Abstract
Hospital-associated infections caused by methicillin-resistant Staphylococcus aureus (MRSA) are a global health burden dominated by a small number of bacterial clones. The pandemic EMRSA-16 clone (ST36-II) has been widespread in UK hospitals for 20 y, but its evolutionary origin and the molecular basis for its hospital association are unclear. We carried out a Bayesian phylogenetic reconstruction on the basis of the genome sequences of 87 S. aureus isolates including 60 EMRSA-16 and 27 additional clonal complex 30 (CC30) isolates, collected from patients in three continents over a 53-y period. The three major pandemic clones to originate from the CC30 lineage, including phage type 80/81, Southwest Pacific, and EMRSA-16, shared a most recent common ancestor that existed over 100 y ago, whereas the hospital-associated EMRSA-16 clone is estimated to have emerged about 35 y ago. Our CC30 genome-wide analysis revealed striking molecular correlates of hospital- or community-associated pandemics represented by mobile genetic elements and nonsynonymous mutations affecting antibiotic resistance and virulence. Importantly, phylogeographic analysis indicates that EMRSA-16 spread within the United Kingdom by transmission from hospitals in large population centers in London and Glasgow to regional health-care settings, implicating patient referrals as an important cause of nationwide transmission. Taken together, the high-resolution phylogenomic approach used resulted in a unique understanding of the emergence and transmission of a major MRSA clone and provided molecular correlates of its hospital adaptation. Similar approaches for hospital-associated clones of other bacterial pathogens may inform appropriate measures for controlling their intra- and interhospital spread.
Keywords: nosocomial, epidemiology
The Gram-positive bacterium Staphylococcus aureus is a component of the normal flora of about 30% of the human population, but is capable of causing severe infections of immunocompromised patients in hospitals and healthy humans in the community (1). Hospital-associated methicillin-resistant S. aureus (HA-MRSA) is represented by a small number of clones that rarely cause disease outside of the health-care setting and that are characterized by resistance to β-lactam antibiotics in addition to other front-line antimicrobials (2). During the last 60 y, the S. aureus clonal complex 30 (CC30) has had a profound impact on global human health by giving rise to three pandemic waves and the toxic shock syndrome (TSS) epidemic (3, 4). Furthermore, one study indicated S. aureus isolates from life-threatening endocarditis infections are more likely to belong to CC30 than to other S. aureus lineages (5). The first CC30 pandemic was caused by the methicillin-sensitive phage type 80/81 clone in the 1950s and 1960s, which spread from hospitals causing a significant disease burden in the community and was characterized by resistance to penicillin and production of the Panton-Valentine leukocidin (PVL) toxin (6–9). The Southwest Pacific clone (SWP) is a contemporary PVL+ community-associated MRSA clone, which has spread to several continents and which largely causes skin and soft tissue infections of otherwise healthy individuals (10, 11). In contrast to phage type 80/81 and SWP, the EMRSA-16 (ST36) clone appears to be restricted to the hospital setting and has reduced virulence due in part to low levels of expression of cytolytic toxins (12). Along with the EMRSA-15 (ST22) clone, EMRSA-16 has been endemic in UK hospitals for over 20 y and has also been reported less commonly in other European countries, Southeast Asia, South Africa, Australia, and North America (3, 13–17). The high rate of MRSA infections and the rapid spread of HA-MRSA between UK hospitals led the UK government to introduce stringent infection control legislation from 2003, resulting in a decrease in rates of nosocomial MRSA infection (18, 19). Of note, EMRSA-16 prevalence has declined more rapidly than that of EMRSA-15, implying the existence of unknown strain-dependent factors that confer increased susceptibility to hospital infection prevention and control measures (18, 19).
Despite its clinical importance, the evolution of the EMRSA-16 clone, in addition to the molecular basis for its success are poorly understood. Here we use a phylogenomic approach to examine the diversity of EMRSA-16 relative to other CC30 pandemic clones. The results represent a high resolution insight into the emergence, expansion, and transmission of a major clone of MRSA, revealing unique molecular correlates for its hospital association.
Results and Discussion
Phylogenetic and Dating Analysis of CC30 Pandemics.
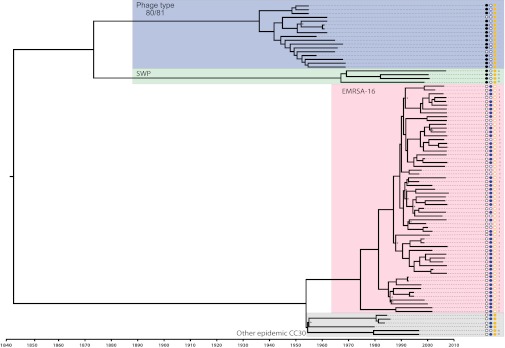
The core genome of the 87 CC30 isolates examined consisted of 2,381,276 bases (82% of the MRSA252 reference genome), containing a total of 4,499 high-confidence single nucleotide polymorphisms (SNPs). We applied a Bayesian coalescent method using a relaxed molecular clock model to infer the phylogeny and the rate of molecular evolution of the CC30 lineage and its major clades. The phylogeny indicates the existence of three major clades within the CC30 lineage, representative of the major pandemic clones, 80/81, SWP, and EMRSA-16, in addition to an EMRSA-16 sister clade represented by other epidemic CC30 isolates (Fig. 1). There was strong posterior support for the majority of nodes in the tree and parsimony analysis indicates a very low frequency of homoplasies across the phylogenetic tree (consistency index of 0.92 for parsimony informative sites), suggesting there was limited conflicting phylogenetic signal. The mean nucleotide substitution rate within CC30 was 1.42 × 10−6 substitutions per site per year [95% highest posterior densities (HPDs) 1.04 × 10−6–1.80 × 10−6], which varied negligibly depending on the clock model and choice of tree prior and within each pandemic clade. Given the rate of molecular evolution determined for the CC30 lineage, we calculated the time of the most recent common ancestor (MRCA) for each of the three major clades that correspond to the three pandemic CC30 clones. The date for the MRCA of the 80/81 clone was estimated as 1936 (95% HPDs, 1926–1945), the date for the MRCA of the SWP clone was estimated as 1967 (95% HPDs, 1952–1984), and the date for the MRCA of the EMRSA-16 clone was estimated as 1975 (95% HPDs, 1965–1983). The latter date precedes the first reports of EMRSA-16 identification in UK hospitals by about 18 y. Finally, the date for the MRCA of the entire CC30 lineage was estimated as 1842 (95% HPDs, 1765–1909).
Fig. 1.
The CC30 lineage is divided into multiple distinct clades characterized by the presence of different toxin and antibiotic resistance determinants. Bayesian phylogenetic reconstruction of the CC30 lineage using all sites in the core genome. Blue, green, red, and gray shading correspond to the 80/81, Southwest Pacific, EMRSA-16, and other epidemic CC30 clones, respectively. Presence of the pvl locus is denoted by shaded black circles, tst carriage by shaded blue circles, intact crtM gene by shaded yellow circles, and SCCmec type is indicated for methicillin-resistant isolates. Branch lengths are scaled according to time-scale bar. All nodes have posterior probability support >0.8 unless labeled with an asterisk. Tip (isolate no.) and node labels are shown in Fig. S4.
Previously, Robinson et al. used multilocus sequence typing (MLST) and PCR genotyping to examine the evolution of CC30 pandemic clones and inferred that the SWP clone originated from the historic phage type 80/81 clone (3). However, an important recent study by DeLeo et al. based on a comparative genome sequence analysis of nine CC30 isolates demonstrated that the SWP clone is not a direct descendent of the phage 80/81 clone, but that each clone has evolved from a single ancestral strain, which gave rise to all three CC30 pandemics (12). In agreement with this finding, our phylogenetic analysis clearly demonstrates the independent origins of each of the three major CC30 pandemic clones. Furthermore, the larger number of genome sequences included in the current study facilitated the use of a Bayesian coalescent method to estimate the rate of molecular evolution within CC30, the date of emergence of each of the major pandemic clones, and the timing of other important CC30 evolutionary events. Temporal analysis of bacterial epidemics may implicate contemporaneous environmental factors or human practices that promoted their emergence and expansion. For example, the phage type 80/81 clone is predicted to have emerged shortly before a time of intensive penicillin use, which may have provided a selection for clonal expansion after acquisition of a β-lactamase plasmid (3, 20, 21). For each of the CC30 pandemic clones, the predicted date of the MRCA is several years earlier than the time that these clones were first reported as clinical isolates in the literature, consistent with observations for the pandemic ST239 clone of S. aureus (22–24). Early strains of the emergent clones may have existed for some time before acquiring mobile genetic elements, or before encountering selective pressures, which promoted rapid clonal expansion. Furthermore, it is feasible that EMRSA-16 strains were causing low-level clinical infections for several years before they were first described in the literature as a significant clinical problem.
Multiple Independent Acquisitions of pvl Occurred During the Evolution of CC30 Pandemic Clones.
Comparative genomic analysis of closely related isolates from epidemics occupying different niches or associated with different disease manifestations is a powerful means for identifying genetic events that may have contributed to clone emergence and pathogenesis (23, 25–27). PVL has been a marker for community-associated clones of S. aureus associated with skin and soft tissue infections or severe necrotizing pneumonia (28), although increased identification of PVL+ strains associated with nosocomial infections weakens this correlation (29, 30). We examined the genomes of the CC30 isolates included in the current study for the presence of the pvl locus (lukS-PV and lukF-PV) and found it to be present in 19 of 21 isolates belonging to the community-associated phage type 80/81 and SWP clones (Fig. 1). The PVL toxin is encoded by temperate phage, which can be differentiated into distinct morphological groups on the basis of their elongated- or icosahedral-head types (31). Sequence analysis of the small and large terminase subunits of the PVL phage revealed that three of four SWP clone isolates contained the elongated-head phage type, whereas the remaining SWP isolate and the phage type 80/81 isolates had the icosahedral-head phage type. Phylogenetic analysis of the small and large terminase subunit genes indicates the close relatedness of the icosahedral phage heads of the SWP and phage type 80/81 clones (Fig. S1), suggesting that a progenitor of the SWP and phage type 80/81 clones, which existed over 100 y ago, had previously acquired an icosahedral-head phage type encoding PVL (although independent horizontal acquisition cannot be ruled out). Subsequently, the phage was replaced by the elongated-head phage type in some isolates of the SWP clone (Fig. S1). Overall, these data suggest that PVL has a long residency with some community-associated S. aureus clones, having been maintained in CC30 clades since a likely acquisition event that occurred about 137 y ago (95% HPDs, 86–197 y ago). These data are consistent with a central role for PVL in the success of some community-associated S. aureus clones (32).
Single Acquisition of the tst Gene Led to CC30 Strains Responsible for the Toxic Shock Syndrome Epidemic.
Previous studies have demonstrated that the majority of cases of menstrual TSS are caused by a single clone that corresponds to CC30 (33, 34). However, the distribution of the tst gene encoding the toxic shock syndrome toxin 1 (TSST-1) among CC30 subclades has not been previously examined. We determined that the tst gene is harbored by the staphylococcal pathogenicity island 2 (SaPI2) and is present in 55 of 66 (83%) of the isolates in the sister clades represented by EMRSA-16 and other CC30 epidemic isolates (Fig. 1), but is not present in any of the 21 (0%) isolates from the clades represented by SWP and 80/81 clones. These data imply the restriction of the tst gene to specific contemporary CC30 clones and its absence from the phage 80/81 and SWP clones. The high level of sequence identity of SaPI2 among the 55 tst+ isolates (five or fewer SNPs in 14.7-kb SaPI2) strongly suggests that a single acquisition event occurred in an ancestor that existed 10–140 y before the TSS epidemic of the 1970/80s (Fig. 1). Of note, previous studies have provided evidence that the TSS epidemic was caused by already widely disseminated TSST-1+ strains rather than rapid clonal expansion of a single strain that had acquired a fitness mutation (e.g., SaPI2 acquisition) (33).
Identification of Mutations That Correlate with the Hospital Habitat of EMRSA-16.
The hospital setting provides a unique array of insults that bacteria must overcome, including numerous classes of antibiotics used in treatment and routine use of disinfectants and antiseptics. However, in contrast to the community setting, the hospital provides a continuous supply of immunocompromised human hosts, who offer plentiful opportunities for infection and transmission. Our CC30 genome-wide analysis indicates that 58 of 60 EMRSA-16 isolates contained the type II staphylococcal cassette chromosome mec element (SCCmecII) (Fig. 1). Acquisition of SCCmecII was a critical genetic event in the evolution of the EMRSA-16 clone as a hospital-associated antibiotic-resistant clone refractory to treatment with β-lactam antibiotics. Of note, the type II SCCmec element has been demonstrated to reduce the toxicity of MRSA CC30 strains in comparison with methicillin-sensitive CC30 strains, by preventing normal stationary phase induction of the agr system, leading to decreased expression of cytolytic toxins (35, 36). It is speculated that this reduction in energy requirement could compensate for the metabolically costly maintenance of a large SCCmec element and its associated methicillin resistance, but which would be likely to lead to reduced fitness outside of the hospital setting (37).
In addition to SCCmecII, we identified a number of nonsynonymous mutations specific to EMRSA-16 in loci previously demonstrated to influence antibiotic resistance, which are likely to have been the result of selective pressures prevalent in hospitals. Specifically, a S84L replacement in DNA gyrase subunit A, and an S80F replacement in DNA topoisomerase IV subunit A have been demonstrated to confer resistance to fluoroquinolones (38–41). Of note, a selection of 16 EMRSA-16 isolates tested were all fluoroquinolone resistant. In addition, nonsynonymous mutations were identified in additional loci implicated in resistance, including genes encoding penicillin binding proteins 2 and 4, the vraD component of the bacitracin resistance pathway, and the gene encoding the multidrug efflux transporter NorA in 50 of 60 EMRSA-16 isolates. Finally, plasmids encoding resistance to quaternary ammonium compounds were found in 7 of 60 EMRSA-16 isolates (Dataset S1).
In their recent study, DeLeo and colleagues included a single EMRSA-16 isolate (MRSA252) and identified mutations in agr and hla genes, which occurred in an ancestor of MRSA252 and other contemporary CC30 isolates leading to reduced virulence in mouse models of infection (12). As proposed by the authors, these mutations may have contributed to the hospital association of contemporary CC30 isolates (4). However, isolates that belong to the sister clade to EMRSA-16, annotated as other epidemic CC30 (Fig. 1), contain the same agr and hla mutations but have been isolated from episodes of severe community-associated infections of healthy humans such as TSS (Fig. 1 and Dataset S2) (42). We therefore suggest that, whereas agr and hla mutations are indeed likely to influence the capacity of CC30 isolates to cause certain types of infection, they are probably not sufficient to explain the hospital restriction of the EMRSA-16 clone since its differentiation from the other epidemic CC30 clade. To investigate this hypothesis further, we screened for mutations that occurred on the branch leading to the EMRSA-16 clone only (Dataset S3). It should be pointed out that some of the mutations are likely to be the result of fixation due to genetic drift before clonal expansion rather than the result of selective pressures encountered in hospitals. However, several of the identified mutations are predicted to have functional consequences that could impact on virulence. For example, a nonsense mutation occurred in the squalene desaturase gene (crtM) leading to pseudogene formation and disruption of the terminal portion of the mevalonate pathway, which leads to staphyloxanthin carotenoid biosynthesis (42). This is consistent with the nonpigmented colony phenotype of EMRSA-16 strains described in the earliest reports of the EMRSA-16 clone (43). Bacterial carotenoids confer resistance to oxidative killing during phagocytosis, and staphyloxanthin-deficient S. aureus mutants have diminished virulence in animal models of infection (42). The loss of metabolically expensive pigment production may in part compensate for the energy cost of maintaining the large SCCmecII element mediating antibiotic resistance (23, 44). In turn, and in combination with the previously described hla and agr mutations, lack of pigment may affect its capacity to cause disease of healthy humans outside of the hospital setting. In addition, nonsynonymous mutations were identified in genes encoding proteins involved in virulence such as the virulence gene regulator CcpA and, for a proportion of EMRSA-16 strains, cell wall-associated proteins Fib, SasH, IsdB, and Ebh (Dataset S2). Whereas the examples discussed are selected on the basis of known or implicated roles in virulence, we cannot dismiss the possibility that some of the other nonsynonymous mutations identified among EMRSA-16 strains may also have had a role in shaping its hospital-specialist lifestyle (Dataset S3). In contrast to DeLeo et al. who neatly demonstrated the virulence-attenuating effects of mutations in agr and hla (12), we have not yet examined the functional consequences of the identified genetic differences. However, the identification of EMRSA-16 clade-specific mutations provides important avenues for future studies examining the hospital specialization of this important clone.
EMRSA-16 Has Spread Within the United Kingdom by Transmission from Hospitals in Major Cities to Regional Centers.
The geographic spread of hospital-associated bacterial clones is not well understood. Whole genome sequencing of large numbers of nosocomial isolates allows the high-resolution tracking of the transmission of strains through space and time (45). Bayesian phylogenetic analysis of a subset of the genome sequence data, which included the UK EMRSA-16 isolates only (54 of 60 EMRSA-16 isolates), resolved several subclades consisting of isolates from proximal UK geographic locations, consistent with the existence of EMRSA-16 strains, which are endemic to particular hospitals or regions (Fig. 2). In particular, EMRSA-16 strains isolated in Aberdeen Royal Infirmary between 2006 and 2007 are more closely related to each other (subclade a) than to other EMRSA-16 isolates (Fig. 2). In addition, subclade b consists largely of isolates from central Scotland, implying the existence of an EMRSA-16 subtype, which is endemic to this region (Fig. 2). However, isolates from London and Glasgow are widely distributed among clusters of closely related isolates from regional hospitals, consistent with hospitals in major population centers acting as reservoirs for EMRSA-16 UK transmissions. To examine this observation further, we used the discrete phylogeographic diffusion model implemented in BEAST, with isolates grouped by geographic region (London, southeast, south, and central England and north, east, and west Scotland), and mean distance between cities calculated. An alternative approach using a matrix of geographic locations based on city of isolation resulted in an overparameterized model. Using the regional groupings, we identified statistical support for London as a source of EMRSA-16 transmission events to south and southeast regions of England (Bayes factors 3.00 and 4.36, respectively). In addition, Glasgow (west Scotland) was identified as a reservoir for transmission of EMRSA-16 to surrounding population centers in the north and east of Scotland (Bayes factors 4.04 and 7.16, respectively). The dataset is limited by the number of isolates included and the relatively small number of hospitals sampled. For example, it would be interesting to examine the dynamics between regional networks within the city of London. Nonetheless, these data provide evidence for transmission routes from hospitals in major cities to UK regions, leading to endemic strains circulating in local hospitals. These findings are consistent with a recent US study that estimated high transmission rates between large hospitals and long-term care facilities using a simulation model (46). It has previously been postulated that an increase in the willingness of patients to travel further for treatment, coupled with the centralization of specialist treatment centers have been contributing factors to the spread of MRSA throughout the United Kingdom (47). These data could inform the design of infection control protocols, such as decolonization of patients before transfer from large hospitals, to limit interhospital transmission as a major driving force for epidemics.
Fig. 2.
EMRSA-16 has been transmitted from hospitals in major population centers to regional centers. (A) Bayesian phylogenetic reconstruction of UK EMRSA-16 isolates. Terminal branches representing London and Glasgow isolates are colored in red and blue, respectively. Black branches depict isolates from other locations. Branches are scaled with time (years). Gray shading indicates examples of geographically restricted subclades a and b. (B) Map of the United Kingdom indicating regions represented in the phylogeographic analysis (colored). Isolates were grouped by geographic region: L, London; SEE, southeast England; SE, south England; CE, central England; NS, north Scotland; ES, east Scotland; WS, west Scotland), and mean distance between cities was calculated. Blue and red shaded regions represent Glasgow (G) and London (L), respectively.
Concluding Comments.
The current study extends the recent findings of DeLeo et al. (12) who revealed the independent origin of phage 80/81 and SWP pandemics and identified mutations leading to reduced virulence among contemporary CC30 strains. The capacity to rapidly sequence the genome of large numbers of bacterial isolates can result in very enhanced insights into bacterial epidemics. By using a high-resolution phylogenomic approach, we have determined a time frame for the emergence and expansion of CC30 pandemics, and identified pathways of transmission of a major hospital-associated MRSA clone, which may be used to inform control methods. In addition, we have identified unique genetic events that correlate with its adaptation to the hospital environment, some of which may help to explain its lack of success in the community setting. The use of a similar approach for other hospital-associated bacteria could lead to the identification of risk factors that promote the emergence and expansion of epidemics, and thereby inform the rational design of methods for controlling their inter- and intrahospital spread.
Methods
Bacterial Isolates.
In addition to seven publicly available complete or draft CC30 genome sequences, a total of 80 S. aureus isolates were selected from a larger collection for genome sequencing to represent the breadth of genotypic diversity within the CC30 lineage sampled through time and space, with an emphasis on EMRSA-16 isolates in the United Kingdom (Dataset S1). The genetic diversity of CC30 isolates was based on molecular typing methods such as pulsed field gel electrophoresis (PFGE), staphylococcal protein A (spA), and MLST, in addition to limited information relating to the distribution of selected virulence and antibiotic resistance determinants. The 80 isolates sequenced for the current study were typed by MLST as ST30 (n = 21) or its single locus variant (slv) ST36 (n = 58), with one isolate identified as ST500, an slv of both ST30 and ST36. Genomic DNA was isolated from S. aureus as previously described (33).
Whole Genome Sequencing, Mapping, and Alignment.
Whole genome sequencing was carried out with the Illumina Genome Analyzer II, or the Roche 454 GS FLX platform. Reads from the previously sequenced ED83, ED84, ED86 (48). and WW2703/97 (49) isolates were included in this study. Adaptor sequences were trimmed from Illumina reads using the ea-utils FASTQ processing tool (50), and low-quality reads were filtered out using the FASTX toolkit (51). Filtered Illumina reads were mapped to the ST36 MRSA252 genome sequence (accession no. NC_002952) using the Burrows–Wheeler short-read aligner (BWA) with the Smith–Waterman alignment of unmapped mates disabled for paired end reads (52). Reads generated using the 454 platform were trimmed using the Biopython SeqIO module and mapped to MRSA252 using the BWA long-read aligner (53). Consensus sequences were called and point mutations and insertions/deletions (indels) identified for sites covered by at least 3 reads, with average mapping and PHRED scores greater than 30. Consensus genomes and whole genomes representative of the EMRSA-16 clone (MRSA252), SWP clone (TCH60, acc. no. CP002110.1), and other epidemic CC30 (MN8, acc. no. CM000952.1) were aligned using the progressiveMauve algorithm and gap positions removed (54).
Bayesian Evolutionary Analysis.
Bayesian analysis of evolutionary rates and divergence times was performed using BEAST v1.6.1 (45) under the Hasegawa–Kishino–Yano model of nucleotide substitution with a gamma correction for rate heterogeneity. The SWP and Phage 80/81 clades were constrained together on the basis of robust phylogenies determined using maximum-likelihood (Fig. S2), neighbor-joining, and maximum parsimony analyses. All isolates were dated on the basis of year and month (when known) of isolation. Before carrying out the Bayesian analysis, the dataset was assessed for temporal signal using a linear regression of root-to-tip branch length against date of isolation for the maximum likelihood phylogeny (Fig. S3). Markov chain Monte Carlo (MCMC) samples from three independent analyses each run for 1.5 × 108 iterations, sampled every 1,000 generations and the first 10% discarded as burn-in, were combined for estimation of posterior probabilities. The log-normal relaxed molecular clock model was used, with a constant coalescent prior (55). For an alignment of the UK isolates of the EMRSA-16 clone, phylogeographic distribution was examined using the discrete diffusion model (56) with distance-informed priors and using the clock rate determined for the whole dataset. Using city of isolation to construct the matrix of geographic locations resulted in an overparameterized model. Therefore, isolates were grouped by geographic region (London, southeast, south, and central England and north, east, and west Scotland), and mean distance between cities was calculated.
Supplementary Material
Acknowledgments
The authors thank R. Goering, H. de Lencastre, and A. Shore for providing S. aureus isolates and ARK-Genomics, Roslin Institute, for sequencing services. The project was supported by a grant from the Chief Scientist’s Office Scotland, a doctoral training grant from the Medical Research Council (United Kingdom), the Biotechnology Biological Sciences Research Council (United Kingdom), Grant GM080602 from the National Institute of General Medical Sciences [National Institutes of Health (NIH)], The Wellcome Trust Grant 098051, and Wellcome Trust funding (B.G.S.) This project has been funded in part with federal funds from the National Institute of Allergy and Infectious Diseases, NIH, Department of Health and Human Services, under Contract HHSN272200900018C.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the National Center for Biotechnology Information (NCBI) sequence read archive database (accession nos. SRA051389, ERA050956, and SRP000757).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1202869109/-/DCSupplemental.
References
- 1.Wertheim HFL, et al. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis. 2005;5:751–762. doi: 10.1016/S1473-3099(05)70295-4. [DOI] [PubMed] [Google Scholar]
- 2.Chambers HF, Deleo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol. 2009;7:629–641. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robinson DA, et al. Re-emergence of early pandemic Staphylococcus aureus as a community-acquired meticillin-resistant clone. Lancet. 2005;365:1256–1258. doi: 10.1016/S0140-6736(05)74814-5. [DOI] [PubMed] [Google Scholar]
- 4.Altemeier WA, et al. Staphylococcus aureus associated with toxic shock syndrome: Phage typing and toxin capability testing. Ann Intern Med. 1982;96:978–982. doi: 10.7326/0003-4819-96-6-978. [DOI] [PubMed] [Google Scholar]
- 5.Nienaber JJC, et al. International Collaboration on Endocarditis-Microbiology Investigators Methicillin-susceptible Staphylococcus aureus endocarditis isolates are associated with clonal complex 30 genotype and a distinct repertoire of enterotoxins and adhesins. J Infect Dis. 2011;204:704–713. doi: 10.1093/infdis/jir389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blair JE, Carr M. Distribution of phage groups of Staphylococcus aureus in the years 1927 through 1947. Science. 1960;132:1247–1248. doi: 10.1126/science.132.3435.1247. [DOI] [PubMed] [Google Scholar]
- 7.Nahmias A, Sakurai N, Blumberg R, Doe Ge A, Sulzer C. The Staphylococcus “80/81 complex: epidemiological and laboratory observations. J Infect Dis. 1961;109:211–222. doi: 10.1093/infdis/109.3.211. [DOI] [PubMed] [Google Scholar]
- 8.Donahue JA, Baldwin JN. Hemolysin and leukocidin production by 80/81 strains of Staphylococcus aureus. J Infect Dis. 1966;116:324–328. doi: 10.1093/infdis/116.3.324. [DOI] [PubMed] [Google Scholar]
- 9.Goldie DJ, Alder VG, Gillespie WA. Changes in the drug resistance of Staphylococcus aureus in a non-hospital population during a 20-year period. J Clin Pathol. 1971;24:44–47. doi: 10.1136/jcp.24.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collignon P, et al. Australian Group on Antimicrobial Resistance Community-acquired meticillin-resistant Staphylococcus aureus in Australia. Lancet. 1998;352:145–146. doi: 10.1016/s0140-6736(98)85051-4. [DOI] [PubMed] [Google Scholar]
- 11.Alesana-Slater J, et al. Methicillin-resistant Staphylococcus aureus, Samoa, 2007-2008. Emerg Infect Dis. 2011;17:1023–1029. doi: 10.3201/eid1706.101083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeLeo FR, et al. Molecular differentiation of historic phage-type 80/81 and contemporary epidemic Staphylococcus aureus. Proc Natl Acad Sci USA. 2011;108:18091–18096. doi: 10.1073/pnas.1111084108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho P-L, Chow K-H, Lo P-Y, Lee K-F, Lai EL. Changes in the epidemiology of methicillin-resistant Staphylococcus aureus associated with spread of the ST45 lineage in Hong Kong. Diagn Microbiol Infect Dis. 2009;64:131–137. doi: 10.1016/j.diagmicrobio.2009.01.030. [DOI] [PubMed] [Google Scholar]
- 14.Jansen van Rensburg MJ, et al. The dominant methicillin-resistant Staphylococcus aureus clone from hospitals in Cape Town has an unusual genotype: ST612. Clin Microbiol Infect. 2011;17:785–792. doi: 10.1111/j.1469-0691.2010.03373.x. [DOI] [PubMed] [Google Scholar]
- 15.Carleton HA, Diep BA, Charlebois ED, Sensabaugh GF, Perdreau-Remington F. Community-adapted methicillin-resistant Staphylococcus aureus (MRSA): Population dynamics of an expanding community reservoir of MRSA. J Infect Dis. 2004;190:1730–1738. doi: 10.1086/425019. [DOI] [PubMed] [Google Scholar]
- 16.Scicluna EA, et al. Characterisation of MRSA from Malta and the description of a Maltese epidemic MRSA strain. Eur J Clin Microbiol Infect Dis. 2010;29:163–170. doi: 10.1007/s10096-009-0834-1. [DOI] [PubMed] [Google Scholar]
- 17.Pérez-Roth E, Lorenzo-Díaz F, Batista N, Moreno A, Méndez-Alvarez S. Tracking methicillin-resistant Staphylococcus aureus clones during a 5-year period (1998 to 2002) in a Spanish hospital. J Clin Microbiol. 2004;42:4649–4656. doi: 10.1128/JCM.42.10.4649-4656.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wyllie DH, et al. Decline of meticillin-resistant Staphylococcus aureus in Oxfordshire hospitals is strain-specific and preceded infection-control intensification. BMJ Open. 2011;1:e000160. doi: 10.1136/bmjopen-2011-000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellington MJ, et al. Decline of EMRSA-16 amongst methicillin-resistant Staphylococcus aureus causing bacteraemias in the UK between 2001 and 2007. J Antimicrob Chemother. 2010;65:446–448. doi: 10.1093/jac/dkp448. [DOI] [PubMed] [Google Scholar]
- 20.Gould JC. Environmental penicillin and penicillin-resistant Staphylococcus aureus. Lancet. 1958;1:489–493. doi: 10.1016/s0140-6736(58)90807-9. [DOI] [PubMed] [Google Scholar]
- 21.Rountree PM, Asheshov EH. Further observations on changes in the phage-typing pattern of phage type 80/81 staphylococci. J Gen Microbiol. 1961;26:111–122. doi: 10.1099/00221287-26-1-111. [DOI] [PubMed] [Google Scholar]
- 22.Smyth DS, et al. Population structure of a hybrid clonal group of methicillin-resistant Staphylococcus aureus, ST239-MRSA-III. PLoS ONE. 2010;5:e8582. doi: 10.1371/journal.pone.0008582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harris SR, et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327:469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gray RR, et al. Testing spatiotemporal hypothesis of bacterial evolution using methicillin-resistant Staphylococcus aureus ST239 genome-wide data within a bayesian framework. Mol Biol Evol. 2011;28:1593–1603. doi: 10.1093/molbev/msq319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mutreja A, et al. Evidence for several waves of global transmission in the seventh cholera pandemic. Nature. 2011;477:462–465. doi: 10.1038/nature10392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Croucher NJ, et al. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331:430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hendriksen RS, et al. Population genetics of Vibrio cholerae from Nepal in 2010: Evidence on the origin of the Haitian outbreak. mBio. 2011;2:e00157–11. doi: 10.1128/mBio.00157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diep BA, et al. Polymorphonuclear leukocytes mediate Staphylococcus aureus Panton-Valentine leukocidin-induced lung inflammation and injury. Proc Natl Acad Sci USA. 2010;107:5587–5592. doi: 10.1073/pnas.0912403107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rossney AS, et al. The emergence and importation of diverse genotypes of methicillin-resistant Staphylococcus aureus (MRSA) harboring the Panton-Valentine leukocidin gene (pvl) reveal that pvl is a poor marker for community-acquired MRSA strains in Ireland. J Clin Microbiol. 2007;45:2554–2563. doi: 10.1128/JCM.00245-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Popovich KJ, Weinstein RA, Hota B. Are community-associated methicillin-resistant Staphylococcus aureus (MRSA) strains replacing traditional nosocomial MRSA strains? Clin Infect Dis. 2008;46:787–794. doi: 10.1086/528716. [DOI] [PubMed] [Google Scholar]
- 31.Ma XX, et al. Two different Panton-Valentine leukocidin phage lineages predominate in Japan. J Clin Microbiol. 2008;46:3246–3258. doi: 10.1128/JCM.00136-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diep BA, Sensabaugh GF, Somboonna N, Carleton HA, Perdreau-Remington F. Widespread skin and soft-tissue infections due to two methicillin-resistant Staphylococcus aureus strains harboring the genes for Panton-Valentine leucocidin. J Clin Microbiol. 2004;42:2080–2084. doi: 10.1128/JCM.42.5.2080-2084.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fitzgerald JR, Sturdevant DE, Mackie SM, Gill SR, Musser JM. Evolutionary genomics of Staphylococcus aureus: Insights into the origin of methicillin-resistant strains and the toxic shock syndrome epidemic. Proc Natl Acad Sci USA. 2001;98:8821–8826. doi: 10.1073/pnas.161098098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peacock SJ, et al. Virulent combinations of adhesin and toxin genes in natural populations of Staphylococcus aureus. Infect Immun. 2002;70:4987–4996. doi: 10.1128/IAI.70.9.4987-4996.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rudkin JK, et al. Methicillin resistance reduces the virulence of healthcare-associated methicillin-resistant Staphylococcus aureus by interfering with the agr quorum sensing system. J Infect Dis. 2012;205:798–806. doi: 10.1093/infdis/jir845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Collins J, et al. Offsetting virulence and antibiotic resistance costs by MRSA. ISME J. 2010;4:577–584. doi: 10.1038/ismej.2009.151. [DOI] [PubMed] [Google Scholar]
- 37.Trong HN, Prunier A-L, Leclercq R. Hypermutable and fluoroquinolone-resistant clinical isolates of Staphylococcus aureus. Antimicrob Agents Chemother. 2005;49:2098–2101. doi: 10.1128/AAC.49.5.2098-2101.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Łeski TA, Tomasz A. Role of penicillin-binding protein 2 (PBP2) in the antibiotic susceptibility and cell wall cross-linking of Staphylococcus aureus: Evidence for the cooperative functioning of PBP2, PBP4, and PBP2A. J Bacteriol. 2005;187:1815–1824. doi: 10.1128/JB.187.5.1815-1824.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Banerjee R, Gretes M, Harlem C, Basuino L, Chambers HF. A mecA-negative strain of methicillin-resistant Staphylococcus aureus with high-level β-lactam resistance contains mutations in three genes. Antimicrob Agents Chemother. 2010;54:4900–4902. doi: 10.1128/AAC.00594-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hiron A, Falord M, Valle J, Débarbouillé M, Msadek T. Bacitracin and nisin resistance in Staphylococcus aureus: A novel pathway involving the BraS/BraR two-component system (SA2417/SA2418) and both the BraD/BraE and VraD/VraE ABC transporters. Mol Microbiol. 2011;81:602–622. doi: 10.1111/j.1365-2958.2011.07735.x. [DOI] [PubMed] [Google Scholar]
- 41.Couto I, Costa SS, Viveiros M, Martins M, Amaral L. Efflux-mediated response of Staphylococcus aureus exposed to ethidium bromide. J Antimicrob Chemother. 2008;62:504–513. doi: 10.1093/jac/dkn217. [DOI] [PubMed] [Google Scholar]
- 42.Liu GY, et al. Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J Exp Med. 2005;202:209–215. doi: 10.1084/jem.20050846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Epidemic methicillin resistant Staphylococcus aureus. Commun Dis Rep CDR Wkly. 1997;7:191. [PubMed] [Google Scholar]
- 44.Beres SB, et al. Molecular complexity of successive bacterial epidemics deconvoluted by comparative pathogenomics. Proc Natl Acad Sci USA. 2010;107:4371–4376. doi: 10.1073/pnas.0911295107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barnes SL, Harris AD, Golden BL, Wasil EA, Furuno JP. Contribution of interfacility patient movement to overall methicillin-resistant Staphylococcus aureus prevalence levels. Infect Control Hosp Epidemiol. 2011;32:1073–1078. doi: 10.1086/662375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murchan S, Aucken HM, O’neill GL, Ganner M, Cookson BD. Emergence, spread, and characterization of phage variants of epidemic methicillin-resistant Staphylococcus aureus 16 in England and Wales. J Clin Microbiol. 2004;42:5154–5160. doi: 10.1128/JCM.42.11.5154-5160.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McAdam PR, Holmes A, Templeton KE, Fitzgerald JR. Adaptive evolution of Staphylococcus aureus during chronic endobronchial infection of a cystic fibrosis patient. PLoS ONE. 2011;6:e24301. doi: 10.1371/journal.pone.0024301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomas JC, Godfrey PA, Feldgarden M, Robinson DA. Candidate targets of balancing selection in the genome of Staphylococcus aureus. Mol Biol Evol. 2012;29:1175–1186. doi: 10.1093/molbev/msr286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aronesty E. 2011. ea-utils. Available at http://code.google.com/p/ea-utils/. Accessed May 16, 2011.
- 51.Gordon A. 2010. FASTX-toolkit. Available at http://hannonlab.cshl.edu/fastx_toolkit/index.html. Accessed Nov 18, 2010.
- 52.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Darling AE, Mau B, Perna NT. progressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE. 2010;5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lemey P, Rambaut A, Drummond AJ, Suchard MA. Bayesian phylogeography finds its roots. PLOS Comput Biol. 2009;5:e1000520. doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.