Abstract
The use of synthetic biological systems in research, healthcare, and manufacturing often requires autonomous history-dependent behavior and therefore some form of engineered biological memory. For example, the study or reprogramming of aging, cancer, or development would benefit from genetically encoded counters capable of recording up to several hundred cell division or differentiation events. Although genetic material itself provides a natural data storage medium, tools that allow researchers to reliably and reversibly write information to DNA in vivo are lacking. Here, we demonstrate a rewriteable recombinase addressable data (RAD) module that reliably stores digital information within a chromosome. RAD modules use serine integrase and excisionase functions adapted from bacteriophage to invert and restore specific DNA sequences. Our core RAD memory element is capable of passive information storage in the absence of heterologous gene expression for over 100 cell divisions and can be switched repeatedly without performance degradation, as is required to support combinatorial data storage. We also demonstrate how programmed stochasticity in RAD system performance arising from bidirectional recombination can be achieved and tuned by varying the synthesis and degradation rates of recombinase proteins. The serine recombinase functions used here do not require cell-specific cofactors and should be useful in extending computing and control methods to the study and engineering of many biological systems.
Keywords: DNA inversion, synthetic biology, genetic engineering, standard biological parts
Most engineered genetic data storage systems use auto- or cross-regulating bistable systems of transcription repressors or activators to define and hold state via continuous gene expression (1–4). Such epigenetic storage systems can be subject to evolutionary counter selection due to resource burdens placed on the host cell or spontaneous switching due to putatively stochastic fluctuations in cellular processes, including gene expression. Moreover, heterologous expression-based systems are difficult to redeploy given differences in gene regulatory mechanisms across organisms.
Another approach for storing data inside organisms is to code extrinsic information within genetic material (5). Nucleic acids have undergone natural selection to serve as heritable data storage material in organismal lineages. Moreover, DNA provides attractive features in terms of data storage robustness, scalability, and stability (6). In addition, engineered transmission of DNA molecules could support data exchange between organisms as needed to implement higher-order multicellular behaviors within programmed consortia (6, 7).
Practically, researchers have begun to use enzymes that modify DNA, typically site-specific recombinases, to study and control engineered genetic systems. For example, recombinases can catalyze strand exchange between specific DNA sequences and enable precise manipulation of DNA in vitro and in vivo (8). Depending on the relative location or orientation of recombination sites, three distinct recombination outcomes, integration, excision or inversion, can be realized.
From such knowledge, several natural recombination systems have been reapplied to support research in cell and developmental biology (9, 10). However, all in vivo DNA-based control or data storage systems implemented to date are “single-write” systems (11–13). Consequently, the amount of information such systems are able to store is linearly proportional to the number of implemented elements (for example, a “thermometer-code” counter capable of recording N events given N data storage elements) (13).
Single-write architectures are limiting if many of the uses for genetic data storage are considered in detail. For example, studies of replicative aging in yeast or human fibroblasts typically track at least 25 or 45 cell division events prior to the onset of senescence, respectively (14). Lineage mapping during worm development frequently tracks at least 10 differentiation events (15), while research with mouse and human systems considers up to several hundred cell divisions (16). In situations where the same signal is being recorded over multiple occurrences (for example, a series of cell division events), reliably rewritable elements are needed to realize geometric increases in data storage capacity (for example, combinatorial counters capable of recording 2N events given N storage elements).
Among the recombinase family of DNA-modifying enzymes, phage integrases are unique in that the directionality of the recombination reaction can be influenced by an excisionase cofactor (17). In natural systems, a phage integrase alone typically catalyzes site-specific recombination between an attachment site on the infecting phage chromosome (attP) and an attachment site encoded within the host chromosome (attB). The resulting integration reaction inserts the phage genome within the host chromosome bracketed by newly formed attL and attR (LR) sites. Upon induction leading to lytic growth, the prophage coexpresses integrase and excisionase that together restore an independent phage genome and the original attB and attP (BP) sites (18).
Early work with the r32 polar mutations of bacteriophage lambda revealed that integrase mediated recombination of antiparallel BP sites could also lead to the inversion of the intervening DNA (19, 20). Subsequent studies on DNA supercoiling used phage integrases to invert recombinant DNA sequences flanked by opposing BP sites (11, 21). Further in vitro work has since demonstrated that an integrase excisionase complex can revert a DNA sequence flanked by opposing LR sites (22). We thus sought to develop a stable data register that could invert and restore a target DNA sequence in vivo by appropriately controlling the conditional heterologous expression of integrase and excisionase.
Phages integrases are thought to represent two evolutionary and mechanistically distinct recombinase families (23). Tyrosine integrases, such as the bacteriophage lambda integrase, often have relatively long attachment sites (∼200 bp), use a Holliday junction mechanism during strand exchange, and require host specific cofactors. By contrast, serine integrases use a double-strand break mechanism during recombination and can have shorter attachment sites (∼50 bp). In addition, some serine integrases do not require host cofactors, a feature that has led to their successful reuse across a range of organisms (24). We thus chose to explore the engineering of rewritable genetic data storage systems using a bacteriophage serine integrase.
Bacteriophage Bxb1 now provides the best characterized serine integrase excisionase system (25–28). Bxb1 gp35 is a serine integrase that catalyzes integration of the Bxb1 genome into the GroEL1 gene of Mycobacterium smegmatis (25). Bxb1 gp47 is an excisionase that mediates excision in vivo and has been shown to control recombination directionality in vitro with high efficiency (27). Minimal attB, attP, attL, and attR sites have been defined for the Bxb1 system (25–27). The Bxb1 excisionase does not bind DNA independently and, from in vitro studies, is thought to control integrase directionality in a stoichiometric manner (27). From these and other studies several models have been proposed for how Bxb1 excisionase controls integrase directionality, but it is not yet clear how excisionase-mediated recombination proceeds or is regulated in vivo (27, 29).
Results
Architecture and Model for a RAD Module.
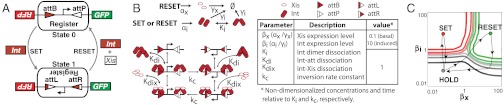
We developed a RAD module based on a two-state latch architecture that switches between states in response to distinct inputs and stores the last state recorded in the absence of either input signal. Here, our RAD module consists of an inducible “set” generator producing integrase, an inducible “reset” generator producing integrase and excisionase, and a DNA data register (Fig. 1A). Briefly, production of integrase alone should set a DNA register sequence flanked by oppositional attB and attP sites, thereby producing an inverted sequence flanked by attL and attR sites (State “1”). A second independent transcriptional input drives the simultaneous production of integrase and excisionase and should reset the register sequence to its original orientation and flanking sequences (State “0”).
Fig. 1.
Architecture, mechanisms, and operation of a recombinase addressable data (RAD) module. (A) The DNA inversion RAD module is driven by two generic transcription input signals, set and reset. A set signal drives expression of integrase that inverts a DNA element serving as a genetic data register. Flipping the register converts flanking attB and attP sites to attL and attR sites, respectively. A reset signal drives expression of integrase and excisionase and restores both register orientation and the original flanking attB and attP sites. The register itself encodes a constitutive promoter which initiates strand-specific transcription. Following successful set or reset operations, mutually exclusive transcription outputs “1” or “0” are activated, respectively. For the RAD module developed here, a “1” or “0” register state produces red or green fluorescent protein, respectively. (B) Elementary chemical reactions, molecular species, and kinetic parameters used to model the RAD module. Molecular concentrations are normalized to the integrase dimer dissociation constant (Ki). Kinetic rates are normalized to the integrase-mediated recombination rate (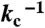 ). (C) Simulated phase diagram detailing pseudoequilibrium operating regimes for a RAD module experiencing sustained integrase and excisionase expression levels for 200/kc. The red, green, and gray lines represent, with decreasing intensity, 95, 75, and 55% switching (or hold) efficiencies (main text and SI Appendix).
). (C) Simulated phase diagram detailing pseudoequilibrium operating regimes for a RAD module experiencing sustained integrase and excisionase expression levels for 200/kc. The red, green, and gray lines represent, with decreasing intensity, 95, 75, and 55% switching (or hold) efficiencies (main text and SI Appendix).
We built a chemical kinetic model to better understand the potential behavior and failure modes of a DNA inversion RAD module (Fig. 1B). Our model reflects available knowledge of the mechanics and kinetics of the Bxb1 recombinase system, specifically (27, 30, 31). We used the model to estimate the operational phase diagram of our latch at pseudoequilibrium (SI Appendix). We found three distinct latch operating regions as a function of integrase and excisionase expression levels, corresponding to expected “set,” “reset,” or “hold” operations (Fig. 1C). One complete latch cycle requires the dynamic adjustment of integrase and excisionase expression through a “set, hold, reset, hold” pattern. These operations are realized in practice by cycling the transcription signals that define latch set and reset inputs and by tuning the specific genetic elements that provide fine control over integrase and excisionase synthesis and degradation.
Unidirectional DNA Inversion and Data Storage.
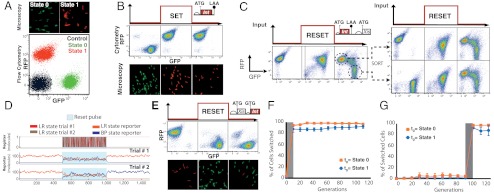
We first implemented a data storage register via a DNA fragment encoding fluorescent reporter proteins and Bxb1 recombinase recognition sites flanking a constitutive promoter on the chromosome of Escherichia coli DH5αZ1 (32) (Fig. 1A). We then confirmed via microscopy and cytometry that the state of the register could be assayed reliably (Fig. 2A). We next established that the register could set and hold state via a pulse of integrase expression within cells containing a single coding sequence for integrase. To do this, we built integrase driven “set” switches by cloning Bxb1 integrase under the control of an inducible promoter (32, 33) and a ribosome binding site library (Materials and Methods). We transformed the set-encoding vectors into cells containing the chromosomal BP register and isolated cells that only switched when induced; many variants switch spontaneously in the absence of an input signal or do not switch when induced (SI Appendix, Fig. S4; Table 1). We were able to isolate set functions that switch with greater than 95% efficiency at the single-cell level and that hold state following inducer removal (Fig. 2B).
Fig. 2.
Independent set and reset operations plus long-term data storage and switching in vivo. (A) Microscopy and flow cytometry data showing two distinguishable states for an invertible data register integrated in the E. coli chromosome driving red (RFP) or green (GFP) fluorescent proteins, and also a control sample in which cells express neither reporter. (B) Data register inversion via expression of integrase. Growing cells (doubling time ∼90 min) start in state “0” expressing GFP and, following a 16-h set input pulse, switch to and hold state “1” expressing RFP. (C) Bidirectionally of the integrase-excisionase reaction. Cells were transformed with plasmids containing the LR DNA data register and a bidirectional reset element on a plasmid and pulsed with arabinose (main text and SI Appendix). During a pulse, cells entered an intermediate state where both GFP and RFP are expressed. After inducer removal, cells split into two major populations corresponding to BP and LR states. We sorted split BP and LR populations by FACS and pulsed these sorted cells with arabinose again. The same behavior was observed regardless of the initial register state. (D) Stochastic simulation of bidirectional DNA inversion for a single-copy DNA register (top row) before, during (blue shaded area), and after a reset pulse. BP to LR and LR to BP recombination propensities are assumed to be equal. Two independent time-course stochastic simulations (middle and bottom rows) of expected GFP and RFP expression levels given the depicted (top row) BP and LR states. Fluorescent reporter degradation propensities modeled as tenfold slower than recombination propensities. (E) Data register restoration via expression of integrase and excisionase. Growing cells (doubling time ∼90 min) start in state “1” expressing RFP and, following a 16-h reset input pulse, return to and hold state “0” expressing GFP. (F) Stable long-term data storage. Cells were serially propagated without input signals for 100 generations following data register set (orange) or reset (blue). The fraction of individual cells maintaining register state was assayed by cytometry (see SI Appendix). (G) Long-term functionality of data register. Cells were serially propagated without input signals for 90 generations and then exposed to set (orange) or reset (blue) input signals. The fraction of individual cells switching state was assayed by cytometry.
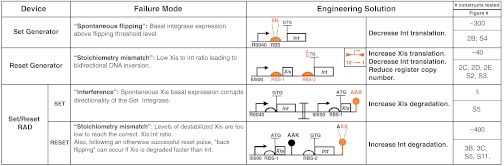
Table 1.
Failure modes and engineering solutions for set and reset operations. Putative failure causes as noted. Expression cassette schematics highlight (orange) regulatory regions targeted for reengineering with element-specific redesign goals. 6N is a full six nucleotide library within a Shine-Dalgarno ribosome binding site (RBS) core. RBS-1 and RBS-2 are collections of “standard” or computationally designed RBSs. AXX is a peptide library sampling 12 biochemically representative amino acids. The number of independent clonal constructs tested in each case plus main text or supplementary figures as stated. Details via SI Appendix.
Bidirectionality of Excisionase-Mediated DNA Inversion in Vivo.
We next determined if Bxb1 integrase and excisionase could mediate DNA inversion from an LR to BP state efficiently and unidirectionally in vivo. Previous in vitro experiments show that Bxb1 integrase and excisionase can catalyze LR to BP recombination to near completion (27). Unexpectedly, we found that a reset function mediated by integrase plus excisionase is reversible in vivo. For example, using constructs (15–20 copies per cell) expressing both integrase and excisionase we observed that upon induction using a reset signal (arabinose), both DNA register states are sampled across a mixed population and then a split population arises following reset signaling (Fig. 2C).
We observed bidirectional behavior starting from either initial register state, suggesting that, in the context of our system, expression of integrase plus excisionase results in repeated cycles of register inversion between BP and LR states (Fig. 2C, Right). We postulated that the system enters a bidirectional regime if the concentration of excisionase is too low relative to integrase, as might be needed to completely reverse recombination directionality (27). We therefore termed this type of behavior as “stoichiometry mismatch” failure. Since register flipping should occur faster than fluorescent reporter degradation cells display an intermediate state in which both reporter proteins are expressed. Following the reset pulse, cells resolve to one of two register states randomly and express a single reporter protein. A stochastic simulation of register “coin flipping” recapitulated the observed behavior (Fig. 2D and SI Appendix). From this framing, we were then able to engineer reset controllers that produce a range of weighted outcomes in the final register state by tuning the reset-specific integrase degradation rate (25, 50, 75% BP:LR distributions; SI Appendix, Fig. S11).
Engineering Excisionase-Mediated Inversion Toward Unidirectionality.
We overcame stoichiometry mismatch failures by engineering reset generators that should increase the expressed ratio of excisionase to integrase (Table 1). First, we expressed integrase from a lower copy plasmid (5–10 per cell) while expressing excisionase from a higher copy plasmid (50–70 per cell). Such changes were sufficient to drive excisionase-directed recombination to a BP state at ∼85% efficiency on a single-cell basis (SI Appendix, Fig. S2). We next expressed excisionase and integrase together from a bicistronic operon on the same higher copy plasmid (50–70 per cell), but with a weaker GTG start codon for initiating translation of the integrase. These changes drove excisionase-directed recombination to a BP state at greater than 90% efficiency on a single-cell basis (Fig. 2E).
We also tuned the DNA copy number of the data register in order to optimize the effective stoichiometry of excisionase associated with DNA-bound integrase (Table 1). Hatfull and coworkers proposed that Bxb1 excisionase only interacts with integrase that is complexed with DNA in either the LR or, more strongly, BP state (27). Thus, by reducing the number of DNA binding sites for integrase, we sought to decrease the fraction of integrase available for excisionase binding and therefore increase the effective excisionase-to-integrase ratio. For example, by only reducing the DNA register copy number from 5–10 per cell (Fig. 2C) to 1 per cell (SI Appendix, Fig. S3A, Bottom), we observed an increase in excisionase-mediated recombination directionality from ∼40% to ∼65%. Such observations are consistent with a model that accounts for relative DNA copy number and whether or not excisionase can interact with cytoplasmic integrase (SI Appendix, Fig. S3 B and C).
Stable in Vivo Data Storage over Many Generations.
For in vivo data storage systems to be most useful, they must be able to store state over an extended period of time or many cell doublings. Thus, to study the temporal and evolutionary stability of our RAD module, we repetitively grew and diluted E. coli cells every day for 10 days and monitored data storage at the single-cell level by measuring the continuous expression of fluorescent reporters. We established that starting from either state, the register could switch and hold state for 100+ cell doublings (Fig. 2F) or could hold state and then switch reliably following 90+ cell doublings (Fig. 2G). Taken together, these data demonstrate the practical stability and long-term operational reliability of a RAD register in the absence of feedback-mediated latch bistability.
Composing Opposing Recombinase Functions.
We next sought to operate a full set/reset cycle within a single cell. However, we found that most combinations of set and reset functions that work independently fail when used in combination within the same cell (SI Appendix, Fig. S5). For example, we observed that stand-alone reset functions, which encode for high levels of excisionase synthesis, result in spontaneous accumulation of excisionase sufficient to corrupt set functions (SI Appendix, Fig. S5). We termed such behavior set/reset “interference” failure.
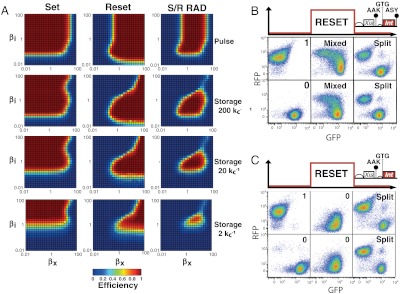
More generally, we eventually reasoned that the ranges of integrase and excisionase synthesis and degradation rates within which set and reset functions operate reliably together and for which a RAD module holds state are increasingly distinct with decreasing input pulse lengths, leading to increasingly stringent expression requirements (Fig. 3A, top to bottom). For example, when we destabilized the reset excisionase peptide in attempting to overcome interference failures (Table 1), we found that reset generators could be corrupted in two ways. First, we returned to stoichiometry mismatch failures during reset induction (Fig. 3B). Second, we found that if insufficient excisionase is maintained during the relaxation period following an apparently reliable reset, then split state populations can still emerge (Fig. 3C and SI Appendix, Fig. S6).
Fig. 3.
Functional composition, expected operable ranges, and reset failure modes for a RAD module. (A) Simulated phase diagrams detailing the expected operation of RAD module functions in response to dynamic pulses of integrase and excisionase across a range of expression levels (main text). Top row: switching efficiencies (colormap) for set and reset functions when combined in a RAD module. Second, third, and fourth rows: combined switching and storage efficiencies (colormap) for set and reset alone and for the integrated RAD module across decreasing input pulse widths (200/kc, 20/kc, and 2/kc, respectively) (main text and SI Appendix). (B) Reset failure during a reset pulse due to stoichiometric mismatch-mediated bidirectional register switching. Cells containing a chromosomal DNA data register starting in either state “1” (top row) or “0” (bottom row) were exposed to a 16-h reset pulse, producing a mixed state population during signal input and a split population thereafter (see also SI Appendix, Fig. S6). (C) Reset failure immediately following a reset pulse due to stoichiometric mismatch-mediated setting to state “1”. Cells containing a chromosomal DNA data register starting in either state “1” (top row) or “0” (bottom row) were exposed to a 16-h reset pulse, producing a single state “0” population during signal input but a split population thereafter.
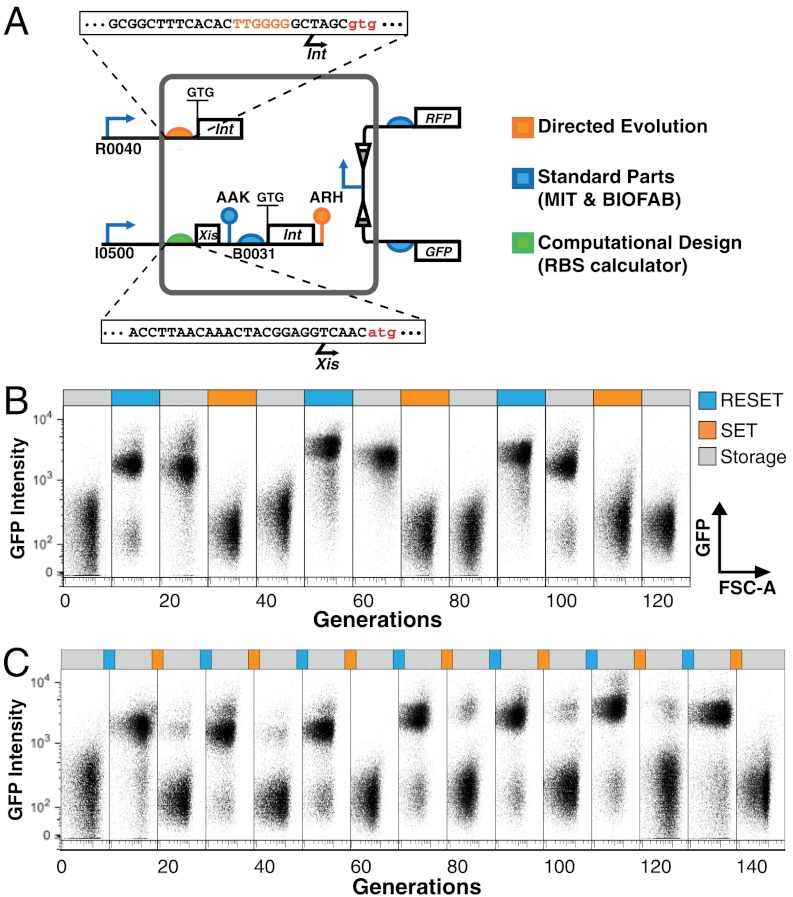
We ultimately screened ∼400 clones encoding a library of destabilized reset-encoded integrase peptides in order to obtain a reset cassette that functions reliably without also corrupting set functionality (Table 1). The working RAD module that resulted from this overall process uses weaker GTG start codons for both set and reset integrase coding sequences and encodes distinct nonconsensus ssrA proteolysis tags (34) on the reset integrase and excisionase peptides (Fig. 4A and SI Appendix, Fig. S4). Once assembled, we demonstrated that the RAD module can be cycled repeatedly and reliably in response to transition and hold inputs lasting ∼10 cell doublings (∼900 min) (Fig. 4B). We also demonstrated that the module functions in response to shorter transition inputs (∼240 min) (Fig. 4C).
Fig. 4.
Optimized genetic elements and reliable multicycle operation of a DNA-inversion RAD module. (A) Details of an integrated DNA inversion RAD module optimized for reliable set, reset, and storage functions. Specific genetic regulatory elements controlling protein synthesis and degradation were obtained from standard biological parts collections, via computational design, or via random mutagenesis and screening, as noted (main text and SI Appendix). (B) Experimental RAD module operation over multiple duty cycles. Growing cells (doubling time ∼90 min) starting in state “1” were cycled through a “reset, hold, set, hold” input pattern, with each step lasting ∼10 generations. Cell state was assayed via multicolor cytometry. Population distribution GFP (y axis) and forward scatter (x axis) levels are shown for samples taken from the generation number following the input step given directly above each scatter plot (for example, the first scatter plot shows the population distribution at generation 10) (main text and SI Appendix). (C) Multicycle RAD module operation driven by shorter SET and RESET input pulses. As in B but with set and reset pulses lasting for ∼2.5 cell doublings (∼4 h).
Discussion
Our experimental results are not inconsistent with a model in which inversion of DNA by Bxb1 integrase alone is unidirectional but inversion of DNA by integrase plus excisionase is bidirectional. However, biased directionality of Bxb1 excisionase-mediated recombination can be realized at a system level by controlling the excisionase-to-integrase ratio and dynamics, and also the integrase-to-DNA target site ratio. By carefully tuning and integrating the expression of competing recombinase functions, we were able to develop a first reliable and rewritable DNA inversion-based data storage system that works in vivo.
The process by which we eventually engineered a working RAD module warrants consideration. Practically, we needed to surmount challenges associated with enacting control over the relative levels and timing of expression for three proteins within E. coli. Initially, we did not know what specific quantitative levels or expression timing would drive DNA inversion in vivo. However, even during this first stage of the project, we lacked tools, whether working standard biological parts or computational design methods, sufficient to rationally engineer gene expression cassettes to provide qualitative control over switchable gene expression. For example, a series of computationally designed translation control elements produced qualitative distinct phenotypes during set operations across a range of expected protein synthesis rates, and also for repeated design attempts at the same target translation rate (SI Appendix, Fig. S4). The lack of genetic control elements that can be reliably composed so as to enable precise expression control with novel heterologous coding sequences, and the precision limits of computational tools that optimize control elements for specific genetic contexts, forced us to empirically validate gene expression on a gene-by-gene and gene combination-by-combination basis.
Our use of conditional control over recombination directionality to implement a repeatedly rewritable DNA data storage element likely only partially aligns with the natural contexts in which integrase and excisionase performance have been selected. For example, integrase alone naturally mediates integration of a phage genome into a host chromosome under circumstances in which the phage will not destructively lyse the host cell. Such integration reactions are likely under positive selection to be fast and efficient, given that failure to integrate prior to host chromosome replication and cell division could result in loss of the phage from a daughter lineage. Integration reactions are also likely under negative selection to be irreversible, since integration followed by immediate excision could result in an abortive infection. Both selective pressures would align well with our performance requirements for integrase during a set operation. However, in nature, when integrase plus excisionase excise a prophage, we suspect that there are not similarly strong selective pressures against recombination bidirectionality. For example, prophage induction via integrase plus excisionase is typically associated with the expression of phage factors leading to irreversible lytic growth. Such an evolutionary context is different from the performance requirements of a reset operation for a genetic data storage system in which both the recombination products and the host cell must continue to exist. Searching for phage-host systems in which prophage induction is followed by a period of delayed lysis or even host cell reproduction may help to identify natural excisionase-mediated recombination systems that are fully unidirectional.
The DNA inversion RAD module developed here should be translatable to applications requiring stable long-term data storage (for example, replicative aging) or under challenging conditions (for example, clinical or environmental contexts requiring in situ diagnosis or ex post facto reporting via PCR or DNA sequencing). Given the natural phage recombination functions from which the latch is implemented, (35) we believe that reliable operation with less than 30-min switching times should be obtainable via continued optimization of integrase and excisionase synthesis and degradation rates. Further improvements to latch speed or reliability might also be realized by thresholding- or closed-loop control architectures that produce system-level bistability.
A typical architecture for an 8-bit synchronous counter capable of recording a series of 256 input pulses (36) would require 16 recombinases recognizing distinct DNA sequences or the multiplexing of recombinase activity across repeating DNA recognition sites. Additional biochemically independent RAD modules could likely be identified from the increasing set of known natural recombinases, (37–39) or perhaps by engineering synthetic integrase excisionase pairs with altered DNA recognition specificity (40–42). Such work, along with puzzles of integrating dozens of competing biochemical functions, suggest that engineering increased capacity in vivo data storage systems will help define and challenge the limits of synthetic biology.
Materials and Methods
Plasmids.
The set element (pBAD-driven-integrase generator, Fig. 2 B,D, and E) was cloned in pSB3K1 plasmid (p15A origin; 15–20 copies). The reset element (pBAD-driven-excisionase+integrase generator, Fig. 2 C–E) was cloned on J64100 plasmid. The full RAD module (PLtet-O1 driven integrase generator and pBAD-driven-excisionase+integrase generator, Fig. 4 and SI Appendix, Fig. S9) was cloned in J64100. Additional details are given in the SI Appendix.
Cell Culture and Experimental Conditions.
All experiments were performed in E. coli DH5alphaZ1. For each experiment, three colonies were inoculated in supplemented M9 medium [M9 salts (Sigma), 1 mM thiamine hydrochloride (Sigma), 0.2% casamino acids (Across Organics), 0.1 M MgSO4 (EMD reagents), 0.5 M CaCl2 (Sigma) with glycerol (0.4%, Fisher Scientific) added as a carbon source] and appropriate antibiotics, and grown for approximately 18 h at 37 °C. Antibiotics used were carbenicillin (25 μg/mL), kanamycin (30 μg/mL), and chloramphenicol (25 μg/mL) (Sigma). L-arabinose (Calbiochem) was used at a final 0.5% wt/vol concentration; anhydrotetracycline (Sigma) was used at a final concentration of 20 ng/mL.
For long inputs, a saturated culture was diluted 1∶2,000 in media with inducer. For evolutionary stability experiments, cultures were diluted 1∶2,000 every day in media without inducer to achieve ∼10 generations per day [log 2 2,000 = 10.96, (43)] Cells were centrifuged and washed before each dilution step. For short inputs, overnight grown cultures were diluted 1∶100 in media with inducer, grown for 4 h, at which point cells were washed, diluted 1∶2,000, and grown for an additional 16 h.
Measurement and Data Analysis.
Flow cytometry was performed on a LSRII cytometer (BD-Bioscience). For each data point, GFP and RFP fluorescence was measured for 30,000 cells. Data were analyzed using the FlowJo software (Treestar, Inc.). State distributions were quantified by plotting GFP and RFP intensities against each other and gating according to control cells containing only the chromosomal LR or BP DNA registers. No gate was applied to the cell population before quantification.
Supplementary Material
Acknowledgments.
We thank Roger Brent, Christina Smolke, Alfonso Farrugio, Michele Calos, Francois St-Pierre, members of the Endy and Smolke labs, and the Stanford Shared FACS Facility for discussion and assistance. Support was provided by the NSF Synthetic Biology Engineering Research Center and Stanford University. J.B. is a recipient of the Stanford Center for Longevity (SCL) postdoctoral fellowship. P.S. is a recipient of the Stanford BioX bioengineering graduate fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1202344109/-/DCSupplemental.
References
- 1.Toman Z, Dambly-Chaudiere C, Tenenbaum L, Radman M. A system for detection of genetic and epigenetic alterations in Escherichia coli induced by DNA-damaging agents. J Mol Biol. 1985;186:97–105. doi: 10.1016/0022-2836(85)90260-8. [DOI] [PubMed] [Google Scholar]
- 2.Gardner TS, Cantor CR, Collins JJ. Construction of a genetic toggle switch in Escherichia coli. Nature. 2000;403:339–342. doi: 10.1038/35002131. [DOI] [PubMed] [Google Scholar]
- 3.Ajo-Franklin CM, et al. Rational design of memory in eukaryotic cells. Genes Dev. 2007;21:2271–2276. doi: 10.1101/gad.1586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burrill DR, Silver PA. Making cellular memories. Cell. 2010;140:13–18. doi: 10.1016/j.cell.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bancroft C. Long-term storage of information in DNA. Science. 2001;293:1763c–1765. doi: 10.1126/science.293.5536.1763c. [DOI] [PubMed] [Google Scholar]
- 6.Ham TS, Lee SK, Keasling JD, Arkin AP. Design and construction of a double inversion recombination switch for heritable sequential genetic memory. PLoS One. 2008;3:e2815. doi: 10.1371/journal.pone.0002815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abelson H, et al. Amorphous computing. Commun ACM. 2000;43:74–82. [Google Scholar]
- 8.Grindley NDF, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu Rev Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 9.Sauer B. Site-specific recombination: Developments and applications. Curr Opin Biotechnol. 1994;5:521–527. doi: 10.1016/0958-1669(94)90068-x. [DOI] [PubMed] [Google Scholar]
- 10.Branda CS, Dymecki SM. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev Cell. 2004;6:7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- 11.Podhajska AJ, Hasan N, Szybalski W. Control of cloned gene expression by promoter inversion in vivo: Construction of the heat-pulse-activated att-nutL-p-att-N module. Gene. 1985;40:163–168. doi: 10.1016/0378-1119(85)90038-1. [DOI] [PubMed] [Google Scholar]
- 12.Ham TS, Lee SK, Keasling JD, Arkin AP. A tightly regulated inducible expression system utilizing the fim inversion recombination switch. Biotechnol Bioeng. 2006;94:1–4. doi: 10.1002/bit.20916. [DOI] [PubMed] [Google Scholar]
- 13.Friedland AE, et al. Synthetic gene networks that count. Science. 2009;324:1199–1202. doi: 10.1126/science.1172005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steinkraus KA, Kaeberlein M, Kennedy BK. Replicative aging in yeast: The means to the end. Annu Rev Cell Dev Biol. 2008;24:29–54. doi: 10.1146/annurev.cellbio.23.090506.123509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- 16.Frumkin D, Wasserstrom A, Kaplan S, Feige U, Shapiro E. Genomic variability within an organism exposes its cell lineage tree. PLoS Comput Biol. 2005;1:e50. doi: 10.1371/journal.pcbi.0010050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nash HA. Integration and escision of bacteriophage lambda: The mechanism of conservative site specific recombination. Annu Rev Genet. 1981;15:143–167. doi: 10.1146/annurev.ge.15.120181.001043. [DOI] [PubMed] [Google Scholar]
- 18.Ptashne M. A Genetic Switch. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2004. [Google Scholar]
- 19.Fiandt M, Szybalski W, Malamy MH. Polar mutations in lac, gal and phage lambda consist of a few IS-DNA sequences inserted with either orientation. Mol Gen Genet. 1972;119:223–231. doi: 10.1007/BF00333860. [DOI] [PubMed] [Google Scholar]
- 20.Reyes O, Gottesman M, Adhya S. Formation of lambda lysogens by IS2 recombination: Gal operon-lambda pR promoter fusions. Virology. 1979;94:400–408. doi: 10.1016/0042-6822(79)90470-7. [DOI] [PubMed] [Google Scholar]
- 21.Mizuuchi K, Fisher LM, O’Dea MH, Gellert M. DNA gyrase action involves the introduction of transient double-strand breaks into DNA. Proc Natl Acad Sci USA. 1980;77:1847–1851. doi: 10.1073/pnas.77.4.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pollock TJ, Nash HA. Knotting of DNA caused by a genetic rearrangement. Evidence for a nucleosome-like structure in site-specific recombination of bacteriophage lambda. J Mol Biol. 1983;170:1–18. doi: 10.1016/s0022-2836(83)80224-1. [DOI] [PubMed] [Google Scholar]
- 23.Groth AC, Calos MP. Phage integrases: Biology and applications. J Mol Biol. 2004;335:667–678. doi: 10.1016/j.jmb.2003.09.082. [DOI] [PubMed] [Google Scholar]
- 24.Keravala A, et al. A diversity of serine phage integrases mediate site-specific recombination in mammalian cells. Mol Genet Genomics. 2006;276:135–146. doi: 10.1007/s00438-006-0129-5. [DOI] [PubMed] [Google Scholar]
- 25.Kim AI, et al. Mycobacteriophage Bxb1 integrates into the Mycobacterium smegmatis groEL1 gene. Mol Microbiol. 2003;50:463–473. doi: 10.1046/j.1365-2958.2003.03723.x. [DOI] [PubMed] [Google Scholar]
- 26.Ghosh P, Kim AI, Hatfull GF. The orientation of mycobacteriophage Bxb1 integration is solely dependent on the central dinucleotide of attP and attB. Mol Cell. 2003;12:1101–1111. doi: 10.1016/s1097-2765(03)00444-1. [DOI] [PubMed] [Google Scholar]
- 27.Ghosh P, Wasil LR, Hatfull GF. Control of phage Bxb1 excision by a novel recombination directionality factor. PLoS Biol. 2006;4:e186. doi: 10.1371/journal.pbio.0040186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mediavilla J, et al. Genome organization and characterization of mycobacteriophage Bxb1. Mol Microbiol. 2000;38:955–970. doi: 10.1046/j.1365-2958.2000.02183.x. [DOI] [PubMed] [Google Scholar]
- 29.Savinov A, Pan J, Ghosh P, Hatfull GF. The Bxb1 gp47 recombination directionality factor is required not only for prophage excision, but also for phage DNA replication. Gene. 2011 doi: 10.1016/j.gene.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghosh P, Pannunzio NR, Hatfull GF. Synapsis in phage Bxb1 integration: Selection mechanism for the correct pair of recombination sites. J Mol Biol. 2005;349:331–348. doi: 10.1016/j.jmb.2005.03.043. [DOI] [PubMed] [Google Scholar]
- 31.Ghosh P, Bibb LA, Hatfull GF. Two-step site selection for serine-integrase-mediated excision: DNA-directed integrase conformation and central dinucleotide proofreading. Proc Natl Acad Sci USA. 2008;105:3238–3243. doi: 10.1073/pnas.0711649105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee NL, Gielow WO, Wallace RG. Mechanism of araC autoregulation and the domains of two overlapping promoters, Pc and PBAD, in the L-arabinose regulatory region of Escherichia coli. Proc Natl Acad Sci USA. 1981;78:752–756. doi: 10.1073/pnas.78.2.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andersen JB, et al. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl Environ Microbiol. 1998;64:2240–2246. doi: 10.1128/aem.64.6.2240-2246.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ringrose L, et al. Comparative kinetic analysis of FLP and cre recombinases: Mathematical models for DNA binding and recombination. J Mol Biol. 1998;284:363–384. doi: 10.1006/jmbi.1998.2149. [DOI] [PubMed] [Google Scholar]
- 36.Horowitz P, Hill W. The Art of Electronics. Cambridge, UK: Cambridge Univ Press; [Google Scholar]
- 37.Hendrix RW. Evolutionary relationships among diverse bacteriophages and prophages: All the world’s a phage. Proc Natl Acad Sci USA. 1999;96:2192–2197. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lewis JA, Hatfull GF. Control of directionality in integrase-mediated recombination: Examination of recombination directionality factors (RDFs) including Xis and Cox proteins. Nucleic Acids Res. 2001;29:2205–2216. doi: 10.1093/nar/29.11.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith MCM, Thorpe HM. Diversity in the serine recombinases. Mol Microbiol. 2002;44:299–307. doi: 10.1046/j.1365-2958.2002.02891.x. [DOI] [PubMed] [Google Scholar]
- 40.Buchholz F, Stewart AF. Alteration of Cre recombinase site specificity by substrate-linked protein evolution. Nat Biotechnol. 2001;19:1047–1052. doi: 10.1038/nbt1101-1047. [DOI] [PubMed] [Google Scholar]
- 41.Ashworth J, et al. Computational redesign of endonuclease DNA binding and cleavage specificity. Nature. 2006;441:656–659. doi: 10.1038/nature04818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gaj T, Mercer AC, Gersbach CA, Gordley RM, Barbas CF. Structure-guided reprogramming of serine recombinase DNA sequence specificity. Proc Natl Acad Sci USA. 2011;108:498–503. doi: 10.1073/pnas.1014214108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Canton B, Labno A, Endy D. Refinement and standardization of synthetic biological parts and devices. Nat Biotechnol. 2008;26:787–793. doi: 10.1038/nbt1413. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.