Abstract
Toll-like receptor (TLR) 3 is an endosomal TLR that mediates immune responses against viral infections upon activation by its ligand double-stranded RNA, a replication intermediate of most viruses. TLR3 is expressed widely in the body and activates both the innate and adaptive immune systems. However, little is known about how TLR3 intracellular trafficking and maturation are regulated. Here we show that newly synthesized endogenous TLR3 is transported through the ER and Golgi apparatus to endosomes, where it is rapidly cleaved. TLR3 protein expression is up-regulated by its own ligand, leading to the accumulation of its cleaved form. In agreement with its proposed role as a transporter, UNC93B1 expression is required for TLR3 cleavage and signaling. Furthermore, TLR3 signaling and cleavage are sensitive to cathepsin inhibition. Cleavage occurs between aa 252 and 346, and results in a functional receptor that signals upon activation. A truncated form of TLR3 lacking the N-terminal 345 aa also signals from acidic compartments in response to ligand activation. Screening of the human cathepsin family by RNA interference identified cathepsins B and H as key mediators of TLR3 processing. Taken together, our data indicate that TLR3 proteolytic processing is essential for its function, and suggest a mechanism of tight control of TLR3 signaling and thus immunity.
Keywords: Toll-like receptor trafficking, proteolysis, proteases, poly(I:C)
Germ-line–encoded pattern recognition receptors can be divided into cytosolic and transmembrane receptors (1). Toll-like receptors (TLRs) belong to the latter group and are key sensors of microorganisms. This sensing leads to the production of cytokines and chemokines that activate the innate and adaptive immune responses. A subset of TLRs, the endosomal TLRs 3, 7, 8, and 9, have evolved to sense the abnormal presence of nucleic acids in the endosomal pathway, a hallmark of viral infections (2).
In contrast to TLR7, TLR8, and TLR9, which are expressed mainly in immune system cells, TLR3 expression is nearly ubiquitous. TLR3 detects double-stranded RNA, which is produced during the replication cycle of most viruses, leading to activation of the transcription factors NF-κB and interferon regulatory factor (IRF) 3, and rapid and massive production of type I interferons (IFNs) and proinflammatory cytokines, such as IL-6 and IL-8 (3). Activation of the TLR3 pathway can also induce apoptosis in tumor cells via the activation of caspases 8 and 9 (4).
Human genetic studies in children carrying an autosomal dominant TLR3 deficiency have established that TLR3 is crucial for the defense against herpes simplex virus 1 (HSV-1) encephalitis (5). This was the first human TLR immunodeficiency detected, and its discovery established that, in contrast to TLR7 and TLR9 responses that are redundant during host defense, TLR3 is vital for natural immunity to HSV-1 in the central nervous system (5). Nonetheless, data to date suggest that TLR3 is redundant in host defense against other microbes. The importance of TLR3-mediated antiviral immune responses is underscored by the fact that several viruses such as hepatitis C virus, influenza, and West Nile virus have evolved specific mechanisms to bypass TLR3 detection (6).
Despite the fact that TLR3 mediates a fundamental defense against viral pathogens in most cell types and may represent an important therapeutic target in cancer, little is known about how TLR3 is regulated within the cell or initiates signaling in response to its ligand. Most of our current understanding on endosomal TLRs derives from studies of TLR9, which is synthesized in the endoplasmic reticulum (ER) as a full-length precursor, and transported to the endocytic pathway in a UNC93B1-dependent manner (7). There, TLR9 is subjected to two steps of proteolytic processing that involve asparagine endopeptidase (AEP) and cathepsins (8). TLR9 requires such processing to initiate signaling in response to its ligand CpG DNA (8–10). A recent study using transfected mouse cells expressing tagged forms of TLR7 or TLR3 observed that these receptors are also subjected to proteolytic cleavage (8). However, it remains unclear whether TLR3 cleavage is required for signaling and downstream events such as cytokine and chemokine production. Here we show that TLR3 protein expression is regulated by dsRNA, that TLR3 activity is tightly controlled by proteolytic cleavage, and that UNC93B1 is required for TLR3 cleavage and signaling.
Results
poly(I:C) Triggers a TLR3-Specific Response in RPE1 Cells via NF-κB and IRF3 Nuclear Translocation.
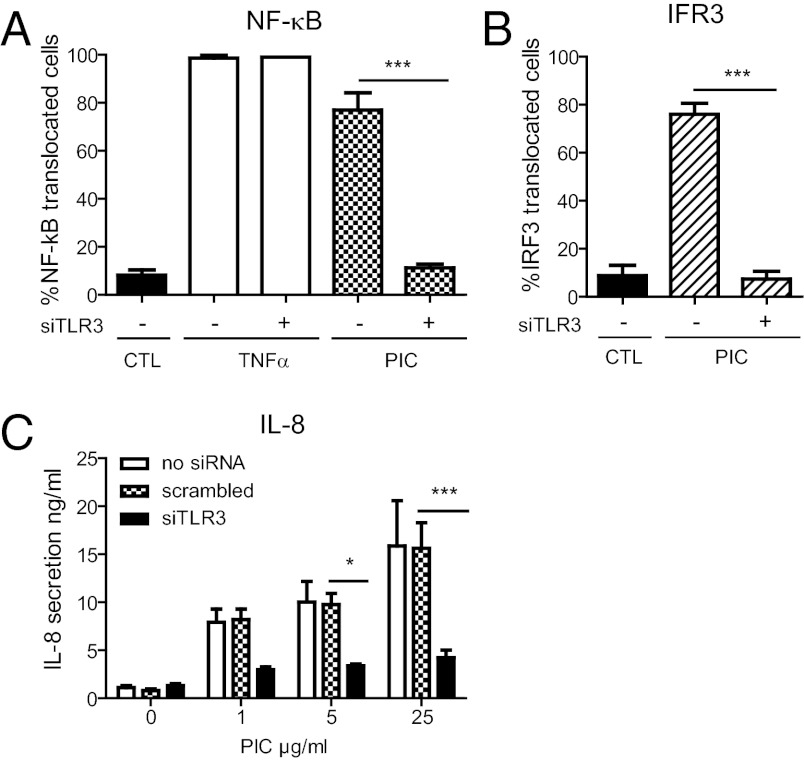
We studied TLR3 signaling in the human retinal epithelial cell line RPE1 that has been immortalized but not transformed (11). Exposure of RPE1 cells to poly(I:C) induced translocation of both NF-κB and IRF3 into the nucleus (Fig. S1 A and B). TLR3 activation was determined throughout this study by automated quantification of nuclear translocation of these transcription factors using algorithms based on ImageJ (National Institutes of Health) (Fig. S1C). Induction of NF-κB translocation by poly(I:C) was dose dependent (Fig. S1D and E) and peaked after 75 min (Fig. S1F). poly(I:C)-induced IRF3 translocation also peaked after 75 min (Fig. S1G). Depletion of TLR3 by RNAi blocked poly(I:C)-induced NF-κB and IRF3 translocation but did not affect NF-κB translocation induced by TNFα (Fig. 1 A and B). Similarly, overnight exposure of RPE1 cells to poly(I:C) led to secretion of IL-8 in a dose- and TLR3-dependent manner (Fig. 1C). poly(I:C) exposure thus triggers a TLR3-specific response in RPE1 cells and does not activate cytosolic sensors such as RIGI and MDA5 (12).
Fig. 1.
poly(I:C) triggers a TLR3-specific response in RPE1 cells via NF-κB and IRF3 nuclear translocation. (A and B) Translocation of NF-κB (A) and IRF3 (B) into the nucleus. RPE1 cells transfected or not transfected with siRNA specific for TLR3 were incubated with poly(I:C) (PIC) for 75 min, TNFα for 30 min or unstimulated (CTL), stained for NF-κB (A) or IRF3 (B) and assessed by epifluorescence microscopy (Fig. S1 A and B). Histograms represent percentage of cells with NF-κB or IFR3 translocated into the nucleus calculated from three independent experiments. The method used to generate these histograms is explained in Fig. S1 C–E. (C) Cells transfected or not transfected with TLR3-specific siRNA or scrambled siRNA were incubated with poly(I:C) for 15 h, and IL-8 present in the supernatant was assessed by ELISA. Data presented are from three independent experiments. One-way ANOVA, Tukey HSD: *P < 0.05; **P < 0.01; ***P < 0.001.
TLR3 Signaling Requires a Proteolytic Activity Sensitive to Cathepsin Inhibitors.
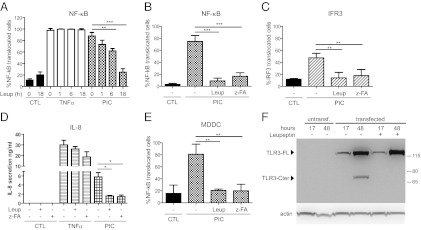
As serine/cysteine proteases are involved in TLR9 processing, we assessed the involvement of leupeptin-sensitive proteases in TLR3 signaling and cleavage. Leupeptin pretreatment led to a gradual decrease in poly(I:C)-induced NF-κB translocation with complete inhibition at 18 h without affecting TNFα signaling (Fig. 2A). Exposure to leupeptin or to the cathepsin inhibitor z-FA-fmk did not affect cell survival (Fig. S2A), but inhibited poly(I:C)-induced TLR3 signaling as observed by NF-κB and IRF3 nuclear translocation (Fig. 2 B and C) or IL-8 secretion (Fig. 2D). Similar findings were observed in primary human monocyte-derived dendritic cells (MDDC), with exposure to poly(I:C) leading to nuclear translocation of NF-κB, except in cells pretreated with leupeptin or z-FA-fmk (Fig. 2E).
Fig. 2.
TLR3 signaling requires a proteolytic activity sensitive to cathepsin inhibitors. (A) RPE1 cells were treated with leupeptin for different time periods before stimulation with poly(I:C) or TNFα (B, C, and E). Cells were treated with leupeptin or z-FA-fmk (z-FA) for 18 h before stimulation with poly(I:C) and assessed by (B) NF-κB pathway, (C) IRF3 pathway, and (E) NF-κB pathway in monocyte-derived dendritic cells (MDDC) from three different donors. (D) Cells were treated with leupeptin or z-FA-fmk (z-FA) for 18 h before stimulation with poly(I:C) for 15 h. IL-8 present in the supernatant was assessed by ELISA. All analyses were conducted from at least three independent experiments. (F) HEK-293 cells transiently transfected with TLR3-4HA were exposed or not exposed to leupeptin, and analyzed at 17 or 48 h posttransfection by HA-specific immunoblot. Data are representative of three experiments.
These results potentially indicate that proteolysis of TLR3 itself has to occur for proper signaling, in a manner similar to TLR9 (10). To analyze the effect of protease inhibitor treatment on TLR3 cleavage, we made a human TLR3-4HA construct in which four HA tags were added in tandem at the C terminus of the TLR3 protein. HEK-293 cells, which do not express endogenous TLR3, were transiently transfected with this construct and exposed to leupeptin. HA-specific immunoblot analysis of the cell lysates revealed a 130-kDa band corresponding to the full-length TLR3, which accumulated with time posttransfection, and a shorter fragment of 70 kDa corresponding to the C terminus cleaved form of TLR3, which appeared at 48 h and was absent from cells exposed to leupeptin (Fig. 2F). Similar results were obtained when immunoprecipitations were carried out on lysates from cells stably expressing TLR3-4HA (Fig. S2B). Notably, the shorter TLR3 fragment appeared to accumulate in stably expressing cells (Fig. S2B). These results suggest that leupeptin-sensitive proteolytic cleavage of TLR3 occurs with time and is necessary for signaling.
UNC93B1-Dependent Pool of Cleaved TLR3 Is Present in Endosomes Ready to Signal.
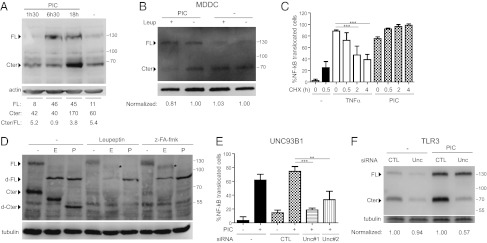
Analysis of the endogenous TLR3 protein revealed that, in the absence of stimuli, TLR3 is found as both a full-length protein of 130 kDa and a shorter form of 70 kDa (Fig. 3A). Exposure to poly(I:C) resulted in increased expression over time of both the full-length TLR3 and the 70 kDa fragment (Fig. 3A). The increased expression of full-length TLR3 observed upon poly(I:C) exposure peaked after 6 h 30 min, whereas the increase in the 70-kDa fragment was most pronounced after 18 h, possibly suggesting a time course for de novo synthesis of TLR3 followed by processing into the cleaved form that accumulates in the cell.
Fig. 3.
A UNC93B1-dependent pool of cleaved TLR3 is present in endosomes ready to signal. (A and B) Immunoblot analysis of endogenous TLR3 expression. (A) RPE1 cells were incubated for different time periods with poly(I:C), or (B) MDDCs were treated or not treated with leupeptin in the presence or absence of poly(I:C) for 18 h before lysis. Cter; TLR3-C-terminal 70 kDa fragment; FL, TLR3 full-length. Bands corresponding to the full length and the C-terminal fragment were quantified. The ratio of the 70-kDa TLR3 C-terminal form over the full-length form is shown for each condition or normalized (Norm) to nonleupeptin in each group. Actin is shown as a control for protein loading. Data are representative of three experiments. (C) RPE1 cells were treated with cycloheximide (CHX) for different time periods as indicated before stimulation with poly(I:C) or TNFα. Data are from three independent experiments. (D) Immunoblot analysis of endogenous TLR3 expression. Cells were treated with leupeptin or z-FA-fmk in the presence of poly(I:C) for 24 h. Cell lysates were incubated with EndoH or PNGase for 2 h, as indicated. *Position of full-length EndoH resistant TLR. degly, Deglycosylated TLR3. Tubulin is shown as a control for protein loading. (E and F) RPE1 cells were transfected with UNC93B1 (U) or scrambled (CTL) siRNAs for 3 d before stimulation with poly(I:C) for (E) 75 min, and assessed by NF-κB translocation assay for three independent experiments, or (F) for 18 h and lysates assessed by immunoblot analysis for endogenous TLR3. The ratio of the 70 kDa TLR3 C-terminal form over the full-length form is normalized to CTL siRNA in each group. Tubulin is shown as a control for protein loading. Data are representative of three independent experiments.
Similar biochemical analysis was performed on primary MDDCs with similar results. At steady state, both the full-length and shorter forms of endogenous TLR3 were present (Fig. 3B). Exposure to poly(I:C) led to increased levels of TLR3, whereas leupeptin treatment reduced levels of the 70-kDa fragment (Fig. 3B). Stimulation by IFNγ instead of poly(I:C) led to similar regulation of TLR3 expression and sensititivity to leupeptin (Fig. S3A). We conclude that the regulation of expression of TLR3 and of its 70-kDa fragment are similar in RPE1 cells and in primary MDDCs.
As TLR3 expression levels were increased by poly(I:C) exposure, we investigated whether protein neosynthesis was required for signaling. Inhibition of protein synthesis by cycloheximide treatment did not inhibit NF-κB translocation in response to poly(I:C) exposure, although it did inhibit translocation in response to TNFα (Fig. 3C). This result suggests that at steady state there is a cellular pool of TLR3 ready to signal.
To study TLR3 trafficking from the ER, we monitored its N-glycosylation status. Studies from TLR3-transfected cells have shown that N-glycans account for roughly one third of its molecular weight (4). RPE1 cell lysates were analyzed by immunoblot for endogenous TLR3 after pretreatment with either endoglycosidase H (Endo H), which is unable to digest complex sugars added as proteins passage through the Golgi apparatus, or peptide N-glycosidase (PNGase), which digests all sugars including those found on post-Golgi proteins. Full-length TLR3 is heavily glycosylated but remains Endo H sensitive (Fig. 3D). In contrast, the TLR3 70 kDa fragment was partially Endo H resistant, as observed by comparison with the PNGase treatment (Fig. 3D), suggesting that the cleaved form of TLR3 is the predominant form found in post-Golgi compartments. Exposure to leupeptin or z-FA-fmk prevented the generation of the 70-kDa fragment and enabled the visualization of a faint band corresponding to partially Endo H–resistant full-length TLR3 of ∼100 kDa, (Fig. 3D, star). This suggests that full-length TLR3 is also able to traffic through the Golgi apparatus. Even so, full-length TLR3 appears to be unable to signal, as activation of TLR3 upon poly(I:C) exposure was blocked by overnight exposure to either leupeptin or z-FA-fmk (Fig. 2 B–D). Thus, the generation of the cleaved form of TLR3 correlates with the signaling capacity upon exposure to poly(I:C).
Endosomal TLRs are thought to be transported from the ER to the endosomes in a UNC93B1-dependent manner (7, 13). We would therefore anticipate that in the absence of UNC93B1, TLR3 transport, and hence subsequent cleavage and signaling in the endosomes would be impaired. To test this hypothesis, RPE1 cells were silenced for UNC93B1 expression using two different siRNAs (Fig. S3B) and were exposed to poly(I:C). Compared with nontransfected cells or to cells transfected with a control siRNA, UNC93B1-silenced cells exhibited a strong inhibition of poly(I:C)-dependent NF-κB translocation (Fig. 3E). Moreover, we observed in the same cells decreased levels of the endogenous TLR3 70-kDa fragment compared with control cells (Fig. 3F).
Taken together, our results indicate that TLR3 is synthesized in the ER as a full-length protein, exits from the ER in an UNC93B1-dependent manner, and is trafficked rapidly through the Golgi apparatus to reach the endocytic compartments, where it is cleaved by cathespins, and accumulates there ready to signal.
Endosomal Acidification Has a Direct Impact on TLR3 Signaling.
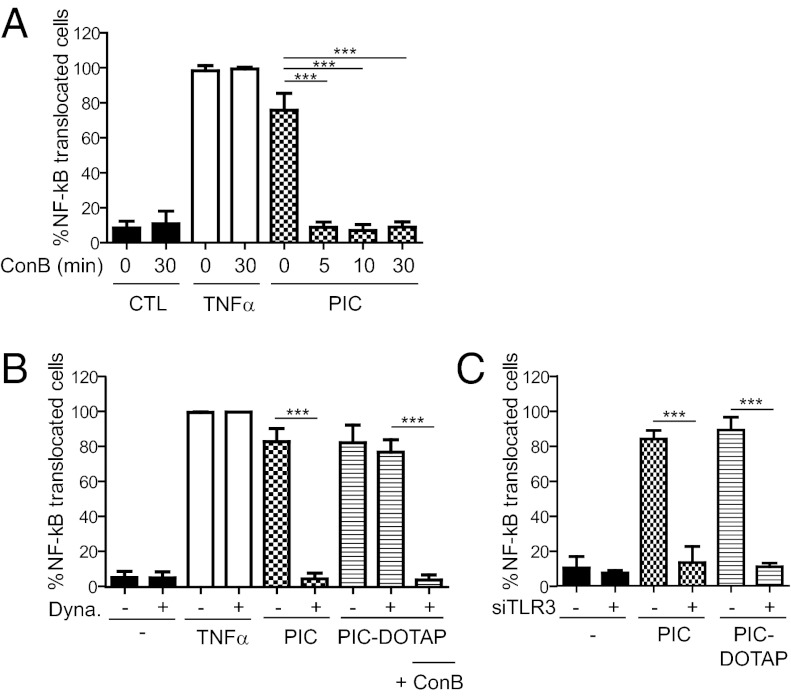
Endosomal TLRs were first identified because of their sensitivity to chloroquine treatment, which neutralizes pH in the endocytic pathway (14). To confirm that TLR3 function was dependent on acidification of the endosomes, we treated cells with concanamycin B (ConB), a potent inhibitor of the v-ATPase that is responsible for endosomal acidification. Indeed, neutralization of endosomes by ConB leads to inhibition of poly(I:C)-induced TLR3 activation but had no effect on translocation of NF-κB in response to TNFα (Fig. 4A). Strikingly, ConB-induced inhibition was observed within 5 min of exposure to poly(I:C) (Fig. 4A). Furthermore, inhibition of dynamin-dependent endocytosis using the drug Dynasore blocked poly(I:C)-induced TLR3 activation; however, such inhibition could be overcome by complexing poly(I:C) with the lipofectant DOTAP, which allows direct delivery of ligand to endosomes (Fig. 4B). Nonetheless, ConB treatment still prevented TLR3 activation, indicating that mere delivery of ligand to endosomes was insufficient for signaling (Fig. 4B). To test whether cytosolic sensors might be involved when poly(I:C) is delivered in the presence of DOTAP, cells silenced for TLR3 expression by RNAi were exposed to poly(I:C) mixed or not mixed with DOTAP. In all cases, NF-κB translocation was inhibited by TLR3 silencing (Fig. 4C), ruling out the involvement of cytosolic sensors in mediating poly(I:C) responses in this system.
Fig. 4.
Acidification in endosomes has a direct impact on TLR3 signaling. RPE1 cells were treated with either (A) ConcanamycinB (ConB) for different time periods, before stimulation with 5 μg/mL poly(I:C); or (B) Dynasore (Dyna) or Dynasore and ConB for 30 min and then exposed to poly(I:C), TNFα, or poly(I:C) complexed with DOTAP (PIC-DOTAP), as indicated. (C) Cells were transfected with TLR3 siRNAs for 3 d before stimulation with poly(I:C) or PIC-DOTAP, as indicated. TLR3 activation in all cases is represented as percentage of cells with NF-κB translocated to the nucleus of at least three independent experiments.
These results suggest that acidic pH most likely regulates the affinity of TLR ligand-binding and/or induces a conformational change required for downstream signaling from the pool of cleaved TLR3.
A C-Terminal Truncated Version of TLR3 Can Signal in Response to poly(I:C).
To identify the TLR3 cleavage site, we performed large-scale immunoprecipitation and separation by SDS/PAGE of TLR3 from HEK-293 cells expressing TLR3-4HA. Bands corresponding to the full-length and the 70 kDa forms of TLR3 were excised, trypsin-digested and subjected to tandem mass spectrometry. Several TLR3 peptides were identified in both bands. Analysis of the positioning of these peptides suggested a 90-aa-long region of TLR3 that contains the probable cleavage site (Fig. 5A). Notably, this region includes the leucine-rich repeat 12 (LRR12) loop, which represents a possible target for proteases because it protrudes from the protein structure and may therefore be more accessible for proteolysis (10, 15).
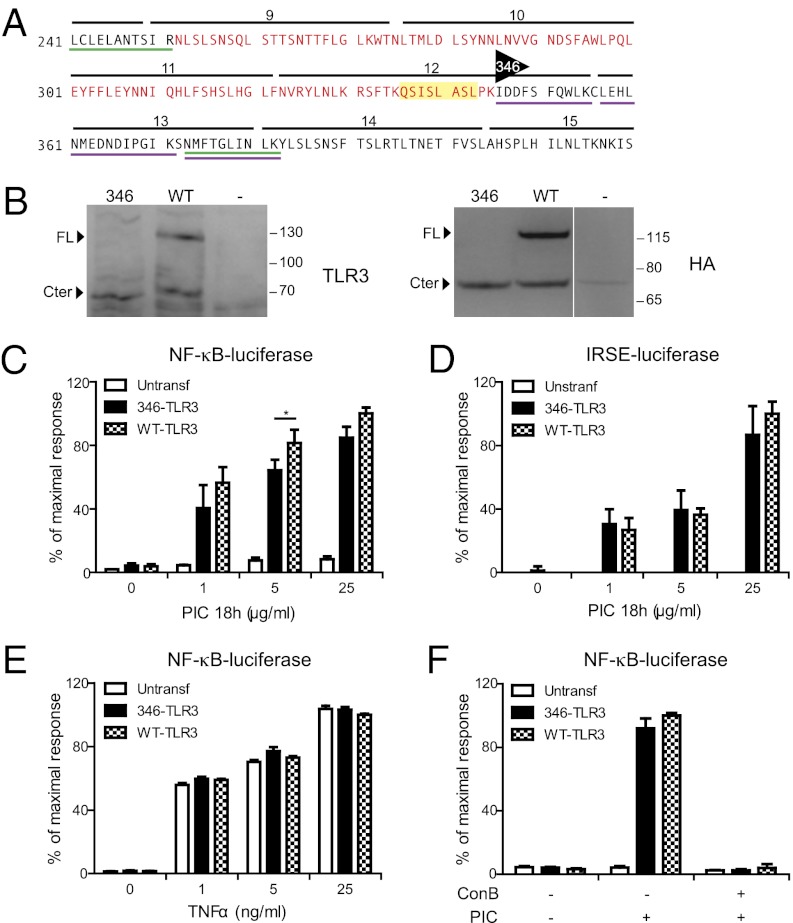
Fig. 5.
Truncated TLR3 can transduce signal in response to poly(I:C). (A) TLR3 amino acid sequence from position 241–370. Peptides identified by mass spectrometry in the full-length TLR3 band (green) and in the 70-kDa TLR3 (purple) are underlined. Leucine-rich repeats are indicated by black lines, the protruding loop by yellow, and the putative region of cleavage by red. The first amino-terminal position of the truncated form of TLR3 is indicated by the black triangle. (B) Cell lysates from nontransfected (−), or 346-TLR3-4HA (346) or WT TLR3-4HA (WT) stably transfected HEK-293-luc cells were immunoblotted for TLR3 or HA. (C, E, and F) Stably transfected HEK-293-luc cells were assessed by NF-κB luciferase reporter assay after (C) 18 h exposure to poly(I:C); (E) 2 h exposure to TNFα; or (F) 18 h exposure to poly(I:C) before pretreatement with ConB for 30 min. (D) Stably transfected HEK-293-IRSE luc cells were assessed by IRSE luciferase reporter assay after 18 h exposure to poly(I:C). Untransf, untransfected cells. Data are from at least three independent experiments.
To test our hypothesis that cleavage of TLR3 was required for signaling, we generated a truncated form of TLR3 by removing the first 345 aa from the N-terminal ectodomain (346-TLR3; Fig. 5A). This position was chosen at the C-terminal end of the 90-aa region identified above, which corresponds to the C-terminal end of the LRR12 loop. After transfection into HEK-293 cells, we verified by immunoblot against TLR3 and HA that the construct was correctly expressed and had the predicted molecular weight (Fig. 5B).
The response of the 346-TLR3 to poly(I:C) was functionally tested in HEK-293 cells stably transfected with either an NF-κB- or an IRSE-reporter construct, both driving the expression of the luciferase gene (HEK-293-luc and HEK-293 IRSE-luc, respectively). In both reporter systems, cells transfected with either WT TLR3 or 346-TLR3 exhibited similar luciferase activity in response to 18 h of exposure to poly(I:C) (Fig. 5 C and D), indicating that the truncated receptor is fully functional. However, at 6 h, we observed notably less poly(I:C)-induced activation in cells expressing the 346-TLR3 construct compared with the WT (Fig. S4 A and B), indicating some differences in kinetics between the WT and truncated receptor. As a control, exposure to TNFα induced similar levels of luciferase activity in the different cell populations in a dose-dependent manner (Fig. 5E). Notably, poly(I:C)-induced signaling was still inhibited by ConB treatment, indicating that endosomal acidic pH was necessary for signaling even if the receptor was already cleaved (Fig. 5F and Fig. S4B). These data confirm that the cleaved form of TLR3 can signal from acidic endosomes when cells are exposed to poly(I:C).
Cathepsins B and H Are Involved in TLR3 Cleavage and Signaling.
As TLR3 cleavage and signaling was inhibited when cells were exposed to leupeptin or to the cathepsin inhibitor z-FA-fmk (Figs. 2 B–F and 3 B and D), we sought to identify the cathepsin(s) involved using an RNA interference screen. RPE1 cells were transfected with siRNA specific for each of the 15 human cathepsins (four different siRNA/gene; Table S2). Three days later, poly(I:C)-induced translocation of NF-κB was quantified. Results from three independent experiments identified cathepsins B, H, and V as inhibiting poly(I:C)-induced signaling with at least one of the siRNA tested in all replicates and with a Z factor inferior to −3 (P < 0.05; Table S1). Although cathepsin V (also called L2) was a hit, we could not detect this protein in RPE1 cells by immunoblot and thus did not study it further (Fig. S5B).
Because cathepsins possess broad substrate specificity (16), there is likely redundancy among these enzymes. The NF-κB translocation assay was thus repeated with siRNAs specific for cathepsin B or H used either alone or in combination (Fig. 6A). Cathepsin L was used as a negative control, as none of the four siRNA tested in the first screen affected TLR3 activation (Table S1). The efficiency of the depletions induced by RNAi was verified in parallel by immmunoblot analysis (Fig. 6B and Fig. S5). Knockdown of cathepsin B or H expression using individual siRNA led to a substantial inhibition of poly(I:C)-induced NF-κB translocation (Fig. 6A). The extent of inhibition varied between siRNA and correlated with the efficiency of protein depletion. Mixtures of siRNAs specific for each cathepsin led to a stronger inhibition of translocation, suggesting that both cathepsin B and H activities control signaling via TLR3.
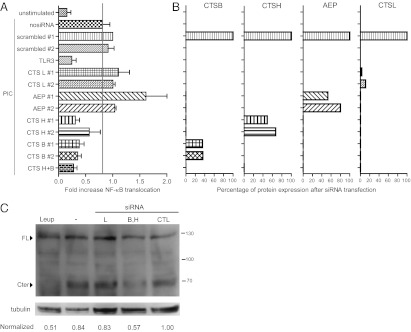
Fig. 6.
Cleavage of TLR3 by Cathepsin B and H is required for signaling. (A) RPE1 cells were transfected with the indicated siRNA for 3 d and then exposed to poly(I:C). Fold increase of NF-κB translocation in poly(I:C)-activated cells is represented, normalized to scrambled siRNA #1. Unstimulated: pooled data of cells transfected with each siRNA without stimuli. Gray line defines the limit under which the siRNA targeted genes are considered as hits (1 SD of scrambled siRNA). Data are from three independent experiments. (B) Percentage change in protein expression assessed by immunoblot after 3 d of the indicated siRNA transfection, normalized to scrambled #1 siRNA. (C) Immunoblot analysis of endogenous TLR3 present in the indicated RPE1 cell lysates. Cells were either transfected with the indicated siRNA or exposed to leupeptin (Leup) in the presence of poly(I:C) for 24 h. Tubulin is shown as a control for protein loading. Bands corresponding to the full-length and the C-terminal fragment were quantified. The ratios of the 70 kDa TLR3 C-terminal form over the full-length form are shown for each condition normalized to the scrambled siRNA. CTL, scrambled siRNA. Data are representative of three experiments.
Simultaneous knockdown of cathepsins B and H led to a 43% reduction of the level of the endogenous 70 kDa TLR3 fragment, as assessed by immunoblot, compared with control cells transfected with a scrambled siRNA (Fig. 6C). These results show that cathepsins B and H are involved in TLR3 cleavage, and provide further evidence that proteolytic cleavage is required for signal transduction.
We also included in our screen siRNAs specific for Asparagine endopeptidase (AEP), which is involved in TLR9 cleavage (8, 9). However, none of the four siRNA tested had any effect on poly(I:C)-induced NF-κB translocation (Table S1 and Fig. 6A), even though AEP expression was reduced (Fig. 6B).
Discussion
The activity of endosomal TLRs is tightly regulated through a combination of specific intracellular trafficking and proteolysis-dependent maturation. Comparison of TLR3 with TLR9 reveals significant differences with respect to the regulation of their expression and cleavage. Several studies indicate that TLR9 is retained in the ER at steady state and that exposure to CpG DNA ligand promotes trafficking of TLR9 to the endosomal pathway (17). In contrast, we show that TLR3 traffics to the endosome via a classical-secretion pathway, where cleavage occurs and the C-terminal fragment accumulates in the absence of ligand. Our data further indicate that it is this cleaved form of TLR3 that signals from endosomes upon ligand induction in nonimmune (epithelial) and immune (MDDC) cells. As with other endosomal TLRs, TLR3 trafficking, and thus subsequent cleavage and signaling, depend on the transporter protein UNC93B1. Acidification of endosomes is required not only for cleavage of TLR3 but also for activation of cleaved TLR3, likely by regulating its affinity for binding ligand or by inducing a conformational change required for signaling. The former possibility would correlate with previous studies showing that TLR3 binding to dsRNA was optimal at acidic pH (14, 18). Although cathepsins responsible for TLR3 cleavage would also be inhibited by neutralization of the endosomes, the rapidity of ConB-induced inhibition of TLR3 signaling (<5 min) along with the fact that a pool of cleaved TLR3 exists at steady state in the endosomes indicate that the change in pH is directly affecting either ligand binding or induction of signaling. Such sensitivity to pH makes sense, as it will serve to restrict the activity of TLR3 to endosomal compartments and to prevent inappropriate activation of antiviral and apoptotic pathways in the cell.
Importantly, we observed that exposure to poly(I:C) led to increased expression of TLR3, indicating that a positive feedback loop regulates the expression of TLR3 in both epithelial and monocyte-derived dendritic cells. This is relevant because it would allow the cell to maintain an antiviral response throughout infection.
Upon reaching the endosome, TLR9 is processed by two cleavage events: the first by AEP (or cathepsins if AEP is absent); and the second a trimming by cathepsins (8). The identity of the cathepsins involved in these processes is yet to be established. In contrast, TLR3 cleavage appears to be AEP independent in epithelial and human dendritic cells, as both are sensitive to leupeptin, which does not inhibit AEP activity (8, 9). Our results further indicate that in epithelial cells TLR3 cleavage depends on cathepsins B and H, as poly(I:C)-dependent TLR3 signaling is inhibited and cleavage is reduced in cells silenced for both cathepsins. This does not preclude the possibility that AEP, or indeed other cathepsins, could play a role in TLR3 maturation in dendritic or other cell types.
The cleavage sites for TLR9, TLR7, and TLR3 all remain to be determined. For TLR9, the predicted cleavage site has been suggested to be between amino acids 378 and 475 of the ectodomain (10) that comprises a region (441–470) that is part of a flexible loop that contains the potential AEP (9) and cathepsin cleavage sites (10). For TLR3, our data predict a cleavage site within amino acids 252–346. We know from the 3D structure of TLR3 that this region encompasses the LRR12 flexible loop that protrudes from the lateral face of the ectodomain and that is potentially well exposed to proteases (10, 15). Cathepsins B and H possess broad substrate specificity (16), so it is not yet possible to further clarify the cleavage site.
Although our model suggests that full-length TLR3 does not signal from endosomes, we know from its 3D structure that full-length TLR3 can bind to dsRNA as a dimer (19). Structural studies of the TLR3 ectodomain predict two putative ligand binding sites: one located at the N-terminal end within LRR-NT to LRR3, and the other at the C terminus within LRR19 to LRR21 (19). Mutational analysis has shown that both sites contain residues required for poly(I:C)-dependent NF-κB and IRF3 activation (19). However, in our system, the N-terminal site is dispensable for signaling, as both the cleaved form of TLR3 that accumulates in endosomes and our truncated 346-TLR3 construct only retain the C-terminal of these binding sites and can respond to poly(I:C) ligand by inducing NF-κB and IRF3 activation. Similarly, analysis of TLR9 cleavage has shown that a truncated form of TLR9 has levels of activation similar to those of the the WT receptor (10), despite also missing an N-terminal binding site thought to be required for activation (20).
The processing of TLR3 by endosomal proteases may reflect a common mechanism of regulation for all endosomal TLRs (3, 7, 9) to restrict their capacity to signal only from the endosomal compartments where they sense nucleic acids (8). The pH sensitivity of cleaved TLR3 would also serve to restrict the ability of TLR3 to signal to endosomes. The evolutionary importance of these mechanisms may be to restrain the activity of endosomal TLRs to the cellular compartments where foreign nucleic acids are more likely to be found.
Materials and Methods
hTLR3-4HA (WT-TLR3), cDNA encoding four repetitions of the hemmaglutinin (HA) epitope (5′TACCCATACGATGTTCCAGATTACGCT3′) was inserted at the 3′ end of TLR3 in pUNO-hTLR3 (Invivogen). The 346-TLR3-4HA construct (346-TLR3) was generated from hTLR3-4HA by PCR cloning (sense sequence: 5′ATTGATGATTTTTCTTTTCAGTGGC3′). More information is provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank S. Amigorena, S. Suchting, S. Lebecque, and A. Vathaman for helpful discussions; and M. Desdouits for assistance. We thank the Nikon Imaging Centre (F. Waharte) and the Mass Spectrometry and Proteomics platform (D. Loew, F. Dingli, and B. Lombard) of the Curie Institute. This work was supported by Grant ANR-09-MIE-021 from L'Agence Nationale de la Recherche (to P.B.) and by a fellowship from Fondation pour la Recherche Médicale (to A.G.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1115091109/-/DCSupplemental.
References
- 1.Medzhitov R, Janeway CA., Jr Decoding the patterns of self and nonself by the innate immune system. Science. 2002;296:298–300. doi: 10.1126/science.1068883. [DOI] [PubMed] [Google Scholar]
- 2.Barton GM. Viral recognition by Toll-like receptors. Semin Immunol. 2007;19:33–40. doi: 10.1016/j.smim.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Takeda K, Akira S. TLR signaling pathways. Semin Immunol. 2004;16:3–9. doi: 10.1016/j.smim.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Sun R, et al. Toll-like receptor 3 (TLR3) induces apoptosis via death receptors and mitochondria by up-regulating the transactivating p63 isoform alpha (TAP63alpha) J Biol Chem. 2011;286:15918–15928. doi: 10.1074/jbc.M110.178798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang SY, et al. TLR3 deficiency in patients with herpes simplex encephalitis. Science. 2007;317:1522–1527. doi: 10.1126/science.1139522. [DOI] [PubMed] [Google Scholar]
- 6.Wilson JR, de Sessions PF, Leon MA, Scholle F. West Nile virus nonstructural protein 1 inhibits TLR3 signal transduction. J Virol. 2008;82:8262–8271. doi: 10.1128/JVI.00226-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim YM, Brinkmann MM, Paquet ME, Ploegh HL. UNC93B1 delivers nucleotide-sensing toll-like receptors to endolysosomes. Nature. 2008;452:234–238. doi: 10.1038/nature06726. [DOI] [PubMed] [Google Scholar]
- 8.Ewald SE, et al. Nucleic acid recognition by Toll-like receptors is coupled to stepwise processing by cathepsins and asparagine endopeptidase. J Exp Med. 2011;208:643–651. doi: 10.1084/jem.20100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sepulveda FE, et al. Critical role for asparagine endopeptidase in endocytic Toll-like receptor signaling in dendritic cells. Immunity. 2009;31:737–748. doi: 10.1016/j.immuni.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 10.Park B, et al. Proteolytic cleavage in an endolysosomal compartment is required for activation of Toll-like receptor 9. Nat Immunol. 2008;9:1407–1414. doi: 10.1038/ni.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bodnar AG, et al. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 12.Yoneyama M, Fujita T. Structural mechanism of RNA recognition by the RIG-I-like receptors. Immunity. 2008;29:178–181. doi: 10.1016/j.immuni.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 13.Brinkmann MM, et al. The interaction between the ER membrane protein UNC93B and TLR3, 7, and 9 is crucial for TLR signaling. J Cell Biol. 2007;177:265–275. doi: 10.1083/jcb.200612056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Bouteiller O, et al. Recognition of double-stranded RNA by human toll-like receptor 3 and downstream receptor signaling requires multimerization and an acidic pH. J Biol Chem. 2005;280:38133–38145. doi: 10.1074/jbc.M507163200. [DOI] [PubMed] [Google Scholar]
- 15.Choe J, Kelker MS, Wilson IA. Crystal structure of human toll-like receptor 3 (TLR3) ectodomain. Science. 2005;309:581–585. doi: 10.1126/science.1115253. [DOI] [PubMed] [Google Scholar]
- 16.Turk V, Turk B, Turk D. Lysosomal cysteine proteases: Facts and opportunities. EMBO J. 2001;20:4629–4633. doi: 10.1093/emboj/20.17.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukui R, et al. Unc93B1 biases Toll-like receptor responses to nucleic acid in dendritic cells toward DNA- but against RNA-sensing. J Exp Med. 2009;206:1339–1350. doi: 10.1084/jem.20082316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukuda K, et al. Analysis of the interaction between human TLR3 ectodomain and nucleic acids. Nucleic Acids Symp Ser (Oxf) 2006;50:249–250. doi: 10.1093/nass/nrl124. [DOI] [PubMed] [Google Scholar]
- 19.Liu L, et al. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science. 2008;320:379–381. doi: 10.1126/science.1155406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peter ME, Kubarenko AV, Weber AN, Dalpke AH. Identification of an N-terminal recognition site in TLR9 that contributes to CpG-DNA-mediated receptor activation. J Immunol. 2009;182:7690–7697. doi: 10.4049/jimmunol.0900819. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.