding memory B-cell clones have required high-throughput techniques (5). In contrast, stem-reactive Abs were readily found following the pH1N1 2009 vaccine, implying that these Abs were induced more frequently as a consequence of the major change in epitopes from the HA head while the stem was conserved. In addition, although two stem-specific mAbs came from one subject and one stem-specific mAb came from another, several subjects had none, suggesting that the propensity for developing cross-reactive Abs might be due to the underlying B-cell repertoire or previous antigenic history.
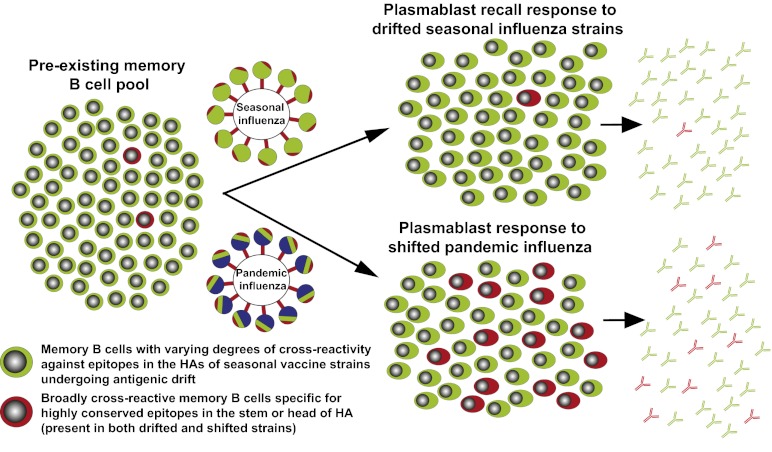
Why does the pandemic influenza vaccine induce more broadly cross-reactive antibody responses compared with the seasonal vaccine? It should be noted that both vaccines induced a predominantly B-cell recall response. Thus, a possible reason for the observed differences in the breadth of the response could be due to competition between the large pool of influenza-virus-specific memory B cells that recognize dominant but variable regions of the HA and the much rarer memory B cells that recognize conserved but subdominant HA epitopes. A model based on this competition is described in Fig. 6. Annual vaccination with the seasonal influenza virus strains that differ by only a few mutations from year to year (antigenic drift) will continue to recruit similar memory B cells that recognize dominant epitopes in the globular head of the HA and comprise the major pool of the memory cell population. Under these conditions, there is minimal chance of activating the rare memory B cells that recognize conserved and subdominant epitopes in the HA stem (or head). However, following immunization or infection with the pandemic strain that contains a dramatically altered HA (antigenic shift), there is now a greater probability of activating the rare memory B cells that recognize conserved HA epitopes. Under these conditions, there is much less competition because the shifted HA is missing many of the dominant and variable epitopes recognized by the majority of preexisting influenza-specific memory B cells.
Fig. 6.
A model contrasting the antibody response induced after infection or vaccination with seasonal versus pandemic influenza virus strains. The preexisting influenza-specific B-cell pool primarily consists of memory cells that recognize epitopes from recent seasonal strains in the globular head of HA that change relatively little (drift) year to year (shown in green). These memory B cells are repeatedly expanded following infection or vaccination with the seasonal influenza virus strains, whereas the less frequent memory B cells specific for highly conserved epitopes in the stem and head of HA (shown in red) are crowded out. With a pandemic strain, many epitopes in the HA head are replaced with new epitopes (depicted in blue), whereas conserved epitopes in the stem and head remain. Cross-reactive memory B cells specific for the conserved epitopes now have a greater chance of being recruited into the response. Naive responses to the novel epitopes are likely also induced but, based on the Ig isotype and the rapid appearance of the plasmablast response, it is unlikely that these newly induced naive B cells make up a significant proportion of the overall response.
These findings might offer an explanation as to why the preceding seasonal H1N1 strain almost completely disappeared following the emergence of the pH1N1 2009 virus (18–20). Our studies in individuals infected or vaccinated with pH1N1 2009 have shown that, in either situation, large numbers of cross-reactive Abs with activity against A/Brisbane/59/07 are generated (7). Thus, most individuals who have encountered the pH1N1 2009 strain will also have developed protective immunity against A/Brisbane/59/07, leading to a rapid decrease in the number of susceptible hosts.
Our data show that broadly cross-reactive stem-binding Abs can be induced by the pH1N1 2009 vaccine, thus demonstrating that productive infection is not necessary. However, the frequency of these stem-binding Abs following the pH1N1 2009 vaccine was low and could not be detected in all vaccinees. In order for a truly universal vaccine to provide robust heterosubtypic immunity, it must induce cross-reactive Abs to a high level in all recipients. Current vaccines are therefore inadequate, and the design of a vaccine with improved immunogenicity that preferentially stimulates protective cross-reactive responses remains one of the great challenges ahead.
Materials and Methods
All studies were approved by the Emory University institutional review board. Twenty-four healthy adult volunteers were given the monovalent pH1N1 2009 vaccine and were compared with data from 2008/09 and 2009/10 seasonal TIV vaccinees. Viral stocks were provided by R. J. Webby (St. Jude Children’s Hospital, Memphis, TN) or the Centers for Disease Control (CDC, Atlanta, GA). Recombinant HA proteins were provided by the CDC and by the Biodefense and Emerging Infections research repository. Direct ELISPOT to enumerate total and HA-specific plasmablasts were performed as described (7). Analytical and cell sorting flow cytometry analysis was performed as described (11). Monoclonal Abs were generated and analyzed as described (11, 12, 21). Whole virus, recombinant HA, and vaccine-specific ELISA, as well as HAI, neutralization, and competition assays were performed as described (11). For more details, see SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Becky Gherkin, Robert Karaffa, and Sommer Durham for technical support. This work was funded in parts by National Institutes of Health (NIH)/National Institute of Allergy and Infectious Diseases (NIAID) Grant U19-AI057266 with American Recovery and Reinvestment Act Supplement U19 AI057266-06S2 (to R.A. and P.C.W.); NIH/NIAID Awards HHSN266200700006C Center of Excellence for Influenza Research and Surveillance (to R.A. and P.C.W.), HHSN266200500026C (to P.C.W.), and 5U19AI062629-05 (to P.C.W.); and the Intramural Research Program of the NIAID (J.W.Y.).
Footnotes
Conflict of interest statement: R.A., J.W., and P.C.W. have a licensing agreement with MedImmune on the influenza-virus-specific human monoclonal antibodies.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. JX027379–JX027476).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118979109/-/DCSupplemental.
References
- 1.Beigel JH. Influenza. Crit Care Med. 2008;36:2660–2666. doi: 10.1097/CCM.0b013e318180b039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Throsby M, et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS ONE. 2008;3:e3942. doi: 10.1371/journal.pone.0003942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ekiert DC, et al. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009;324:246–251. doi: 10.1126/science.1171491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sui J, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corti D, et al. Heterosubtypic neutralizing antibodies are produced by individuals immunized with a seasonal influenza vaccine. J Clin Invest. 2010;120:1663–1673. doi: 10.1172/JCI41902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corti D, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333:850–856. doi: 10.1126/science.1205669. [DOI] [PubMed] [Google Scholar]
- 7.Wrammert J, et al. Broadly cross-reactive antibodies dominate the human B cell response against 2009 pandemic H1N1 influenza virus infection. J Exp Med. 2011;208:181. doi: 10.1084/jem.20101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang TT, et al. Broadly protective monoclonal antibodies against H3 influenza viruses following sequential immunization with different hemagglutinins. PLoS Pathog. 2010;6:e1000796. doi: 10.1371/journal.ppat.1000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang TT, et al. Vaccination with a synthetic peptide from the influenza virus hemagglutinin provides protection against distinct viral subtypes. Proc Natl Acad Sci USA. 2010;107:18979–18984. doi: 10.1073/pnas.1013387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hancock K, et al. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N Engl J Med. 2009;361:1945–1952. doi: 10.1056/NEJMoa0906453. [DOI] [PubMed] [Google Scholar]
- 11.Wrammert J, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith K, et al. Rapid generation of fully human monoclonal antibodies specific to a vaccinating antigen. Nat Protoc. 2009;4:372–384. doi: 10.1038/nprot.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papenburg J, et al. Household transmission of the 2009 pandemic A/H1N1 influenza virus: Elevated laboratory-confirmed secondary attack rates and evidence of asymptomatic infections. Clin Infect Dis. 2010;51:1033–1041. doi: 10.1086/656582. [DOI] [PubMed] [Google Scholar]
- 14.Whittle JR, et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci USA. 2011;108:14216–14221. doi: 10.1073/pnas.1111497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khurana S, et al. MF59 adjuvant enhances diversity and affinity of antibody-mediated immune response to pandemic influenza vaccines. Sci Transl Med. 2011;3 doi: 10.1126/scitranslmed.3002336. 85ra48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ledgerwood JE, et al. DNA priming and influenza vaccine immunogenicity: Two phase 1 open label randomised clinical trials. Lancet Infect Dis. 2011;11:916–924. doi: 10.1016/S1473-3099(11)70240-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pica N, et al. Hemagglutinin stalk antibodies elicited by the 2009 pandemic influenza virus as a mechanism for the extinction of seasonal H1N1 viruses. Proc Natl Acad Sci USA. 2012;109:2573–2578. doi: 10.1073/pnas.1200039109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahmed R, Oldstone MB, Palese P. Protective immunity and susceptibility to infectious diseases: Lessons from the 1918 influenza pandemic. Nat Immunol. 2007;8:1188–1193. doi: 10.1038/ni1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dormitzer PR, et al. Influenza vaccine immunology. Immunol Rev. 2011;239:167–177. doi: 10.1111/j.1600-065X.2010.00974.x. [DOI] [PubMed] [Google Scholar]
- 21.Wardemann H, et al. Predominant autoantibody production by early human B cell precursors. Science. 2003;301:1374–1377. doi: 10.1126/science.1086907. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.