Abstract
The derivation of germ-line competent avian primordial germ cells establishes a cell-based model system for the investigation of germ cell differentiation and the production of genetically modified animals. Current methods to modify primordial germ cells using DNA or retroviral vectors are inefficient and prone to epigenetic silencing. Here, we validate the use of transposable elements for the genetic manipulation of primordial germ cells. We demonstrate that chicken primordial germ cells can be modified in vitro using transposable elements. Both piggyBac and Tol2 transposons efficiently transpose primordial germ cells. Tol2 transposon integration sites were spread throughout both the macro- and microchromosomes of the chicken genome and were more prevalent in gene transcriptional units and intronic regions, consistent with transposon integrations observed in other species. We determined that the presence of insulator elements was not required for reporter gene expression from the integrated transposon. We further demonstrate that a gene-trap cassette carried in the Tol2 transposon can trap and mutate endogenous transcripts in primordial germ cells. Finally, we observed that modified primordial germ cells form functional gametes as demonstrated by the generation of transgenic offspring that correctly expressed a reporter gene carried in the transposon. Transposable elements are therefore efficient vectors for the genetic manipulation of primordial germ cells and the chicken genome.
Keywords: poultry, transgenesis, transposition
Primordial germ cells (PGCs) are specified in the early embryo and are the lineage-restricted stem cells for the germ cell population. In the chicken, segregation of PGCs from somatic cells occurs during the early stages of blastoderm formation (1–3). These cells accumulate in the germinal crescent region anterior to the forming head at day 1 of incubation [stage 4 Hamburger and Hamilton (HH)] and subsequently migrate via the circulatory system to the forming gonads on day 3 of incubation (stage 15 HH) (4). Chicken PGCs can be isolated from embryonic blood at this developmental stage and propagated indefinitely in culture (5). When returned to the circulatory system of an equivalent stage host embryo, the cultured PGCs migrated to and populated the forming embryonic gonad. Breeding from the resulting germ-line chimeras demonstrated that the cultured PGCs formed functional gametes by production of offspring derived from these cells. Germ-line transmission of cultured PGCs has now been demonstrated for three different chicken breeds (5–7).
This in vitro system for the long-term propagation of chicken PGCs is the basis of a unique stem cell model for both the investigation of germ cell differentiation and the generation of genetically modified chickens. Thus far, chicken PGCs have proved highly resistant to genetic modification. Stable transfection of PGCs has been achieved by electroporation of plasmid DNA, at a frequency of ∼1 in 106, but expression from integrated reporter constructs depended on the presence of flanking insulator sequences, suggesting that epigenetic silencing of the introduced transgenes occurred. Stable integration frequencies were improved 20-fold by incorporation of an attB site in the transgene plasmid and expression of ϕC31 integrase to promote chromosomal integration (8). The apparently low frequencies of stable integration may be due to transcriptional repression of the integrated transgenes by the genome-wide epigenetic modifications present in the PGCs of many organisms (9–12).
DNA transposons are naturally occurring mobile genetic elements that transpose by a “cut and paste” process involving the mobilization of a transposon from one genomic location and its reintegration at a novel site within the host genome (reviewed in ref. 13). A transposon-encoded transposase enzyme recognizes the inverted terminal repeats (ITRs) flanking a transposon and catalyzes the transposition of the element. Transposition of a number of transposons has been shown to require only the transposase enzyme and the ITR sequences of its respective transposon. This transposition has formed the basis of development of simple integrating gene transfer vectors, where the vector consists of the ITR sequences flanking a transgene of choice and the transposase is supplied in trans, as mRNA or from an expression vector.
Several transposons have been developed for use in genetic modification, including Sleeping Beauty, reactivated from a salmonid genome (14); piggyBac, isolated from the genome of the cabbage-looper moth, Trichoplusia ni (15); and the Tol2 transposable element, isolated from the genome of medaka fish (16). The piggyBac and Sleeping Beauty transposons have been shown to integrate preferentially into transcriptional units in several cell types (17–19). Similarly, the Tol2 transposon frequently integrates into intragenic regions (20, 21). All three of these transposable elements are functional in a wide range of host species, including chickens (22, 23), and have been used to modify the germ line of many species (mouse, reviewed in ref. 13; zebrafish, ref. 24; insects, ref. 25; Xenopus, ref. 26; rat, ref. 27; and pig, refs. 28–30). Genome-wide mutational screens using gene-trap versions of these vectors have also been carried out successfully in several vertebrate species (20, 31–34).
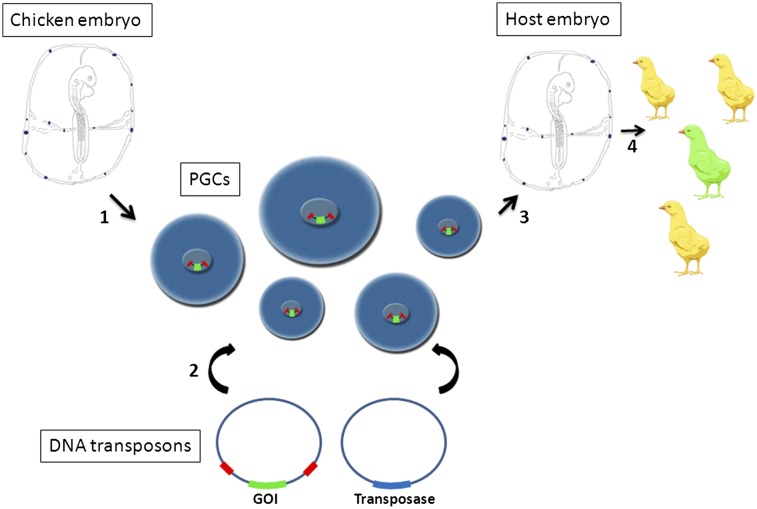
The aim of this study was to evaluate whether DNA transposons can be used to increase the efficiency of the genetic modification of chicken PGCs. We compared the efficiencies of stable integration of piggyBac and Tol2 transposons in both a chicken embryonic cell line and PGCs. We chose these two transposons as they have been used extensively in nonmammalian systems (24–26). Using a selectable reporter transgene, we observed that both transposon vectors transposed efficiently in chicken PGCs. The apparent frequency of integration of the transposon was not increased by the presence of insulator sequences flanking the reporter gene. Mapping of Tol2 integration sites revealed that integrations were predominantly located in transcriptional units and introns of PGCs. We also found that endogenous PGC transcripts could be trapped and mutated using a gene-trap construct contained in the Tol2 vector. Chicken PGCs containing integrated transposons colonized the gonad of host embryos and formed functional gametes and offspring. PiggyBac and Tol2 vectors are thus useful for the genetic modification of PGCs and the production of transgenic chickens.
Results
Stable Transfection of Chicken Embryonic Cells Using piggyBac and Tol2 Vectors.
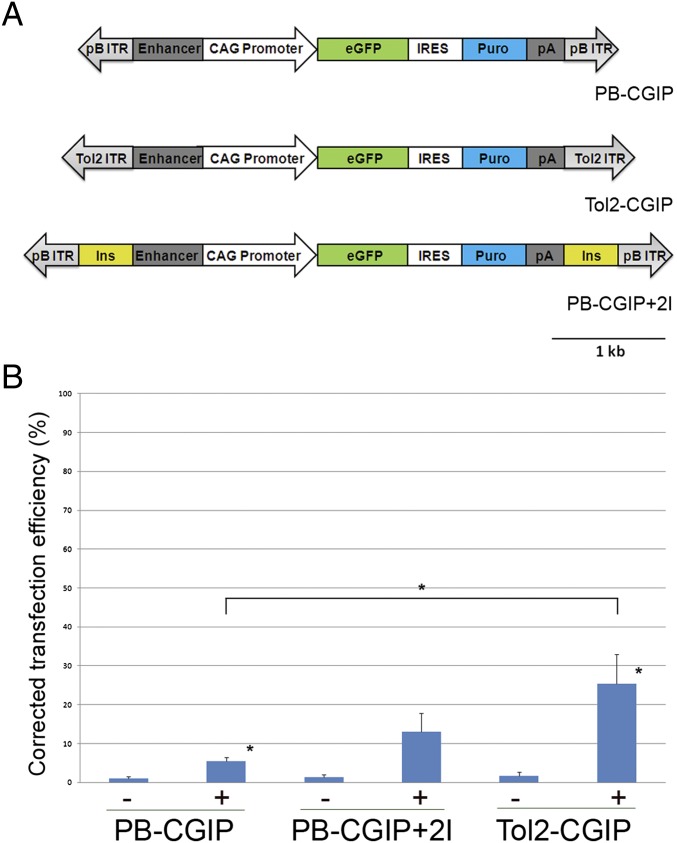
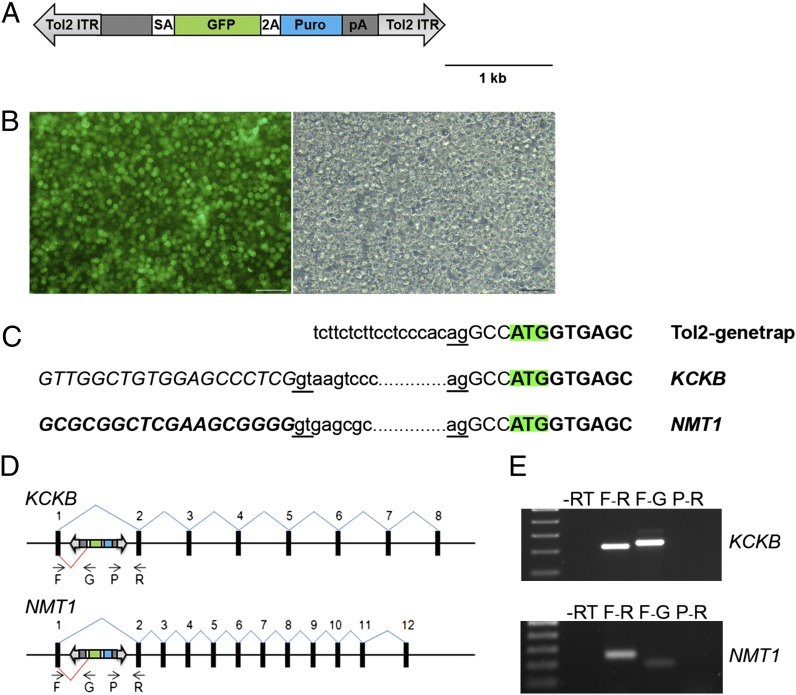
The piggyBac and Tol2 vectors used in this study contain ITRs of 236 bp and 550 bp, respectively (35, 36). To directly compare the integration efficiency of the piggyBac and Tol2 transposons in chicken PGCs, an identical reporter cassette was cloned between the ITRs of these vectors (Fig. 1A). The reporter cassette comprised the CAG promoter, a hybrid enhancer/promoter containing the CMV-IE enhancer fused to the chicken β-actin promoter and first intron (37), driving expression of a dual reporter, GFP-IRES–puromycin (CGIP). The reporter cassette allowed the visualization of GFP to measure stable genomic integration and expression from the transposable element and selection for puromycin resistance to ablate cells that do not express the reporter construct. In addition, two copies of the insulator element, HS4, from the β-globin locus were inserted into the piggyBac transposon adjacent to each ITR and flanking the reporter cassette to generate PB-CGIP+2I (Fig. 1A) to shield the integrated transposon from epigenetic silencing events (38).
Fig. 1.
DNA transposon vectors and stable transfection rates into a chicken cell line. (A) Diagram of the piggyBac and Tol2 vectors used in this study. Tol2-CGIP and PB-CGIP contain identical reporter cassettes between the ITRs. The CAG enhancer/promoter drives expression of a GFP-IRES–puromycin reporter construct. Flanking insulator elements (Ins) from the chicken β-globin locus were inserted adjacent to the ITRs in the piggyBac construct, PB-CGIP+2I. (B) DF-1 cells were transfected with transposon constructs with (+) or without (−) transposase and assayed 3 wk after transfection for GFP expression by flow cytometry. Data represent three independent experiments. The stable transfection efficiencies are corrected for the initial rate of transfection. Error bars, SEM. *P < 0.05.
We assayed the transposition efficiencies of the piggyBac and Tol2 transposons carrying the reporter cassette in the immortalized embryonic chicken fibroblast cell line, DF-1 (39). Cells were transfected with the transposons in the presence or absence of the appropriate transposase and assayed 3 wk posttransfection for the expression of GFP. In the absence of transposase a low frequency, ∼1%, of stably transfected cells was observed (Fig. 1B). The addition of transposase increased the frequency of stably transfected cells significantly: 5.4% of cells transfected with the piggyBac transposon and 25.5% of cells transfected with the Tol2 transposon expressed GFP. To determine whether the presence of insulator elements flanking the reporter cassette increased the frequency of GFP-expressing cells, the experiment was repeated using piggyBacCGIP+2I. The frequency of GFP-expressing cells 3 wk posttransfection was 13%, a 2.4-fold increase. We next determined whether increasing the amount of transposase increased the frequency of stable transfection. We found that increasing amounts of Tol2 transposase, from 0.5 (transposase):1 (transposon) to 5:1 increased the frequency of stable transfection (Fig. S1). Similarly, increasing the ratio of piggyBac transposase to transposon vector increased the frequency of stable transfection up to a ratio of 2.5:1. These results demonstrate that both piggyBac and Tol2 vectors can be used for stable transfection of chicken cells, presumably as a result of transposase-mediated integration.
Efficient Stable Transfection of Primordial Germ Cells Using piggyBac and Tol2 Transposons.
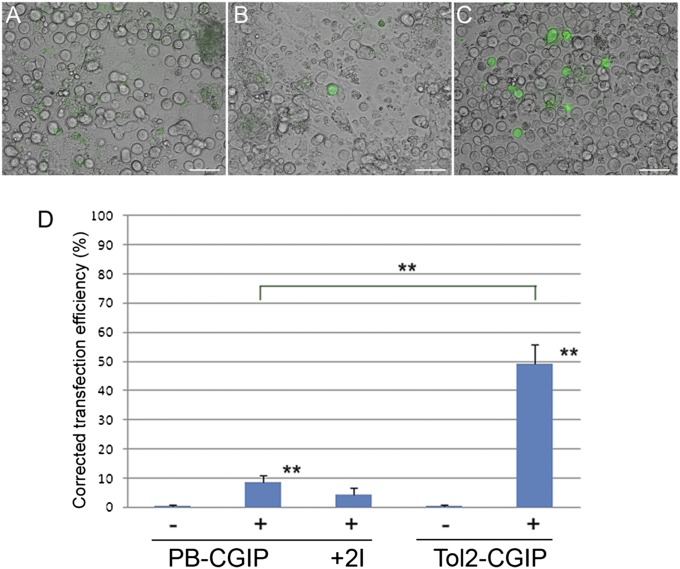
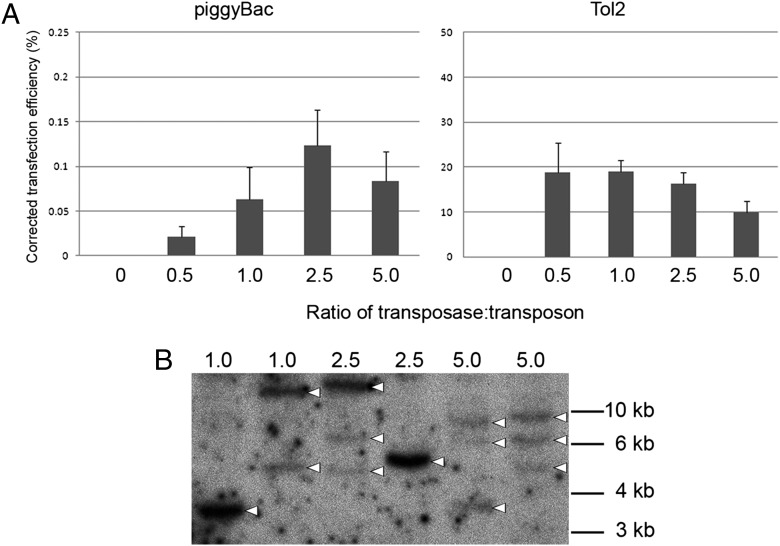
Previous results indicated that stable genetic modification of chicken PGCs could be achieved only at very low frequencies (8). To determine whether transposon vectors can be used to increase the efficiency of stable transfection of PGCs, we carried out a series of transfections, on the basis of the results described above, using three established chicken PGC lines (7). PGCs were transfected with either the piggyBac or the Tol2 vectors in the presence or absence of the appropriate transposase. The transfected cells were observed 3 d and 3 wk posttransfection for the expression of GFP and cell counts were taken to calculate the frequency of stable transfection. In contrast to DF-1 cells, no GFP expression was observed at 3 wk posttransfection in the absence of transposase (Fig. 2A). Cotransfection with the transposase plasmid resulted in stable transfection, as measured by persistent GFP expression, for both the piggyBac and the Tol2 vectors (Fig. 2 B and C). The frequency of GFP-expressing PGCs observed was significantly higher for the Tol2 vector in comparison with the piggyBac vector (45.2% vs. 10.5%), as was observed in DF-1 cells (Fig. 2D). We next tested the stable transfection efficiency of the piggyBac vector flanked by HS4 insulator elements. Notably, no increase in stable transfection rates was observed (4.5%; Fig. 2D). Finally, we determined whether increasing the ratio of transposase to transposon increased the frequency of stable transfection. In contrast to the results observed in DF-1 cells, increasing the ratio of Tol2 transposase to transposon had little effect on the stable transfection rate in PGCs (Fig. 3A). Increasing ratios of piggyBac transposase to transposon (up to 2.5:1) increased the stable transfection efficiencies of chicken PGCs. A ratio higher than this lowered the stable transfection frequency.
Fig. 2.
Stable genetic modification of primordial germ cells using piggyBac and Tol2 vectors. Chicken PGCs were transfected with transposon constructs with (+) or without (−) transposase and visualized a minimum of 2 wk after transfection for GFP expression. (A) Minus transposase control; (B) PB-CGIP vector; (C) Tol2-CGIP vector. (Scale bar, 50 μm.) (D) Comparison of stable transfection rates of piggyBac and Tol2 transposons in PGCs. Data are corrected for transfection efficiency and represent a minimum of four independent experiments, using two lines of PGCs. Error bars, SEM. **P < 0.01.
Fig. 3.
Analysis of the affect of increasing the amount of transposase on stable transfection and genomic integration events in primordial germ cells. (A) Chicken PGCs were transfected with 1.0 μg of a PB-CGIP or a Tol2-CGIP vector and increasing amounts (micrograms) of the appropriate DNA transposase and visualized a minimum of 2 wk after transfection for GFP expression. All transfection contained equal total amounts of plasmid DNA. The data are from four independent experiments. The stable transfection efficiencies are corrected for the initial rate of transfection. (B) Genomic DNA from pools of stably transfected PGCs was analyzed for presence and copy number of the piggyBac transposon by digestion with BamHI to generate junction fragments and hybridized with a probe to the GFP sequence. Arrowheads, junctional fragments.
The chicken PGCs transfected with increasing amounts of piggyBac transposase were selected with puromycin and propagated in culture, and genomic DNA was isolated for Southern blot analysis (Fig. 3B). This analysis detected between one and three piggyBac vector integration events in the pools of stably transfected PGCs.
Analysis of Tol2 Chromosomal Integration Sites in Primordial Germ Cells.
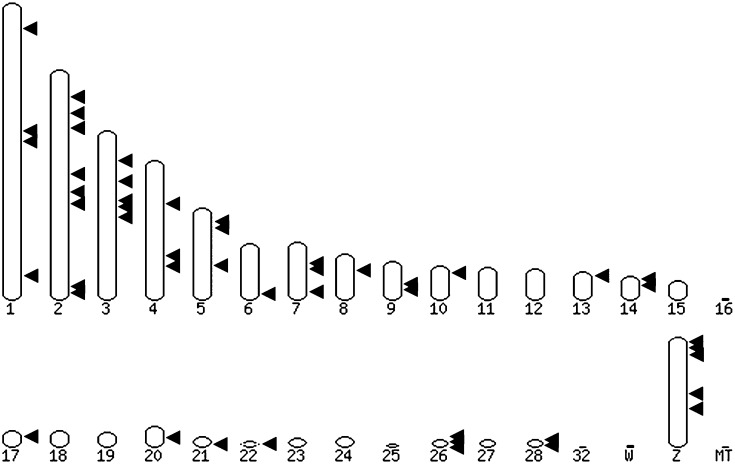
The results above indicate that the Tol2 vector was more efficient than the piggyBac vector for the genetic modification of PGCs. Tol2 vectors have proved useful for a range of genome modifications, including the integration of novel transgenes, mutagenesis by transposon integration, and gene-trap screens (reviewed in ref. 40). The effectiveness of these applications depends on the distribution of Tol2 integrations in the chicken genome in relation to coding and noncoding sequences. To determine the chromosomal integration sites of the Tol2 transposon in stably transfected chicken PGCs and to verify that the integrations were due to a transposition event, the flanking region surrounding the transposon termini was mapped using inverse PCR. We were able to assign 50 integration sites to specific chromosomal sites in the chicken genome of 55 integration sites identified by inverse PCR (Table S1). The remaining 5 insertion sites either did not identify sequences within the current chicken genome assembly or were ambiguous due to integration into a repetitive DNA element. We found that the majority of Tol2 integration sites were within transcriptional units (27/50; 54%). Of these integrations, 25 were located in introns and 2 were in exons. For the remaining integrations, 14 (14/50; 34%) were located within 9 kb of DNA flanking transcriptional units, commonly classified as integrations into gene regulatory regions. The other 9 integrations (9/50; 18%) were located farther than 9 kb from a defined transcriptional unit and were classified as intergenic integrations. All integration sites contained the Tol2 ITR directly followed by genomic DNA, indicating that transposition of the transposon vector from the delivery plasmid backbone into genomic DNA had occurred. The junction fragments for 5 integration events were sequenced on both sides of the Tol2 transposon and are shown in Fig. S2A. An 8-bp duplication of genomic DNA on either side of the transposon was found in each case, indicative of a Tol2 transposition event (24). Analysis of all of the integration sites sequenced did not reveal any nucleotide preferences in the site of integration (Fig. S2B). The integration sites were dispersed over both the macro- and the microchromosomes of the chicken (Fig. 4).
Fig. 4.
Distribution of genomic integration sites of the Tol2 transposon in primordial germ cells. The integration sites for 50 Tol2-CGIP integrations were identified using inverse PCR and the transposon–genomic junction fragments were mapped to the chromosomes of the chicken genome.
Gene Trap of Endogenous Transcripts in PGCs.
Gene-trap screens using transposable elements have been carried out successfully in many species both to mark the expression of endogenous genetic loci and to mutate the corresponding mRNA transcripts (reviewed in ref. 41). We carried out preliminary experiments to demonstrate that it was possible to trap and disrupt genes in chicken PGCs. We hypothesized that gene trapping of transcripts in PGCs would be possible as 54% of the Tol2 integration sites were located within intronic regions of the chicken genome. We constructed a modified Tol2 vector (Tol2–gene trap) containing a splice acceptor site followed by the coding sequence for GFP and puromycin resistance, separated by a self-cleaving peptide sequence and followed by a poly(A) sequence (Fig. 5A). When the gene-trap vector is integrated into an intron of a gene transcribed in PGCs, a truncated transcript and protein are produced that encode the GFP–puromycin dual reporter or GFP fused to the C terminus of the endogenous protein. The Tol2–gene-trap vector was transfected into chicken PGCs, the PGCs were subjected to puromycin selection, and the surviving PGCs were propagated in culture. The selected PGCs expressed high levels of GFP (Fig. 5B). 5′-RACE-PCR analysis was carried out on mRNA from these PGCs, using primers specific to the GFP gene to identify fusion transcripts between GFP and endogenous exons. Two independent gene-trap events were identified. The trapped transcripts contained the endogenous sequence from the first exons of the trapped genes fused to the splice acceptor site upstream of the GFP gene in the gene-trap vector (Fig. 5C). In one case the 5′ exon of the trapped gene contained the start codon fused in frame to the coding sequence of GFP. The structures of the fusion transcripts were verified using RT-PCR and gene-specific primers (Fig. 5 D and E).
Fig. 5.
Gene trap of endogenous transcripts in primordial germ cells. (A) A minimal Tol2 transposable element containing a β-globin splice acceptor (SA) followed by a SA-GFP–puromycin reporter gene containing a self-cleaving peptide sequence and a poly(A) sequence. (B) GFP and brightfield images of a representative PGC culture that was transfected with the gene-trap vector and the transposase expression plasmid and selected with puromycin. (Scale bar, 100 μm.) (C) Fusion transcripts identified by RACE PCR analysis. Amino acids coding sequences are indicated in boldface type and the initiation codon for GFP is highlighted. The splice donor sites of the trapped exons and the gene-trap acceptor sites are underlined. The endogenous exon sequences fused to the transposon sequence are shown in italics. KCKB, ENSGALT00000018766; NMT1, ENSGALT00000001256. (D) A schematic representation of transposon integration sites in the trapped loci and the primers used to verify the fusion transcripts. (E) Independent PCRs were performed on cDNA isolated from the gene-trap PGCs with the indicated primers to verify the fusion transcripts. A primer located in the puromycin gene did not produce a PCR product, suggesting that a truncated transcript was produced.
Germ-Line Competency of PGCs Containing Integrated Transposons.
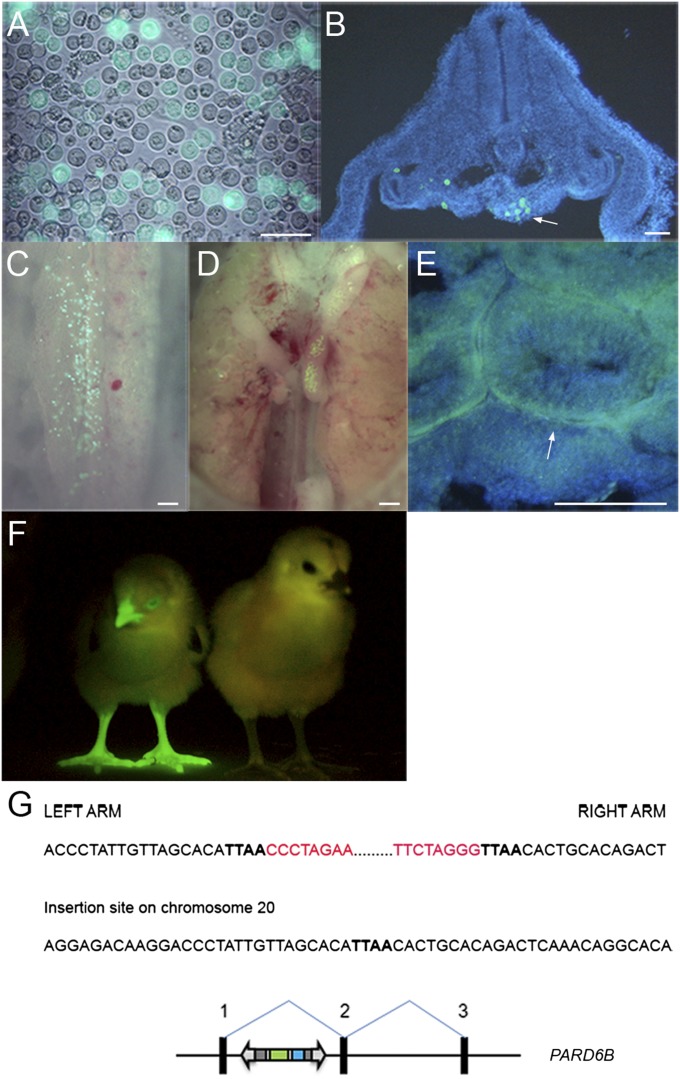
Chicken PGCs propagated in culture for prolonged periods can be tested for the developmental potential to form functional gametes by injection into host chicken embryos, rearing of the manipulated embryos to sexual maturity, and screening offspring to identify chicks derived from the cultured PGCs. To assess germ-line competency of transposon-modified PGCs, cells were stably transfected with a piggyBac vector expressing membrane-localized GFP and a puromycin resistance gene (PB-CmyrGIP). PGCs were selected with puromycin, propagated in culture, and injected into stage 16HH host embryos (Fig. 6A). Host embryos were examined for GFP-expressing PGCs. On day 1, GFP-expressing cells could be seen in the genital ridge (Fig. 6B). GFP-expressing cells at later time points were visible in the forming gonad (Fig. 6 C–E). To determine whether the PGCs formed functional gametes, GFP-expressing PGCs were injected into 16 host embryos and incubated until hatching. Three embryos survived until sexual maturity, two females and one male. The rooster was bred to stock hens, their offspring were screened for GFP fluorescence, and a GFP-expressing chick was obtained (Fig. 6F). Southern blot analysis of genomic DNA from this bird showed that a single transposition event into the chicken had occurred (Fig. S3). The genomic sequences flanking the piggyBac vector ITRs were isolated using inverse PCR and the integration site was mapped to the first intron of the PARD6B gene on chromosome 20 (Fig. 6G).
Fig. 6.
Primordial germ cells modified with transposon vectors are germ-line competent. (A) A piggyBac vector containing a CAGmyrGFP-IRES-Puro reporter gene was transfected into PGCs and selected with puromycin. (Scale bar, 50 μm.) (B) Cross-section of a day 3 embryo, 1 d after injection of GFP-expressing PGCs, contained GFP+ cells in the genital ridge. Arrow, left genital ridge. (C and D) GFP+ cells in the gonads in day 5 (C) and day 10 (D) chicken embryos. (E) Cross-section of seminiferous tubules of an adult cockerel hatched after injection with GFP-expressing PGCs. GFP+ cells are located adjacent to the basement membrane of the tubules. [Scale bars (B–E), 100 μm.] (F) G1 offspring of the adult cockerel injected with GFP-expressing PGCs and visualized for GFP expression. A nontransgenic sibling is on the right. (G) A schematic representation of the piggyBac insertion site in the first intron of the PARD6B gene of the G1 offspring (ENSGALT00000012978). Red, piggyBac ITRs; boldface type, genomic DNA.
Discussion
The results described here demonstrate that DNA transposons can be used to directly modify chicken PGCs with high efficiencies. Leighton et al. (8) previously investigated the efficiency of stable transfection of cultured chicken PGCs. They found that the frequency of stable transfection by electroporation was 100-fold lower than that obtained in chicken ES cells and suggested that the reporter transgene was silenced on chromosomal integration in PGCs. By introducing HS4 insulator elements into their constructs flanking the reporter transgene, to protect the transgene from epigenetic gene silencing, frequencies of stable transfection of ∼1 colony per 106 cells (0.0001%) were observed. Here we have shown that frequencies of stable transfection of PGCs of 10.5% and 45.2% can be obtained using piggyBac and Tol2 transposon vectors. In addition, we evaluated the effect of flanking the reporter transgene in a piggyBac vector by HS4 insulator elements and found that these elements were not necessary for reporter gene expression. Our results suggest that transgene expression from integrated piggyBac and Tol2 transposons is not affected by epigenetic silencing mechanisms as was observed for plasmid vectors in chicken PGCs (5, 8).
The integration sites of over 50 Tol2 transposons were mapped and shown to be distributed throughout the chicken genome, with many of the integrations into the microchromosomes. A large proportion of integrations were within introns or in close proximity to transcribed regions of the genome. This pattern of integration is similar to that in the Tol2 integration sites in other cell types and species (19, 21), suggesting that integration site selection by the Tol2 transposase in chicken PGCs does not greatly differ from integration site selection in other cell types. We have also shown that chicken PGCs transfected with a piggyBac vector and selected for puromycin resistance to select for transposition events can be introduced into the circulatory system of early chicken embryos, the stage at which the endogenous PGCs are migrating to the forming gonads, and form functional spermatozoa in the resulting adult rooster. These results demonstrate that the use of transposon vectors will greatly increase the efficiency of stable genetic modification of PGCs, facilitating the use of genetic modification of PGCs as a method for the generation of transgenic chickens.
The transgenes that can be carried by DNA transposon vectors have been reported to be generally less than 13 kb in length (18, 35). However, recent reports have shown that much larger transgenes can be transposed. Tol2 vectors have been used to mobilize a bacterial artificial chromosomes (BAC) into the genome of early vertebrate embryos (42, 43). Similarly, a piggyBac vector has been used to integrate sequences of up to 100 kb in length (44). On the basis of these advances it may be possible to integrate larger genetic constructs into PGCs, using transposons.
This high efficiency of genetic modification using the piggyBac and Tol2 transposon vectors has the potential to enable additional applications, beyond standard genetic modification. Here, we have shown that endogenous transcripts can be trapped in chicken PGCs, using a Tol2 gene-trap construct. This initial demonstration, coupled with the germ-line transmission of transposon-modified chicken PGCs, validates the prospect of conducting a large-scale mutational screen of gene function in chickens. Large-scale mutational gene-trap screens have been carried out in many species (20, 31, 32). Such an analysis in chickens would be a valuable and alternative model to the mutational screens being carried out in the mouse. The chicken genome is relatively small (1.05 Gbp) in comparison with the human, mouse, and zebrafish genomes and contains less repetitive DNA (<15%) (45, 46). A targeted germ cell library could be produced in PGCs and cryopreserved for later use in the production of knockout animals. It needs to be ascertained whether the trapped transcripts will be specific to the transcriptome of PGCs or will represent the entire chicken transcriptome.
These results also suggest that the modification and genetic screening of other avian model systems (quail, zebrafinch, starling, and sparrow) and potentially mammals could be carried out using PGCs once permissive culture conditions were established. As yet, PGCs from mammals can be cultured only for short periods before either undergoing apoptosis or forming embryonic germ cells, a pluripotent cell displaying many characteristics in common with the ES cell (47–49). Potentially, germ cells from later developmental stages, for example spermatogonial stem cells (SSCs), could also be used as alternative systems for transgenesis and mutagenesis screens as has been shown for rat SSCs (50).
The production of transgenic chickens has wide-ranging applications in academic research, biotechnology, and agriculture (51–53). Transgenic birds are model organisms for the study of developmental biology, as bioreactors for the production of therapeutic proteins, and as models of disease resistance to minimize losses in agriculture. Our results show that chicken PGCs can be efficiently manipulated using transposon vectors, thus providing a cell-based tool for transgenesis.
Materials and Methods
DNA Transposon Vectors.
A CAG-piggyBac transposase (pCyL43) and minimal transposon (pCyL50) were provided by the Wellcome Trust Sanger Institute (Hinxton, United Kingdom) (36). The CAG-GFP-IRES–puromycin-poly(A) fragment from pCGIP (kind gift of A. Smith, Cambridge, United Kingdom) was cloned into the EcoRI site of pCyL50 to produce PB-CGIP. The GFP fragment was removed from PB-CGIP by digestion with EcoRI and replaced with a myrGFP fragment to generate PB-CmyrGIP (54). A KpnI 2 × 250-bp core HS4 insulator element from pNI-CD (38) was directionally cloned into the PstI site and the SpeI site of PB-CGIP to generate PB-CGIP+2I.
Tol2-CGIP was constructed by cloning an IRES–puromycin-poly(A) fragment from pCGIP into the XhoI site of pT2K-CAGGS-EGFP (22). The CAG-Tol2 transposase was described in ref. 22. The PB-SAGFP-2APuro gene-trap construct was produced by recloning Tol2-CGIP (EcoRI-BamHI fragment) into pGEM-T (Promega) to generate pEntryTol2-CGIP. The CMV β-actin promoter/splice donor was deleted from this construct by digesting with SalI/BlpI and religation of the vector. The GFP-IRES–puromycin was removed by digestion with EcoRI/ClaI and replaced with a SA-GFP-2A–puromycin fragment (BamHI/ClaI) from OCT4GFPpuroDonor1 (55).
Culture of PGCs and DF-1 Cells.
PGC cultures were derived and maintained as outlined in ref. 7. Briefly, blood (2–4 μL) isolated from stage 15–16 (HH) embryos of ISA Brown chickens was placed in 300 μL of PGC culture medium on a culture dish containing irradiated Sandoz inbred mouse-derived thioguanine-resistant and ouabain-resistant (STO) feeder cells (3.0 × 104 per well). Every 2 d one-third of the medium was removed and replaced with fresh medium. Once culture outgrowth was observed, the total volume of medium would be changed with similar frequency. The PGC culture medium contained 50% (vol/vol) buffalo rat liver (BRL)-conditioned medium in KO-DMEM (Invitrogen) and 10% (vol/vol) FBS (ES cell tested; PAA Laboratories), 2.5% (vol/vol) chicken serum (Biosera), 2 mM GlutaMax (Invitrogen), 1× NEAA (Invitrogen), 0.1 mM β-mercaptoethanol (Invitrogen), 1× nucleosides (Invitrogen), 1 mM pyruvate (Invitrogen), 1× penicillin–streptomycin (Sigma), and 2 ng/mL human recombinant bFGF (R&D Biosystems). Two male PGC lines were isolated and used for this work (line 08-08-08 and line 193-3). DF-1 cells were grown in 10% (vol/vol) FBS, 2 mM GlutaMax, 1× NEAA, 1× penicillin–streptomycin.
Cell Transfections.
DF-1 cells were plated in six-well plates 24 h before transfection to produce a monolayer. The cells were transfected at ∼60–70% confluence, using FuGene transfection reagent (Roche) with 2.0 μg each of either piggyBac or Tol2 vectors and 2.0 μg of the appropriate transposase expression vector or empty expression vector. Cells were passaged in culture and fluorescent cells were quantified using flow cytometry at 3 d after transfection to determine initial rates of transfection and at 3 wk posttransfection to determine the frequency of stable integration.
PGCs (25,000–50,000 cells) were transfected using 6 μL of DMRIE transfection reagent (Invitrogen) with 2.0 μg each of either piggyBac or Tol2 vector and 2.0 μg of the appropriate transposase vector or empty expression vector. Cells were incubated with transfection reagent for 5 h in suspension and then replated on fresh feeder cells. PGCs were passaged in culture and cells were visually assayed for GFP fluorescence at 2 d and 3 wk posttransfection to determine the rate of transfection and the rate of integration. Between 4 and 11 independent transfections were carried out for each condition. PGCs were selected over a 2-wk period with 0.1 μg/mL of puromycin (Invitrogen) and then expanded in culture for several weeks before injection into host embryos or used to isolate genomic DNA for insertion site analysis. For the transposase titration experiments, DF-1 cells and PGCs were transfected with a total of 6.0 μg of DNA containing 1.0 μg of transposon and either empty plasmid or increasing amounts of the appropriate transposase (0.5–5.0 μg).
P values were determined using Student’s t test with two-tailed distribution comparing + and − transposase conditions.
Inverse PCR.
PGCs were selected with puromycin and grown in culture to 100,000 cells. Genomic DNA was purified from the cells, using a Micro DNA prep kit (Qiagen). Tol2 integration sites were mapped by inverse PCR using the nested primers and conditions of ref. 20, with the following modifications: 1.0 μg of DNA was digested overnight with AluI, HaeIII, or PsiI to map 5′-junction fragments or with HaeIII and HindIII to map 3′-junction fragments. Digested DNA was heat treated and ligated overnight at 15 °C, using T4 DNA ligase. First-round PCR was carried out at 94 °C for 30 min, 60 °C for 30 min, and 72 °C for 60 min for 32 cycles, using Rapid TAQ (Roche), on 0.5 μg of ligated DNA in a volume of 25 μL. One microliter of the first reaction was used for the second-round PCR, using the above PCR conditions. Reactions were electrophoresed on agarose gels. DNA fragments were isolated and subcloned into the pGEM-T Easy (Promega) vector and sequenced. The piggyBac integration site in the generation 1 (G1) chicken was mapped by digesting genomic DNA with SpeI and using nested primers: outer primers GCAAGAGAGCAGAGAGGATA, GCGCGCCGTCGACATTGATT and inner primers GCGATGACGAGCTTGTTGGC, TCATCGTCTAAAGAACTACC. PCR conditions for both rounds were 94 °C for 30 min, 50 °C for 30 min, and 72 °C for 60 min for 30 cycles. Integration sites were determined using the BLAT algorithm at the University of California, Santa Cruz (UCSC) genome browser (http://www.genome.ucsc.edu) on the chicken genome (version WUGSC2.1/galGal3). Individual integration sites were validated by verifying that the restriction site used for inverse PCR was present in the chicken genome at the correct distance from the putative transposon integration site and by reisolation of the same junctional fragment from an independent inverse PCR (n = 7) or mapping both the 5′- and the 3′-junctional fragments for a given integration (n = 5). The 8-bp Tol2 integration sites were analyzed using WebLogo 3 to identify a consensus sequence (56). The piggyBac integration site was verified using specific primers to amplify and sequence both junctional fragments of the insert.
RACE PCR.
PGCs were transfected with the gene-trap construct, selected with puromycin, and expanded in culture to 200,000 cells. The sample was divided and total RNA or genomic DNA was purified using either a RNA Easy Kit (Promega) or a Micro DNA prep kit (Qiagen). RACE PCR was carried out using the Roche 5′3′ Race Kit and nested reverse primers specific to the GFP gene (57). The hybrid messages were verified using the gene-specific primers [Kbrc forward (For), CCGCTGAGGTCCTTACGTT; Kbrc reverse (Rev), CATCATCCAGCGTAAATCCA; NMT1 For, GGACGACAGTGAGACAGCAG; NMT1 Rev, CTTCATTGGCTGGTCCTGAG] or the gene-trap GFP reverse primer TAGGTCAGGGTGGTCACGAG and sequenced. The genomic integration sites were verified using the gene-specific forward primers and the inner primers from the inverse PCR.
Southern Blot Hybridization Analysis.
Ten micrograms of genomic DNA isolated from blood samples or stably transfected PGCs (∼1,000,000 cells) was digested overnight, using the appropriate restriction enzymes. The DNA digests were resolved by gel electrophoresis and transferred via capillary action to Hybond N membrane (GE Healthcare). PB-CGIP vector was digested to isolate a 1.0-kb fragment encoding GFP, which was then labeled with [α-32P]dCTP using the DIG High Prime DNA Labeling and Detection Kit II (Roche) and used to hybridize the Southern blot.
Generation of Chicken Germ-Line Chimeras.
PGCs were transfected with PB-CmyrGIP, selected with puromycin, and grown in culture for several weeks. Two hundred to 400 cells were injected into stage 16 HH host embryos and incubated until hatching (58). The PGCs had been derived and proliferated in culture for a total of 248 d before injection into founder host embryos. The hatched chicks were raised to sexual maturity and genomic DNA extracted from semen of an adult rooster was screened by semiquantitative PCR to quantify the GFP transgene in the semen (59). This rooster was crossed to wild-type hens and the offspring were screened for GFP fluorescence to identify germ cell-derived offspring (1 of 518). Animal experiments were approved and conducted under a UK Home Office license.
Supplementary Material
Acknowledgments
We thank the members of the transgenic chicken facility (M. Hutchison, R. Mitchell, and F. Thomson) for care and breeding of the chickens and D. McBride for help with the Southern blot analysis. This research was funded by the Biotechnology and Biological Sciences Research Council.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 8803 (volume 109, number 23).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118715109/-/DCSupplemental.
References
- 1.Karagenç L, Cinnamon Y, Ginsburg M, Petitte JN. Origin of primordial germ cells in the prestreak chick embryo. Dev Genet. 1996;19:290–301. doi: 10.1002/(SICI)1520-6408(1996)19:4<290::AID-DVG2>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 2.Kagami H, et al. The developmental origin of primordial germ cells and the transmission of the donor-derived gametes in mixed-sex germline chimeras to the offspring in the chicken. Mol Reprod Dev. 1997;48:501–510. doi: 10.1002/(SICI)1098-2795(199712)48:4<501::AID-MRD11>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 3.Tsunekawa N, Naito M, Sakai Y, Nishida T, Noce T. Isolation of chicken vasa homolog gene and tracing the origin of primordial germ cells. Development. 2000;127:2741–2750. doi: 10.1242/dev.127.12.2741. [DOI] [PubMed] [Google Scholar]
- 4.Nieuwkoop PD, Sutasurya LA. Primordal Germ Cells in the Chordates. Cambridge, UK: Cambridge Univ Press; 1979. [Google Scholar]
- 5.van de Lavoir MC, et al. Germline transmission of genetically modified primordial germ cells. Nature. 2006;441:766–769. doi: 10.1038/nature04831. [DOI] [PubMed] [Google Scholar]
- 6.Choi JW, et al. Basic fibroblast growth factor activates MEK/ERK cell signaling pathway and stimulates the proliferation of chicken primordial germ cells. PLoS ONE. 2010;5:e12968. doi: 10.1371/journal.pone.0012968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Macdonald J, Glover JD, Taylor L, Sang HM, McGrew MJ. Characterisation and germline transmission of cultured avian primordial germ cells. PLoS ONE. 2010;5:e15518. doi: 10.1371/journal.pone.0015518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leighton PA, van de Lavoir MC, Diamond JH, Xia C, Etches RJ. Genetic modification of primordial germ cells by gene trapping, gene targeting, and phiC31 integrase. Mol Reprod Dev. 2008;75:1163–1175. doi: 10.1002/mrd.20859. [DOI] [PubMed] [Google Scholar]
- 9.Seydoux G, Dunn MA. Transcriptionally repressed germ cells lack a subpopulation of phosphorylated RNA polymerase II in early embryos of Caenorhabditis elegans and Drosophila melanogaster. Development. 1997;124:2191–2201. doi: 10.1242/dev.124.11.2191. [DOI] [PubMed] [Google Scholar]
- 10.Seki Y, et al. Cellular dynamics associated with the genome-wide epigenetic reprogramming in migrating primordial germ cells in mice. Development. 2007;134:2627–2638. doi: 10.1242/dev.005611. [DOI] [PubMed] [Google Scholar]
- 11.Hajkova P, et al. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008;452:877–881. doi: 10.1038/nature06714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Venkatarama T, et al. Repression of zygotic gene expression in the Xenopus germline. Development. 2010;137:651–660. doi: 10.1242/dev.038554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ivics Z, et al. Transposon-mediated genome manipulation in vertebrates. Nat Methods. 2009;6:415–422. doi: 10.1038/nmeth.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ivics Z, Hackett PB, Plasterk RH, Izsvák Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 15.Cary LC, et al. Transposon mutagenesis of baculoviruses: Analysis of Trichoplusia ni transposon IFP2 insertions within the FP-locus of nuclear polyhedrosis viruses. Virology. 1989;172:156–169. doi: 10.1016/0042-6822(89)90117-7. [DOI] [PubMed] [Google Scholar]
- 16.Kawakami K, Koga A, Hori H, Shima A. Excision of the tol2 transposable element of the medaka fish, Oryzias latipes, in zebrafish, Danio rerio. Gene. 1998;225:17–22. doi: 10.1016/s0378-1119(98)00537-x. [DOI] [PubMed] [Google Scholar]
- 17.Wilson MH, Coates CJ, George AL., Jr PiggyBac transposon-mediated gene transfer in human cells. Mol Ther. 2007;15:139–145. doi: 10.1038/sj.mt.6300028. [DOI] [PubMed] [Google Scholar]
- 18.Nakazawa Y, et al. Optimization of the PiggyBac transposon system for the sustained genetic modification of human T lymphocytes. J Immunother. 2009;32:826–836. doi: 10.1097/CJI.0b013e3181ad762b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang X, et al. Gene transfer efficiency and genome-wide integration profiling of Sleeping Beauty, Tol2, and piggyBac transposons in human primary T cells. Mol Ther. 2010;18:1803–1813. doi: 10.1038/mt.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kotani T, Nagayoshi S, Urasaki A, Kawakami K. Transposon-mediated gene trapping in zebrafish. Methods. 2006;39:199–206. doi: 10.1016/j.ymeth.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 21.Kondrychyn I, Garcia-Lecea M, Emelyanov A, Parinov S, Korzh V. Genome-wide analysis of Tol2 transposon reintegration in zebrafish. BMC Genomics. 2009;10:418. doi: 10.1186/1471-2164-10-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sato Y, et al. Stable integration and conditional expression of electroporated transgenes in chicken embryos. Dev Biol. 2007;305:616–624. doi: 10.1016/j.ydbio.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 23.Kong BW, Carlson DF, Fahrenkrug SC, Foster DN. Application of the Sleeping Beauty transposon system to avian cells. Anim Genet. 2008;39:180–186. doi: 10.1111/j.1365-2052.2008.01702.x. [DOI] [PubMed] [Google Scholar]
- 24.Kawakami K, Shima A, Kawakami N. Identification of a functional transposase of the Tol2 element, an Ac-like element from the Japanese medaka fish, and its transposition in the zebrafish germ lineage. Proc Natl Acad Sci USA. 2000;97:11403–11408. doi: 10.1073/pnas.97.21.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Handler AM. Use of the piggyBac transposon for germ-line transformation of insects. Insect Biochem Mol Biol. 2002;32:1211–1220. doi: 10.1016/s0965-1748(02)00084-x. [DOI] [PubMed] [Google Scholar]
- 26.Hamlet MR, et al. Tol2 transposon-mediated transgenesis in Xenopus tropicalis. Genesis. 2006;44:438–445. doi: 10.1002/dvg.20234. [DOI] [PubMed] [Google Scholar]
- 27.Jang CW, Behringer RR. 2007. Transposon-mediated transgenesis in rats. Cold Spring Harb Protoc 2007:pdb.prot4866.
- 28.Garrels W, et al. Germline transgenic pigs by Sleeping Beauty transposition in porcine zygotes and targeted integration in the pig genome. PLoS ONE. 2011;6:e23573. doi: 10.1371/journal.pone.0023573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carlson DF, et al. Strategies for selection marker-free swine transgenesis using the Sleeping Beauty transposon system. Transgenic Res. 2011;20:1125–1137. doi: 10.1007/s11248-010-9481-7. [DOI] [PubMed] [Google Scholar]
- 30.Jakobsen JE, et al. Pig transgenesis by Sleeping Beauty DNA transposition. Transgenic Res. 2011;20:533–545. doi: 10.1007/s11248-010-9438-x. [DOI] [PubMed] [Google Scholar]
- 31.Horie K, et al. A homozygous mutant embryonic stem cell bank applicable for phenotype-driven genetic screening. Nat Methods. 2011;8:1071–1077. doi: 10.1038/nmeth.1739. [DOI] [PubMed] [Google Scholar]
- 32.Bonin CP, Mann RS. A piggyBac transposon gene trap for the analysis of gene expression and function in Drosophila. Genetics. 2004;167:1801–1811. doi: 10.1534/genetics.104.027557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruf S, et al. Large-scale analysis of the regulatory architecture of the mouse genome with a transposon-associated sensor. Nat Genet. 2011;43:379–386. doi: 10.1038/ng.790. [DOI] [PubMed] [Google Scholar]
- 34.Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–226. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- 35.Urasaki A, Morvan G, Kawakami K. Functional dissection of the Tol2 transposable element identified the minimal cis-sequence and a highly repetitive sequence in the subterminal region essential for transposition. Genetics. 2006;174:639–649. doi: 10.1534/genetics.106.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang W, et al. Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc Natl Acad Sci USA. 2008;105:9290–9295. doi: 10.1073/pnas.0801017105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 38.Recillas-Targa F, et al. Position-effect protection and enhancer blocking by the chicken beta-globin insulator are separable activities. Proc Natl Acad Sci USA. 2002;99:6883–6888. doi: 10.1073/pnas.102179399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Himly M, Foster DN, Bottoli I, Iacovoni JS, Vogt PK. The DF-1 chicken fibroblast cell line: Transformation induced by diverse oncogenes and cell death resulting from infection by avian leukosis viruses. Virology. 1998;248:295–304. doi: 10.1006/viro.1998.9290. [DOI] [PubMed] [Google Scholar]
- 40.Clark KJ, Urban MD, Skuster KJ, Ekker SC. Transgenic zebrafish using transposable elements. Methods Cell Biol. 2011;104:137–149. doi: 10.1016/B978-0-12-374814-0.00008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ivics Z, Izsvák Z. The expanding universe of transposon technologies for gene and cell engineering. Mob DNA. 2010;1:25. doi: 10.1186/1759-8753-1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suster ML, Sumiyama K, Kawakami K. Transposon-mediated BAC transgenesis in zebrafish and mice. BMC Genomics. 2009;10:477. doi: 10.1186/1471-2164-10-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bussmann J, Schulte-Merker S. Rapid BAC selection for tol2-mediated transgenesis in zebrafish. Development. 2011;138:4327–4332. doi: 10.1242/dev.068080. [DOI] [PubMed] [Google Scholar]
- 44.Li MA, et al. Mobilization of giant piggyBac transposons in the mouse genome. Nucleic Acids Res. 2011;39:e148. doi: 10.1093/nar/gkr764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Epplen JT, Leipoldt M, Engel W, Schmidtke J. DNA sequence organisation in avian genomes. Chromosoma. 1978;69:307–321. doi: 10.1007/BF00332134. [DOI] [PubMed] [Google Scholar]
- 46.International Chicken Genome Sequencing Consortium Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 47.Matsui Y, Zsebo K, Hogan BL. Derivation of pluripotential embryonic stem cells from murine primordial germ cells in culture. Cell. 1992;70:841–847. doi: 10.1016/0092-8674(92)90317-6. [DOI] [PubMed] [Google Scholar]
- 48.Resnick JL, Bixler LS, Cheng L, Donovan PJ. Long-term proliferation of mouse primordial germ cells in culture. Nature. 1992;359:550–551. doi: 10.1038/359550a0. [DOI] [PubMed] [Google Scholar]
- 49.Leitch HG, et al. Embryonic germ cells from mice and rats exhibit properties consistent with a generic pluripotent ground state. Development. 2010;137:2279–2287. doi: 10.1242/dev.050427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ivics Z, Izsvák Z, Chapman KM, Hamra FK. Sleeping Beauty transposon mutagenesis of the rat genome in spermatogonial stem cells. Methods. 2011;53:356–365. doi: 10.1016/j.ymeth.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Song G, Han JY. Avian biomodels for use as pharmaceutical bioreactors and for studying human diseases. Ann N Y Acad Sci. 2011;1229:69–75. doi: 10.1111/j.1749-6632.2011.06087.x. [DOI] [PubMed] [Google Scholar]
- 52.Lillico SG, McGrew MJ, Sherman A, Sang HM. Transgenic chickens as bioreactors for protein-based drugs. Drug Discov Today. 2005;10:191–196. doi: 10.1016/S1359-6446(04)03317-3. [DOI] [PubMed] [Google Scholar]
- 53.Mozdziak PE, Petitte JN. Status of transgenic chicken models for developmental biology. Dev Dyn. 2004;229:414–421. doi: 10.1002/dvdy.10461. [DOI] [PubMed] [Google Scholar]
- 54.Rhee JM, et al. In vivo imaging and differential localization of lipid-modified GFP-variant fusions in embryonic stem cells and mice. Genesis. 2006;44:202–218. doi: 10.1002/dvg.20203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hockemeyer D, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo:A sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kawakami K, et al. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell. 2004;7:133–144. doi: 10.1016/j.devcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 58.Perry MM. A complete culture system for the chick embryo. Nature. 1988;331:70–72. doi: 10.1038/331070a0. [DOI] [PubMed] [Google Scholar]
- 59.McGrew MJ, et al. Efficient production of germline transgenic chickens using lentiviral vectors. EMBO Rep. 2004;5:728–733. doi: 10.1038/sj.embor.7400171. [DOI] [PMC free article] [PubMed] [Google Scholar]