Abstract
The Notch signaling pathway is a key determinant in keratinocyte differentiation and growth cycle arrest, and has been reported to have a tumor suppressor function in skin. The papillomavirus life cycle is intricately linked to the differentiation status of keratinocytes. Papillomaviruses are associated with benign proliferative epithelial lesions in their respective hosts. Although human papillomaviruses (HPVs) associated with genital tract lesions have been extensively studied, studies of the cutaneous HPVs are more limited. In particular, it is well established that the E6 proteins of high-risk HPVs of the α-genus such as HPV16 and HPV18 mediate the degradation of p53 by its association with the ubiquitin ligase E6AP. In contrast, less is known about the cellular activities of the cutaneous HPVs of the β-genus. By using an unbiased proteomic approach, we identify MAML1 and other members of the Notch transcription complex as high-confidence cellular interacting proteins of E6 proteins of the β-genus HPVs and of the bovine papillomavirus type 1 associated with cutaneous fibropapillomas. We show that bovine papillomavirus type 1 and β-HPV E6 repress Notch transcriptional activation, and that this repression is dependent on an interaction with MAML1. Finally, we show that the expression levels of endogenous Notch target genes are repressed by β-HPV E6 proteins. These findings elucidate a mechanism of viral antagonism of Notch signaling, and suggest that Notch signaling is an important epithelial cell pathway target for the β-HPVs.
Keywords: Notch1, proteomics, skin cancer
Papillomaviruses infect a variety of mammalian hosts in a species-specific manner and have been associated with a number of epithelial malignancies in their respective hosts (1). The bovine papillomavirus type 1 (BPV-1) induces the formation of cutaneous fibropapillomas and has been a useful tool for studying the molecular biology of these viruses. In humans, more than 120 different HPVs have been described, and the most frequently studied have been those of the α-genus that are associated with mucosal genital tract and upper airway lesions. The low risk α-HPV types, such as HPV6 and HPV11, cause benign genital warts, whereas the high-risk α-HPV types such as HPV16 and HPV18 cause lesions that progress to cancer (1). In contrast, the β-genus HPVs cause cutaneous nongenital skin lesions. This group of viruses was first described in cancers and precancerous lesions of patients with epidermodysplasia verruciformis (2). DNA from these viruses is also found in nonmelanoma skin cancer of immunosuppressed patients and in the general population (3), as well as some normal skin samples (4). Thus, an etiological role for the β-HPV types in the cancers with which they are associated remains unclear.
The E6 oncoproteins encoded by papillomaviruses do not have any reported enzymatic activity, and exert their function by binding cellular proteins. High-risk α-HPV E6 proteins have been extensively investigated, and a major oncogenic activity involves the proteolytic degradation of p53 by complexing with the cellular ubiquitin ligase E6AP (5, 6). In addition, they have a unique PDZ-binding domain at their C terminus that binds proteins with PDZ domains and facilitates their ubiquitylation by E6AP (7, 8). In contrast, BPV-1 and β-HPV E6 proteins do not stimulate p53 degradation (9–11) and lack a PDZ-binding domain. Less is known about the cellular binding partners of E6 for BPV-1 and the β-HPV types. Hence, the activities of BPV-1 and the β-HPV E6 proteins are poorly understood at present compared with the high-risk α-HPV E6 proteins.
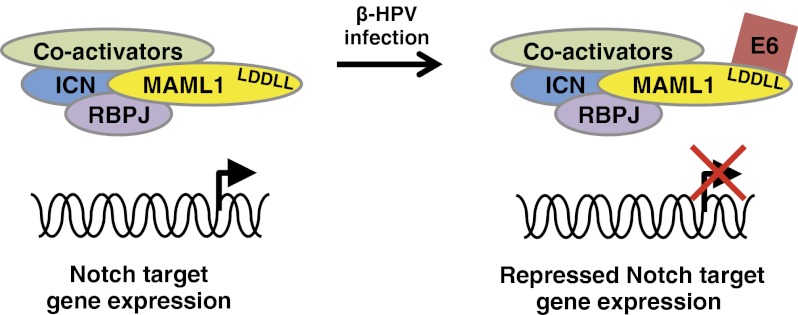
To further our understanding of the cellular activities of the E6 proteins of papillomavirus types other than the high-risk α-HPVs, we initiated studies to identify the cellular interacting proteins of E6 of the other papillomavirus types. By using an unbiased proteomic approach, we identify Mastermind-like 1 (MAML1) and other members of the Notch transcription complex as cellular interacting proteins of the E6 protein encoded by BPV-1 and the β-HPVs. MAML1 is a core component of the transcriptional activation complex that mediates the effects of the canonical Notch signaling pathway (12). We map the interaction of BPV-1 and β-HPV E6 with MAML1 to an LDDLL motif in the C-terminal acidic domain of MAML1. Finally, we show that BPV-1 and β-HPV E6 repress Notch-mediated transcriptional activation of Notch-responsive reporter genes and the expression levels of endogenous Notch genomic target genes.
The Notch signaling pathway is crucial in cell-fate determination and cell proliferation during vertebrate development (13). Notch receptors participate in a signaling pathway in which ligand binding to the extracellular domain of Notch receptors triggers a series of proteolytic cleavages that allow the intracellular domain of Notch to translocate to the nucleus, where additional coactivators such as MAML1 are recruited to activate gene expression (14).
Notch-dependent transcriptional programs are critical in the differentiation and cell cycle arrest of keratinocytes (15, 16). In addition, inactivating Notch pathway mutations have been recently reported in squamous cell carcinomas of the head and neck (17, 18) and the skin (19), suggesting that a significant role of the Notch signaling pathway as a tumor suppressor in squamous epithelial cells.
Results
BPV-1 E6 Protein Interacts with MAML1 and Components of Notch Transcription Complex.
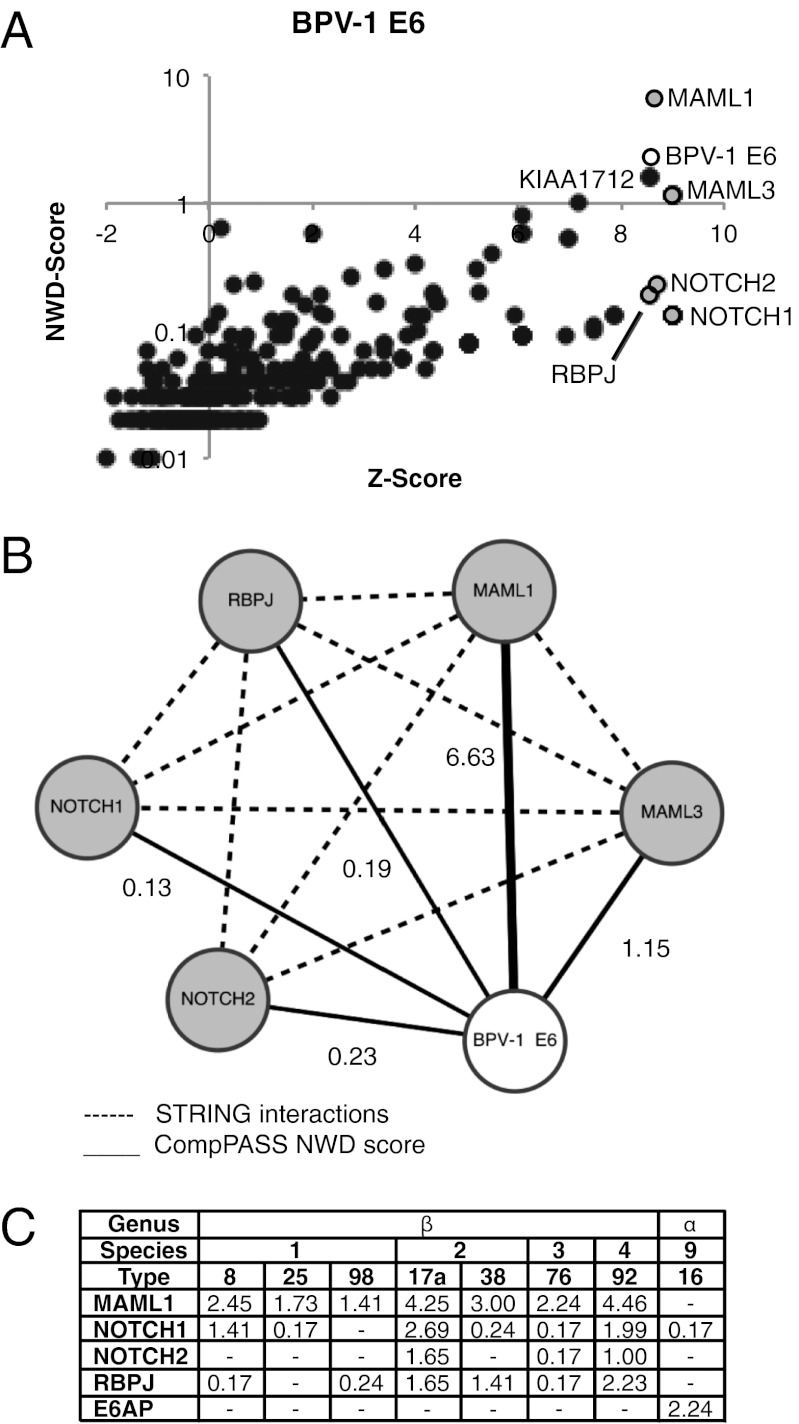
We undertook an unbiased proteomic approach to identify cellular interacting proteins of the BPV-1 E6 protein. HA-tagged BPV-1 E6 was stably expressed in 293T cells via retroviral transduction. After HA immunoprecipitation (IP) and processing of the cell lysates, E6-associated proteins were identified by liquid chromatography/tandem MS, and the data were analyzed with the proteomics software Comparative Proteomics Analysis Software Suite (CompPASS) (20). The CompPASS algorithm compares the abundance (total spectral counts), specificity, and reproducibility of interacting proteins with analogous complexes from dozens of unrelated bait proteins to assign a normalized weighted D-score (NWD score), as well as a z-score based solely on peptide abundance. Proteins with NWD scores of at least 1 are considered high-confidence candidate interacting proteins (HCIPs). Proteins with NWD lower than 1 as a result of very low total spectral counts, but that are nevertheless very specific to the bait in question, can also be identified by z-scores greater than 5, providing additional candidate interacting proteins. Among the proteins with NWD scores of at least 1 for BPV-1 E6 were components of the Notch transcriptional complexes MAML1 and MAML3 as well as a poorly characterized ORF KIAA1712 (not pursued further here; Fig. 1 A and B and Dataset S1). Additional components of the Notch transcriptional complex, including Notch1, Notch2, and RBP-J had NWD scores lower than 1 but were highly specific for BPV-1 E6 (z-score >8; Fig. 1A and Dataset S1).
Fig. 1.
MAML1 is an HCIP of BPV-1 E6 and the β-HPV E6 proteins. (A) Plot of NWD score against z-score of candidate interacting proteins of BPV-1 E6. The z-score reflects the abundance of the protein whereas the NWD score is calculated by CompPASS software and takes into account its uniqueness and reproducibility. (B) Interaction map of BPV-1 E6 with components of the Notch transcription complex identified by CompPASS. The thickness of the solid lines is proportional to the indicated NWD score of that interactor determined through the CompPASS software. Dashed lines denote known interactions from the STRING database. The NWD score for each interactor is as labeled. In this analysis, an HCIP has an NWD score of at least 1. (C) NWD scores of MAML1, other components of the Notch transcription complex, and E6AP identified by CompPASS for different HPV E6 proteins. The genus and species of each E6 protein is indicated.
MAML1 Is Also an Interactor of Cutaneous β-HPV E6 Proteins.
Our laboratory has also initiated a systematic analysis of the cellular interactors of several key proteins encoded by a diverse group of HPVs (21). As part of this ongoing analysis, the binding partners of an array of HA-tagged papillomavirus E6 proteins have been identified in low-passage N/Tert-1 human keratinocytes stably expressing these proteins. A preliminary analysis of the data for the HPV E6 proteins identified MAML1 as an HCIP for seven β-HPV E6s (Fig. 1C and Dataset S2). Other members of the Notch transcription complex were also identified as interactors in this data set. In contrast, the ubiquitin ligase E6AP was not identified as an interactor of any of the β-HPV E6s but was identified as an HCIP for the α-HPV16 E6 protein, consistent with the known interaction between E6-AP and HPV16 E6 (Fig. 1C and Dataset S2). Interestingly, MAML1 was not detected in complex with the α-genus HPV16 E6 (Fig. 1C and Dataset S2), although it should be noted that a previous study has reported an interaction between HPV16 E6 and MAML1 in a yeast two-hybrid screen (12).
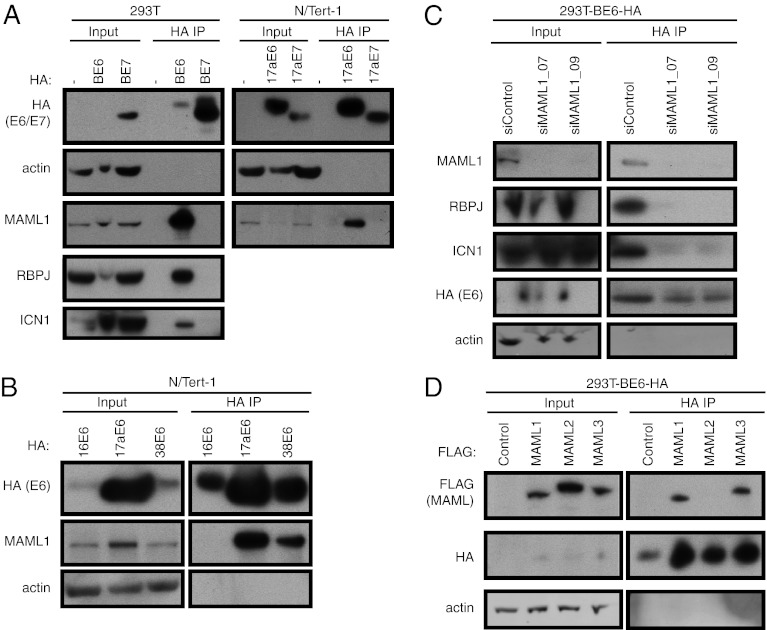
To confirm the interaction of MAML1 with BPV-1 and β-HPV E6, we used 293T cells stably expressing HA-tagged BPV-1 E6 or E7 proteins and N/Tert-1 cells stably expressing HA-tagged HPV17a E6 or E7 proteins to perform HA IP followed by Western blotting with an antibody against endogenous MAML1. We observed specific binding of endogenous MAML1 to the E6 proteins from BPV-1 and the β-genus HPV17a, but not to the E7 proteins (Fig. 2A). In addition, we confirmed the interaction of BPV-1 E6 with two other members of the Notch transcription complex identified in our MS dataset, intracellular Notch1 (ICN1) and RBPJ (Fig. 2A). An interaction with MAML1 was also observed for the β-genus HPV38 E6 protein, but not for the α-genus HPV16 E6 protein (Fig. 2B). These results confirmed that MAML1 is a specific interactor of BPV-1 and β-genus HPV E6 proteins, but not an interactor of the α-genus HPV16 E6.
Fig. 2.
MAML1 and MAML3 interact with BPV-1 and β-HPV E6 proteins. (A) MAML1 interacts with BPV-1 and the β-HPV17a E6. Lysates from 293T cells and 293T cells stably expressing BPV-1 E6 (BE6) and BPV-1 E7 (BE7; Left) were subjected to HA IP and blotted for HA, MAML1, ICN1, RBPJ, and actin. Right: Lysates from N/Tert-1 cells and N/Tert-1 cells stably expressing HPV17a E6 and E7 were similarly subjected to HA IP and blotted for HA, MAML1, and actin. In each experiment, 5% of cell lysate used for IP is loaded as input and 50% of the IP samples is loaded as IP. (B) MAML1 interacts with the β-HPV (HPV17a and HPV38) E6 proteins but not with the α-HPV16 E6 protein. Lysates from N/Tert-1 cells stably expressing the indicated HPV E6 proteins were subjected to HA IP and blotted as in A. (C) BPV-1 E6 does not interact with ICN1 and RBPJ in the absence of MAML1. Lysates from stable 293T-BE6-HA cells transfected with control or MAML1-specific siRNAs were subjected to HA IP and blotted for HA, MAML1, ICN1, RBPJ, and actin. (D) BPV-1 E6 interacts with MAML1 and MAML3, but not MAML2. Lysates from stable 293T-BE6-HA cells transfected with FLAG-tagged MAML1, MAML2, and MAML3 were subjected to HA IP and blotted for HA, FLAG, and actin.
Considering the high NWD score for MAML1 and the relatively lower NWD scores for the other Notch transcription complex components in the BPV-1 E6 CompPASS analysis, we speculated that E6 directly bound MAML1, which in turn mediated the interaction with the other Notch transcription complex components. To test this hypothesis, we examined the binding of E6 to MAML1, RBPJ, and Notch1 in 293T-BE6-HA cells in which MAML1 had been knocked down with siRNAs. Using two different siRNAs directed to MAML1 that each effectively reduced MAML1 levels, we observed reduced binding of BPV-1 E6 to both RBPJ or ICN1 (Fig. 2C). Thus, we conclude that the interaction of E6 to the components of the Notch transcriptional complex is mediated through its interaction with MAML1.
MAML1 belongs to a family of three Notch transcriptional coactivators. MAML3, but not MAML2, was identified as an interactor of BPV-1 E6 in the IP-MS/MS experiment (Fig. 1B). To confirm the binding of BPV-1 E6 to MAML3 and to determine whether it could also bind MAML2, we examined the interactions by using transiently expressed FLAG-tagged MAML1, MAML2, and MAML3 in 293T cells stably expressing BPV-1 E6 (293T-BE6-HA). HA IP of cell lysates followed by Western blotting with FLAG and HA antibodies showed that BPV-1 E6 interacts only with MAML1 and MAML3, and not with MAML2 (Fig. 2D). In addition, in this and subsequent experiments, we observed an apparent increase in BPV-1 E6 levels in cells expressing exogenous MAML proteins that interact with BPV-1 E6.
C-Terminal Acidic Domain of MAML1 Is Necessary for Its Interaction with BPV-1 and β-HPV E6 Proteins.
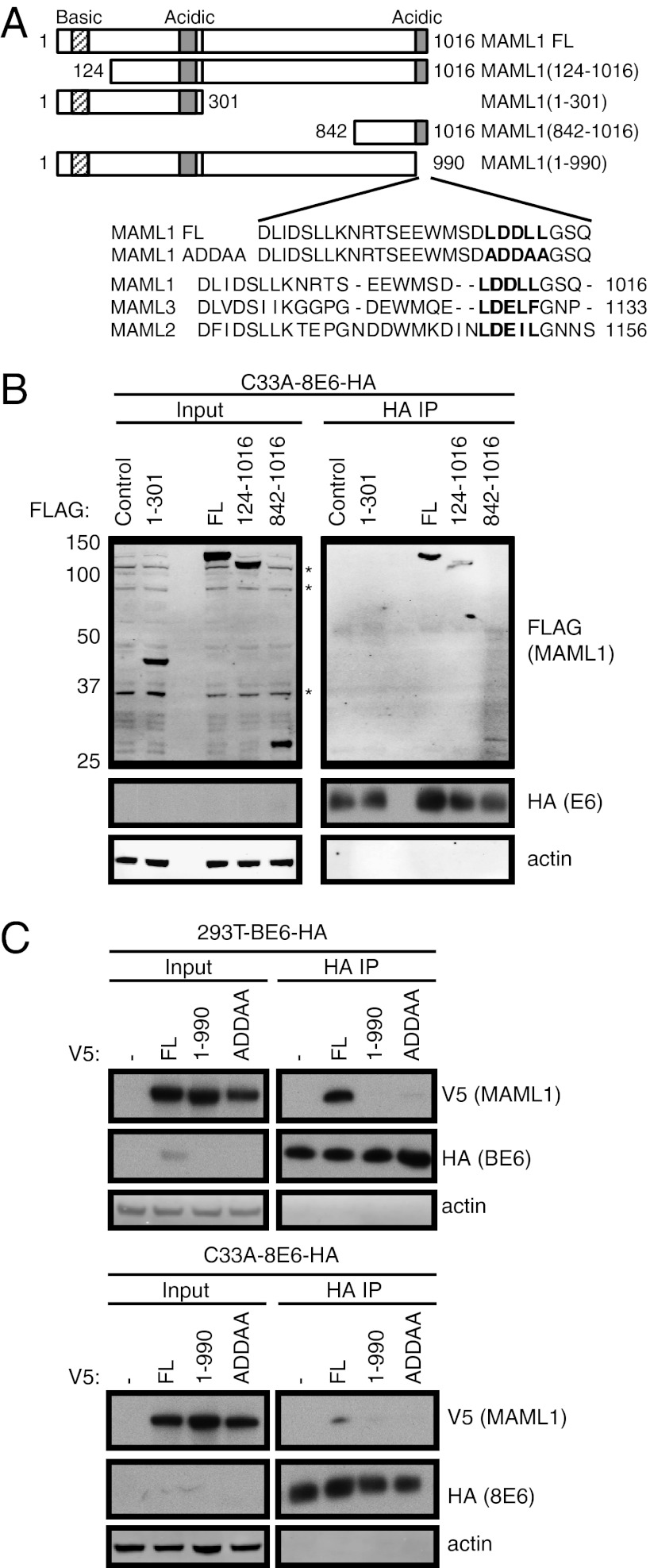
To further characterize the interaction between MAML1 and the papillomavirus E6 proteins, we mapped the domain on MAML1 that mediates its interaction with BPV-1 E6. We expressed various FLAG-tagged full-length (FL) MAML1 or MAML1 fragments (Fig. 3A, Upper) in C33A-8E6-HA cells, and performed HA IP of the cell lysates followed by Western blotting with FLAG and HA antibodies. We found that that a C-terminal fragment of MAML1, MAML1(842-1016), is sufficient for its interaction with β-HPV8 E6 (Fig. 3B). An examination of the amino acid sequence of this fragment of MAML1 revealed an acidic domain with an LXXLL motif. Such acidic α-helical motifs have been shown to be important for interacting with papillomavirus E6 proteins (22, 23), and are found in other E6-binding proteins such as E6AP, paxillin, and IRF3 (24–26). To test whether this acidic domain was necessary for binding to the BPV-1 and β-HPV8 E6 proteins, a truncation mutant of MAML1 missing this acidic domain, MAML1(1-990) was generated (Fig. 3A, Lower). When transiently expressed in 293T-BE6-HA cells or C33A-8E6-HA cells, this MAML1(1-990) mutant was unable to bind BPV-1 or HPV8 E6 protein (Fig. 3C). These results demonstrate that the C-terminal acidic domain of MAML1 is necessary for its interaction with BPV-1 and β-HPV E6. To further elucidate the motif necessary for the interaction, we constructed a MAML1 mutant (MAML1-ADDAA) with leucine-to-alanine mutations in the LDDLL motif (Fig. 3A, Lower). This mutant was found to be deficient in binding BPV-1 and HPV8 E6 proteins (Fig. 3C). Taken together, these results indicate that the LDDLL motif in the C-terminal acidic domain of MAML1 is necessary for its interaction with BPV-1 and the β-HPV E6 proteins.
Fig. 3.
The C-terminal LDDLL motif of MAML1 is necessary for its interaction with E6. (A) Schematic of the N-terminally tagged FLAG- or V5-tagged MAML1 constructs, together with an alignment of the C-termini of MAML1, MAML2, and MAML3. (B) A C-terminal fragment (842–1016) of MAML1 is sufficient for its interaction with BPV-1 E6. 293T-BE6-HA cell lysates transiently expressing the indicated FLAG-tagged fragments of MAML1 were subjected to HA IP and blotted for HA, FLAG, and actin. (C) C-terminal LDDLL motif is necessary for interaction of MAML1 with BPV-1 and the β-HPV8 E6. Lysates from stable 293T-BE6-HA or C33A-8E6-HA cells expressing the indicated V5-tagged MAML1 plasmids were subjected to HA IP and blotted for HA, V5, and actin.
An examination of the C-terminal acidic domains of the MAML family of proteins (Fig. 3A, Lower) reveals a closer conservation for MAML1 and MAML3 than with MAML2, with an additional two amino acid residues proximal to the motif that might alter the structure of this domain MAML2 and perhaps explain its inability to bind BPV-1 E6 as observed in Fig. 2D.
BPV-1 and β-HPV E6 Repress Notch Transcriptional Activation.
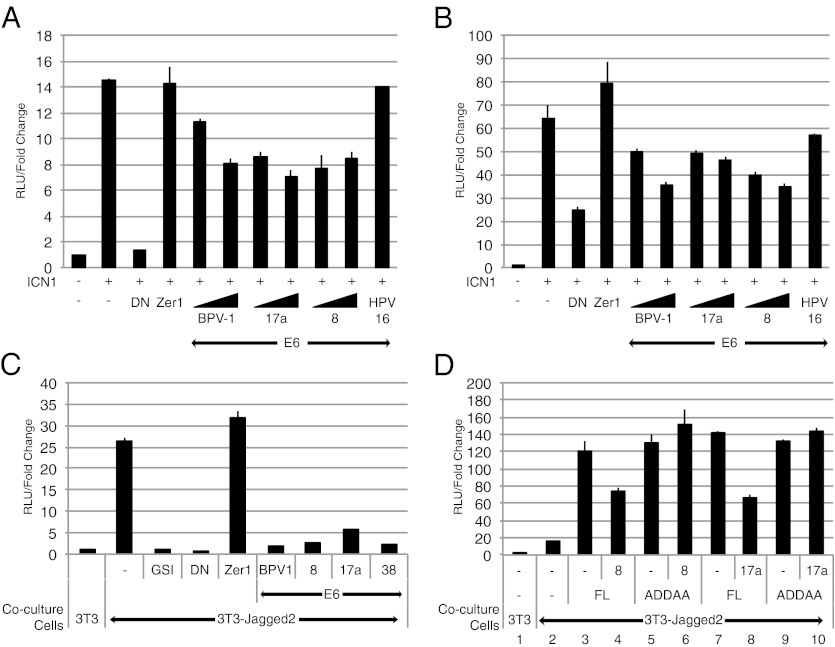
MAML1 functions as a transcriptional coactivator in the Notch signaling pathway (12). We postulated that E6 might affect Notch signaling through its interaction with MAML1. We tested whether the E6 proteins from various papillomavirus types affected Notch transcriptional activation using a Notch-responsive luciferase plasmid HES1-luc. C33A (Fig. 4A) and U2OS (Fig. 4B) cells were cotransfected with the HES1-luc reporter plasmid and the control Renilla luciferase plasmid pRL-tk-luc, along with different E6 expression plasmids. Notch-dependent transcription was activated by the coexpression of ICN1. The total amount of DNA transfected for each experiment condition was normalized by the addition of the appropriate amount of noncoding control plasmid. Luciferase activity was measured 48 h after transfection, and the firefly luciferase readings were normalized to their respective Renilla luciferase readings. The fold change in reporter activity of each experiment condition relative to the nonactivated control is depicted in Fig. 4. BPV-1 E6 and two different β-HPV E6 proteins (HPV8 and HPV17a) exhibited a dose-dependent repression of ICN1-mediated Notch transcriptional activation in C33A and U2OS cells (Fig. 4 A and B). In contrast, the expression of an unrelated control cellular protein Zer1 had no effect on Notch transcription activation. In addition, the expression, even at the highest dose of input plasmid, of α-HPV16 E6 did not repress the activation of transcription from the reporter plasmid. To further validate this finding, we performed additional studies in which we activated Notch signaling by using its ligand Jagged2, which presumably yields a more physiologic level of activated Notch. After cotransfecting U2OS cells with the Notch-responsive TP1-luc and pRL-tk-luc, the cells were cocultured with 3T3 or 3T3-Jagged2 cells (27). 3T3-Jagged2 cells stably express the Notch ligand Jagged2 and activate Notch signaling by binding the Notch receptors on the surface of the transfected U2OS cells (Fig. 4C). Ligand-induced Notch signaling can be abrogated by addition of γ-secretase inhibitors (GSI) such as compound E or the coexpression of the dominant negative (DN) MAML1 fragment MAML1(13-74) (28), which retains the N-terminal ICN/RBPJ interaction domain but lacks the C-terminal transactivation domain (TAD) of MAML1 (12). Like GSI and DN MAML1, coexpression of BPV-1 E6 protein or the E6 proteins from three different β-HPVs (HPV8, HPV17a, and HPV38) led to a strong repression of Notch transcription (Fig. 4C).
Fig. 4.
β-HPV E6 repression of Notch transcriptional activation requires their interaction with MAML1. (A and B) BPV-1 E6 and β-HPV (HPV8 and HPV17a17a) E6 repress ICN1-mediated transcriptional activation in a dose-dependent manner. C33A (A) or U2OS (B) cells were cotransfected with the Notch-responsive firefly luciferase plasmid (300 ng), a control Renilla luciferase plasmid (6 ng), a plasmid encoding activated Notch1 ICN1 (50 ng), and plasmids encoding the following proteins: BPV-1 or β-HPV E6 (300 ng or 500 ng); Zer1 or HPV16 E6 (500 ng); or DN MAML1, a specific inhibitor of the Notch transcription complex (50 ng). The luciferase activity is reported as relative luciferase units (RLUs), as calculated by normalizing the firefly luciferase reading with its corresponding internal Renilla luciferase control. The luciferase activity is expressed as fold activation relative to cells transfected only with empty control plasmid. (C) BPV-1 and β-HPV (i.e., HPVs 8, 17a, and 38) E6 proteins repress Jagged2-mediated transcriptional activation of a Notch reporter gene. U2OS cells were cotransfected with a Notch-responsive firefly luciferase reporter plasmid and a control Renilla luciferase plasmid, and 300 ng of the indicated plasmids (except for 50 ng in the case of DN MAML1). Transfected U2OS cells were then plated onto control 3T3 or Notch-activating 3T3-Jagged2 cells 24 h after transfection. A GSI, compound E, was used at a 1-μM concentration as an additional Notch pathway inhibitor control in the indicated cocultures. Dual-luciferase assays were then performed on the lysates of cocultured cells 48 h after transfection. The relative luciferase activity was calculated as in A. (D) β-HPV (HPV8 and HPV17a) E6 repression of MAML1-mediated Notch transcriptional activation is dependent on interaction with MAML1. U2OS cells were cotransfected with luciferase reporter plasmids as in C, and 16 ng of full-length MAML1 (FL), 50 ng of the non–E6-binding mutant MAML1(ADDAA), or 300 ng of HPV8 or HPV17a E6 were cotransfected into the U2OS cells as indicated. Dual-luciferase assays were then performed on the lysates of cocultured cells 48 h after transfection. The luciferase activity was calculated as in A.
β-HPV E6-Mediated Repression of Notch-Dependent Transcriptional Activation Requires Their Interaction with MAML1.
To investigate whether the repression of Notch activity produced by the different β-HPV E6 proteins depends on its interaction with MAML1, we assayed FL MAML1 and the non–E6-binding MAML1-ADDAA mutant in U2OS cells by using the 3T3-Jagged2 coculture luciferase assay. Ligand-mediated Notch signaling was enhanced approximately sevenfold by FL MAML1 or MAML1-ADDAA, indicating that the LDDLL domain of MAML1 is not critical for MAML1’s transcriptional activation function (Fig. 4D, lanes 3, 5, 7, and 9 vs, lane 2). HPV8 E6 or HPV17a E6 repressed MAML1-mediated transcriptional activation (Fig. 4D, lanes 4 and 8). In contrast, the β-HPV E6 proteins do not repress MAML1-ADDAA–mediated transcriptional activation (Fig. 4D, lanes 6 and 10), indicating that the mutation of the β-HPV E6 binding site on MAML1 rescues it from E6-mediated repression. This result shows that the interaction between MAML1 and the β-HPV E6 proteins is required for β-HPV E6–mediated repression of Notch transcriptional activation.
β-HPV E6 Represses Expression and Activation of Notch Target Genes.
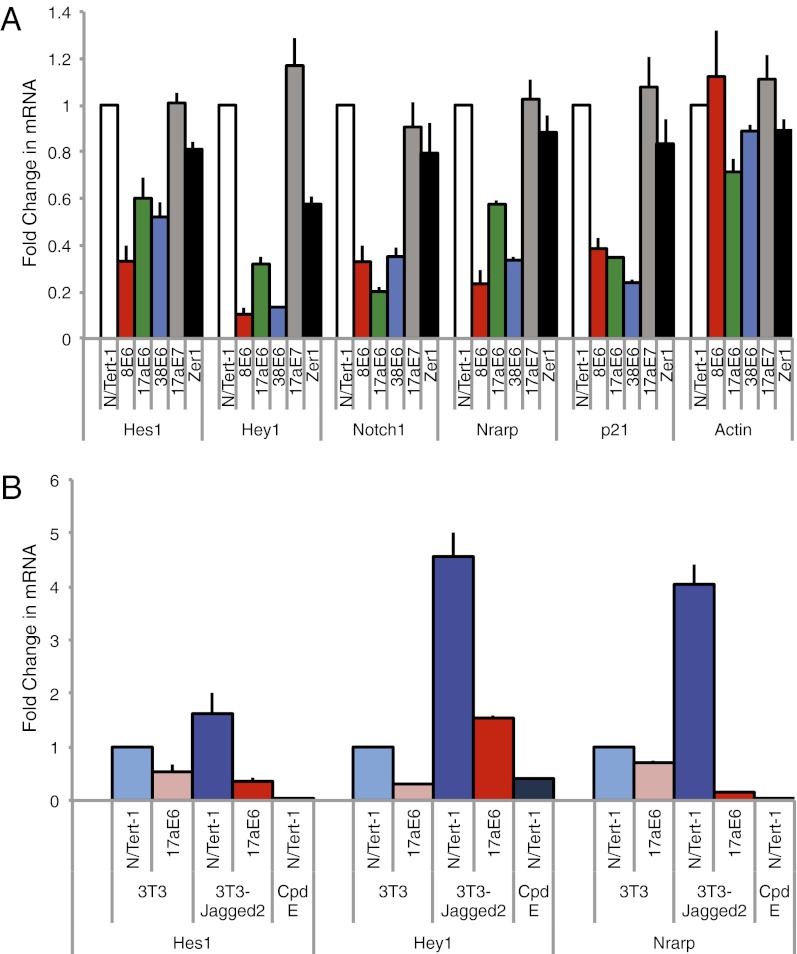
The luciferase reporter assay results suggested that β-HPV E6 proteins should also repress the expression and activation of endogenous Notch target genes. To test the effect of β-HPV E6 proteins on the basal expression of Notch target genes, we assessed levels of Notch target gene expression in N/Tert-1 cells stably expressing a panel of E6 proteins from different β-HPV types (HPV8, HPV17a, or HPV38). As controls, we used N/Tert-1 cells stably expression HPV17aE7 or the cellular protein Zer1. As predicted, the N/Tert-1 cells expressing the various β-HPV E6 proteins had two- to fivefold lower mRNA levels of the Notch-responsive genes HES1, HEY1, NOTCH1, and NRARP, compared with control N/Tert-1 cells (Fig. 5A). In contrast, the control N/Tert-1 cells expressing HPV17aE7 and Zer1 had similar levels of expression of these genes to the control N/Tert-1 cells.
Fig. 5.
β-HPV E6 represses expression of endogenous Notch-responsive genes. (A) Endogenous basal Notch-responsive gene expression is repressed by β-HPV E6s. qRT-PCR analysis of Notch-responsive gene expression in the indicated stable N/Tert-1 cell lines. Reverse transcription was performed, followed by qRT-PCR. Transcript levels were normalized to the housekeeping gene Rad21 and then presented relative to levels in the control N/Tert-1 sample. (B) Notch-responsive gene expression is repressed by HPV17a E6 even in the presence of ligand stimulation of Notch signaling. The N/Tert-1 cells were cocultured with control 3T3 cells or 3T3-Jagged2 cells for 48 h before harvesting for RNA extraction. qRT-PCR analysis of Notch-responsive gene expression was performed and analyzed as in A.
Notch activation has previously been reported to induce p21 expression and cell cycle arrest in keratinocytes (16). We observed a three- to fivefold lower levels of p21 mRNA in N/Tert-1 cells expressing the various β-HPV E6 proteins. To test whether the β-HPV E6 proteins are able to repress Jagged2-mediated activation of Notch target gene expression, we compared the levels of Notch target gene expression in control N/Tert-1 cells with N/Tert-1 cells stably expression HPV17a E6 in the presence and absence of Notch ligand stimulation. As expected, we observed an activation of the Notch target genes (HES1, HEY1, and NRARP) when the control N/Tert-1 cells were stimulated by coculture with 3T3-Jagged2 cells (Fig. 5B). In addition, we observed repression of Notch target gene expression in N/Tert-1 cells stably expressing HPV17a E6, even in the presence of Notch stimulation by Jagged2. These results demonstrate that β-HPV E6 proteins are specifically disrupting Notch target gene expression as part of their cellular activity.
Discussion
The papillomavirus E6 proteins do not have any known intrinsic enzymatic activity, and hence exert their cellular functions by binding host proteins. Although the oncogenic activity of high-risk α-genus HPV E6 proteins have been extensively investigated, less is known about the cellular activities of the BPV-1 or the β-genus HPV E6 proteins.
To further our understanding of the cellular activities of BPV-1 and β-genus HPV E6 proteins, we undertook an unbiased proteomic screen to identify their unique cellular interactors. Through this approach, we have identified MAML1 and other components of the Notch transcription complex as interacting proteins of BPV-1 and β-HPV E6 (Fig. 1). While we were preparing this manuscript for submission, a manuscript from Brimer et al. was published that also independently showed that BPV-1 and HPV8 E6 proteins associate with MAML1 and affect Notch signaling (29). Our study establishes that binding to the MAML coactivators is required for inhibiting Notch signaling in keratinocytes and that this is a general property of the HPV E6 proteins of the β-genus and not shared by the HPV E6 proteins of the α-genus.
MAML1 is a core component of the canonical Notch signaling pathway (12). The N-terminal basic domain of MAML1 interacts with the ankyrin repeats of the intracellular domain of Notch receptors and the DNA-binding factor RBPJ (also known as CSL) (30, 31). MAML1 also contains two TADs that are believed to be involved in the recruitment of additional cellular factors contributing to target gene activation. Forms of MAML1 that lack these C-terminal domains act as DN proteins and induce cutaneous squamous cell carcinomas in mice (32), implicating the Notch/RBPJ/MAML1 complex in suppression of squamous carcinogenesis.
Mapping the interaction between the papillomavirus E6 proteins and MAML1 revealed the importance of the C-terminal acidic domain of MAML1 for its interaction with papillomavirus E6 proteins (Fig. 3). This acidic domain is part of TAD2 of MAML1, which is required for transcription in vivo and has been proposed to interact with additional unknown cellular proteins necessary for Notch transcriptional activation (33). In addition, we show that an LDDLL motif in the acidic domain is necessary for its interaction with papillomavirus E6 proteins.
The Notch signaling pathway plays important roles in cell-fate determination and cell proliferation during vertebrate development (13). Notch receptors participate in a signaling pathway in which ligand binding to the extracellular domain of Notch receptors triggers a series of proteolytic cleavages that allow the intracellular domain of Notch to translocate to the nucleus and activate gene expression. In mouse models, Notch can function as an oncogene or a tumor suppressor, depending on the context (34). Furthermore, whereas activating mutations in the NOTCH1 gene are frequently found in human T-cell acute lymphoblastic leukemia (35), the Notch pathway has tumor-suppressive activity in mammalian epithelial cells (32, 36). Recently, inactivating Notch pathway mutations have been reported in squamous cell carcinomas of the head and neck (17, 18) and the skin (19). Taken together, these findings suggest that the Notch signaling pathway may have a significant role as a tumor suppressor in squamous epithelial cells.
In keratinocytes, the activation of Notch signaling induces differentiation and cell cycle arrest (15, 16). Considering that the virus life cycle of the papillomaviruses are closely linked to the differentiation state of squamous epithelial cells, the Notch pathway represents a potentially important target for papillomaviruses. Indeed, HPV16 E6 has been reported to target the Notch signaling pathway by decreasing Notch1 expression levels in a p53-dependent manner (37, 38). This could lead to a downstream effect on Notch signaling, but such an effect for HPV16 has not yet been reported to our knowledge.
In contrast, neither BPV-1 E6 nor any of the β-HPV E6s induce p53 degradation, nor have they been previously reported to have an effect on Notch signaling. Here we report a repression of Notch transcriptional activation by BPV-1 and β-HPV E6 (Fig. 4). In particular, this repression of Notch signaling is dependent on their interaction with MAML1 (Fig. 4). This represents a unique mode of perturbing Notch signaling.
Finally, to look at the cellular outcome of this perturbation, we examined the expression of Notch target genes in keratinocytes expressing β-HPV E6s. Consistent with the repression of Notch transcription we observed in our luciferase assays, we found a repression of Notch target gene expression in cells expressing β-HPV E6s, even in the presence of ligand stimulation of Notch signaling (Fig. 5). A decrease in HES1, HEY1, Notch1, and Nrarp expression levels was observed in each of the β-HPV E6–expressing cell lines tested.
This work identifies the Notch signaling pathway as a target for a variety of papillomaviruses. Although the perturbation of the Notch signaling pathway via binding of viral proteins encoded by EBV and Kaposi sarcoma-associated herpesvirus to the transcription factor RBPJ has been reported (39–41), perturbation and disruption of Notch signaling by papillomavirus proteins has not been reported. In particular, the repression of Notch transcription by BPV-1 E6 and β-HPV E6s through their interaction with MAML1 represents a unique mechanism of targeting Notch signaling.
The papillomavirus life cycle is closely linked to the differentiation state of squamous epithelial cells (1), making the keratinocyte differentiation program an ideal target for viral perturbation. Indeed, the α-HPV E7 proteins have been reported to alter the differentiation program of keratinocytes via an interaction with p21 (42, 43). Considering the integral role of Notch signaling in keratinocyte differentiation (15, 16), we propose that the β-HPV E6s perturb the Notch signaling pathway to alter the keratinocyte differentiation program and maintain a conducive cellular environment for viral replication.
In light of the importance of the Notch pathway in keratinocyte differentiation and the recent discovery of additional tumor suppressor functions of the Notch pathway, our work lays the foundation to further understanding of the Notch signaling pathway and its perturbation by papillomaviruses. Although an etiologic role for the β-HPVs in the nonmelanoma skin cancers in which they have been found has not been firmly established, the identification of the Notch pathway as a target of E6 through interaction with MAML coactivators provides a potential mechanism by which this group of viruses might contribute to cancer progression in epithelial cells.
Materials and Methods
Cells and Viruses.
293T, C33A, U2OS, 3T3 and 3T3-Jagged2 (27) cells were cultured in DMEM (Invitrogen) supplemented with 10% (vol/vol) FBS (SH3008803; HyClone) and 1% penicillin-streptomycin (Gibco/Invitrogen). N/Tert-1 cells were cultured as described previously (44).
The stable 293T and N/Tert-1 cell lines used for the HA IP-MS/MS analysis were made by using retroviruses generated in 293 Phoenix cells per a standard protocol. Briefly, 293 Phoenix cells were cotransfected with the retroviral vectors encoding HA-tagged versions of the various papillomavirus E6 proteins, along with expression vectors encoding the retrovirus proteins Gag-Pol and vesicular stomatitis virus G protein. 293T or N/Tert-1 cells were then infected with the produced viruses by using a standard protocol and selected for 10 d in DMEM with 0.75 or 0.5 μg/mL puromycin, respectively.
Plasmids.
The various papillomavirus E6 and E7 ORFs were PCR-amplified from genomic plasmids for BPV-1 (45), HPV8 (46), HPV17a (47), and HPV38 (48), and cloned into retroviral expression vectors by using Gateway recombination cloning technology (Invitrogen).
The FLAG-tagged MAML constructs have been described (12), and were gifts of Lizi Wu (University of Florida, Gainesville, FL). The V5-tagged MAML1 constructs were generated by using standard mutagenesis protocols and cloned into lentiviral expression vectors by using Gateway recombination cloning technology (Invitrogen).
The Notch luciferase reporter plasmids HES1-luc (49) and TP1-luc (41) have been described. The firefly luciferase readings were normalized for transfection efficiency by cotransfection with pRL-TK (Promega) that encodes Renilla luciferase as described previously (50). The DN MAML1 and intracellular domain of Notch1 (i.e., ICN1) expression plasmids have been previously described (28).
DNA Transfection.
Cells were seeded the day before transfection to achieve 70% confluence on the day of transfection. C33A and U2OS cells were transfected with the indicated plasmids using a 3:1 FuGENE (in μL)/DNA (in μg) ratio, as recommended in the manufacturer’s protocol (Roche). 293T/Phoenix cells were transfected using TransIT-293 (Mirus) with the indicated plasmids using a 3:1 TransIT-293 reagent (in μL)/DNA (in μg) ratio, per the manufacturer’s protocol.
siRNA Transfection.
siRNAs were obtained from Dharmacon. 293T cells were seeded the day before transfection to achieve 80% confluence on the day of transfection, and transfected using DharmaFECT1, as recommended in the manufacturer’s protocol (Dharmacon/Thermo Scientific). Medium was replaced 24 h after transfection, and transfection efficiency was monitored by using siGLO Red (Dharmacon). Cells were harvested 48 h after transfection for IP experiments.
Protein IP.
HA IP for MS and CompPASS analysis were performed on 293T or N/Tert-1 cells stably expressing the respective HA-tagged proteins as described previously (20). Briefly, four 15-cm plates of 90% confluent cells were harvested in lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% Nonidet P-40, 1 mM EDTA) supplemented with a protease inhibitor mixture (Roche). Cell lysates were sonicated at 30% amplitude for 8 s with a Branson sonifier, and clarified by spinning at 16,100 × g for 10 min and passing through a 0.20-μm filter (model 431218; Corning). The lysate was then incubated with anti–HA-agarose resin (A2095; Sigma) overnight at 4 °C. After five washes with lysis buffer, the resin was exchanged into PBS and eluted with 250 μg/mL HA peptide (Sigma) at room temperature. The eluted proteins were concentrated by trichloroacetic acid (Sigma) precipitation, and washed with acetone (Sigma).
HA IPs for Western blotting were performed as described earlier, except the bound proteins were eluted by boiling in sample buffer for 10 min. V5 IPs for Western blotting were performed like HA IPs but using anti-V5 agarose (A7345; Sigma).
Western Blotting.
Proteins were separated by using NuPAGE (Invitrogen) gels and transferred onto PVDF membranes. After blocking in 5% (wt/vol) milk in Tris-buffered saline solution, pH 7.4, with 0.05% Tween-20, blots were incubated with the following primary antibodies: actin (Millipore), V5 (R960-25; Invitrogen), FLAG (F7425; Sigma), MAML1 [A300-673A (Bethyl) or 4608S (Cell Signaling)], RBPJ (5442S; Cell Signaling), and Notch1 (51). HA was probed directly by using an HA-peroxidase antibody (no. 12013819001; Roche).
Bound primary antibodies were incubated with donkey anti-rabbit and sheep anti-mouse IgG horseradish peroxidase-linked antibodies (NA934 and NA931; Amersham/GE Healthcare). All proteins were detected by using Western Lightning chemiluminescent substrate and visualized on film.
MS and CompPASS Analysis.
HA IPs were performed on lysates from 293T or N/Tert-1 cells stably expressing N-terminal HA-tagged papillomavirus E6 proteins and were prepared for MS (liquid chromatography/tandem MS) as previously described (20). The data were measured using an LTQ Orbitrap Velos (Thermo Scientific). All peptide identifications were made by using Sequest, and the resulting data were analyzed by using CompPASS against a database containing IP-MS/MS data for 74 unrelated bait proteins. Interaction network analysis was performed by using the STRING databases (http://string-db.org/) with software included in CompPASS and visualized by using Cytoscape software (www.cytoscape.org).
Dual-Luciferase Reporter Assay.
U2OS or C33A cells were plated at 70% confluence in 12-well plates 1 d before transfection. The total amount of transfected plasmid DNA was kept constant by adding appropriate amount of control plasmids. In the coculture experiments, the transfected U2OS cells were replated on top of 3T3 or 3T3-Jagged2 cells 24 h after transfection in media supplemented with 1 μM of the GSI compound E (Enzo Life Sciences) or DMSO vehicle. Cells were harvested for luciferase analysis 48 h after transfection as described earlier.
Luciferase activity was determined by using the Dual-Luciferase Reporter Assay System (Promega), per the manufacturer’s protocol. Luciferase readings were measured by using the SpectraMax L Luminescence Microplate Reader (Molecular Devices). The firefly luciferase readings were normalized to relative luciferase units by using the respective Renilla luciferase readings.
Quantitative Real-Time PCR (QRT-PCR).
For quantitative real-time PCR (qRT-PCR), total RNA was isolated from the various N/Tert-1 cell lines using the NucleoSpin RNA II kit (Clontech) per the manufacturer's instructions. The concentration of each sample was determined by UV spectrophotometry (NanoDrop), and equal amounts of RNA were reverse-transcribed by using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Gene expression levels was detected with an Applied Biosystems ABI 7500 Fast Sequence Detection System using TaqMan Fast Universal PCR Master Mix (Applied Biosystems) and the TaqMan Gene Expression assays (Applied Biosystems) for HES1 (assay ID, Hs00172878_m1), HEY1 (assay ID, Hs00232618_m1), Notch1 (assay ID, Hs01062014_m1), Nrarp (assay ID, Hs01104102_s1), and Actin (assay ID, Hs99999903_m1).
The relative amounts of cDNA in each sample were calculated based on a standard curve prepared by using serial dilutions of the reference cDNA. The cDNA amount was then normalized to the housekeeping gene, Rad21 (assay ID, Hs00366726_m1). The fold change in transcription of the gene was calculated by comparison with control N/Tert-1 cells.
Supplementary Material
Acknowledgments
We thank Stephen Blacklow (Dana Farber Cancer Institute) as well as members of the P.M.H., J.W.H., J.C.A., and Blacklow laboratories for critical discussions and suggestions; Lizi Wu (University of Florida) for providing the FLAG-tagged MAML plasmids and James Rheinwald (Brigham and Women’s Hospital) for providing the N/Tert-1 cells; Ethel-Michele de Villiers (Deutsche Krebsforschungszentrum) for providing the genomic clones for HPV8 and HPV38; and Michel Favre (Institut Pasteur) for providing the genomic clone for HPV17a. This work was supported by a scholarship from the Agency of Science, Technology, and Research, Singapore (to M.J.A.T.); National Institutes of Health (NIH) National Research Service Award F32AI080075 (to E.AW.); a Roche Postdoctoral Fellowship (to E.AW.); and NIH Grants 1RC1 CA145188 (to P.M.H. and J.W.H.), R01 GM054137 (to J.W.H.), and P01 CA50661 (to P.M.H.).
Footnotes
The authors declare no conflict of interest.
See Author Summary on page 8804 (volume 109, number 23).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1205991109/-/DCSupplemental.
References
- 1.Howley PM, Lowy DR. Papillomaviruses. In: Knipe D, Howley P, editors. Fields Virology. 5th Ed. Philadelphia: Lippincott Williams and Wilkins; 2007. pp. 2299–2354. [Google Scholar]
- 2.Jablonska S, Orth G. Epidermodysplasia verruciformis. Clin Dermatol. 1985;3:83–96. doi: 10.1016/0738-081x(85)90052-5. [DOI] [PubMed] [Google Scholar]
- 3.Akgül B, Cooke JC, Storey A. HPV-associated skin disease. J Pathol. 2006;208:165–175. doi: 10.1002/path.1893. [DOI] [PubMed] [Google Scholar]
- 4.Antonsson A, et al. Prevalence and type spectrum of human papillomaviruses in healthy skin samples collected in three continents. J Gen Virol. 2003;84:1881–1886. doi: 10.1099/vir.0.18836-0. [DOI] [PubMed] [Google Scholar]
- 5.Huibregtse JM, Scheffner M, Howley PM. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 1991;10:4129–4135. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huibregtse JM, Scheffner M, Howley PM. Cloning and expression of the cDNA for E6-AP, a protein that mediates the interaction of the human papillomavirus E6 oncoprotein with p53. Mol Cell Biol. 1993;13:775–784. doi: 10.1128/mcb.13.2.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gardiol D, et al. Oncogenic human papillomavirus E6 proteins target the discs large tumour suppressor for proteasome-mediated degradation. Oncogene. 1999;18:5487–5496. doi: 10.1038/sj.onc.1202920. [DOI] [PubMed] [Google Scholar]
- 8.Nakagawa S, Huibregtse JM. Human scribble (Vartul) is targeted for ubiquitin-mediated degradation by the high-risk papillomavirus E6 proteins and the E6AP ubiquitin-protein ligase. Mol Cell Biol. 2000;20:8244–8253. doi: 10.1128/mcb.20.21.8244-8253.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper B, et al. Requirement of E6AP and the features of human papillomavirus E6 necessary to support degradation of p53. Virology. 2003;306:87–99. doi: 10.1016/s0042-6822(02)00012-0. [DOI] [PubMed] [Google Scholar]
- 10.Jackson S, Harwood C, Thomas M, Banks L, Storey A. Role of Bak in UV-induced apoptosis in skin cancer and abrogation by HPV E6 proteins. Genes Dev. 2000;14:3065–3073. doi: 10.1101/gad.182100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caldeira S, et al. p53 mutations are common in human papillomavirus type 38-positive non-melanoma skin cancers. Cancer Lett. 2004;209:119–124. doi: 10.1016/j.canlet.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 12.Wu L, et al. MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat Genet. 2000;26:484–489. doi: 10.1038/82644. [DOI] [PubMed] [Google Scholar]
- 13.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: Cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 14.Nam Y, Aster JC, Blacklow SC. Notch signaling as a therapeutic target. Curr Opin Chem Biol. 2002;6:501–509. doi: 10.1016/s1367-5931(02)00346-0. [DOI] [PubMed] [Google Scholar]
- 15.Lowell S, Jones P, Le Roux I, Dunne J, Watt FM. Stimulation of human epidermal differentiation by delta-notch signalling at the boundaries of stem-cell clusters. Curr Biol. 2000;10:491–500. doi: 10.1016/s0960-9822(00)00451-6. [DOI] [PubMed] [Google Scholar]
- 16.Rangarajan A, et al. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 2001;20:3427–3436. doi: 10.1093/emboj/20.13.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stransky N, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agrawal N, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang NJ, et al. Loss-of-function mutations in Notch receptors in cutaneous and lung squamous cell carcinoma. Proc Natl Acad Sci USA. 2011;108:17761–17766. doi: 10.1073/pnas.1114669108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White EA, et al. Systematic identification of interactions between host cell proteins and E7 oncoproteins from diverse human papillomaviruses. Proc Natl Acad Sci USA. 2012;109:E260–E267. doi: 10.1073/pnas.1116776109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen JJ, Hong Y, Rustamzadeh E, Baleja JD, Androphy EJ. Identification of an alpha helical motif sufficient for association with papillomavirus E6. J Biol Chem. 1998;273:13537–13544. doi: 10.1074/jbc.273.22.13537. [DOI] [PubMed] [Google Scholar]
- 23.Vande Pol SB, Brown MC, Turner CE. Association of bovine papillomavirus type 1 E6 oncoprotein with the focal adhesion protein paxillin through a conserved protein interaction motif. Oncogene. 1998;16:43–52. doi: 10.1038/sj.onc.1201504. [DOI] [PubMed] [Google Scholar]
- 24.Tong X, Howley PM. The bovine papillomavirus E6 oncoprotein interacts with paxillin and disrupts the actin cytoskeleton. Proc Natl Acad Sci USA. 1997;94:4412–4417. doi: 10.1073/pnas.94.9.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huibregtse JM, Scheffner M, Howley PM. Localization of the E6-AP regions that direct human papillomavirus E6 binding, association with p53, and ubiquitination of associated proteins. Mol Cell Biol. 1993;13:4918–4927. doi: 10.1128/mcb.13.8.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ronco LV, Karpova AY, Vidal M, Howley PM. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 1998;12:2061–2072. doi: 10.1101/gad.12.13.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo B, Aster JC, Hasserjian RP, Kuo F, Sklar J. Isolation and functional analysis of a cDNA for human Jagged2, a gene encoding a ligand for the Notch1 receptor. Mol Cell Biol. 1997;17:6057–6067. doi: 10.1128/mcb.17.10.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weng AP, et al. Growth suppression of pre-T acute lymphoblastic leukemia cells by inhibition of notch signaling. Mol Cell Biol. 2003;23:655–664. doi: 10.1128/MCB.23.2.655-664.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brimer N, Lyons C, Wallberg AE, Vande Pol SB. Cutaneous papillomavirus E6 oncoproteins associate with MAML1 to repress transactivation and NOTCH signaling. Oncogene. 2012 doi: 10.1038/onc.2011.589. 31, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nam Y, Sliz P, Song L, Aster JC, Blacklow SC. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell. 2006;124:973–983. doi: 10.1016/j.cell.2005.12.037. [DOI] [PubMed] [Google Scholar]
- 31.Nam Y, Weng AP, Aster JC, Blacklow SC. Structural requirements for assembly of the CSL.intracellular Notch1.Mastermind-like 1 transcriptional activation complex. J Biol Chem. 2003;278:21232–21239. doi: 10.1074/jbc.M301567200. [DOI] [PubMed] [Google Scholar]
- 32.Proweller A, et al. Impaired notch signaling promotes de novo squamous cell carcinoma formation. Cancer Res. 2006;66:7438–7444. doi: 10.1158/0008-5472.CAN-06-0793. [DOI] [PubMed] [Google Scholar]
- 33.McElhinny AS, Li JL, Wu L. Mastermind-like transcriptional co-activators: emerging roles in regulating cross talk among multiple signaling pathways. Oncogene. 2008;27:5138–5147. doi: 10.1038/onc.2008.228. [DOI] [PubMed] [Google Scholar]
- 34.Radtke F, Raj K. The role of Notch in tumorigenesis: Oncogene or tumour suppressor? Nat Rev Cancer. 2003;3:756–767. doi: 10.1038/nrc1186. [DOI] [PubMed] [Google Scholar]
- 35.Weng AP, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 36.Nicolas M, et al. Notch1 functions as a tumor suppressor in mouse skin. Nat Genet. 2003;33:416–421. doi: 10.1038/ng1099. [DOI] [PubMed] [Google Scholar]
- 37.Lefort K, et al. Notch1 is a p53 target gene involved in human keratinocyte tumor suppression through negative regulation of ROCK1/2 and MRCKalpha kinases. Genes Dev. 2007;21:562–577. doi: 10.1101/gad.1484707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yugawa T, et al. Regulation of Notch1 gene expression by p53 in epithelial cells. Mol Cell Biol. 2007;27:3732–3742. doi: 10.1128/MCB.02119-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang Y, Chang J, Lynch SJ, Lukac DM, Ganem D. The lytic switch protein of KSHV activates gene expression via functional interaction with RBP-Jkappa (CSL), the target of the Notch signaling pathway. Genes Dev. 2002;16:1977–1989. doi: 10.1101/gad.996502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grossman SR, Johannsen E, Tong X, Yalamanchili R, Kieff E. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the J kappa recombination signal binding protein. Proc Natl Acad Sci USA. 1994;91:7568–7572. doi: 10.1073/pnas.91.16.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zimber-Strobl U, et al. Epstein-Barr virus nuclear antigen 2 exerts its transactivating function through interaction with recombination signal binding protein RBP-J kappa, the homologue of Drosophila Suppressor of Hairless. EMBO J. 1994;13:4973–4982. doi: 10.1002/j.1460-2075.1994.tb06824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones DL, Alani RM, Münger K. The human papillomavirus E7 oncoprotein can uncouple cellular differentiation and proliferation in human keratinocytes by abrogating p21Cip1-mediated inhibition of cdk2. Genes Dev. 1997;11:2101–2111. doi: 10.1101/gad.11.16.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Funk JO, et al. Inhibition of CDK activity and PCNA-dependent DNA replication by p21 is blocked by interaction with the HPV-16 E7 oncoprotein. Genes Dev. 1997;11:2090–2100. doi: 10.1101/gad.11.16.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dickson MA, et al. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol Cell Biol. 2000;20:1436–1447. doi: 10.1128/mcb.20.4.1436-1447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heilman CA, Engel L, Lowy DR, Howley PM. Virus-specific transcription in bovine papillomavirus-transformed mouse cells. Virology. 1982;119(1):22–34. doi: 10.1016/0042-6822(82)90061-7. [DOI] [PubMed] [Google Scholar]
- 46.Pfister H, Nürnberger F, Gissmann L, zur Hausen H. Characterization of a human papillomavirus from epidermodysplasia verruciformis lesions of a patient from Upper-volta. Int J Cancer. 1981;27:645–650. doi: 10.1002/ijc.2910270511. [DOI] [PubMed] [Google Scholar]
- 47.Kremsdorf D, et al. Molecular cloning and characterization of the genomes of nine newly recognized human papillomavirus types associated with epidermodysplasia verruciformis. J Virol. 1984;52:1013–1018. doi: 10.1128/jvi.52.3.1013-1018.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scheurlen W, Gissmann L, Gross G, zur Hausen H. Molecular cloning of two new HPV types (HPV 37 and HPV 38) from a keratoacanthoma and a malignant melanoma. Int J Cancer. 1986;37:505–510. doi: 10.1002/ijc.2910370406. [DOI] [PubMed] [Google Scholar]
- 49.Jarriault S, et al. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 50.Aster JC, et al. Essential roles for ankyrin repeat and transactivation domains in induction of T-cell leukemia by notch1. Mol Cell Biol. 2000;20:7505–7515. doi: 10.1128/mcb.20.20.7505-7515.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hasserjian RP, Aster JC, Davi F, Weinberg DS, Sklar J. Modulated expression of notch1 during thymocyte development. Blood. 1996;88:970–976. [PubMed] [Google Scholar]