Abstract
Cellular senescence is a tumor-suppressive program that involves chromatin reorganization and specific changes in gene expression that trigger an irreversible cell-cycle arrest. Here we combine quantitative mass spectrometry, ChIP deep-sequencing, and functional studies to determine the role of histone modifications on chromatin structure and gene-expression alterations associated with senescence in primary human cells. We uncover distinct senescence-associated changes in histone-modification patterns consistent with a repressive chromatin environment and link the establishment of one of these patterns—loss of H3K4 methylation—to the retinoblastoma tumor suppressor and the H3K4 demethylases Jarid1a and Jarid1b. Our results show that Jarid1a/b-mediated H3K4 demethylation contributes to silencing of retinoblastoma target genes in senescent cells, suggesting a mechanism by which retinoblastoma triggers gene silencing. Therefore, we link the Jarid1a and Jarid1b demethylases to a tumor-suppressor network controlling cellular senescence.
Keywords: H3K4me3, histone demethylase
Cellular senescence is an antiproliferative stress-response program that acts as a potent tumor-suppressor mechanism (1). Senescent cells acquire distinctive features, including a stable cell-cycle arrest, senescence-associated β-galactosidase (SA-β-gal) activity, and marked alterations in higher-order chromatin organization that are associated with dramatic changes in gene expression (2). In the cancer context, senescence can be triggered by telomere attrition, aberrant proliferative signals, or by some cytotoxic chemotherapeutic drugs, in each case providing a barrier to tumor initiation or progression (3). Beyond cancer, senescent cells can be observed in aged and damaged tissues, where they contribute to the resolution of some wound-healing responses (4) and can promote organismal aging (5).
Senescence involves a complex interplay between the p53 and retinoblastoma (RB) pathways. p53 acts to induce various cell-cycle inhibitory proteins, whereas RB acts to repress E2F-driven transcription (6). Although the RB protein family has overlapping and compensatory functions in cell-cycle control, we have shown that RB is specifically required for the repression of E2F target genes during senescence (7). RB is also required for the formation of senescence-associated heterochromatic foci (SAHF), potential centers of gene repression (8). Accordingly, SAHF are enriched in H3K9me3 (a histone modification associated with heterochromatin), are devoid of methylated H3K4me3 (a histone modification associated with gene activation), and exclude sites of active transcription (8). Such RB-mediated changes in chromatin modifications and structure may contribute to its tumor-suppressive role; however, the underlying mechanisms remain unclear.
To better understand the relationship between chromatin changes and gene expression during cellular senescence, we took an unbiased approach to identify global changes in histone modifications specific to senescent cells. Among the changes we observed, we linked one—loss of H3K4 methylation—to the RB tumor suppressor and to the RB-binding proteins and H3K4 demethylases Jarid1a and Jarid1b. Our results implicate histone demethylation as an important regulatory mechanism used by RB to mediate gene repression in senescent cells.
Results
Senescence-Specific Loss of H3K4 Methylation.
We used quantitative mass spectrometry (qMS) to characterize the global changes in histone modifications occurring during Ras-induced senescence of IMR90 cells, a well-characterized system where senescence is enforced by the combined action of the RB and p53 tumor suppressor pathways (7). Senescence was triggered in these cells by the activation of ER:H-RasG12V (9) and monitored by the detection of senescence markers, such as changes in morphology, increased expression of p16 and p21, silencing of proliferation genes (e.g., Cyclin A), and an increase in SA-β-gal activity (Fig. S1 A–C). Histone modifications were quantified in growing and senescent cells using a “one pot” approach, whereby total acid extracted histones were subjected to qMS (10).
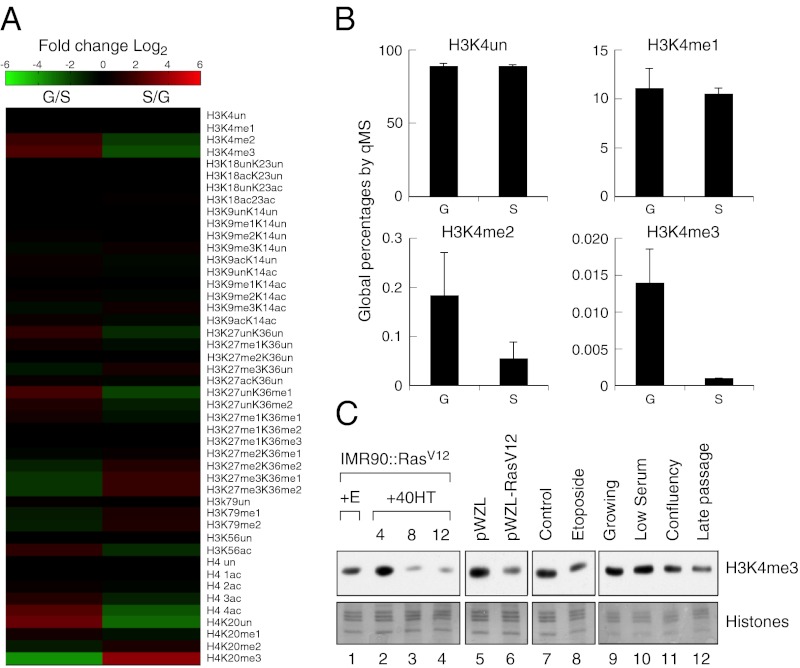
Consistent with heterochromatin formation during senescence (8), we detected decreased H3K9 acetylation and moderate enrichment of H3K9me3 in senescent cells (Fig. S1D and Dataset S1). Senescent cells also displayed a marked increase in the heterochromatic modifications H3K27me3 and H4K20me3, a decrease in several acetylation marks on H3 and H4 (e.g., H3K27ac and H3K56ac), and a striking loss of H3K4me2 and H3K4me3 (Fig. 1 A and B, Fig. S1E, and Dataset S1). qMS can also measure combinatorial histone modification changes, allowing us to detect, for example, a senescence-associated loss of acetylation in multiple lysine residues of histone H4 (H4 4ac: K5, K8, K12, and K16) (Fig. 1A, Fig. S1E, and Dataset S1). These changes, which also occurred in cells triggered to senescence by replicative exhaustion and treatment with etoposide, were confirmed by immunoblotting with modification-specific antibodies (Fig. S1 E and F). Collectively, these data indicate that the chromatin in senescent cells exists predominantly in a repressive state.
Fig. 1.
Senescence-specific global loss of H3K4 methylation. (A) Heatmap representing the relative levels (log2) of the different histone modifications in growing (G) vs. senescent (S) cells. (B) Histograms representing the global percentages of the different methylation states of H3K4 as determined by qMS. H3K4un is unmodified H3K4. The error bars represent the SD from three technical replicates. (C) Immunoblots using H3K4 methylation specific antibodies showing global loss of H3K4me3 in senescent but not quiescent cells. IMR90 cells treated with ethanol (lane 1) or with 4OHT for the indicated time (lanes 2–4), infected with a vector control (lane 5) or a vector expressing activated Ras (lane 6), treated with DMSO (lane 7) or 50 μM etoposide (lane 8), growing (lane 9), low serum quiescence (lane 10), confluency-induced quiescence (lane 11), and replicative senescence (lane 12). Core histones were used as a loading control.
Because H3K4 methylation is associated with actively transcribed genes, the marked loss of this modification in senescent cells suggested that active histone demethylation might contribute to gene repression during senescence. Supporting this idea, we detected a substantial decrease in H3K4me2 and H3K4me3 (Fig. 1 A and B), modifications associated with actively transcribed genes, but not changes in H3K4me1 (Fig. 1B), a modification that delineates developmental enhancers (11). The loss of H3K4me3 was confirmed by immunoblotting chromatin from cells triggered to senesce by oncogenic Ras, etoposide, and replicative exhaustion (Fig. 1C). No consistent changes in H3K4me3 were detected in quiescent cells (Fig. 1C), indicating that loss of H3K4me3 is a unique characteristic of senescent cells.
Cell-Cycle Genes Are the Main Targets for the Loss of H3K4me3.
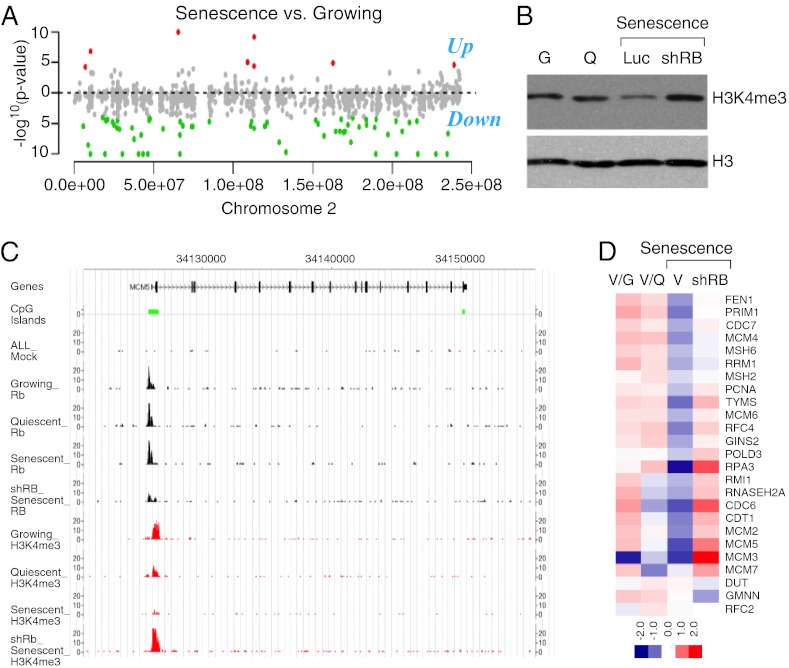
To identify the genomic regions affected by loss of H3K4 methylation, we performed ChIP using anti-H3K4me3 antibody in growing, quiescent, and senescent IMR90 cells, followed by deep sequencing (ChIP-seq). Consistent with previous studies (12), we detected H3K4me3 at the transcriptional start sites of highly expressed genes in each IMR90 cell population (Fig. S2A). Because our qMS analyses detected a global loss of H3K4me3 during senescence, we focused our informatic analyses toward identifying specific chromosomal regions affected by changes in H3K4me3 between growing and senescent cells across each chromosome (Dataset S2). The number of ChIP-seq reads located in each H3K4me3-enriched region in growing or senescent cells were counted, followed by χ2 analysis to identify regions that showed significantly different read counts between the two samples. Consistent with our qMS analysis, we observed a significant loss of H3K4me3 at many loci in senescent cells compared with growing cells (Fig. 2A and Dataset S3).
Fig. 2.
The RB tumor suppressor is required for the senescence-associated global and gene-specific loss of H3K4me3. (A) Shown is the log10 of the P values of the difference in H3K4me3 across chromosome 2 of growing vs. senescent cells (green) or senescent vs. growing cells (red). (B) Immunoblots from chromatin-bound fractions of growing (G), quiescent (Q), senescent (shLuc), and RB- deficient senescent cells (shRB) using an antibody that specifically recognizes H3K4me3 or total histone H3. (C) Genome-browser view documenting the loss of H3K4me3 at the MCM5 gene in senescent cells but not growing, or RB-deficient (shRB) senescent cells. (D) Heatmap representing the relative enrichment for H3K4me3 at the promoter of a subset of RB-regulated genes in growing (G), quiescent (Q), senescent (V), and RB-deficient senescent (shRB) cells.
The genes that lost H3K4me3 were subjected to Gene Ontology (GO) analysis to identify the processes they may control, as well as promoter motif analysis to gain insights into their regulation. These analyses revealed that genes controlling cell-cycle progression and DNA replication were enriched among the set of genes showing loss of H3K4me3 in senescence (Fig. S2B) (P = 3.0e-30 and 1.0e-13, respectively). Interestingly, genes that show loss of H3K4me3 frequently contained E2F binding sites in their promoters, whereas genes that showed either no loss or gain of H3K4me3 during senescence did not (binomial P value < 2.5e-11) (Fig. S2C). Given that RB binds with higher affinity to a subset of E2F target genes as cells exit the cycle into senescence (S2D) (7), we examined the overlap between genes bound by RB and genes that show loss of H3K4me3. Indeed, our ChIP-Seq data revealed a significant correlation (P = 2.6E-16) between RB binding and H3K4me3 loss during cellular senescence (Fig. S2E). Thus, RB target genes involved in cell-cycle control and DNA replication are the most prominent targets for loss of H3K4me3 during cellular senescence.
RB Is Required for H3K4me3 Demethylation During Senescence.
To test if the loss of H3K4me3 in senescent cells is RB-dependent, we examined the effect of suppressing RB on the global levels of H3K4me3 through immunoblotting and ChIP-seq in IMR90 cells triggered to senesce by oncogenic Ras. We have previously identified shRNAs capable of potently suppressing RB and shown that these prevent SAHF formation and suppression of a subset of E2F targets (herein defined as “RB-regulated” genes), but despite an initial delay, these cells eventually arrest because of a secondary barrier involving p21 (7). Here, these same RB shRNAs (Fig. S2F) prevented the senescence-associated global loss of H3K4me3 (Fig. 2B).
By further examining H3K4me3 at specific promoters in growing, quiescent, and senescent cells, we observed that RB suppression abolished the reduction in H3K4me3 that occurs on RB-regulated genes in senescent cells (Fig. 2 C and D, and Fig. S2G). In contrast, the total level of H3K4me3 in quiescent cells remained unchanged relative to growing cells (Fig. 2B), and only a modest reduction was observed on the promoters of RB-regulated genes (Fig. 2 C and D). These results further confirm that marked loss of H3K4me3 is specific for the senescent state and strongly link RB to the regulation of H3K4me3 during cellular senescence.
RB Associates with Jarid1a and Is Required for Jarid1a-Induced Senescence.
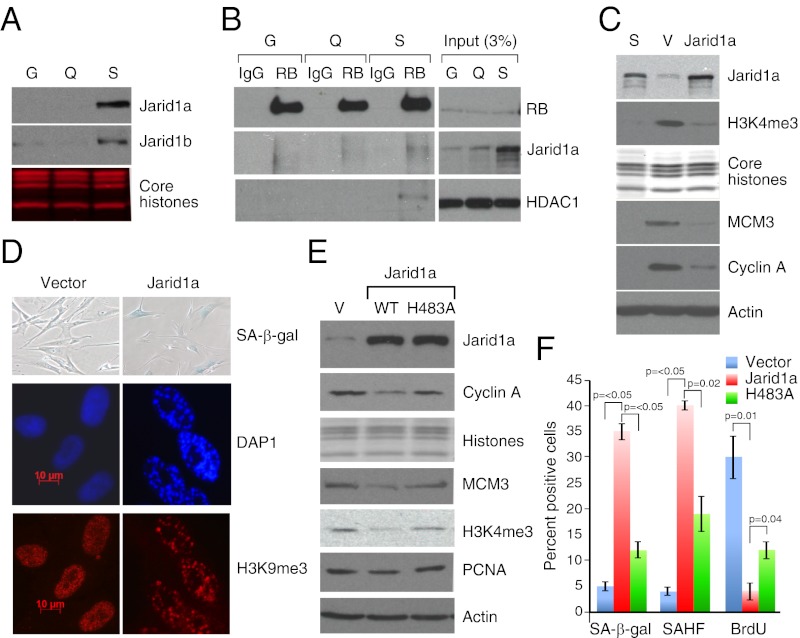
Although RB itself does not possess any known histone demethylase activity, it can associate with the Jarid1a and Jarid1b H3K4me2/3 demethylases (13–15). Like RB, both Jarid1a and Jarid1b accumulate in the chromatin fraction during senescence (Fig. 3A and Fig. S3A), implicating their role in the removal of H3K4me3. To determine whether RB binds the Jarid1 proteins in senescent cells, a physiological setting in which their interaction has not been previously examined, we carried out reciprocal coimmunopreciptiation experiments on chromatin-enriched fractions using anti-RB and anti-Jarid1a antibodies. Although this association can be difficult to detect (16), we reproducibly identified endogenous Jarid1a protein in RB immunoprecipitates from senescent-cell chromatin (Fig. 3B). Although weak, the signal was specific (i.e., not observed in IgG immunoprecipitations) and similar to that observed for the well-established RB binding protein, HDAC1 (17). Conversely, we also detected endogenous RB protein in Jarid1a immunoprecipitations (Fig. S3B). Although detectable in senescent mouse embryo fibroblasts (MEFs) (18), we were unable to detect an association between RB and endogenous Jarid1b in senescent IMR90 cells; whether this reflects a reagent issue or the lack of an interaction remains unclear. In addition, suppression of RB did not prevent chromatin association of the Jarid1 proteins, implying that Jarid1 targeting to chromatin may not be entirely RB-dependent or that Jarid1a has RB-independent targets in senescent cells.
Fig. 3.
Jarid1a associates with RB and induces premature senescence. (A) Immunoblots from chromatin-bound fractions of growing (G), quiescent (Q), and senescent (S) cells using antibodies that specifically recognize Jarid1a or Jarid1b. Coomassie blue staining of core histone proteins is shown as evidence of equal loading. (B) Immunoblots showing Jarid1a coimmunoprecipitation with RB. Note the specific Jarid1a signal in the in the senescent immunoprecipitation, but those from growing and quiescent immunoprecipitates show a nonspecific lower molecular weight smear. (C) Immunoblots documenting expression of Jarid1a, levels of H3K4me3, and levels of MCM3 and Cyclin A in ras-senescent (S), empty vector (V) infected, and Jarid1a-infected IMR90 cells. Parallel blots were probed with an anti-Actin antibody as a protein quantification control. (D) Micrographs showing the senescence markers SA-β-gal and SAHF in Jarid1a-infected IMR90 cells. (E) Immunoblots documenting expression of wild-type or catalytic inactive mutant (H483A) Jarid1a, levels of H3K4me3, and levels of MCM3, Cyclin A, and PCNA. Parallel blots were probed with an anti-Actin antibody as a protein quantification control. (F) Quantification of senescence markers. Values represent the ± SE of at least three independent experiments.
The above data raise the possibility that Jarid1 proteins contribute to senescence by facilitating the silencing of some RB targets by promoting H3K4 demethylation. Consistent with this possibility, enforced Jarid1a expression at levels similar to those observed physiologically during senescence triggered a global reduction of H3K4me3, which was associated with reduced expression of key RB , such as MCM3 and Cyclin A (Fig. 3C). Furthermore, enforced Jarid1a expression triggered cell-cycle arrest in IMR90 cells (Fig. 3F), which was associated with the accumulation of senescence markers, such as SA-β-gal and SAHF (Fig. 3 D and F). Such effects were significantly reduced when using a catalytic inactive mutant of Jarid1a (Fig. 3 E and F) or a C-terminal deletion that lacks the RB binding domain (Fig. S3 C–E), implying that both the demethylase activity of Jarid1a and its ability to bind RB are required for its role in senescence.
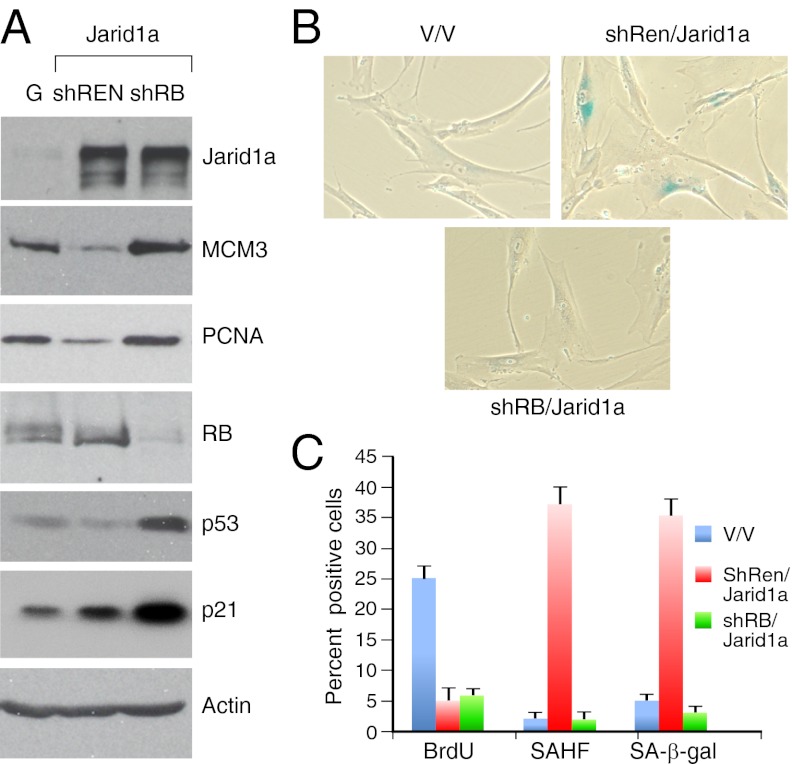
To determine whether RB was required for Jarid1a-mediated repression of key E2F target genes, IMR90 cells expressing either a control or RB shRNA were transduced with Jarid1a. In cells with RB knockdown, Jarid1a was unable to repress the E2F target genes examined (Fig. 4A). Consistent with this finding, suppression of RB prevented the accumulation of SAHF and SA-β-gal in cells expressing Jarid1a (Fig. 4 B and C). However, these cells eventually underwent cell-cycle arrest (Fig. 4C), likely from the same p21-dependent proliferation barrier observed in RasV12-senescent IMR90 cells expressing RB shRNAs (Fig. 4A) (7). Collectively, these data support a model whereby the association between RB and Jarid1a can facilitate H3K4 demethylation and subsequent silencing of E2F targets genes during senescence.
Fig. 4.
RB is required for Jarid1a-induced silencing of E2F target genes. (A) Immunoblots documenting the effect of suppressing RB on Jarid1a-induced silencing of E2F target genes. Parallel blots were probed with an anti-Actin antibody as a protein quantification control. (B). Micrographs showing the senescence marker SA-β-gal. Magnification: 20×. (C). Quantification of senescence markers. Values represent the ± SE of at least three independent experiments.
Jarid1 Proteins Act Redundantly to Demethylate H3K4me3 and Promote Silencing During Senescence.
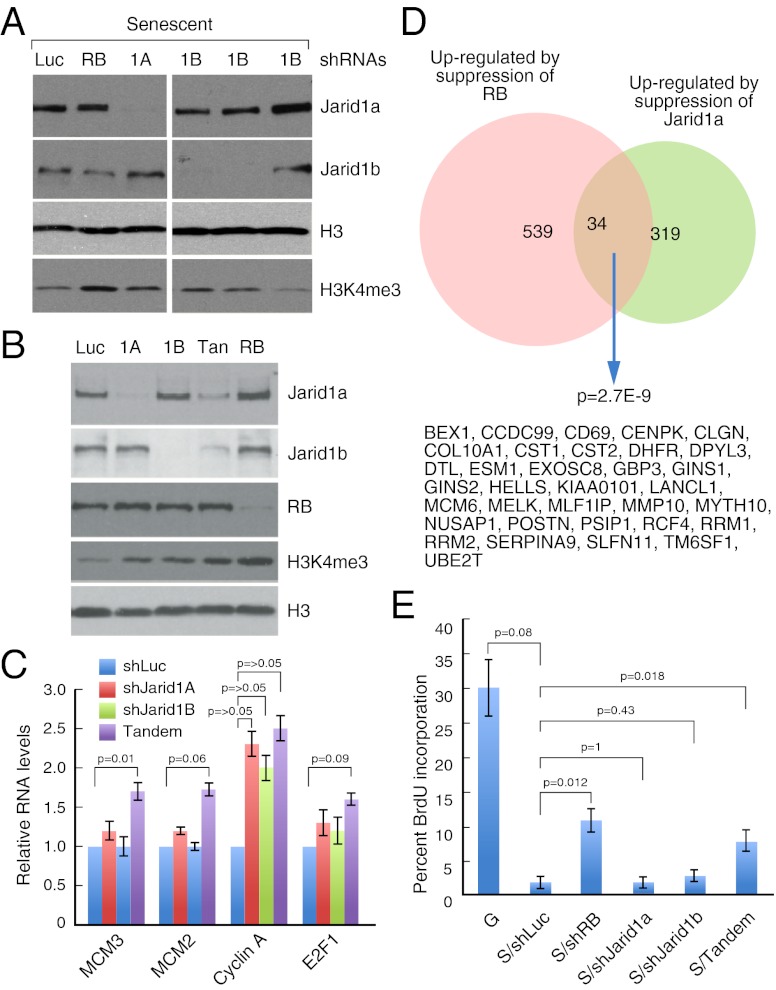
We next assessed whether the Jarid1 proteins are indeed required for H3K4me3 demethylation during oncogene-induced senescence. We generated multiple mir-30–based shRNAs targeting either Jarid1a or Jarid1b and identified several shRNAs that effectively suppressed their respective targets (Fig. 5A and Fig. S4A). Suppression of either Jarid1a or Jarid1b alone in IMR90 cells only partially prevented the senescence-associated global H3K4me3 demethylation (Fig. 5A and Fig. S4A). Although this partial effect could not be explained by the ability of the Jarid1 proteins to compensate for each other at the protein level (Fig. 5B), tandem shRNAs capable of cosuppressing Jarid1a and Jarid1b cooperated to prevent the demethylation of H3K4me3, similarly to suppression of RB (Fig. 5B). These data suggest that Jarid1a and Jarid1b play a redundant role in promoting H3K4 demethylation during senescence.
Fig. 5.
Redundant role for Jarid1 and Jarid1b in the senescence-associated demethylation of H3K4me3. (A) Immunoblots documenting the effect of suppressing Jarid1a (1A) or Jarid1b (1B) on the levels of H3K4me3 in senescent cells. Cells expressing RB shRNAs are used as a control. (B) Immunoblots from senescent cells expressing the indicated shRNA showing that cosuppression of Jarid1a and Jarid1b (Tan), like suppression of RB, prevents the loss of H3K4me3. (C) RT-qPCR analysis documenting the relative expression of the indicated E2F target genes in senescent cells expressing shRNAs targeting the Jarid1 proteins. Values represent the ± SE of at least three independent experiments using two different shRNA for each gene. (D) Venn diagram illustrating the overlap between RB-regulated and Jarid1a regulated genes. The list of genes in the overlap is indicated below the Venn diagram. (E) Histogram documenting BrdU incorporation of cells undergoing senescence at PS3 in the presence of the indicated shRNA. Values represent the ± SE of at least three independent experiments using two different shRNA for each gene.
To evaluate the role of the Jarid1 proteins in the control of RB target-gene expression in senescent cells, we suppressed Jarid1a, Jarid1b, or both and quantified the mRNA levels of known RB target genes by quantitative RT-PCR (RT-qPCR). This analyses showed that suppression of either Jarid1a or Jarid1b alone in senescent cells is insufficient to influence the expression of most cell cycle genes tested (e.g., MCM2, MCM3, E2F1), with the exception of Cyclin A, the expression of which is up-regulated in cells expressing shRNA targeting either Jarid1a or Jarid1b (Fig. 5C). Although the effects were relatively modest, cosuppression of Jarid1a/b cooperated to prevent the repression of these E2F target genes (Fig. 5C), indicating that the Jarid1 proteins were collectively required for their proper silencing. Accordingly, ChIP assays demonstrated that Jarid1a binds to these genes in senescent cells and that suppression of Jarid1a/b partially inhibits the senescence-associated demethylation of H3K4me3 (Fig. S4 B and C). These results are consistent with ChIP-chip data showing that Jarid1a binds and regulates a subset of E2F targets during differentiation (19). Although the Jarid1 proteins have been suggested to have pro-oncogenic functions (20–22), their requirement for E2F target-gene silencing suggests that the Jarid1 proteins are not simply a trigger of oncogene induced senescence, but rather participate in a downstream facet of this program.
To extend this analysis to a genome-wide level, we performed microarray-based gene-expression profiling to identify genes impacted by the Jarid1 proteins in senescent cells. As controls, we also examined the effect of suppressing Jarid1a, Jarid1b, or both demethylases on gene expression in growing, quiescent, and senescent cells [at two time points, postselection (PS) 3 and PS7]. Using a cut-off for significance (1.4-fold) based on the qPCR results above (Fig. 5C), we detected changes in gene expression between vector control-infected cells and cells expressing either the single or tandem shRNAs in all three conditions (Dataset S4).
Although growing and quiescent cells showed similar numbers of up- and down-regulated genes, senescent cells expressing either the single or tandem Jarid1 shRNAs showed a clear bias for up-regulated genes (Dataset S4). Such genes include those that are incompletely repressed during senescence [e.g., proliferating cell nuclear antigen (PCNA), Cyclin A], indicating that the Jarid1 proteins contribute to gene repression in the context of senescence. GO analysis showed that although the up-regulated genes in senescent cells were enriched in cell-cycle genes (P = 8.56E-04 for PS3 with tandem shRNA, P = 1.66E-06 for PS7 with tandem shRNA, and P = 3.3e-20 for PS3 with Jarid1a shRNA), the genes up-regulated in growing and quiescent cells showed no enrichment for this group of genes (Dataset S5). In contrast, there was no specificity in the ontology categories of down-regulated genes under any conditions. Taken together, these observations indicate that the Jarid1 proteins repress expression of a subset of cell-cycle genes specifically in senescent cells.
To further test our working model that the Jarid1 proteins contribute to RB-directed silencing of E2F target genes through demethylation of H3K4, we integrated our gene-expression profiling datasets with our ChIP-seq datasets for RB and H3K4me3. As expected, we detected a significant overlap (Hyper geometric test P < 2.2e-29, 68 genes) between those genes that show loss of H3K4me3 in senescent cells and those that were not effectively repressed in senescent cells expressing Jarid1a shRNAs. Although the effect was less pronounced, genes up-regulated after suppression of Jarid1b and suppression of both family members also showed a correlation with those displaying loss of H3K4me3 [P < 3.3e-3 for shJarid1b (S3), 10 genes, and P < 1.0e-9 for shTan (S3), 23 genes]. Furthermore, unsupervised hierarchical clustering of RB-regulated genes (genes repressed by RB in cells undergoing senescence) (Dataset S6) indicated that suppression of Jarid1a, Jarid1b, or both attenuates repression of many of these genes (Fig. S4D and Dataset S7). Specifically, we observed a significant overlap (P = 2.7E-9) between RB-regulated genes and Jarid1a-regulated genes (Fig. 5D) but not between Jarid1a-regulated genes and p53-regulated (P = 0.052, 2 genes) or NF-κB-regulated (P = 0.23, 6 genes) genes, two key senescence regulators for which we also have gene-expression profiling data (23). These data further support a model whereby Jarid1 proteins cooperate with RB to repress E2F target genes by facilitating H3K4 demethylation.
Jarid1 Proteins Contribute to Cell-Cycle Arrest During Oncogene-Induced Senescence.
We next examined the effect of suppressing Jarid1a and Jarid1b on the ability of cells to properly exit the cell cycle following expression of oncogenic Ras. Previous work indicates that suppression of RB in cells undergoing senescence delays the exit from the cell cycle as cells continue to incorporate BrdU 2–3 d after control cells (7). We therefore measured BrdU incorporation at PS3, a time point at which control shRNA-expressing cells have completely exited the cell cycle. Similar to cells expressing RB shRNAs (7), cells expressing the tandem shRNA continued to incorporate BrdU at PS3, but cells expressing shRNA targeting only Jarid1a or Jarid1b showed no BrdU incorporation, similar to shLuc controls (Fig. 5E). Taken together our study identifies a crucial role for the specific loss of H3K4me3 in establishment of the senescent state. Furthermore, we provide evidence that this occurs by the coordinated action of RB and the H3K4 demethylases Jarid1a and Jarid1b.
Discussion
Although senescence is associated with microscopically visible alterations in chromatin structure, the molecular events leading to these changes and the factors controlling these molecular events remain largely uncharacterized. By taking an integrative approach toward understanding the impact of chromatin changes in senescence, we demonstrate that senescent cells acquire a highly repressive chromatin state. We further show that demethylation of H3K4me3 can be directed in part through an RB-dependent effect of Jarid1 demethylases on a subset of E2F target promoters. Collectively, our study supports a model where Jarid1a and Jarid1b contribute to RB-mediated gene repression by promoting H3K4 demethylation on a subset of E2F target genes during senescence.
Our genome-wide analyses revealed that genes undergoing H3K4 demethylation during senescence are highly enriched in E2F target genes. Jarid1a binds to E2F target genes examined and enforced expression of wild-type Jarid1a, but not a catalytically inactive mutant, triggers H3K4me3 demethylation and silences key E2F target genes. Conversely, cosuppression of both Jarid1a and Jarid1b impairs H3K4 demethylation of some E2F target genes and blocks the down-regulation of these gene targets during senescence. Although we cannot exclude the possibility that other H3K4 demethylases play an active role in this process, our results indicate that the Jarid1 proteins contribute to the repressive chromatin environment of senescent cells and provide strong evidence that demethylation of H3K4 contributes to silencing of E2F target genes as cells exit the cell cycle into a senescent state.
Our results solidify a link between RB and H3K4 demethylation and thus provide insights into RB action in a context where it plays a tumor-suppressive role (7). Hence, we observe that: (i) genes that bind RB and display H3K4 demethylation during senescence are strongly correlated (P = 2.6E-16); (ii) endogenous RB associates with the H3K4 demethylase Jarid1a specifically in senescent cells; (iii) the RB and the Jarid1 proteins are required for the repression of an overlapping subset of E2F target genes; (iv) suppression of RB blocks Jarid1a-induced silencing of E2F target genes; and (v) conversely, a Jarid1a mutant unable to bind RB is defective at promoting senescence.
The overlap between Jarid1a- and b-regulated genes and those controlled by RB is significant (P = 2.7E-9) and specific (i.e., does not occur with p53- and NF-κB–regulated genes) but is by no means complete. This finding is likely because of the fact that suppression of Jarid1a/b is not as effective as RB knockdown in preventing the demethylation of H3K4me3 (Fig. 5A) and that suppression of Jarid1a/b only partially prevents loss of H3K4me3 at E2F target genes (Fig. S4C). In principle, the knockdown of the Jarid1 proteins might not reduce Jarid1 proteins enough to prevent their action on some target genes, or there could be redundancy in promoting gene repression by other H3K4 demethylases. Alternatively, RB could recruit unrelated histone-modifying enzymes that cooperate with the Jarid1 proteins in the RB-mediated gene repression. Consistent with this model, we detect an association between RB and HDAC1 in senescent cells (Fig. 3B), find substantial loss of histone acetylation in senescent cells (Fig. S1) (8), and see a much larger overlap between genes that show loss of H3K4me3 and Jarid1a-regulated genes (68 genes, P < 2.2e-29) than we detect for Jarid1-regulated and RB-target genes. Although multiple mechanisms undoubtedly contribute to RB-mediated gene repression, our results are consistent with a recent RNAi screen performed by Nijwening et al. that identified Jarid1b as a gene that, when repressed, can bypass RB-mediated senescence in MEFs (18). Although neither study excludes the possibility that the Jarid1 proteins also function independently of RB, these collective data support a model whereby the association between RB and Jarid1 proteins can facilitate H3K4 demethylation and silencing of RB targets genes during senescence.
The action of Jarid1 proteins during senescence suggests an antiproliferative or tumor-suppressor role for the Jarid1 proteins. However, recent reports have attributed oncogenic properties to these factors (16, 20–22). For example, Jarid1a and Jarid1b have been implicated in the maintenance of a slow-growing population of cancer cells that are required for continuous tumor growth and that are resistant to cytotoxic and targeted therapy (20, 21), and Jarid1a is required for the tumor initiation and progression in Rb+/− and Men1-defective mice (22). In principle, such pro-oncogenic functions may be revealed by other mutations common to cancer cells; for example, disruption of tumor-suppressor pathways that are intact in cells undergoing senescence. In the case of melanomas, which frequently inactivate the RB pathway gene INK4a, or developing tumors lacking RB, Jarid1 proteins would be unable to cooperate with RB to facilitate the silencing of E2F-target promoters, enabling other normal or aberrant proproliferative functions to predominate. As such, the biological output of chromatin-modifying factors in general will be heavily context-dependent, an important factor to be considered for effective targeting of these activities as therapeutics in cancer or other diseases.
Materials and Methods
Vectors, Cell Culture, and Gene Transfer.
Culture and transduction of IMR90 cells were performed as previously described (8). For information on the retroviral vectors used, please see SI Materials and Methods.
Senescence Assays and Gene Expression.
All of the assays, including detection of SA-β-gal, BrdU incorporation, detection of SAHF, and measuring gene-expression changes were done as previously described (8). Isolation of chromatin-bound proteins was performed as previously described (7) and subjected to immunoprecipitation with Jarid1a (Cell Signaling) or RB-specific (Santa Cruz) antibodies and IgG control. Immunoblotting was carried out as previously described (8). See SI Materials and Methods for a complete list of antibodies used.
Gene-expression changes were measured using RT-qPCR as previously described (7). For the analysis of the expression microarray, the Affymetrix Human Gene ST 1.0 GeneChip data were preprocessed using Robust Multichip Analysis (RMA) function in Bioconductor. An RMA value of 6 or less (log2 scale) was considered to be below detection. The probsets with more than 1.4-fold changes compared with vector control sample were treated as differentially expressed. The enrichment of GO terms in a particular gene list was performed using the online tool DAVID (24). P values reported for this study were corrected using the Benjamini-Hochberg approach (25). Gene-expression values were clustered and visualized using the Cluster and TreeView package (26).
Mass Spectrometry.
qMS analysis was carried out as previously described (27). In brief, bulk acid extracted histones were derivatized by treatment with propionyl anhydride (10). Histones were subsequently labeled with stable isotope using d10-propionic anhydride (Cambridge Isotope Laboratories). Online HPLCy separation of peptides was followed by LC–MS/MS using an LTQ-Orbitrap mass spectrometry (Thermo Fisher Scientific) as previously described (10). All data were manually inspected for quantification and MS/MS interpretation. Three technical replicates were performed for each sample.
ChIP-Seq Analysis.
The ChIP experiments were done as previously described (7). The immunoprecipitated DNA was prepared for Illumina sequencing as previously described (7). The Solexa sequencing tags were mapped to the unmasked human reference genome (NCBI v36, hg18) using the Illumina Eland program. Further details can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank E. Benevolenskaya for providing us with Jarid1a antibodies and J. Duffy for help in making the figures. This work was supported by National Science Foundation of China Grants 60905013, 91019016, and 31061160497 (to X.W. and M.Q.Z); Postdoctoral Fellowship 5F32AG027631 from the National Institutes of Health (NIH) (to A.C.); Department of Defense Predoctoral Fellowship W81XWH-11-1-0019 (to A.K.); Grant DP2OD007447 from the Office of the Director of the NIH; a National Science Foundation career award and New Jersey Cancer Commission on Research seed grant (to B.A.G.); The Ellison Medical Foundation New Scholar Award and NIH R21CA150117-01 (to E.B.); and NIH Grants HG001696 (to M.Q.Z.) and AG16379 (to S.W.L.). S.W.L. is an investigator in the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1119836109/-/DCSupplemental.
References
- 1.Collado M, Serrano M. Senescence in tumours: Evidence from mice and humans. Nat Rev Cancer. 2011;10:51–57. doi: 10.1038/nrc2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campisi J, d’Adda di Fagagna F. Cellular senescence: When bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 3.Prieur A, Peeper DS. Cellular senescence in vivo: A barrier to tumorigenesis. Curr Opin Cell Biol. 2008;20:150–155. doi: 10.1016/j.ceb.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Krizhanovsky V, et al. Senescence of activated stellate cells limits liver fibrosis. Cell. 2008;134:657–667. doi: 10.1016/j.cell.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker DJ, et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479:232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Courtois-Cox S, Jones SL, Cichowski K. Many roads lead to oncogene-induced senescence. Oncogene. 2008;27:2801–2809. doi: 10.1038/sj.onc.1210950. [DOI] [PubMed] [Google Scholar]
- 7.Chicas A, et al. Dissecting the unique role of the retinoblastoma tumor suppressor during cellular senescence. Cancer Cell. 2010;17:376–387. doi: 10.1016/j.ccr.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Narita M, et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113:703–716. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 9.Young AR, et al. Autophagy mediates the mitotic senescence transition. Genes Dev. 2009;23:798–803. doi: 10.1101/gad.519709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plazas-Mayorca MD, et al. One-pot shotgun quantitative mass spectrometry characterization of histones. J Proteome Res. 2009;8:5367–5374. doi: 10.1021/pr900777e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rada-Iglesias A, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 13.Christensen J, et al. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007;128:1063–1076. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 14.Klose RJ, et al. The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell. 2007;128:889–900. doi: 10.1016/j.cell.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 15.Xiang Y, et al. JARID1B is a histone H3 lysine 4 demethylase up-regulated in prostate cancer. Proc Natl Acad Sci USA. 2007;104:19226–19231. doi: 10.1073/pnas.0700735104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benevolenskaya EV, Murray HL, Branton P, Young RA, Kaelin WG., Jr Binding of pRB to the PHD protein RBP2 promotes cellular differentiation. Mol Cell. 2005;18:623–635. doi: 10.1016/j.molcel.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 17.Brehm A, et al. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- 18.Nijwening JH, Geutjes EJ, Bernards R, Beijersbergen RL. The histone demethylase Jarid1b (Kdm5b) is a novel component of the Rb pathway and associates with E2f-target genes in MEFs during senescence. PLoS ONE. 2011;6:e25235. doi: 10.1371/journal.pone.0025235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopez-Bigas N, et al. Genome-wide analysis of the H3K4 histone demethylase RBP2 reveals a transcriptional program controlling differentiation. Mol Cell. 2008;31:520–530. doi: 10.1016/j.molcel.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roesch A, et al. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141:583–594. doi: 10.1016/j.cell.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma SV, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin W, et al. Loss of the retinoblastoma binding protein 2 (RBP2) histone demethylase suppresses tumorigenesis in mice lacking Rb1 or Men1. Proc Natl Acad Sci USA. 2011;108:13379–13386. doi: 10.1073/pnas.1110104108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chien Y, et al. Control of the senescence-associated secretory phenotype by NF-κB promotes senescence and enhances chemosensitivity. Genes Dev. 2011;25:2125–2136. doi: 10.1101/gad.17276711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dennis G, Jr, et al. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- 25.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc, B. 1995;57:289–300. [Google Scholar]
- 26.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kapoor A, et al. The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature. 2010;468:1105–1109. doi: 10.1038/nature09590. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.