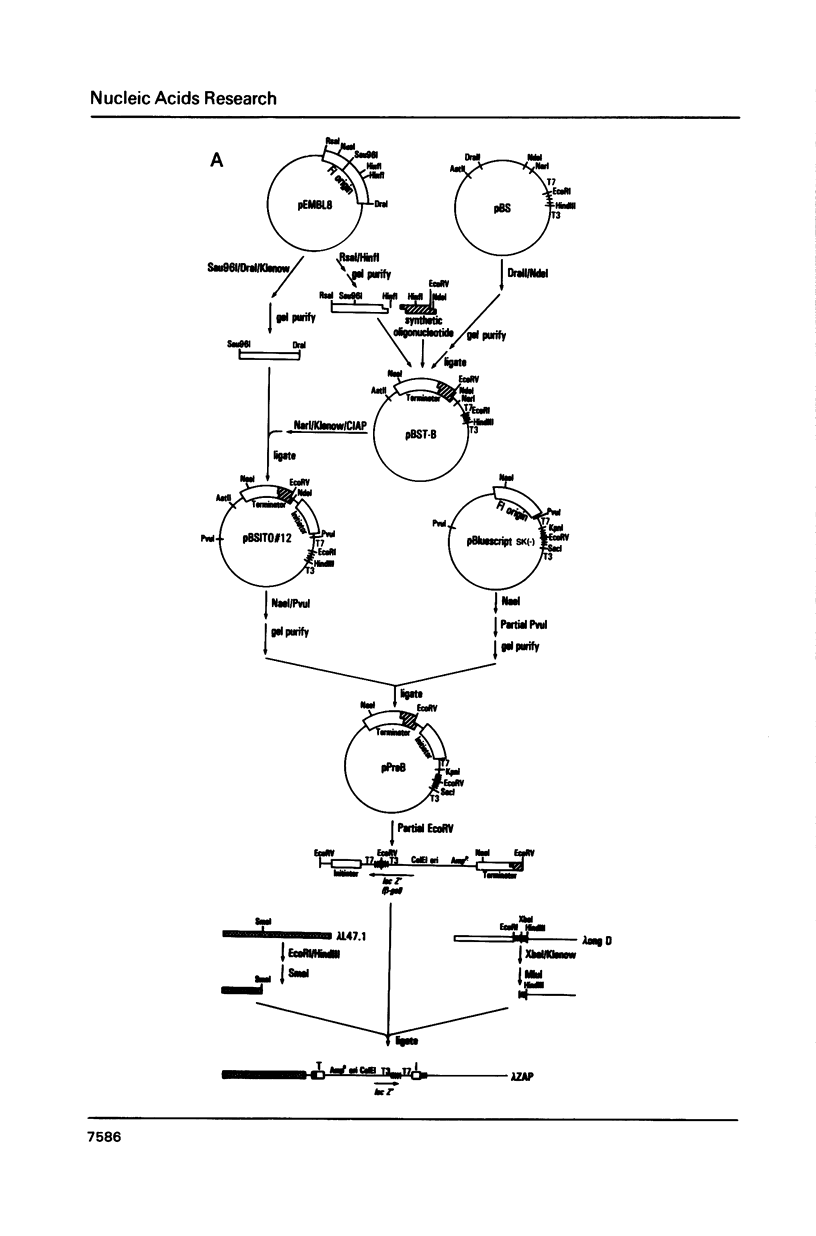
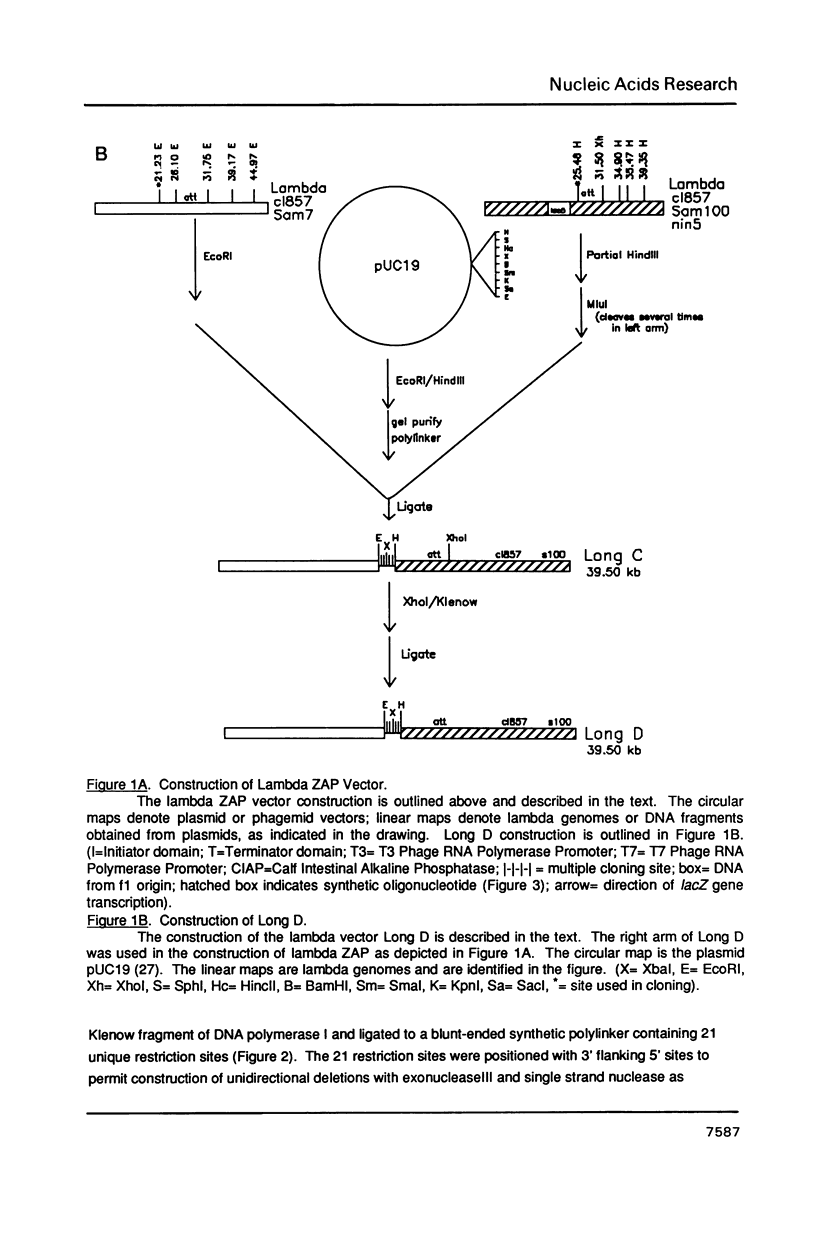
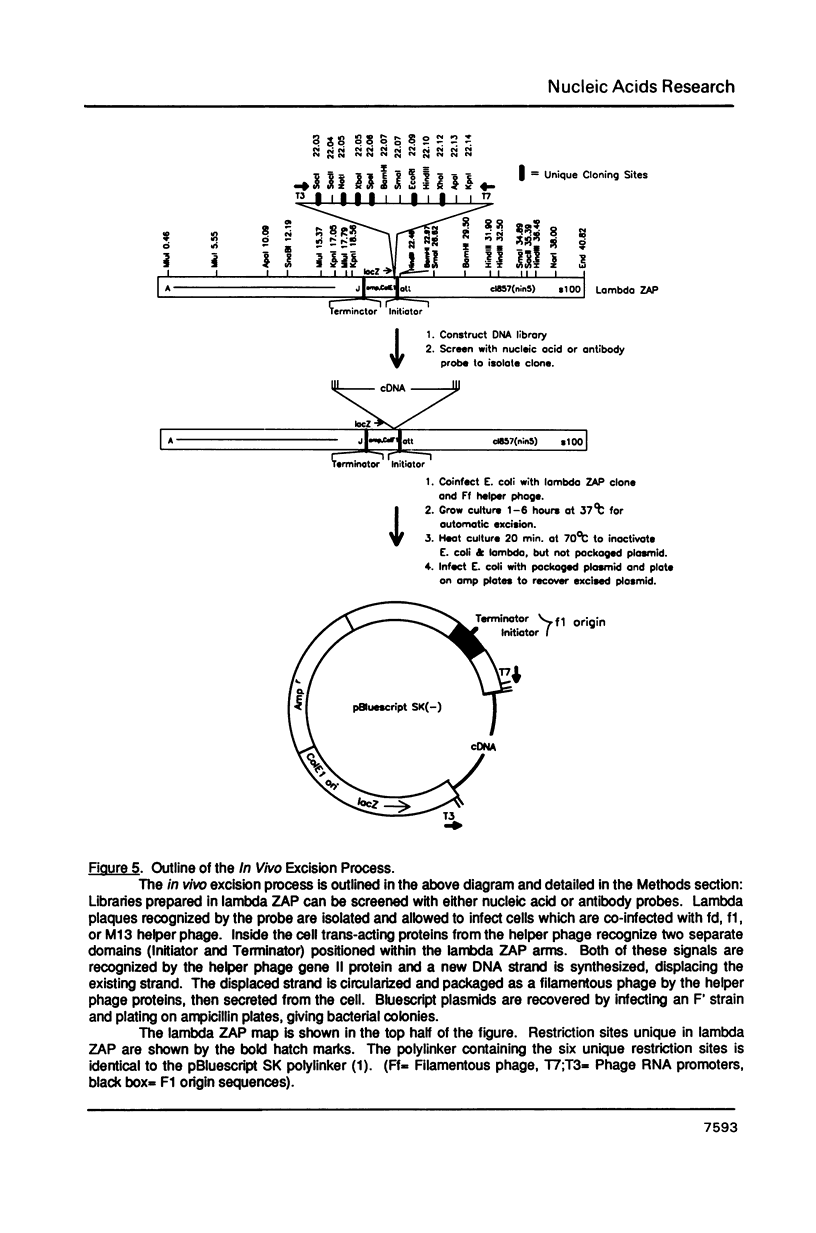
Abstract
A lambda insertion type cDNA cloning vector, Lambda ZAP, has been constructed. In E. coli a phagemid, pBluescript SK(-), contained within the vector, can be excised by f1 or M13 helper phage. The excision process eliminates the need to subclone DNA inserts from the lambda phage into a plasmid by restriction digestion and ligation. This is possible because Lambda ZAP incorporates the signals for both initiation and termination of DNA synthesis from the f1 bacteriophage origin of replication (1). Six of 21 restriction sites in the excised pBluescript SK polylinker, contained within the NH2-portion of the lacZ gene, are unique in lambda ZAP. Coding sequences inserted into these restriction sites, in the appropriate reading frame, can be expressed from the lacZ promoter as fusion proteins. The features of this vector significantly increase the rate at which clones can be isolated and analyzed. The lambda ZAP vector was tested by the preparation of a chicken liver cDNA library and the isolation of actin clones by screening with oligonucleotide probes. Putative actin clones were excised from the lambda vector and identified by DNA sequencing. The ability of lambda ZAP to serve as a vector for the construction of cDNA expression libraries was determined by detecting fusion proteins from clones containing glucocerbrosidase cDNA's using rabbit IgG anti-glucocerbrosidase antibodies.
Full text
PDF













Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bailey J. N., Klement J. F., McAllister W. T. Relationship between promoter structure and template specificities exhibited by the bacteriophage T3 and T7 RNA polymerases. Proc Natl Acad Sci U S A. 1983 May;80(10):2814–2818. doi: 10.1073/pnas.80.10.2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck E., Zink B. Nucleotide sequence and genome organisation of filamentous bacteriophages fl and fd. Gene. 1981 Dec;16(1-3):35–58. doi: 10.1016/0378-1119(81)90059-7. [DOI] [PubMed] [Google Scholar]
- Brenner S., Cesareni G., Karn J. Phasmids: hybrids between ColE1 plasmids and E. coli bacteriophage lambda. Gene. 1982 Jan;17(1):27–44. doi: 10.1016/0378-1119(82)90098-1. [DOI] [PubMed] [Google Scholar]
- Chang K. S., Zimmer W. E., Jr, Bergsma D. J., Dodgson J. B., Schwartz R. J. Isolation and characterization of six different chicken actin genes. Mol Cell Biol. 1984 Nov;4(11):2498–2508. doi: 10.1128/mcb.4.11.2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary J. M., Ray D. S. Replication of the plasmid pBR322 under the control of a cloned replication origin from the single-stranded DNA phage M13. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4638–4642. doi: 10.1073/pnas.77.8.4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dente L., Cesareni G., Cortese R. pEMBL: a new family of single stranded plasmids. Nucleic Acids Res. 1983 Mar 25;11(6):1645–1655. doi: 10.1093/nar/11.6.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotto G. P., Enea V., Zinder N. D. Functional analysis of bacteriophage f1 intergenic region. Virology. 1981 Oct 30;114(2):463–473. doi: 10.1016/0042-6822(81)90226-9. [DOI] [PubMed] [Google Scholar]
- Dotto G. P., Horiuchi K., Jakes K. S., Zinder N. D. Replication origin of bacteriophage f1. Two signals required for its function. J Mol Biol. 1982 Dec 5;162(2):335–343. doi: 10.1016/0022-2836(82)90530-7. [DOI] [PubMed] [Google Scholar]
- Dotto G. P., Horiuchi K. Replication of a plasmid containing two origins of bacteriophage. J Mol Biol. 1981 Nov 25;153(1):169–176. doi: 10.1016/0022-2836(81)90532-5. [DOI] [PubMed] [Google Scholar]
- Dotto G. P., Horiuchi K., Zinder N. D. Initiation and termination of phage f1 plus-strand synthesis. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7122–7126. doi: 10.1073/pnas.79.23.7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotto G. P., Horiuchi K., Zinder N. D. The functional origin of bacteriophage f1 DNA replication. Its signals and domains. J Mol Biol. 1984 Feb 5;172(4):507–521. doi: 10.1016/s0022-2836(84)80020-0. [DOI] [PubMed] [Google Scholar]
- Enea V., Zinder N. D. Interference resistant mutants of phage f1. Virology. 1982 Oct 15;122(1):222–226. doi: 10.1016/0042-6822(82)90395-6. [DOI] [PubMed] [Google Scholar]
- Geider K., Beck E., Schaller H. An RNA transcribed from DNA at the origin of phage fd single strand to replicative form conversion. Proc Natl Acad Sci U S A. 1978 Feb;75(2):645–649. doi: 10.1073/pnas.75.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geider K., Hohmeyer C., Haas R., Meyer T. F. A plasmid cloning system utilizing replication and packaging functions of the filamentous bacteriophage fd. Gene. 1985;33(3):341–349. doi: 10.1016/0378-1119(85)90242-2. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Horiuchi K. Interaction between gene II protein and the DNA replication origin of bacteriophage f1. J Mol Biol. 1986 Mar 20;188(2):215–223. doi: 10.1016/0022-2836(86)90306-2. [DOI] [PubMed] [Google Scholar]
- Horiuchi K. Origin of DNA replication of bacteriophage f1 as the signal for termination. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5226–5229. doi: 10.1073/pnas.77.9.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost T. A., Theodorakis N., Hughes S. H. The nucleotide sequence of the chick cytoplasmic beta-actin gene. Nucleic Acids Res. 1983 Dec 10;11(23):8287–8301. doi: 10.1093/nar/11.23.8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson A., Silver D., Seed B. Minimal size plasmids containing an M13 origin for production of single-strand transducing particles. J Mol Appl Genet. 1984;2(6):507–517. [PubMed] [Google Scholar]
- Loenen W. A., Brammar W. J. A bacteriophage lambda vector for cloning large DNA fragments made with several restriction enzymes. Gene. 1980 Aug;10(3):249–259. doi: 10.1016/0378-1119(80)90054-2. [DOI] [PubMed] [Google Scholar]
- McAllister W. T., Carter A. D. Regulation of promoter selection by the bacteriophage T7 RNA polymerase in vitro. Nucleic Acids Res. 1980 Oct 24;8(20):4821–4837. doi: 10.1093/nar/8.20.4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer T. F., Geider K. Bacteriophage fd gene II-protein. II. Specific cleavage and relaxation of supercoiled RF from filamentous phages. J Biol Chem. 1979 Dec 25;254(24):12642–12646. [PubMed] [Google Scholar]
- Meyer T. F., Geider K., Kurz C., Schaller H. Cleavage site of bacteriophage fd gene II-protein in the origin of viral strand replication. Nature. 1979 Mar 22;278(5702):365–367. doi: 10.1038/278365a0. [DOI] [PubMed] [Google Scholar]
- Morris C. E., Klement J. F., McAllister W. T. Cloning and expression of the bacteriophage T3 RNA polymerase gene. Gene. 1986;41(2-3):193–200. doi: 10.1016/0378-1119(86)90098-3. [DOI] [PubMed] [Google Scholar]
- Poustka A., Pohl T. M., Barlow D. P., Frischauf A. M., Lehrach H. Construction and use of human chromosome jumping libraries from NotI-digested DNA. Nature. 1987 Jan 22;325(6102):353–355. doi: 10.1038/325353a0. [DOI] [PubMed] [Google Scholar]
- Roychoudhury R., Wu R. Novel properties of Escherichia coli exonuclease III. J Biol Chem. 1977 Jul 25;252(14):4786–4789. [PubMed] [Google Scholar]
- Russel M., Kidd S., Kelley M. R. An improved filamentous helper phage for generating single-stranded plasmid DNA. Gene. 1986;45(3):333–338. doi: 10.1016/0378-1119(86)90032-6. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen P., Huang H. V. Homologous recombination in Escherichia coli: dependence on substrate length and homology. Genetics. 1986 Mar;112(3):441–457. doi: 10.1093/genetics/112.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorge J., West C., Westwood B., Beutler E. Molecular cloning and nucleotide sequence of human glucocerebrosidase cDNA. Proc Natl Acad Sci U S A. 1985 Nov;82(21):7289–7293. doi: 10.1073/pnas.82.21.7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt V. M., Ingles C. J., Urdea M. S., Rutter W. J. Homology requirements for recombination in Escherichia coli. Proc Natl Acad Sci U S A. 1985 Jul;82(14):4768–4772. doi: 10.1073/pnas.82.14.4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K. R., Alberts B. M., Benzinger R., Lawhorne L., Treiber G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology. 1970 Mar;40(3):734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Young R. A., Davis R. W. Efficient isolation of genes by using antibody probes. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1194–1198. doi: 10.1073/pnas.80.5.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]