Abstract
The lux operon derived from Photorhabdus luminescens incorporated into bacterial genomes, elicits the production of biological chemiluminescence typically centered on 490 nm. The light-producing bacteria are widely used for in vivo bioluminescence imaging. However, in living samples, a common difficulty is the presence of blue-green absorbers such as hemoglobin. Here we report a characterization of fluorescence by unbound excitation from luminescence, a phenomenon that exploits radiating luminescence to excite nearby fluorophores by epifluorescence. We show that photons from bioluminescent bacteria radiate over mesoscopic distances and induce a red-shifted fluorescent emission from appropriate fluorophores in a manner distinct from bioluminescence resonance energy transfer. Our results characterizing fluorescence by unbound excitation from luminescence, both in vitro and in vivo, demonstrate how the resulting blue-to-red wavelength shift is both necessary and sufficient to yield contrast enhancement revealing mesoscopic proximity of luminescent and fluorescent probes in the context of living biological tissues.
Keywords: quantum dot, optical molecular imaging, Escherichia coli, Staphyloccus aureus, ex vivo
Light-based optical methods are emerging at the cutting edge of molecular imaging technologies. However, many challenges remain including significant practical limitations, and especially the poor propagation of blue-green light in living tissues (1, 2). Peak photon absorption by mammalian tissues is mainly determined by the presence of oxyheamoglobin, deoxyheamoglobin, and melanin, which dramatically reduce light propagation, deteriorating efficient photon detection (3, 4). Numerous studies have aimed at the development of red-shifted optical probes that emit in the range 700–900 nm where absorption of photons is minimal, reducing signal loss and allowing for deep tissue imaging in vivo. However, although there has been considerable success with efforts to red-shift fluorescent chemical and genetically engineered probes (4, 5), there has been rather less progress toward the goal of red-shifting chemiluminescent and bioluminescent probes (6, 7). For example, methods like random mutagenesis of Renilla reniformis luciferase have achieved red-emission shifts of only 547 nm (4, 8), a peak luminescence wavelength far from the optimal target range of 700–900 nm. As an alternative approach, some studies have proposed red shift of blue bioluminescence using bioluminescence resonance energy transfer (BRET). For example, So et al. (9) demonstrated “self-illuminating quantum dots,” where blue luminescence from a mutant of Renilla reniformis luciferase was red-shifted by covalently coupling to carboxylate-functionalized quantum dots (QDs), providing a new emission maximum at 655 nm. Likewise, a modified Photinus pyralis luciferase was closely bound to the commercially available Alexa Fluor 750, resulting in light emission of 780 nm (10). Similarly, far-red bioluminescent protein was constructed from Cypridina luciferase conjugated to an indocyanine dye, giving a red shift to 675 nm (11). In another study, BRET3 used a red-shifted Renilla luciferase conjugated to a mutant red fluorescent protein (mOrange, 564 nm emission; ref. 12). In all these cases, the mechanism of red-shifted bioluminescent probe functioning was defined as BRET, implying that two fundamental criteria were satisfied. First, a spectral overlap between a bioluminescent-donor and fluorescent-acceptor; second, that the distance between these moieties should be 10 nm, or less. Satisfying these conditions, a dipole–dipole interaction may occur between the moieties prerequisite to nonradiative resonant energy transfer between the donor and the acceptor resulting in a desirable red shift of the emission light (9–12).
We previously reported a luminescence red-shift phenomenon constituted in a simple, robust, and reproducible manner using blue bioluminescent RT57 bacteria (a nonpathogenic Escherichia coli strain expressing the lux operon on a plasmid, see Materials and Methods) and red-emission quantum dots (QD705) (13). There we provided some evidence demonstrating a detectable red shift occurred, apparently independent from covalent conjugation of the moieties, and hypothesized that the phenomenon arose from a conventional excitation–emission process between the luminescent light source and the fluorophore. In the current study, we report a characterization “fluorescence by unbound excitation from luminescence” (FUEL), a method that utilizes this conventional epifluorescence phenomenon.
We show that FUEL is a conventional radiating, excitation–emission process occurring under experimental conditions entirely precluding BRET. Further, we demonstrate that FUEL is preserved in vivo under standard imaging conditions, and that its spectral characteristics make it highly exploitable therein. Our results suggest FUEL is an important and overlooked component of BRET phenomena, potentially altering the empirical, quantitative, and experimental interpretation of such studies.
Results
Fluorescence Excitation by Bacterial Luminescence in Vitro.
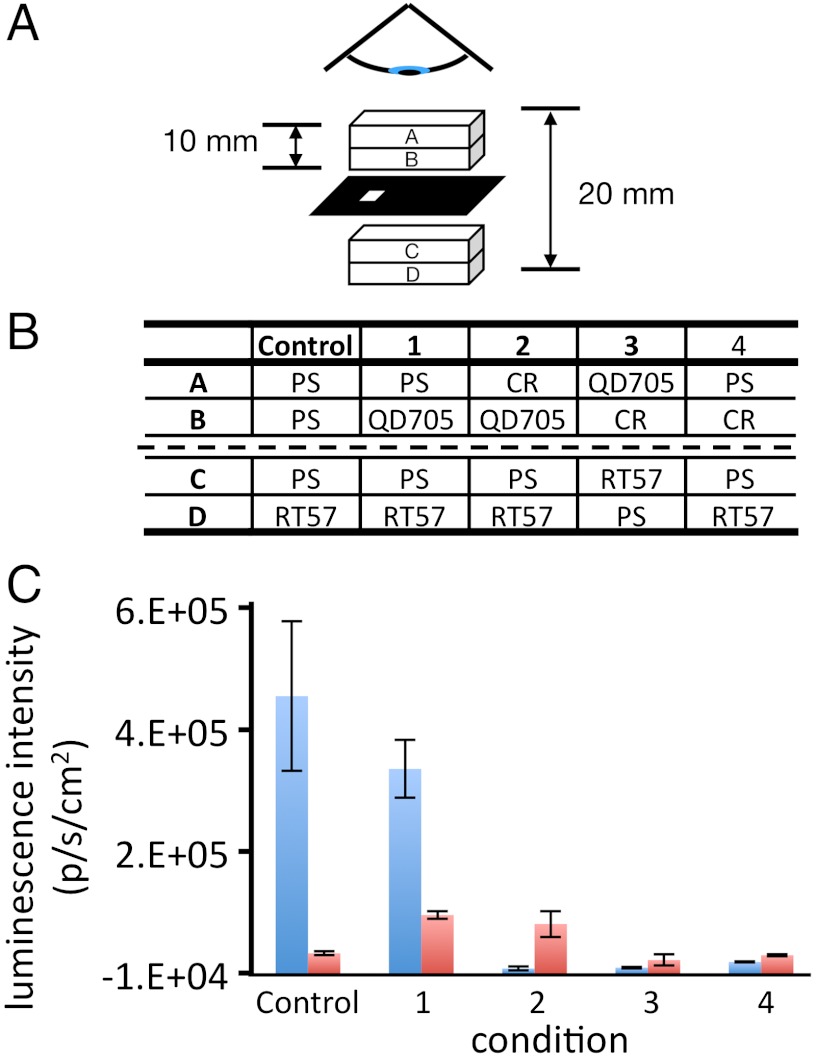
For the purposes of the current study, we sought an experimental design that would recapitulate our previous experimental conditions without mixing the RT57 and QD705 or employing a common substrate. Dilute suspensions of RT57 and QD705 were pipetted separately into high-quality quartz cuvettes. The cuvettes were then physically closely juxtaposed, separated only by a thin, opaque, black paper mask that included a small hole to allow the blue bioluminescent light from the bacteria to pass into the QD705 compartment (Fig. 1A). We next established five conditions (Fig. 1B) wherein bioluminescence was measured concomitantly through the blue (470–490 nm), and then the red (695–770 nm) emission filters respectively (Fig. 1C). Under these conditions, a luminescent red shift was clearly observed, evidenced by the detection of a strongly enhanced red-signal specific to the 695–770 nm filter, dependent upon the introduction of QD705 between the RT57 cuvette compartment and the detector (compare condition 1 and the control, Fig. 1C). This approximately eightfold enhancement of red-light measured in condition 1 suggested that the QD705 isolated in compartment B were directly excited by blue-luminescent light from the bacteria at a physical distance of 1 cm in compartment D of the cuvette setup. To verify this, we examined in parallel the effect of Congo red (CR), a red dye with strong blue absorption closely resembling that of biological absorbers such as whole blood (Figs. S1 and S2). The presence of CR isolated in the compartment A of the cuvette setup, closest to the detector, blocked blue luminescence completely while the red-emission enhancement remained unchanged (Fig. 1C; condition 2). Further, when CR filled the compartment B of the cuvette setup, thereby physically situated between the RT57 luminescent source (compartment C) and the QD705 fluorophore (compartment A) this reduced both red and blue light compared with conditions 1 and 2 (Fig. 1C; condition 3). Interestingly, the level of red photon production detected in condition 3 was comparable to that measured through the red filter in the presence of bacteria alone (control), or when the CR was placed between the RT57 and the detector, in the absence of QD705 (condition 4). This result is probably due to the fact that CR did not block low levels of red light in any condition, even the low level of red-light component comprising the broad spectrum of bacterial lux derived luminescence from bacteria alone. These results strongly suggest QD705 are excited when illuminated by weak, unfocused blue light from bioluminescent bacteria. Under these conditions, there was no possibility of any physical molecular interaction occurring between the luminescent source (RT57) and the fluorophore (QD705) other than incident radiating photons traveling via the water-glass-water route. As such, these results rule out any possibility of circuitous dipole–dipole, molecular resonance energy transfer processes such as BRET (14–17), pointing instead to the occurrence of an extremely low-light level epifluorescent excitation–emission process. This experiment demonstrated low-light level bioluminescence acting over macroscopic distances as an illumination source capable of evoking detectable fluorescent emission from a fluorophore.
Fig. 1.
In vitro demonstration of FUEL. (A) Schematic representation of the cuvette orientation. Two tandem quartz cuvettes, each with a path length of 1 cm, were separated by a piece of black paper, cut to allow light to pass between the cuvettes through a 2-mm square hole. The contents of the cuvette compartments annotated A, B, C, and D correspond to the same letters in the table (B) below, describing each of the five conditions numbered 1–4 plus a control with bacteria alone. Luminescent intensity [photon·s-1·cm-2 (p·s-1·cm-2)] was observed from above the sample through both a 470–490 nm (blue) and a 695–770 nm (red) filter. The quantitative results from these experiments in the graph (C) show the averaged p·s-1·cm-2 corresponding to the five conditions from the table above. For clarity, the p·s-1·cm-2 data were systematically converted to positive integers by addition of 10,000 counts throughout. All conditions were repeated three times using different RT57 cultures. Error bars represent SEM.
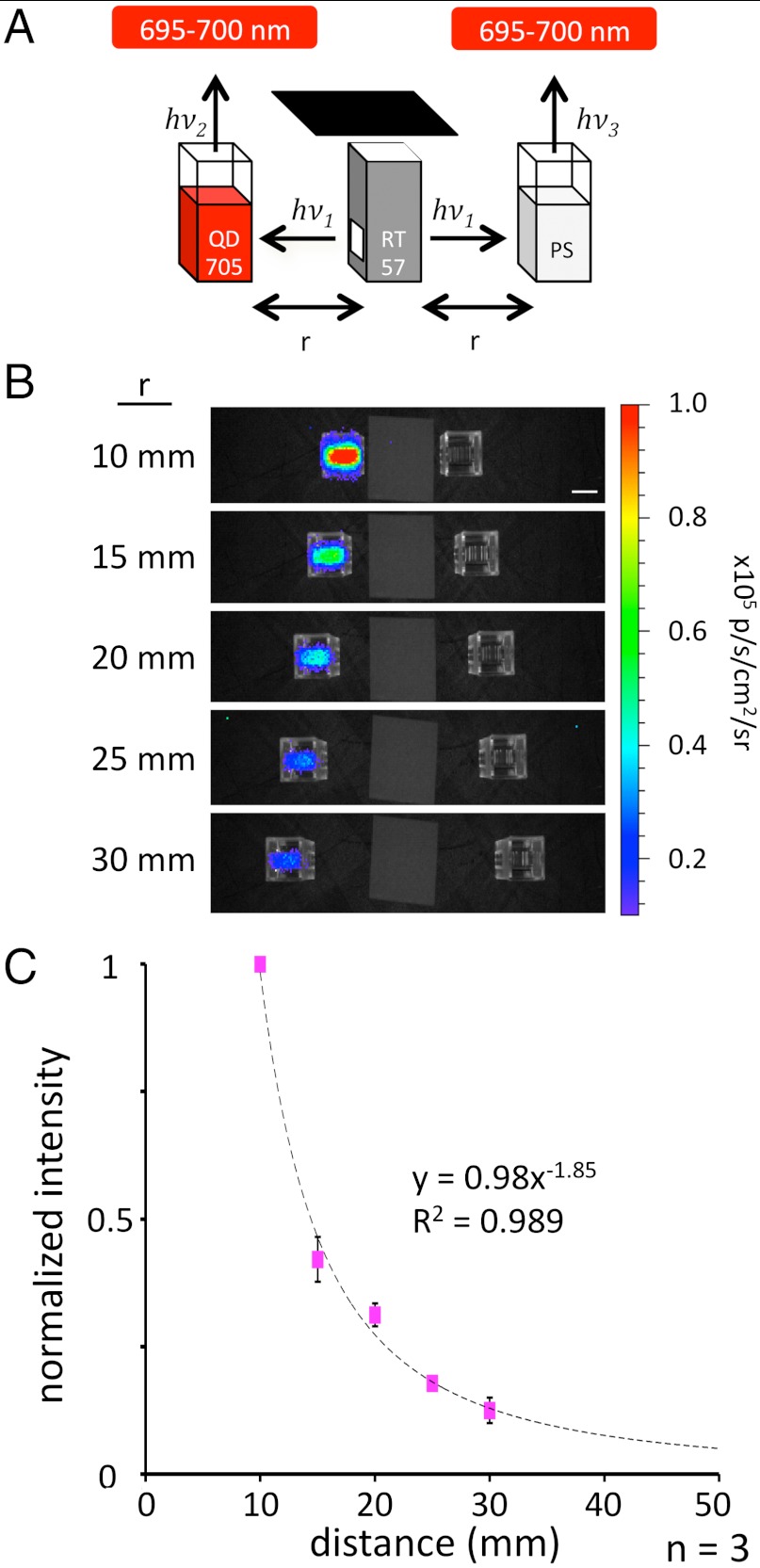
Occurring under conditions entirely precluding BRET, we hypothesized that the FUEL phenomenon arose from simple excitation–emission processes and should therefore display distance dependence characteristics of a radiating light source. To directly examine the dependence of FUEL on distance, we used an experimental design again based on cuvettes, as shown in Fig. 2. Briefly, we filled three cuvettes with either an aliquot of freshly prepared RT57 bacteria, a solution of QD705 in physiological saline (PS), or just PS. The three cuvettes were oriented such that the cuvette containing the RT57 was centrally placed and equidistant between the other two (Fig. 2A), ensuring that luminescence from the contained bacteria emanated symmetrically, unaffected by refraction, and thereby illuminated the two cuvettes placed either side, simultaneously, and in exactly the same manner. As can be seen in Fig. 2, the FUEL effect was strongly reconstituted under these conditions, evidenced by the significant photon flux detected uniquely in the cuvettes containing QD705, observed with the 695–770 nm filter. Taking full advantage of this macroscopic effect, we quantified the fluorescence emission and correlated the averaged values to the measured distances between the cuvettes (Fig. 2B). We observed that the measured intensity of QD705 fluorescence diminished according to the reciprocal of the distance squared (1/r2), entirely consistent with a mechanism of radiating fluorescent excitation–emission occurring between the blue-luminescent bacteria and the QD705 fluorophore.
Fig. 2.
Distance dependence of the FUEL effect. As illustrated in the schematic (A), an aliquot of bioluminescent E. coli (“RT57”) filled a cuvette between two cuvettes filled with either QD705 solution (Left) or PS (Right). The resulting luminescence was observed at varying distances through the 695–770 nm emission filter set. The resulting luminescence decreased as a function of distance from the central cuvette (B). Images show black and white photos of the cuvette in low light overlaid with pseudocolor indicating the bioluminescence signal intensity [photon·s-1·cm-2 (p·s-1·cm-2)] measured through a 695—770 nm filter. The averaged intensities were then normalized to the most intense signal and then plotted as a function of distance, resulting in a nearly 1/r2 relationship shown in the graph (C). Each distance was repeated in triplicate using different RT57 cultures. (Scale bar: 10 mm.) The error bars show the SEM.
Fluorescence Excitation by Bacterial Luminescence in Vivo.
Among the broad gamut of bioluminescence applications in vivo molecular imaging is widespread and provides a valuable tool for life science studies. It was therefore important to establish whether FUEL may occur, and to what extent it is detectable in vivo. Inasmuch as ex vivo experiments revealed that whole blood increased the FUEL contrast comparable to the effect of CR (Fig. S2), we next chose to address these questions using a standard in vivo model for studies on bacterial infection: subcutaneous injection of luminescent bacteria into a mouse. Specifically, we designed in vivo experiments using either bioluminescent Staphylococcus aureus Xen 36 (18) or bioluminescent RT57.
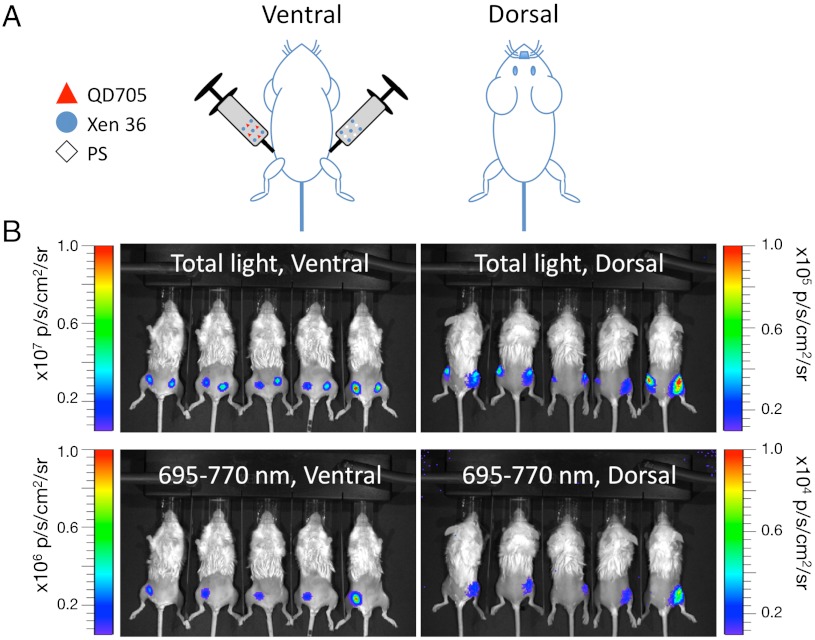
In the first experimental regime (Fig. 3), Xen 36 were freshly harvested from culture in liquid media and diluted to a final concentration of 1.67 × 109 bacteria/mL. A solution containing 30 μL of bacteria stock and 20 μL of PS was subcutaneously injected into one thigh while a solution containing 30 μL of bacteria stock, 15 μL of PS, and 5 μL QD705 stock was subcutaneously injected into the other thigh. The mice were then immediately visualized using a bioluminescence in vivo imaging system with both total light and 695–770 nm emission settings, from both ventral and dorsal sides (Fig. 3). Under these experimental conditions, the FUEL phenomenon was evidenced by detection of a characteristic red signal exclusive to the bacteria/QD705 injections, and distinguished by observation of the ventral side through the 695–770 nm filter (Fig. 3B), wherein it represented a 15-fold increase in relative signal enhancement compared to the control (bacteria alone; see Table 1). Having established that the red FUEL signal could be detected in vivo by observation through the shallow tissue depth of a ventral view, it was interesting to know to what extent this signal could be detected through deep tissue. We therefore quantified the detection of total light and red light in the same experiments, but this time turning the animals over and observing them from their dorsal side, wherein a greater depth of tissue obscured the signal. As shown in Table 1, we found that the FUEL signal was clearly distinguished through deep tissue as compared to bacteria alone (Table 1 and Fig. 3), without recourse to red filters. Presumably because the deep tissue served as a red filter, absorbing blue light. These results demonstrate that FUEL may be reconstituted in vivo and that the relative abundance of signal is detectable under normal experimental conditions used for molecular imaging (Table 1).
Fig. 3.
Observation of FUEL under in vivo conditions. To study whether FUEL can occur under relevant experimental conditions in vivo subcutaneous injections of Staphylococcus aureus Xen36 (blue circle) suspended in PS (diamond), with and without QD705 (red triangle) were made into the inner thighs of live anesthetized female BALB/c mice as shown in A. The mice were imaged from both the ventral and dorsal sides either without a filter (total light), or through a 695–770 nm filter (B). Images show black and white photos of the five mice observed from either ventral or dorsal sides as labeled. Localized bioluminescence (shown by pseudocolor overlay) was measured and the spatial luminescence intensity of the detected light signal [photon·s-1·cm-2 (p·s-1·cm-2)] was determined for the total light and the 695–770 nm filter as annotated. Note: the bioluminescence light scale indicated by full-scale deflection across the rainbow pseudocolored lookup table reveals that total light measured ventral≫dorsal (107≫104 p·s-1·cm-2).
Table 1.
Global average p·s-1·cm-2 of subcutaneous injections
| Filter |
Bac |
PS |
QD |
Bac + QD |
| Ventral | ||||
| Total light | 1.39E+7 ± 0.13E+7 | 8.93E+3 ± 2.14E+3 | 1.12E+4 ± 0.11E+4 | 1.39E+7 ± 0.48E+7 |
| 695–770 nm | 9.15E+4 ± 1.04E+4 | 2.6E+3 ± 0.89E+3 | 1.18E+3 ± 0.71E+3 | 1.31E+6 ± 0.48E+6 |
| Dorsal | ||||
| Total light | 1.97E+5 ± 0.56E+5 | 1.32E+4 ± 0.32E+4 | 8.60E+3 ± 3.64E+3 | 3.22E+5 ± 1.12E+5 |
| 695–770 nm | 9.88E+3 ± 1.98E+3 | 1.46E+3 ± 0.54E+2 | 1.82E+3 ± 0.83E+3 | 1.07E+5 ± 0.31E+5 |
A set of five female BALB/c mice were subcutaneously injected into the inner thigh with either bioluminescent S. aureus Xen 36 (Bac) or a suspension of Xen 36 and quantum dot 705 (Bac + QD). In the same manner, a second set of five female BALB/c mice were subcutaneously injected into the inner thigh with either PS or a suspension of QD 705. The total volume of each injection was identical, and the concentrations of the Xen 36 and QD 705 were kept constant throughout. The mice were observed without a filter (total light) and through a 695–770 nm filter, from the ventral and the dorsal side in an IVIS100. Values indicate the mean photon·s-1·cm-2 ± SEM, n = 5.
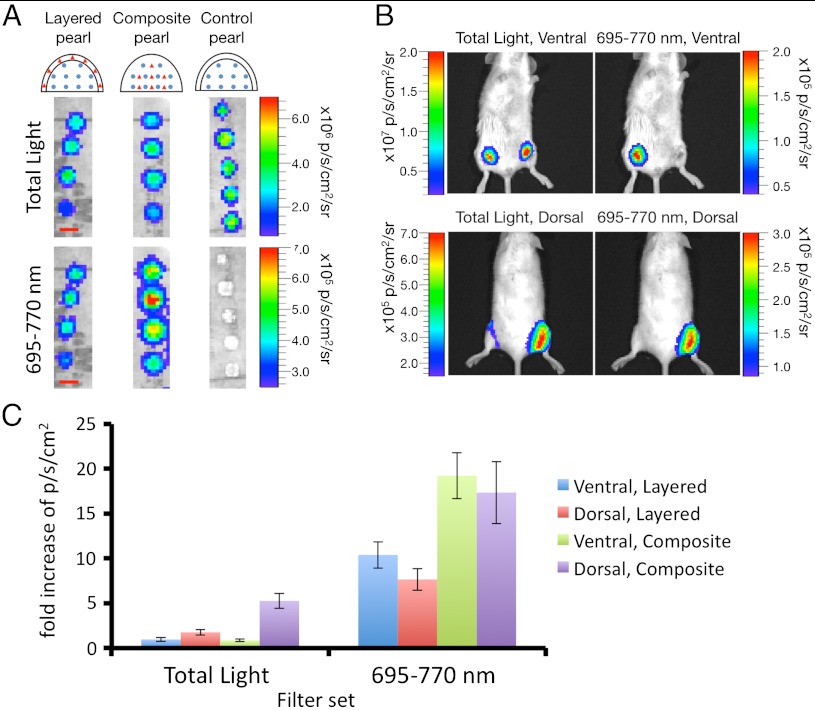
That FUEL was detected in vivo using injections of luminescent bacteria and QD705 was highly consistent with the view that macroscopic coproximity of luminescent and fluorescent moieties limited by diffusion in soft tissue is sufficient for the phenomenon to occur. This property could be of practical value if, for example, FUEL intensity could be interpreted as proportional to spatial proximity in the context of a niche targeting of the moieties, e.g., comparable to the distance dependence of FUEL revealed in vitro. To address this question in vivo, we designed an experimental model using small (5–7 mm) agarose beads (referred hereon as “pearls”) inside which luminescent bacteria and QD705 were suspended. We manufactured two types of pearl, designed to create a niche for either increased, or decreased average distance between luminescent bacteria and the QD705 thereby increasing, or decreasing, the FUEL signal. The first type of pearl was produced by layering two separate agarose droplets, one containing luminescent bacteria and the second containing QD705 in a manner that the two layers remained permanently separated in two juxtaposed layers (“layered” pearls; Fig. 4A). In parallel, a second type of pearl was formed from a single agarose droplet into which both bacteria and QD705 were mixed together and suspended homogenously throughout at the same final concentration as used for layered pearls (“composite” pearls, see Fig. 4A). We reasoned that these two types of pearl served to maintain FUEL moieties (bacteria and QD705) in relatively distant macroscopic proximity (i.e., layered pearls), or relatively close microscopic proximity (i.e., composite pearls). As such the pearls provide a stable FUEL signal reconstituted reproducibly under controlled conditions in a defined, inert, and constant microcosm, protected from in vivo diffusional effects. Consistent with the greater, or lesser, proximity of the FUEL moieties in these two types of pearl, we found in vitro that the composite pearls yielded two-times more red light than layered pearls, agreeing with the hypothesis that the more constrained the proximity of the FUEL moieties, the larger the in vitro FUEL signal revealed through the 695–770 nm filter (Fig. 4A).
Fig. 4.
Reconstitution of in vivo FUEL under constrained conditions. As a means to minimize diffusion of the relevant moieties, bioluminescent E. coli (blue circle) and QD705 (red triangle) were suspended in agar droplets in either a layered, composite, or control pearl geometry as outlined in the schematic. (A) (Scale bar: 5 mm.) The fabricated pearls were observed in the IVIS100 without a filter (total light) and through a 695–770 nm filter. Small incisions were made into the inner thighs of female 6-wk-old BALB/c mice, and the various pearl types inserted, one per thigh. The mice were then observed from the dorsal and ventral sides for total light and through a 695–770 nm filter. Shown is an example of a composite pearl (right thigh) versus a control pearl (left thigh) (B). The photon·s-1·cm-2 (p·s-1·cm-2) for each pearl type through both filter sets and from both sides was then determined, and directly compared to the control pearl. Values are reported as mean fold increase above control ± SEM, n = 5 mice (C).
The layered, and composite pearls were implanted subcutaneously by incision on the inner thighs of mice and visualized from ventral and dorsal points of view, providing insight on shallow (millimeters) and deep (centimeters) tissue performance (Fig. 4 B and C and Table S1). Compared with control pearls loaded with bacteria alone, neither layered, nor composite pearls showed any increase in total photon light output when observed through shallow tissue in vivo (ventral view; see Fig. 4C, total light, and Table S1). By contrast, both layered and composite pearls enhanced the detectable light output by two- and fivefold, respectively, when observed through deep tissue in vivo (dorsal view; see Fig. 4C, total light, and Table S1). That this enhancement of detectable photons through deep tissue was due specifically to FUEL driven spectral red shift was revealed by observing the ventral and dorsal points of view through the 695–770 nm filter (Fig. 4 B and C and Table S1). Layered pearls showed a 10-fold enhancement in red-light signal compared with bacteria alone, and composite pearls a 20-fold enhancement. These results were entirely consistent with the predicted characteristics of bacteria and QD705 in the two types of agarose pearl due to the reduced distance between the luminescent and fluorescent moieties.
Discussion
Our results show unambiguously that extremely low levels of unfocused, radiating bioluminescence can be used as an epifluorescent illumination light source. This conventional process, we term FUEL, has not been reported in previous studies using BRET (19); in particular, those studies aiming at red-shift contrast enhancement (9–12, 20, 21). In all cases, these studies evoked BRET mechanisms implying spectral overlap of bioluminescent-donor and fluorescent-acceptor constrained to molecular proximity of less than 10 nm, thereby facilitating dipole–dipole interaction, and nonradiative resonant energy transfer. Our results show that FUEL may occur entirely distinct from BRET mechanisms and are likely significant under conditions where BRET occurs. Our results suggest the critical importance for careful controls when interpreting BRET experiments. Indeed, we recognize that although BRET and FUEL are discrete processes, they are by no means mutually exclusive. Rather, we suggest that they coexist to a greater or lesser extent in all permissive conditions, reminiscent of Stryer’s first description of FRET (14). The extent to which FUEL and BRET coexist in such experiments needs to be addressed quantitatively. That FUEL has until now been overlooked is most likely due to the fact that quantitative descriptions of BRET have seemed unnecessary because all the studies have focused on achieving qualitative “red-shift” paradigms. That both FUEL and BRET both yield red shift in light output makes them difficult to distinguish without careful controls. Our current results beckon appraisal, whereby BRET/FUEL signals should be quantitatively distinguished in vitro and in vivo.
Our results describing FUEL paired probes may help toward establishing alternative methods to address the practical limitations of poor propagation of blue-green light in living tissues (1–3, 22). It is of significant interest for in vivo imaging that we report FUEL to enhance the detection of bioluminescent signals by an order of magnitude, by virtue of a Stokes shift toward red wavelengths to which living tissue is more transparent. In this context, the use of bioluminescent bacteria is of practical interest considering there already exist a broad range of established applications using the blue-luminescent lux operon transgene to study bacterial infection in animal models (23, 24). A priori the methodology of FUEL may offer itself as a unique regime by which to enhance quantitative deep tissue detection of bacterial infection in vivo. Such approaches to enhance bacterial luminescence in vivo might benefit the use of probes other than inorganic nanoparticles, especially organic fluorophores. However, perhaps even more interesting than the simple ability to enhance bioluminescent light another possible FUEL application might be its use as a means to detect microscopic and macroscopic coproximity of targets porting luminescent and fluorophore moieties in the same tissue (i.e., incident detection of “FUEL-probe-pair” proximity). As a reporter mechanism, this would uniquely distinguish FUEL from nanometer scale molecular proximity reporter methods such as BRET (19), because it offers possibilities for experimental design aimed at detection of physical proximity of functionalized moieties across mesoscopic scales (microscopic to macroscopic).
In conclusion, our results indicate that the FUEL phenomenon is experimentally relevant both in vitro and in vivo, representing a process that cannot be dismissed as trivial. Along with the significant advances in low-light imaging technology, the surprising possibility of observing this process under standard bioluminescent imaging conditions provides exciting future perspective likely enabling the development of imaging paradigms for quantitative molecular imaging both in vitro and in vivo.
Materials and Methods
Reagents.
Bioluminescent Escherichia coli carrying the plasmid expressing the lux operon (pUC18-mini-Tn7T-Gm-lux; referred to as RT57) were provided by José A. Bengoechea (Laboratory Microbial Pathogenesis, Bunyola, Spain) with permission from Herbert P. Schweizer (Department of Microbiology, Immunology, and Pathology, Colorado State University, Fort Collins, CO) (25). Staphylococcus aureus Xen 36 was provided by Xenogen. Qtracker 705 (2 μM, QD705) nontargeted quantum dots were acquired from Invitrogen, and kept in the dark at 4 °C until use. The QD705 were used as received. CR (nvitrogen) was used at 1% wt/vol in water. Physiological saline (0.09% NaCl, PS) was prepared inhouse. For all animal experiments, female 6-wk-old BALB/c mice (Janvier) were used. The day before imaging, an overnight culture of the RT57 was established, providing a typical concentration of 1.2 × 109 bacteria per mL. The following morning, the OD600 was measured and the bacteria diluted to an OD600 = 2 (for RT57 E. coli, an OD600 = 1 is 5 × 108 bacteria per mL).
Bioluminescence Imaging.
An IVIS 100 whole animal imaging system (Xenogen Corporation, Caliper Life Sciences) equipped with an intensified deep-cooled CCD and a filter wheel was used to acquire all bioluminescent and fluorescent images. A 480 nm ± 10 nm band-pass filter was used to collect the RT57-specific photons, termed the “480” filter set. A Cy5.5 band-pass filter (695–770 nm) was used to distinguish red photons. One location within the filter wheel was kept empty for total light detection (Total Light). For each experiment, the CCD was cooled to -105 °C and the stage warmed to 37 °C. For each image, the acquisition settings for the Living Image (Xenogen) software version 3.1 were set as follows: 8× bin, field of view B (20 cm), and f-stop 1. The acquisition time was adjusted as necessary for each experiment (typically 10–60 s). For fluorescence acquisitions, the illumination power was set to low, the bin setting was reduced to 4×, and the acquisition time set to 0.5 s. QD705 fluorescence was observed using a Cy5.5 filter set that included a 615–665 nm excitation filter and a 695–770 nm emission filter.
Demonstration of FUEL.
Tandem quartz cuvettes (Starna Scientific, Ltd.) were used to examine the FUEL effect at controlled distances and variables. The cuvettes were filled as indicated in Fig. 1 (1 mL per reagent), sealed to prevent leaking, and then oriented as shown. For the reagent abbreviations, RT57 indicates an aliquot of stock RT57 from an overnight culture (OD = 2), PS represents physiological saline, CR is Congo red, and QD705 is a solution containing 50 μL of QD705 stock in 950 μL of PS. A piece of black paper with a 2 × 3 mm window was used to separate the two cuvettes. The paper was placed such that the window was near the base of the cuvettes, allowing for visualization to occur away from both the edges of the cuvettes and the meniscus of the reagents. Further, the paper ensured a physical separation existed between the RT57 and the QD705, thus allowing only the FUEL effect to be observed. With this configuration, we estimated the detectable light output of the luminescent bacteria filled cuvette to be 1.84 × 10-13 ± 0.51 × 10-13W/cm2 based on the empirical number of detected photons over time (4.45 × 105 ± 1.23 × 105 p·s-1·cm-2, n = 3; E = hc/λ) at an emission maximum of 490 nm. The luminescence was observed under the 480 and 695–770 nm filter sets as described previously. This was repeated for three different bacterial cultures.
Distance Dependence of FUEL in Vitro.
Black tape was used to create two 2 × 3 mm optical windows on a reduced-volume disposable cuvette (Ratiolab), with the two windows situated on opposite faces of the cuvette, and located near, but away from, the base. This modified cuvette was filled with 1 mL of RT57 stock from an overnight culture (OD600 = 2), covered by a piece of black paper, and placed into the IVIS imaging system. The black paper obscured direct observation of the RT57 bioluminescence. Two unmodified reduced-volume cuvettes were filled with either 1 mL of PS, or 1 mL of the previously described QD705 solution, and then placed on either side of the modified cuvette containing RT57 such that the three cuvettes were axially aligned and at a distance of 10 mm between the central and exterior cuvettes (face to face). The resulting luminescence was observed for the 695–770 nm filter set. After acquisition, the distance between the two unmodified cuvettes and the central cuvette was increased by 5 mm, and the luminescence observed. This procedure was repeated up to a total separation of 30 mm for three different bacterial cultures.
In Vivo Demonstration of FUEL.
A subculture of Xen 36 was established from an overnight culture and allowed to grow for several hours ensuring the bacteria were in exponential phase at the time of use. Prior to the experiment, the OD for the fresh culture was determined and the bacteria diluted to a final concentration of 2.5 × 108 bacterial per mL. Four different solutions were prepared. The first solution consisted of, per mouse, 20 μL of PS and 30 μL of the bacterial stock. The second solution consisted, per mouse, of 5 μL of the QD705 stock, 15 μL PS, and 30 μL of the bacterial stock. The third solution consisted of 50 μL of PS. The final solution consisted of 5 μL of QD705 and 45 μL of PS. After solutions were prepared, five female 6-wk-old BALB/c mice were anesthetized and their hind limbs shaved and epilated using cream. Solutions 1 and 2 were then subcutaneously injected into each inner thigh (both left and right). The mice were then placed into the IVIS100 and visualized from the dorsal and ventral sides under the Total Light and 695–770 nm filter sets. As controls, solutions 3 and 4 were injected into the inner thigh of second set of five female BALB/c mice. Like the initial set of mice, the second set was placed into the IVIS100 and visualized from both sides under both filter sets.
Fabrication of Pearls.
To demonstrate FUEL in vivo, 2% low-melting point agarose pearls consisting of an RT57-based core layered by agarose, or a mixture of agarose and QD705 were constructed. Briefly, to create an individual pearl, 12.5 μL of RT57 stock at an OD of 2 was mixed with 12.5 μL warm 4% agarose and pipetted onto parafilm. After the 25 μL pearl core was allowed to cool, a mixture of either 5 μL of PS or 5 μL QD705 stock and 7.5 μL of 3% agarose was pipetted onto the pearl core, forming the control and layered pearls, respectively. The composite pearls were formed by mixing 12.5 μL of RT57 (OD = 2), 5 μL of QD705, and 17.5 μL of 4% agarose. The suspension was then pipetted onto parafilm and allowed to cool. After construction of the pearls, female 6-wk-old BALB/c mice were chemically anesthetized using Ketamine/Xylazine. The hind limbs, both the dorsal and ventral sides, of all mice were then shaved using an electric razor and epilated using cream. A small incision was made on the interior of both hind limbs, allowing for the subcutaneous placement of the control, layered, or composite pearl. The mice were then placed into the IVIS100 and the luminescence observed from the ventral side for both the Total Light and 695–770 nm filter sets. The mice were then flipped over and the luminescence observed from the dorsal side for both filter sets. All animal experiments were carried out according to national and institutional guidelines.
Supplementary Material
Acknowledgments.
The authors thank Bruno Baron of the Plate-Forme de Biophysique des Macromolécules et de leurs Interactions, Marie-Anne Nicola and Nathalie Aulner of the Plate-Forme d’Imagerie Dynamique for technical support and assistance, José Bengoechea (Laboratory Microbial Pathogenesis, Bunyola, Spain) and Herbert Schweizer (University of Colorado, Fort Collins, CO) for reagents, and Philippe Sansonetti (Institut Pasteur, Paris, France) for use of lab space, reagents, and equipment. Finally, the authors would like to extend their sincerest gratitude to Serge Mostowy for his constructive comments. Joe Dragavon is a Florence Gould Scholar of the Pasteur Foundation Postdoctoral Fellowship Program. The authors would like to extend their gratitude for financial support from the Pasteur Foundation of New York (J.D. and C.S.), the EU-FP7 Program “Automation” (S.L.S.), the Institut Carnot Program 11 (R.T. and S.L.S.), and Project Imnos coordinated by Jean-Christophe Olivo-Marin (Institut Pasteur, Paris, France) (R.T. and S.L.S.), the Conny-Maeve Charitable Foundation (S.L.S.), the European Masters in Molecular Imaging (I.T.), the Region Ile de France programs Modexa (S.L.S.), Sesame (S.L.S.), and DimMalInf (R.T. and S.L.S.), and the Institut Pasteur, Paris.
Footnotes
Conflict of interest statement: J.D., S.B., K.L.R. and S.L.S. contributed to European Patent EP10290158.4.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1204516109/-/DCSupplemental.
References
- 1.Lakowicz JR. Principles of Fluorescence Spectroscopy. 2nd Ed. New York: Kluwer Academia; 1999. pp. 656–658. [Google Scholar]
- 2.Colin M, et al. Haemoglobin interferes with the ex vivo luciferase luminescence assay: Consequence for detection of luciferase reporter gene expression in vivo. Gene Ther. 2000;7:1333–1336. doi: 10.1038/sj.gt.3301248. [DOI] [PubMed] [Google Scholar]
- 3.König K. Multiphoton microscopy in life sciences. J Microsc. 2000;200:83–104. doi: 10.1046/j.1365-2818.2000.00738.x. [DOI] [PubMed] [Google Scholar]
- 4.Loening AM, Wu AM, Gambhir SS. Red-shifted Renilla reniformis luciferase variants for imaging in living subjects. Nat Methods. 2007;4:641–643. doi: 10.1038/nmeth1070. [DOI] [PubMed] [Google Scholar]
- 5.Shu X, et al. Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science. 2009;324:804–807. doi: 10.1126/science.1168683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Branchini BR, et al. Red-emitting luciferases for bioluminescence reporter and imaging applications. Anal Biochem. 2010;396:290–297. doi: 10.1016/j.ab.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 7.Roda A, Guardigli M. Analytical chemiluminescence and bioluminescence: Latest achievements and new horizons. Anal Bioanal Chem. 2011;402:69–76. doi: 10.1007/s00216-011-5455-8. [DOI] [PubMed] [Google Scholar]
- 8.Loening AM, Dragulescu-Andrasi A, Gambhir SS. A red-shifted Renilla luciferase for transient reporter-gene expression. Nat Methods. 2010;7:5–6. doi: 10.1038/nmeth0110-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.So M-K, Xu C, Loening AM, Gambhir SS, Rao J. Self-illuminating quantum dot conjugates for in vivo imaging. Nat Biotechnol. 2006;24:339–343. doi: 10.1038/nbt1188. [DOI] [PubMed] [Google Scholar]
- 10.Branchini BR, Ablamsky DM, Rosenberg JC. Chemically modified firefly luciferase is an efficient source of near-infrared light. Bioconjugate Chem. 2010;21:2023–2030. doi: 10.1021/bc100256d. [DOI] [PubMed] [Google Scholar]
- 11.Wu C, et al. In vivo far-red luminescence imaging of a biomarker based on BRET from Cypridina bioluminescence to an organic dye. Proc Natl Acad Sci USA. 2009;106:15599–15603. doi: 10.1073/pnas.0908594106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De A, Ray P, Loening AM, Gambhir SS. BRET3: A red-shifted bioluminescence resonance energy transfer (BRET)-based integrated platform for imaging protein-protein interactions from single live cells and living animals. FASEB J. 2009;23:2702–2709. doi: 10.1096/fj.08-118919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dragavon J, et al. Validation of method for enhanced production of red-shifted bioluminescent photons in vivo. In: Farkas DL, Nicolau DV, Leif RC, editors. Imaging, Manipulation, and Analysis of Biomolecules, Cells, and Tissues IX. Bellingham, WA: International Society for Optical Engineering; 2011. pp. 790210–790219. [Google Scholar]
- 14.Stryer L. Fluorescence energy transfer as a spectroscopic ruler. Annu Rev Biochem. 1978;47:819–846. doi: 10.1146/annurev.bi.47.070178.004131. [DOI] [PubMed] [Google Scholar]
- 15.Piston DW, Kremers G-J. Fluorescent protein FRET: The good, the bad and the ugly. Trends Biochem Sci. 2007;32:407–414. doi: 10.1016/j.tibs.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 16.Jares-Erijman EA, Jovin TM. FRET imaging. Nat Biotechnol. 2003;21:1387–1395. doi: 10.1038/nbt896. [DOI] [PubMed] [Google Scholar]
- 17.Majoul I, Jia Y, Duden R. Practical fluorescence resonance energy transfer or molecular nanobioscopy of living cells. In: Pawley JB, editor. Handbook of Biological Confocal Microscopy. Third Ed. New York: Springer; 2006. pp. 788–808. [Google Scholar]
- 18.Brand AM, de Kwaadsteniet M, Dicks LMT. The ability of nisin F to control Staphylococcus aureus infection in the peritoneal cavity, as studied in mice. Lett Appl Microbiol. 2010;51:645–649. doi: 10.1111/j.1472-765X.2010.02948.x. [DOI] [PubMed] [Google Scholar]
- 19.Bacart J, Corbel C, Jockers R, Bach S, Couturier C. The BRET technology and its application to screening assays. Biotechnol J. 2008;3:311–324. doi: 10.1002/biot.200700222. [DOI] [PubMed] [Google Scholar]
- 20.Baubet V, et al. Chimeric green fluorescent protein-aequorin as bioluminescent Ca2+ reporters at the single-cell level. Proc Natl Acad Sci USA. 2000;97:7260–7265. doi: 10.1073/pnas.97.13.7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rogers KL, et al. Visualization of local Ca2+ dynamics with genetically encoded bioluminescent reporters. Eur J Neurosci. 2005;21:597–610. doi: 10.1111/j.1460-9568.2005.03871.x. [DOI] [PubMed] [Google Scholar]
- 22.Ntziachristos V. Going deeper than microscopy: The optical imaging frontier in biology. Nat Methods. 2010;7:603–614. doi: 10.1038/nmeth.1483. [DOI] [PubMed] [Google Scholar]
- 23.Hutchens M, Luker GD. Applications of bioluminescence imaging to the study of infectious diseases. Cell Microbiol. 2007;9:2315–2322. doi: 10.1111/j.1462-5822.2007.00995.x. [DOI] [PubMed] [Google Scholar]
- 24.Contag CH, Ross BD. It’s not just about anatomy: In vivo bioluminescence imaging as an eyepiece into biology. J Magn Reson Imaging. 2002;16:378–387. doi: 10.1002/jmri.10178. [DOI] [PubMed] [Google Scholar]
- 25.Choi K-H, Schweizer HP. Mini-Tn7 insertion in bacteria with single attTn7 sites: Example Pseudomonas aeruginosa. Nat Protoc. 2006;1:153–161. doi: 10.1038/nprot.2006.24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.