Abstract
Histone acetyltransferase 1 is the founding member of the histone acetyltransferase superfamily and catalyzes lysine acetylation of newly synthesized histone H4. Here we report a 1.9-Å resolution crystal structure of human histone acetyltransferase 1 in complex with acetyl coenzyme A and histone H4 peptide. The crystal structure reveals that the cofactor and the side chain of lysine 12 of histone H4 peptide are placed in the canyon between the central and C-terminal domains. Histone H4 peptide adopts a well-defined conformation and establishes an extensive set of interactions with the enzyme including invariant residues Glu64 and Trp199, which together govern substrate-binding specificity of histone acetyltransferase 1. Our structure-guided enzyme kinetic study further demonstrates a cumulative effect of the active-site residues Glu187, Glu276, and Asp277 on deprotonation of the ɛ-amino group of reactive Lys12 for direct attack of the acetyl group of the cofactor.
Histone acetylation is the most prominent posttranslational modification leading to activation of gene expression, DNA replication, and DNA repair. It is postulated that histone acetylation, through charge neutralization of the cationic histone tails, weakens nucleosomal electrostatic interactions with anionic DNA, thus destabilizing internucleosomal contacts and nucleosomal structure and facilitating access to the promoter region for RNA polymerase and transcription factors. Histone acetylation also plays a functional role in providing binding sites for regulatory proteins that mediate transcription and other chromatin-based processes.
Histone acetyltransferase 1 (HAT1; EC 2.3.1.48) is the founding member of the family of histone acetyltransferases that catalyze the transfer of an acetyl group from acetyl coenzyme A (AcCoA) to the lysine ε-amino groups on the N-terminal tails of histones (1, 2). HAT1 has a very broad function in many DNA regulatory processes and is found in virtually all eukaryotic organisms examined (3). Originally, HAT1 was identified as a cytosolic enzyme (2) that acetylates newly synthesized histone H4 molecules before their deposition in replicating chromatin and was therefore suspected of being involved in replication-dependent chromatin assembly. In vitro, HAT1 specifically acetylates Lys5 and Lys12 of free (nonnucleosomal) histone H4, and this specificity is entirely consistent with the pattern of acetylation found on newly synthesized histone H4 from many organisms. Biochemical studies revealed that the HAT1 functions as a member of the HAT-B complex (2), which also contains the p46/48 protein (RbAp46 or RBBP7 in humans and HAT2 in yeast). The p46/48 protein is a WD40 repeat protein involved in a wide variety of chromatin-modifying complexes. In yeast, the association of HAT2 with HAT1 increases the catalytic activity of HAT1 by a factor of 10 and appears to function by increasing HAT1 binding to histone H4 (2).
Subsequent studies showed that HAT1 is predominantly localized in the nucleus (3–6) where it forms a trimeric complex with HAT2 and the histone chaperone HIF1. HIF1 is detected exclusively in the nucleus and accompanies newly modified H3-H4 tetramers to facilitate their incorporation into nascent chromatin during DNA synthesis. Deletions of HAT1, HAT2, or HIF1 alone produced no apparent phenotypic effect (1, 2, 6, 7), but when combined with specific mutations in the N terminus of histone H3, caused defects in both telomeric gene silencing and resistance to DNA-damaging agents (7–9). Such defects are reproduced by the substitution of Lys for Arg at position 12 of histone H4, but not at position 5 (8, 9). HAT1 and HAT2 have been found to interact with the origin recognition complex, suggesting a unique role for the HAT1–HAT2 complex at the replication fork and likely a role in replication-dependent chromatin assembly (10).
On the basis of the available results, working models have emerged to describe the activity of HAT1 in the cell (11, 12). In yeast, the HAT1–HAT2 complex binds to and acetylates newly synthesized histone H4 in the cytoplasm and remains associated with histone H4. At a certain point, the newly synthesized histone H3 joins the complex. The HAT1–HAT2 complex is then imported into the nucleus in association with histones H3-H4. Once in the nucleus, the HAT1–HAT2–H3–H4 complex becomes associated with the histone chaperone/chromatin assembly factor HIF1 to form the NuB4 complex. In human cells, the sNASP chaperone binds H3.1–H4 heterodimers and presents the H4 carboxyl domain to RbAp46, which recruits HAT1 activity. After acetylation of histone H4, the complex is stabilized and the histones are transferred to the ASF1B chaperone. ASF1B associates with importin-4, and the histones are then transported into the nucleus (12). These models suggest that HAT1 may not be functioning exclusively in a catalytic capacity and that the association of the HAT1–HAT2 (or HAT1–RbAp46) complex with histones H4 and H3 may be involved in both the import of these histones into the nucleus and their subsequent delivery to chromatin assembly factors.
Despite the fundamental importance of HAT1 in DNA replication and chromatin assembly, we still have a limited understanding of its function and regulation as a HAT. The crystal structure of yeast HAT1 in complex with AcCoA was reported by Dutnall et al. (13) who described an overall fold of the enzyme and its relationship to the structures of other HAT enzymes and AcCoA-binding site. However, how HAT1 recognizes both the substrate and AcCoA at the enzyme active site and how this complex facilitates acetyl group transfer from the latter to the former remains elusive. To address these questions, we solved the crystal structure of human HAT1 in a ternary complex with both AcCoA and substrate histone H4 peptide. The unique HAT1–AcCoA–H4 complex structure offers us detailed atomic-level insights into the enzyme active site and provides mechanistic knowledge of the molecular basis of HAT1 substrate recognition and selectivity. Our structure-guided enzyme kinetic study sheds light on the catalytic mechanism of HAT1.
Results and Discussion
Protein Expression, Purification, and Crystallization.
Human HAT1 is expressed in two isoforms, a and b, which encode proteins of 419 and 334 amino acid residues, respectively. Isoform b lacks an alternate internal exon, compared with isoform a, and uses a downstream start codon. Isoform b has a shorter N terminus (residues 1–85) compared with isoform a. We were not able to express isoform b in soluble form in Escherichia coli cells. However, the full-length protein of isoform a was soluble and stable after purification, although it did not produce crystals. We therefore generated a series of constructs encoding different lengths of the HAT1 protein. We found that a fragment of HAT1 consisting of residues 22–341 was the most soluble when expressed in E. coli cells. Hereafter we describe this fragment used in our study as human HAT1.
Human HAT1 was purified to homogeneity by a Ni chelating column, followed by gel filtration chromatography and ion exchange chromatography as described in SI Materials and Methods. The purified protein was crystallized in complex with AcCoA and histone H4 peptide (residues 1–20). After an initial screen, we used the alkylated enzyme to improve crystal quality. Surface-exposed cysteines Cys27, Cys101, and Cys168 were found to be alkylated during the refinement of the crystal structure. The crystals obtained belong to space group P21212, and the structure was solved by molecular replacement and refined to 1.9-Å resolution (Table S1).
The final model of the human HAT1–AcCoA–H4 ternary complex contains two HAT1 protein molecules per crystallographic asymmetric unit. The two molecules are identical and can be superimposed with root-mean-square deviation (rmsd) of 0.477 Å for all Cα atoms. Each complex consists of residues 23–341 of the HAT1 protein, one AcCoA molecule, and one molecule of histone H4 peptide. The cofactor molecule is clearly defined in the electron density map of the ternary complex with an average B-factor of 30.7 Å2. The HAT1–AcCoA–H4 ternary complex contains a 15-residue histone H4 peptide centered around lysine 12 and containing the sequence Lys5-Gly6-Gly7-Lys8-Gly9-Leu10-Gly11-Lys12-Gly13-Gly14-Ala15-Lys16-Arg17-His18-Arg19. The histone H4 peptide residues 1–4 (Ser-Gly-Arg-Gly) and 20 (Lys) are not visible in the final electron density map. The average B-factor for H4 peptide is 24.8 Å2.
Overall Structure.
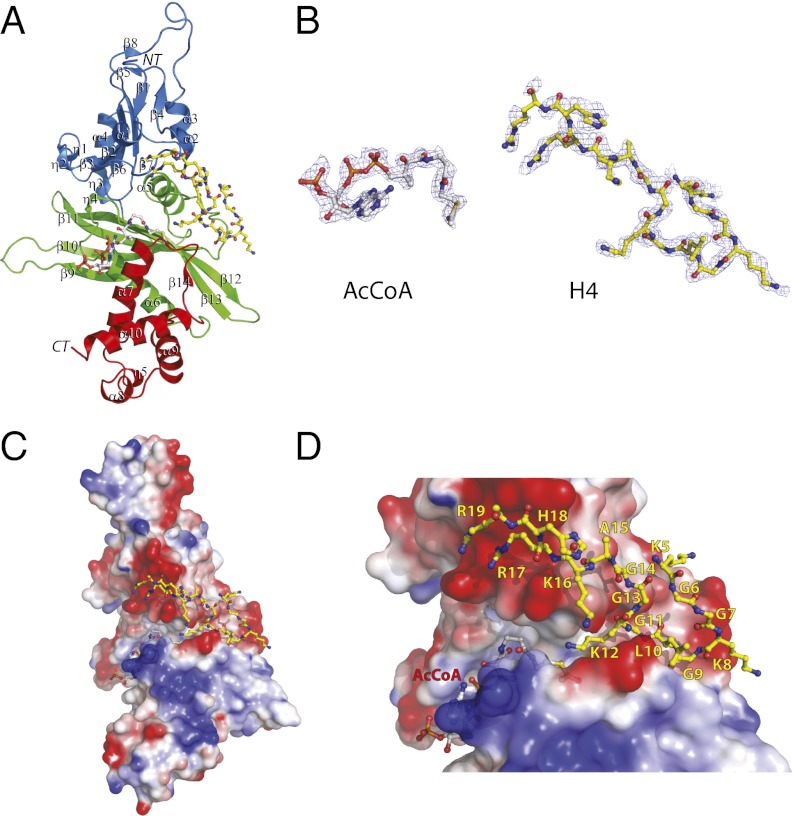
HAT1 structure has an elongated shape with approximate dimensions of 78 Å × 40 Å × 40 Å and can be described in terms of three domains: an N-terminal domain from residues 23–136 (blue in Fig. 1A), the central domain from residues 137–270 (green), and a C-terminal domain from residues 271–341 (red).
Fig. 1.
Crystal structure of human HAT1. (A) Overall fold of HAT1. The structure is shown in cartoon representation with the N, central, and C domains colored in blue, green, and red, respectively. H4 peptide and AcCoA are shown in stick-and-ball representation with carbon atoms in yellow (H4) and gray (AcCoA). (B) Electron density map of AcCoA and H4 peptide. The Fo-Fc maps were computed after simulated annealing with AcCoA and H4 peptide omitted from the atomic model and shown in mesh (contoured to 1.0 σ). AcCoA and H4 are presented in the same orientation as in A. (C) Electrostatic potential surface of the whole HAT1 molecule. Negatively and positively charged surfaces are colored in red and blue, respectively. (D) Close-up view of the substrate binding site of HAT1, with H4 and AcCoA shown in stick-and-ball representation. The figure was generated by using PyMOL (DeLano Scientific).
The N-terminal domain includes eight β-strands, four α-helices, and three 310 (η1–η3) helices. Strands β2, β3, β6, and β7 form a planar β-sheet, with β2 and β6 strands aligned in antiparallel fashion to the central β-strands β3 and β7, which are in parallel orientation to each other. On one side (top) of the β-sheet is a mixed network of helices (α1–α3) and β-strands (β1, β4, β5, and β8). On the other side (bottom) of the β-sheet are helices η1–η3 and α4. The N-terminal domain is connected by the short loop (η3–β9) to the central domain, which contains the twisted β-sheet formed by five antiparallel β-strands (β9–β13) sandwiched between helices η4, α5, and α6. The C-terminal domain is a bundle of helices (α7–α10 and η5) with one short β-strand β14.
Dali analysis (14) performed with individual domains of HAT1 showed no significant similarity with other known protein structures for the N-terminal domain, whereas the central domain showed a considerable structural similarity to the GCN5 and MYST families’ histone acetyltransferases [Protein Data Bank (PDB) ID code 1M1D (15), Z-score 10.6, rmsd 2.3 Å and PDB ID code 2PQ8, Z-score 10.2, rmsd 2.2 Å, respectively], and the C-terminal domain showed little structural similarity to the mating-type protein α-2 homedomain [PDB ID code 1K61 (16), Z-score 4.8, rmsd 2.8 Å].
Remarkably, the central and the C-terminal domains form a canyon whose sides are the inward faces of helix α7, strand β14, and the loop between them (from the C-terminal domain) and strand β13, helix η4, and the η4–α6 loop (from the central domain). The HAT1 canyon is ∼30 Å long and tends to be wider at the AcCoA-binding side (11–12 Å) than at the peptide-binding side (5–6 Å) with ∼25 water molecules solvating its surface. The canyon is highly polar with an electropositive character at the cofactor-binding site and an electronegative one at the peptide-binding site (Fig. 1 C and D).
The overall structure of human HAT1 is similar to yeast HAT1 structure (PDB ID code 1BOB) (13), and the two structures can be superimposed with an rmsd of 1.87 Å for all Cα atoms. The major difference between the two structures is that yeast protein has a longer β12–β13 loop (11 residues), which is disordered in the crystal structure, and strand β12 and the β11–β12 loop of yeast HAT1 are shorter (Figs. S1 and S2).
AcCoA-Binding Site.
The cofactor AcCoA molecule is bound in the canyon between the central and C-terminal domains (Fig. 1 B and D) and buries ∼565 Å2 of the protein-accessible surface area. It adopts a bent conformation with the N6 atom of adenine pointing toward the center of the protein molecule and a significant kink in the PC6-PC7 position of the pantotheine arm. The sugar pucker of AcCoA is in a C3′-endo configuration, and the glycosidic bond is in an anti conformation, placing the 3′-phosphate group exposed to the solvent.
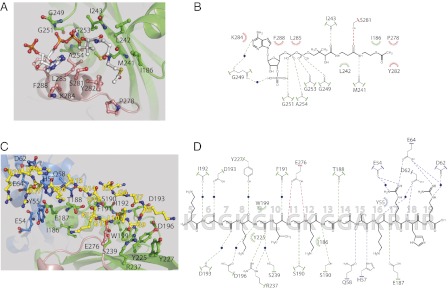
AcCoA makes an extensive set of direct hydrophobic and hydrogen-bonding interactions with residues from helix α7, the β14-α7 loop (C-terminal domain), β-strand β13, helix α6, and the η4-α6 loop (central domain) (Fig. 2 A and B). Only one residue, Gly249, makes water-mediated hydrogen bonds with the 3′-phosphate group of ribose and the N1 of the adenine ring of AcCoA. The adenine ring of AcCoA creates a hydrophobic interaction with the side chain of Lys284, and the ribose ring is in stacking interactions with Phe288. The pyrophosphate group of AcCoA establishes van der Waals contacts with Leu285 and a series of hydrogen bonds with the carbonyl groups of residues Gly249 and Gly253 and with the amide nitrogens of residues Gly251 and Ala254. The pantotheine moiety of AcCoA forms van der Waals contacts with the hydrophobic residue Leu242. These interactions make a kink of the pantotheine arm at the position PC6-PC7. Strikingly, the residue Leu242 is absolutely conserved through HAT1 proteins from different species (Fig. S2) and thus plays an important role in orienting the pantotheine moiety of AcCoA for acetyl transfer. In addition, the carbonyl group of the pantotheine moiety forms a hydrogen bond with the amide nitrogen atom of Ile243. The nitrogen atom and the carbonyl group of the β-mercaptoethylamine moiety of AcCoA establish hydrogen bonds with the carbonyl group of Met241 and side chain of Ser281. The acetyl group of the cofactor is anchored by residues Ile186, Pro278, and Tyr282 through hydrophobic interactions, placing the acetyl group close to the ε-amino group of Lys12 of histone H4 peptide, with a distance of ∼4.3 Å between the carbonyl carbon of the acetyl moiety of AcCoA and ε-amino group of Lys12 (Fig. 1D).
Fig. 2.
Substrate binding sites of HAT1. (A) Close-up view of AcCoA-binding site. (B) Illustration of AcCoA-binding site. (C) Close-up view of H4-binding site. (D) Illustration of H4-binding site. HAT1 structure is shown in cartoon representation. AcCoA, H4, and HAT1 residues interacting with AcCoA and H4 are shown in stick-and-ball representation. The carbon atoms in AcCoA and H4 are in gray and yellow, respectively. In C and D, all of the residues interacting with AcCoA and the H4 peptide are labeled. Hydrogen bonds are shown as dashed lines. Hydrophobic interactions are shown as arcs. Water molecules are shown as blue spheres.
A comparison of AcCoA-binding modes in yeast and human HAT1 revealed that, despite a marked architectural similarity between yeast and human enzymes, their respective modes of AcCoA recognition significantly differ (Fig. S1B). As described earlier, in the human HAT1–AcCoA–H4 ternary complex, the position of ribose and adenine rings is stabilized through interactions with Phe288 and the side chain of Lys284, whereas, in yeast HAT1, the corresponding residues are replaced by Arg267 and Asp263, respectively. In yeast HAT1, Arg267 pushes away the ribose ring of AcCoA and establishes a hydrogen bond with a 3′-phosphate group. The adenine ring of AcCoA in the yeast HAT1–AcCoA complex does not have interactions with the protein and the N6 atom of the ring pointing toward the solvent. Thus, human HAT1 likely has stronger binding to AcCoA than the yeast enzyme due to its interactions with the adenine and ribose rings. Despite these differences, the structures of AcCoA bound to yeast and human HAT1 converge at the pyrophosphate and the pantotheine groups. However, the position of the sulfur atom is different in two structures, suggesting that the reactive Lys12 of the histone H4 peptide plays a certain role in orientation of the acetyl group for the proper transfer (Fig. S1B).
H4 Peptide-Binding Site.
The H4 peptide in the HAT1–AcCoA–H4 ternary complex adopts a well-defined structure with two β-turns at positions Gly7-Lys8-Gly9-Leu10 and Leu10-Gly11-Lys12-Gly13 (Fig. 2 C and D). The conformation of the peptide is stabilized by two backbone hydrogen bonds between carbonyl groups of residues Gly7 and Gly13 with amide nitrogens of residues Leu10 and Lys5, respectively (Fig. 2 C and D). NMR solution studies (17) showed that the N-terminal domain of histone H4 (residues 1–27) prefers an extended backbone conformation at neutral pH; however, upon acetylation, the regions containing lysine residues induce structural transition, having a defined structural form especially at Lys12 position. These results and our observed well-defined structure of histone H4 peptide in the HAT1–AcCoA–H4 ternary complex suggest that the peptide undergoes significant conformation changes upon binding to HAT1 for placing the backbone of residues Gly11 and Lys12 at the entrance and the side chain of Lys12 deep inside the canyon formed between the central and the C-terminal domains. A superimposition of yeast and human HAT1 structures did not reveal any conformation changes that may occur upon histone H4 binding to the enzyme (Fig. S1).
The histone H4-binding site of HAT1 runs across the surface of the protein and comprises residues located in the central domain (α5-β11 loop, β12 and β13 strands) and the N-terminal domain (loop-helix α2-loop) (Fig. 1A). A total of ∼770 Å2 of solvent-exposed surface area of HAT1 is buried upon complex formation, and the majority of the surface where the peptide binds is negatively charged (Fig. 1 C and D).
Fourteen residues of the H4 peptide make interactions with human HAT1 (Fig. 2 C and D). Lys5 of the substrate, even though well-resolved in electron density, does not make any contacts with the protein. The carbonyl and amide groups of Gly6 of H4 form water-mediated hydrogen bonds with the side chain of Asp193 and carbonyl group of Ile192 of HAT1, respectively. The amide nitrogen of Asp193 also makes water-mediated hydrogen bond with the carbonyl group of Gly7 of H4. The side chain of Lys8 (H4 peptide) established van der Waals contacts with residue Tyr225, in addition to the water-mediated hydrogen bond between carbonyl group of Lys8 and side chain of Tyr227. The amide nitrogen of Gly9 of H4 peptide forms a water-mediated hydrogen bond with the side chain of Asp196, and the carbonyl group forms direct and water-mediated hydrogen bonds with the side chains of residues Arg237 and Ser239, respectively. In addition, backbone atoms of Gly9 make van der Waals interaction with Trp199. The residue Trp199 is strictly conserved through HAT1 proteins from different species (Fig. S2) and thus plays a crucial role in orienting H4 peptide and causing a β-turn of H4 peptide at the Gly9 position. Leu10 of H4 peptide has only a water-mediated hydrogen bond between its carbonyl group and the backbone carbonyl of Phe191. The side chain of Glu276 (HAT1) is directly involved in hydrogen bonding with the amide nitrogens of Gly11 and Lys12 of histone H4 peptide. The side chain of Lys12 also makes van der Waals contact with Ile186 of HAT1. The residue Ile186 is highly conserved in HAT1 proteins and is substituted only by valine in Drosophila melanogaster HAT1 at this position, suggesting that hydrophobic contacts are involved in orientation of the side chain of reactive Lys12 of the substrate. The amide nitrogens of Gly13 and Gly14 make direct hydrogen bonds with the side chain of Ser190 and carbonyl group of Thr188, respectively. Residue Ala15 of H4 interacts with side chains of Gln58 and His57 through hydrogen bonding. Lys16 of histone H4 peptide has only a water-mediated hydrogen bond between its carbonyl and the side chain of Asp62, whereas the latter also forms a hydrogen bond with the amide nitrogen of His18 from H4 peptide. The side chains of Arg17 and Arg19 of the histone H4 peptide make an extensive set of hydrogen bonds with the side chain of Glu64. In addition, the side chains of Arg17 and Arg19 interact through water-mediated hydrogen bonds with carbonyl groups of Glu54 and Asp62, respectively. Also, the amide nitrogen of Arg17 is in hydrogen bonding with the side chain of Glu187. This network of hydrogen bonding between Arg17 and Arg19 of H4 peptide and residues Glu54, Asp62, and Glu64 is responsible for anchoring the histone H4 substrate to the HAT1 protein. The residue Glu64 is absolutely conserved in HAT1 proteins (Fig. S2) and therefore plays a crucial role in the substrate-binding specificity of HAT1.
Substrate Recognition and Specificity of HAT1.
Newly synthesized histone H4 is deposited in a di-acetylated form during active DNA replication and chromatin assembly, a modification that was originally described over 30 years ago (18). Subsequent studies have identified that, among all of the lysine residues in the H4 N-terminal tail (lysines 5, 8, 12, 16), lysines 5 and 12 are the exclusive sites of di-acetylation in newly synthesized histone H4, and this pattern is absolutely conserved in organisms ranging from protozoa to humans. Since the identification of HAT1 as an enzyme responsible for this modification, the substrate specificity of HAT1 has been a subject of extensive studies.
HAT1 purified from yeast extracts specifically acetylates only residue Lys12 of histone H4, but recombinantly produced yeast HAT1 also acetylates Lys5 of H4 to a lesser extent and weakly acetylates histone H2A (1, 2). Human HAT1 has been shown to modify Lys5 and Lys12 of histone H4 and weakly modify Lys5 of H2A (3), whereas the maize HAT1 enzyme is specific for H4 at Lys5 and Lys12 (19, 20). On the basis of these studies, a common motif in the vicinity of the acetylated lysine (GxGKxG) has been proposed (2).
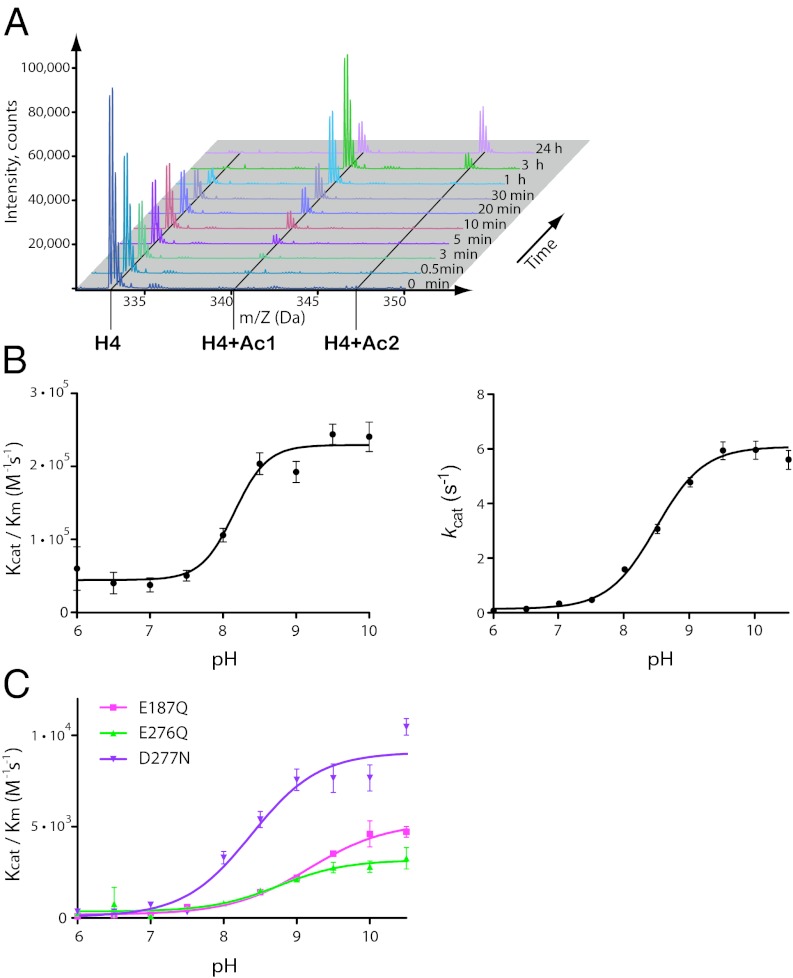
The crystal structure of the human HAT1–AcCoA–H4 ternary complex clearly shows that substrate recognition of HAT1 is achieved by altering H4 conformation and adjusting it from extended to defined conformation. The invariant residue Glu64, together with highly conserved residue Asp62 (substituted by Glu in D. melanogaster HAT1), form a negatively charged surface and attract the positively charged Arg17 and Arg19 of H4 peptide. The residues Trp199 (absolutely conserved) and Tyr225 (substituted by Phe in Caenorhabditis elegans) of HAT1 enzyme “freeze” the orientation of the peptide through interactions with Gly9 and the side chain of Lys8 of the substrate, respectively. Alanine substitutions of these residues resulted in a significant increase in Km values for the H4 peptide—∼6.1-fold for E64A and ∼4.2- and ∼2.9-fold for D62A and W199A mutants, respectively (Table 1). The salient feature of HAT1 structure is the presence and the shape of the canyon between the central and C-terminal domains. The canyon is 5–6 Å wide at the entrance of H4-binding site and can accommodate only residues Gly11 and Lys12 and becomes narrower at side chain of the Lys12. Additionally, the absolutely conserved Glu276 “tunes up” the specificity of the HAT1 enzyme through interactions with Gly11 and Lys12 of histone H4 peptide. The crystal structure of the human HAT1–AcCoA–H4 ternary complex reveals that Lys12 of histone H4 is the highly preferable substrate for HAT1 enzymes. This is also in agreement with our enzyme kinetics study monitored with mass spectrometry (Fig. 3A), which shows that di-acetylated species of H4 peptide start to appear only after the peptide is fully monoacetylated (3 h), and the peptide is not totally converted to di-acetylated form even after 24 h of the reaction. Also, the relative activity of the HAT1 enzyme for H4K12Ac was 20-fold less than that for the H4 peptide (Fig. S3A).
Table 1.
Kinetic properties of HAT1 mutants
| HAT1 | Substrate | kcat (s−1) | Km (μM) | kcat/Km (M−1⋅s−1) |
| WT | AcCoA | 4.14 ± 0.35 | 6.68 ± 0.3 | 619,760 |
| H4 peptide | 4.28 ± 0.01 | 20.77 ± 0.47 | 205,970 | |
| D62A | H4 peptide | 1.99 ± 0.2 | 85.59 ± 4.06 | 23,250 |
| E64A | H4 peptide | 0.50 ± 0.02 | 126.7 ± 28.83 | 3,946 |
| W199A | H4 peptide | 0.41 ± 0.01 | 59.46 ± 3.75 | 4,877 |
| E187Q | H4 peptide | 0.28 ± 0.01 | 197.6 ± 18.4 | 1,415 |
| E276Q | H4 peptide | 0.15 ± 0.01 | 99.8 ± 8.4 | 1,503 |
| D277N | H4 peptide | 0.53 ± 0.02 | 98.9 ± 12.6 | 5,358 |
Fig. 4.
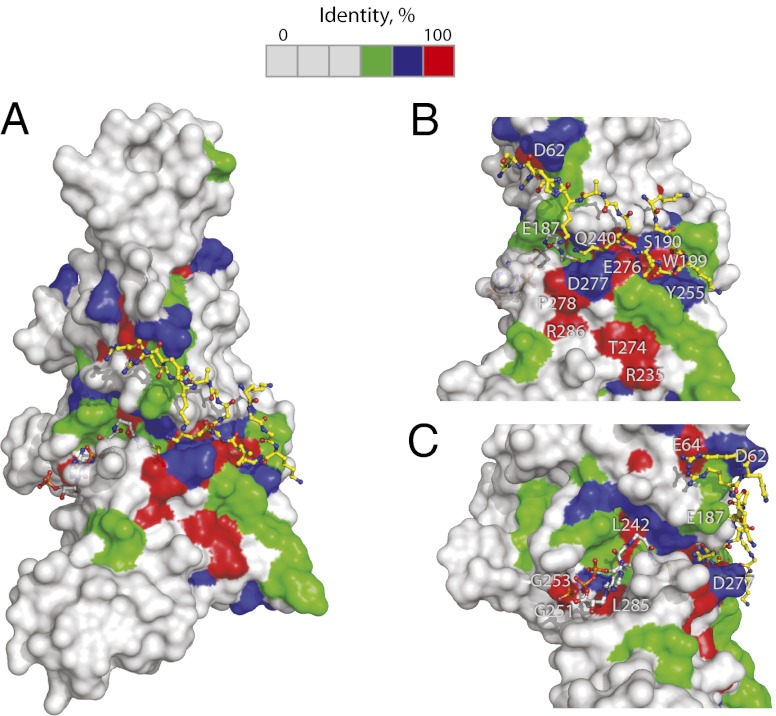
Sequence conservation of HAT1. (A) The human HAT1 structure is shown in surface representation. The identical, very similar, similar, and different residues are in red, green, blue, and gray. AcCoA and H4 peptide are shown in stick-and-ball representation. In B and C, the critical active site residues are labeled.
Implications in the Catalytic Mechanism of HAT1.
Histone lysine residues at physiological pH are normally charged and should be deprotonated by histone acetyltransferase enzymes to facilitate its nucleophilic attack on the acetyl group of AcCoA. Structural studies on GCN5, P/CAF, and ESA1 histone acetyltransferases identified a structurally conserved glutamic acid residue (Glu173 in GCN5, Glu570 in P/CAF, and Glu338 in ESA1) (21–24). This acidic amino acid residue has been shown to function as a general catalytic base to deprotonate the ε-amino group of reactive lysines. The deprotonated lysine can then attack the carbonyl carbon of the acetyl moiety of AcCoA, forming a tetrahedral intermediate, which then collapses to form CoA and acetylated-lysine products (25).
The pH dependence of kcat/Km and of kcat for HAT1 protein showed the plateau-shaped pH profiles (Fig. 3B), and the uprising nature of the plots indicates that a catalytic base required for deprotonation of the positively charged amine and the substrate binding does not change ionizaton of a catalytic base. A detailed view of substrate binding sites of the human HAT1–AcCoA–H4 ternary complex revealed that the closest HAT1 residues to Lys12 of the histone H4 peptide are Glu187, Glu276, and Asp277. The side chains of those residues are 6.7, 7.6, and 6.4 Å away from the ε-amino group of the reactive lysine, respectively. These distances are too far for direct lysine deprotonation by these residues, and probably water molecules mediate deprotonation. However, no ordered water molecules were found between the side chains of these residues and the substrate lysine of H4 during refinement of our crystal structure. The residues Glu187 and Asp277 in HAT1 are not strictly conserved: Glu187 is substituted by aspartic acid in HAT1 proteins from other species, and Asp277 is substituted by serine in C. elegans HAT1. Glu276 is invariant in HAT1 enzymes and also structurally conserved in the GNAT family of histone acetyltransferases as shown in GCN5 and ESA1 structures (13, 21, 24, 26).
Fig. 3.
Enzymatic activity of HAT1. (A) Mass spectrometry results of HAT1 activity at different time points. Peaks are labeled that correspond to unmodified peptide (H4) as well as to mono- or di-acetylated peptide (H4 + Ac1 or H4 + Ac2). pH profile of wild-type (B) and mutant forms of HAT1 (C).
To determine the importance of these residues in HAT1 catalysis, we generated a series of mutants of HAT1 and analyzed their kinetic properties at pH 8.5, which is near the pKa for the wild-type enzyme (pKa = 8.15 ± 0.06) (Fig. 3B). The E276Q mutant showed a 28.5-fold loss of activity (kcat = 0.15 ± 0.01 s−1) compared with the wild-type enzyme (kcat = 4.28 ± 0.01 s−1). The E187Q and D276N mutants were less affected, with ∼15.3- and ∼8.1-fold decreases in activity, respectively (Table 1). The kcat/Km decreased by ∼145- and ∼137-fold for E187Q and E276Q, respectively, and ∼38-fold for the D277N mutant compared with that of the wild-type enzyme. We were not able to determine reliably Km and kcat/Km values for double and triple mutants of HAT1, and hence we present only their relative activities (Fig. S3B)
To further determine which of these residues may act as a catalytic base, we performed pH dependence analysis of enzyme kinetics (i.e., kcat/Km) for the HAT1 mutants. As shown in Fig. 3 B and C, mutation D277N resulted in a slight upward shift of pKa (0.2 pH units) (pKa = 8.35 ± 0.12) compared with the wild-type enzyme (pKa = 8.15 ± 0.06), whereas E187Q and E276Q had a significant increase in pKa to 9.15 ± 0.09 and 8.74 ± 0.15, respectively. Collectively, these results suggest that Glu187 and Glu276 could act as the general catalytic base and together with Asp277 have a cumulative effect on deprotonation of the ε-amino group of substrate Lys12 of the histone H4 peptide, which could be mediated by water molecules.
Conclusion
The high-resolution crystal structure of the wild-type human HAT1 in a ternary complex with both substrate and cofactor reported in this study provides atomic-level understanding of the substrate recognition and specificity and detailed molecular insights into the HAT1 catalytic mechanism. Importantly, the structure also provides a framework for future studies on understanding the functional role of HAT2 in modulating enzymatic activity of HAT1. The interacting partner of the HAT1 enzyme, HAT2, increases the catalytic activity of the enzyme by a factor of 10, and it is believed to function by increasing the interaction between HAT1 and histone H4 (2). Biochemical studies and recent crystal structures of human RbAp46 (also known as RBBP7) and Drosophila RbAp46 (p55 or Nurf55) bound to histone H4 peptide have shown that histone H4 binds to the protein through contacts with the first helix proximal to the N-terminal tail (3, 27, 28). RbAp46 exhibits remarkable sequence conservation, reasonably suggesting that its interaction with the HAT1 enzyme is mediated by highly conserved residues of the two proteins. A survey of absolutely conserved residues of HAT1 proteins whose side chains are exposed to the protein surface reveals that residues Arg286, Thr274, and Arg235 might be very good candidates for these interactions (Fig. 4). These residues are located on the same side of the protein surface, just beneath the residues involved in catalysis. We propose that binding of RbAp46 to HAT1 would cause a conformation change of the β14-α7 loop, which in turn results in close proximity of catalytic residues to the ε-amino group of Lys12 of histone H4. These changes, which would enhance the catalytic activity of HAT1, might also cause a movement of the side chain of Lys12 of histone H4 peptide to a closer proximity to the acetyl group of the cofactor.
Because of the important role of HAT1 in chromatin assembly, a number of studies have begun to link HAT1 to different types of human cancer. The levels of HAT1 have been found to increase substantially in liver tumors (29). Also, HAT1 mRNA and protein levels are elevated in primary and metastatic human colon cancer tissues (30). In addition, immunohistochemical studies show that HAT1 is primarily nuclear in normal cells, but the localization of HAT1 largely shifted to cytoplasm in the tumor tissues (30). The observation of an elevated level of HAT1 is consistent with the proposed role for HAT1 in chromatin assembly (11) and reflects the obvious need to maintain a high level of chromatin assembly capacity in quickly proliferating cells. In light of this, HAT1 becomes an attractive target for the design of specific inhibitors to halt the growth of inappropriately proliferating cells. Therefore, we believe that the crystal structure of the human HAT1–AcCoA–H4 ternary complex will play an instrumental role in the rational design of potent small-molecule inhibitors with potential therapeutic applications.
Materials and Methods
The details of molecular cloning, expression, purification, crystallization, X-ray diffraction data collection, structure determination, and biochemical experiments are described in SI Materials and Methods. In short, HAT1 protein was expressed by using a pET28a-LIC vector that contained an N-terminal hexahistidine tag and thrombin cleavage site. Recombinant protein was purified by using several chromatographic techniques. Diffraction data of HAT1 crystals were collected at the ×12B beamline at the National Synchrotron Light Source, Brookhaven National Laboratory (Upton, NY). The structure of the acetyltransferase domain of HAT1 was solved by molecular replacement using the program MOLREP (31). The crystal structure of yeast HAT1 (PDB ID code 1BOB) (13) was used as the search model. ARP/wARP (32) was used for automatic model building. Refmac (33) was used for structure refinement. Data collection and refinement statistics are summarized in Table S1. The structure factors and atomic coordinates have been deposited in the Protein Data Bank with the PDB ID code 2P0W.
Supplementary Material
Acknowledgments
We thank P. Loppnau for his excellent technical support. This work is supported by the Structural Genomics Consortium, which is a registered charity (1097737) that receives funds from the Canadian Institutes for Health Research, the Canadian Foundation for Innovation, Genome Canada through the Ontario Genomics Institute, GlaxoSmithKline, Karolinska Institutet, The Knut and Alice Wallenberg Foundation, the Ontario Innovation Trust, the Ontario Ministry for Research and Innovation, Merck & Co., the Novartis Research Foundation, the Swedish Agency for Innovation Systems, the Swedish Foundation for Strategic Research, and The Wellcome Trust. The work in the Zhou laboratory has been supported by the research grants from the National Institutes of Health (to M.-M.Z.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.M. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates and structure factors reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2P0W).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1114117109/-/DCSupplemental.
References
- 1.Kleff S, Andrulis ED, Anderson CW, Sternglanz R. Identification of a gene encoding a yeast histone H4 acetyltransferase. J Biol Chem. 1995;270:24674–24677. doi: 10.1074/jbc.270.42.24674. [DOI] [PubMed] [Google Scholar]
- 2.Parthun MR, Widom J, Gottschling DE. The major cytoplasmic histone acetyltransferase in yeast: Links to chromatin replication and histone metabolism. Cell. 1996;87:85–94. doi: 10.1016/s0092-8674(00)81325-2. [DOI] [PubMed] [Google Scholar]
- 3.Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr Biol. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- 4.Imhof A, Wolffe AP. Purification and properties of the Xenopus Hat1 acetyltransferase: Association with the 14-3-3 proteins in the oocyte nucleus. Biochemistry. 1999;38:13085–13093. doi: 10.1021/bi9912490. [DOI] [PubMed] [Google Scholar]
- 5.Lusser A, et al. Analysis of the histone acetyltransferase B complex of maize embryos. Nucleic Acids Res. 1999;27:4427–4435. doi: 10.1093/nar/27.22.4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poveda A, et al. Hif1 is a component of yeast histone acetyltransferase B, a complex mainly localized in the nucleus. J Biol Chem. 2004;279:16033–16043. doi: 10.1074/jbc.M314228200. [DOI] [PubMed] [Google Scholar]
- 7.Ai X, Parthun MR. The nuclear Hat1p/Hat2p complex: A molecular link between type B histone acetyltransferases and chromatin assembly. Mol Cell. 2004;14:195–205. doi: 10.1016/s1097-2765(04)00184-4. [DOI] [PubMed] [Google Scholar]
- 8.Kelly TJ, Qin S, Gottschling DE, Parthun MR. Type B histone acetyltransferase Hat1p participates in telomeric silencing. Mol Cell Biol. 2000;20:7051–7058. doi: 10.1128/mcb.20.19.7051-7058.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin S, Parthun MR. Histone H3 and the histone acetyltransferase Hat1p contribute to DNA double-strand break repair. Mol Cell Biol. 2002;22:8353–8365. doi: 10.1128/MCB.22.23.8353-8365.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suter B, et al. Association with the origin recognition complex suggests a novel role for histone acetyltransferase Hat1p/Hat2p. BMC Biol. 2007;5:38. doi: 10.1186/1741-7007-5-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parthun MR. Hat1: The emerging cellular roles of a type B histone acetyltransferase. Oncogene. 2007;26:5319–5328. doi: 10.1038/sj.onc.1210602. [DOI] [PubMed] [Google Scholar]
- 12.Campos EI, et al. The program for processing newly synthesized histones H3.1 and H4. Nat Struct Mol Biol. 2010;17:1343–1351. doi: 10.1038/nsmb.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dutnall RN, Tafrov ST, Sternglanz R, Ramakrishnan V. Structure of the histone acetyltransferase Hat1: A paradigm for the GCN5-related N-acetyltransferase superfamily. Cell. 1998;94:427–438. doi: 10.1016/s0092-8674(00)81584-6. [DOI] [PubMed] [Google Scholar]
- 14.Holm L, Sander C. Dali: A network tool for protein structure comparison. Trends Biochem Sci. 1995;20:478–480. doi: 10.1016/s0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 15.Poux AN, Cebrat M, Kim CM, Cole PA, Marmorstein R. Structure of the GCN5 histone acetyltransferase bound to a bisubstrate inhibitor. Proc Natl Acad Sci USA. 2002;99:14065–14070. doi: 10.1073/pnas.222373899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aishima J, et al. A Hoogsteen base pair embedded in undistorted B-DNA. Nucleic Acids Res. 2002;30:5244–5252. doi: 10.1093/nar/gkf661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bang E, Lee CH, Yoon JB, Lee DW, Lee W. Solution structures of the N-terminal domain of histone H4. J Pept Res. 2001;58:389–398. doi: 10.1034/j.1399-3011.2001.00941.x. [DOI] [PubMed] [Google Scholar]
- 18.Ruiz-Carrillo A, Wangh LJ, Allfrey VG. Processing of newly synthesized histone molecules. Science. 1975;190:117–128. doi: 10.1126/science.1166303. [DOI] [PubMed] [Google Scholar]
- 19.Kölle D, Sarg B, Lindner H, Loidl P. Substrate and sequential site specificity of cytoplasmic histone acetyltransferases of maize and rat liver. FEBS Lett. 1998;421:109–114. doi: 10.1016/s0014-5793(97)01544-5. [DOI] [PubMed] [Google Scholar]
- 20.Eberharter A, Lechner T, Goralik-Schramel M, Loidl P. Purification and characterization of the cytoplasmic histone acetyltransferase B of maize embryos. FEBS Lett. 1996;386:75–81. doi: 10.1016/0014-5793(96)00401-2. [DOI] [PubMed] [Google Scholar]
- 21.Rojas JR, et al. Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide. Nature. 1999;401:93–98. doi: 10.1038/43487. [DOI] [PubMed] [Google Scholar]
- 22.Tanner KG, et al. Catalytic mechanism and function of invariant glutamic acid 173 from the histone acetyltransferase GCN5 transcriptional coactivator. J Biol Chem. 1999;274:18157–18160. doi: 10.1074/jbc.274.26.18157. [DOI] [PubMed] [Google Scholar]
- 23.Clements A, et al. Crystal structure of the histone acetyltransferase domain of the human PCAF transcriptional regulator bound to coenzyme A. EMBO J. 1999;18:3521–3532. doi: 10.1093/emboj/18.13.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yan Y, Barlev NA, Haley RH, Berger SL, Marmorstein R. Crystal structure of yeast Esa1 suggests a unified mechanism for catalysis and substrate binding by histone acetyltransferases. Mol Cell. 2000;6:1195–1205. doi: 10.1016/s1097-2765(00)00116-7. [DOI] [PubMed] [Google Scholar]
- 25.Berndsen CE, Albaugh BN, Tan S, Denu JM. Catalytic mechanism of a MYST family histone acetyltransferase. Biochemistry. 2007;46:623–629. doi: 10.1021/bi602513x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schuetz A, et al. Crystal structure of a binary complex between human GCN5 histone acetyltransferase domain and acetyl coenzyme A. Proteins. 2007;68:403–407. doi: 10.1002/prot.21407. [DOI] [PubMed] [Google Scholar]
- 27.Murzina NV, et al. Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure. 2008;16:1077–1085. doi: 10.1016/j.str.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song JJ, Garlick JD, Kingston RE. Structural basis of histone H4 recognition by p55. Genes Dev. 2008;22:1313–1318. doi: 10.1101/gad.1653308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pogribny IP, Tryndyak VP, Muskhelishvili L, Rusyn I, Ross SA. Methyl deficiency, alterations in global histone modifications, and carcinogenesis. J Nutr. 2007;137(1, Suppl):216S–222S. doi: 10.1093/jn/137.1.216S. [DOI] [PubMed] [Google Scholar]
- 30.Seiden-Long IM, et al. Transcriptional targets of hepatocyte growth factor signaling and Ki-ras oncogene activation in colorectal cancer. Oncogene. 2006;25:91–102. doi: 10.1038/sj.onc.1209005. [DOI] [PubMed] [Google Scholar]
- 31.Vagin A, Teplyakov A. MOLREP: An automated program for molecular replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 32.Perrakis A, Harkiolaki M, Wilson KS, Lamzin VS. ARP/wARP and molecular replacement. Acta Crystallogr D Biol Crystallogr. 2001;57:1445–1450. doi: 10.1107/s0907444901014007. [DOI] [PubMed] [Google Scholar]
- 33.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.