Abstract
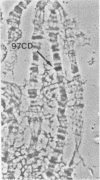
The Tetrahymena histone H2A variant designated hv1 is localized exclusively in the transcriptionally active macronucleus and is absent from the quiescent micronucleus (1). A cDNA clone of the hv1 gene (2) was used to screen a Drosophila cDNA library. A cross-hybridizing clone was recovered and shown by sequence analysis to code for a protein homologous to hv1 as well as to the chicken H2A variant, H2A.F (3), the sea urchin H2A variant, H2A.F/Z (4) and the mammalian H2A variant H2A.Z (5). Southern analysis of Drosophila genomic DNA indicates that the H2AvD (H2A variant Drosophila) gene is present in one copy. In situ hybridization places the locus at 97CD on chromosome 3, while the S-phase regulated histone genes are on chromosome 2 (6). Thus the Drosophila H2A variant should be accessible to genetic analysis, which will enable its function to be determined.
Full text
PDF


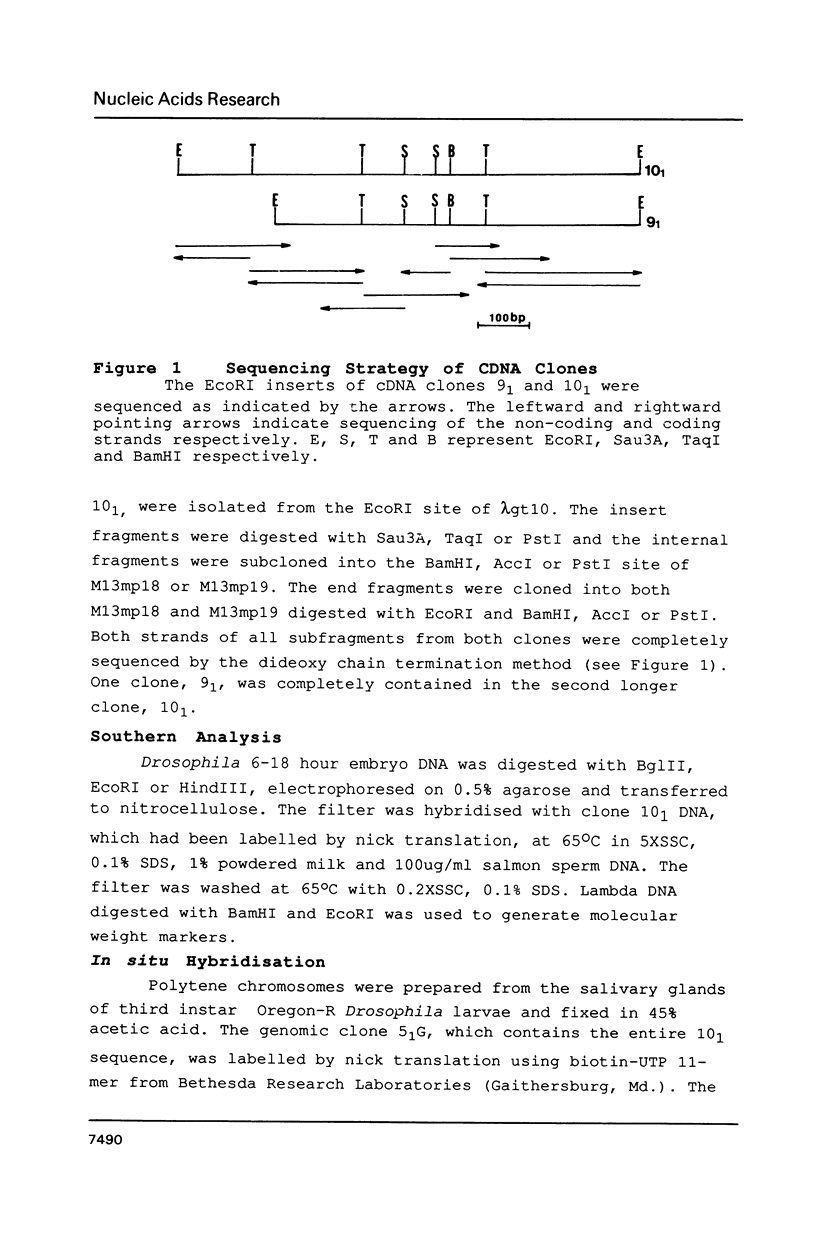







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allis C. D., Glover C. V., Bowen J. K., Gorovsky M. A. Histone variants specific to the transcriptionally active, amitotically dividing macronucleus of the unicellular eucaryote, Tetrahymena thermophila. Cell. 1980 Jul;20(3):609–617. doi: 10.1016/0092-8674(80)90307-4. [DOI] [PubMed] [Google Scholar]
- Allis C. D., Richman R., Gorovsky M. A., Ziegler Y. S., Touchstone B., Bradley W. A., Cook R. G. hv1 is an evolutionarily conserved H2A variant that is preferentially associated with active genes. J Biol Chem. 1986 Feb 5;261(4):1941–1948. [PubMed] [Google Scholar]
- Brush D., Dodgson J. B., Choi O. R., Stevens P. W., Engel J. D. Replacement variant histone genes contain intervening sequences. Mol Cell Biol. 1985 Jun;5(6):1307–1317. doi: 10.1128/mcb.5.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst S. G., Miller H., Brenner C. A., Nocente-McGrath C., Francis S., McIsaac R. Characterization of a cDNA clone coding for a sea urchin histone H2A variant related to the H2A.F/Z histone protein in vertebrates. Nucleic Acids Res. 1987 Jun 11;15(11):4629–4644. doi: 10.1093/nar/15.11.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey R. P., Whiting J. A., Coles L. S., Krieg P. A., Wells J. R. H2A.F: an extremely variant histone H2A sequence expressed in the chicken embryo. Proc Natl Acad Sci U S A. 1983 May;80(10):2819–2823. doi: 10.1073/pnas.80.10.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch C. L., Bonner W. M. Sequence of cDNAs for mammalian H2A.Z, an evolutionarily diverged but highly conserved basal histone H2A isoprotein species. Nucleic Acids Res. 1988 Feb 11;16(3):1113–1124. doi: 10.1093/nar/16.3.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Hardison R. C., Lacy E., Lauer J., O'Connell C., Quon D., Sim G. K., Efstratiadis A. The isolation of structural genes from libraries of eucaryotic DNA. Cell. 1978 Oct;15(2):687–701. doi: 10.1016/0092-8674(78)90036-3. [DOI] [PubMed] [Google Scholar]
- Palmer D., Snyder L. A., Blumenfeld M. Drosophila nucleosomes contain an unusual histone-like protein. Proc Natl Acad Sci U S A. 1980 May;77(5):2671–2675. doi: 10.1073/pnas.77.5.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardue M. L., Kedes L. H., Weinberg E. S., Birnstiel M. L. Localization of sequences coding for histone messenger RNA in the chromosomes of Drosophila melanogaster. Chromosoma. 1977 Aug 25;63(2):135–151. doi: 10.1007/BF00292726. [DOI] [PubMed] [Google Scholar]
- Poole S. J., Kauvar L. M., Drees B., Kornberg T. The engrailed locus of Drosophila: structural analysis of an embryonic transcript. Cell. 1985 Jan;40(1):37–43. doi: 10.1016/0092-8674(85)90306-x. [DOI] [PubMed] [Google Scholar]
- Richmond T. J., Finch J. T., Rushton B., Rhodes D., Klug A. Structure of the nucleosome core particle at 7 A resolution. Nature. 1984 Oct 11;311(5986):532–537. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells D., Kedes L. Structure of a human histone cDNA: evidence that basally expressed histone genes have intervening sequences and encode polyadenylylated mRNAs. Proc Natl Acad Sci U S A. 1985 May;82(9):2834–2838. doi: 10.1073/pnas.82.9.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E. M., Shapiro D. L., Allis C. D., Gorovsky M. A. Sequence and properties of the message encoding Tetrahymena hv1, a highly evolutionarily conserved histone H2A variant that is associated with active genes. Nucleic Acids Res. 1988 Jan 11;16(1):179–198. doi: 10.1093/nar/16.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R. S., Bonner W. M. Separation of basal histone synthesis from S-phase histone synthesis in dividing cells. Cell. 1981 Dec;27(2 Pt 1):321–330. doi: 10.1016/0092-8674(81)90415-3. [DOI] [PubMed] [Google Scholar]