Abstract
From September 2008 to December 2010, 112 Haemophilus parasuis strains were isolated from 536 pigs with clinical signs of Glässer’s disease in South China, for a frequency of 21%. The 112 strains were subjected to serovar analysis by gel diffusion (GD) and indirect hemagglutination (IHA) tests and to genotype analysis by means of pulsed-field gel electrophoresis (PFGE). With a combination of the GD and IHA results, serovars 5 and 4 were found to be the most prevalent, at 23% and 17%, respectively, followed by serovars 2 (8%), 15 (7%), 13 (6%), and 12 (5%); 20% of the strains were nontypeable. The 112 strains were genetically diverse, with 85 genotypes identified (discriminatory index 0.992). The 89 typeable isolates belonged to 15 H. parasuis serovars displaying 63 different PFGE profiles. The 23 nontypeable strains displayed 22 different PFGE profiles. These findings confirmed that 15 serovars and diverse genotypes of H. parasuis were widely distributed in southern China.
Résumé
Entre septembre 2008 et décembre 2010, 112 isolats d’Haemophilus parasuis ont été obtenus à partir de 536 porcs (21 %) provenant du sud de la Chine et présentant des signes cliniques de maladie de Glässer. Les 112 isolats ont été sérotypés par diffusion en gel (GD) et hémagglutination indirecte (IHA) et génotypés par électrophorèse en champs pulsés (PFGE). En combinant les résultats de GD et d’IHA, les sérovars 5 et 4 sont apparus comme les plus prévalents, représentant respectivement 23 % et 17 % des isolats, suivis des sérovars 2 (8 %), 15 (7 %), 13 (6 %), et 12 (5 %); 20 % des isolats étaient non-typables. Les 112 isolats étaient génétiquement diversifiés, avec 85 génotypes identifiés (index de discrimination de 0,992). Les 89 isolats typables appartenaient à 15 sérovars d’H. parasuis et présentaient 63 profiles différent par PFGE. Les 23 isolats non-typables présentaient 22 profiles différents par PFGE. Ces résultats confirment que 15 sérovars et divers génotypes d’H. parasuis sont largement distribués dans le sud de la Chine.
(Traduit par Docteur Serge Messier)
Haemophilus parasuis is a gram-negative bacillus and the causative agent of Glässer’s disease, which affects swine worldwide and is characterized by fibrinous polyserositis, polyarthritis, or meningitis (1,2). Recently H. parasuis was described as one of the most infectious agents affecting swine populations in the world (3).
Traditionally, strains of H. parasuis have been classified by serotyping (4), and most epidemiologic studies of H. parasuis infection have been based on serotyping information (5–8). The currently available H. parasuis vaccines are based on certain serovars. Therefore, knowledge of serovar distribution is essential in evaluating the possible benefits of vaccination, although cross-protection between some serovars of H. parasuis has been demonstrated (9–11). In addition, approximately 15% to 41% of field isolates of H. parasuis are untypeable by current serotyping methods (5,8,12).
To overcome the limitations of serotyping in epidemiologic studies of H. parasuis infection, many molecular-based methods have been developed to assess the heterogeneity of H. parasuis isolates by genotyping (13–17). Pulsed-field gel electrophoresis (PFGE) is a widely used and highly discriminatory molecular typing method based on comparison of patterns of fragments of chromosomal DNA generated by digestion with restriction enzymes (18). It is often considered the gold standard of genomic methods for subtyping many bacterial species owing to its high discriminatory power and reproducibility between laboratories (19–21). Also, this technique has been used to evaluate the genetic relationship between isolates suspected to be epidemiologically related (22–24). We previously developed a modified PFGE method for characterizing H. parasuis isolates (25) and used it to analyze the genetic relatedness of 20 antimicrobial-resistant strains and 6 strains positive for plasmid-mediated quinolone resistance (PMQR) (26). Therefore, PFGE is an excellent approach for epidemiologic studies of H. parasuis outbreaks in addition to being suitable for characterizing unrelated H. parasuis strains.
In the current study the serovars of 112 H. parasuis isolates from pigs in 5 provinces of southern China with clinical signs of Glässer’s disease were defined and the genotypes characterized by means of PFGE. In addition, the serotyping and molecular subtyping approaches were compared.
All the field isolates (Table I) were derived in our diagnostic laboratory between September 2008 and December 2010 from lung, heart, brain, nasal, and synovial samples from the thorax, pericardial sac, peritoneum, and joints of 536 pigs that had presented with fibrinous polyserositis, arthritis, or meningitis characteristic of Glässer’s disease. The samples were cultured on tryptic soy agar (TSA; Difco Laboratories, Detroit, Michigan, USA) containing 10 mg/mL of nicotinamide adenine dinucleotide and 5% bovine serum. The isolates were identified by biochemical tests (27) and diagnostic analysis of fragments of the 16S ribosomal RNA gene amplified by polymerase chain reaction (28). Used as positive controls were 15 H. parasuis strains representing all known serovars kindly provided by Professor Huanchun Chen, College of Veterinary Medicine, Huazhong Agricultural University, Wuhan, Hubei Province, China.
Table I.
Source of the 112 isolates of Haemophilus parasuis from pigs in southern China with clinical signs of Glässer’s disease
| Province and district | Number of isolates | Isolate name | Clinical source |
|---|---|---|---|
| Guangdong | |||
| TaiShan City | 4 | SC001–SC004 | Lung, liver, nose |
| Meizhou City | 7 | SC005–SC011 | Lung, synovium, nose |
| Sanshui City | 10 | SC0012–SC0021 | Lung, synovium, brain, heart |
| Shenzhen City | 7 | SC0067–SC0073 | Lung, synovium, brain |
| Zhongshan City | 6 | SC0074–SC0079 | Lung, heart, nose |
| Sihui City | 7 | SC0080–SC0086 | Lung, synovium |
| Guangzhou City | 7 | SC0027–SC0033 | Lung, synovium |
| Huizhou City | 6 | SC0045, SC0049–SC0053 | Lung, synovium |
| Heyuan City | 2 | SC0091, SC0092 | Lung, brain |
| Jiangmen City | 2 | SC0106, SC0107 | Lung, synovium |
| Yunfu City | 5 | SC0096–SC0100 | Lung, synovium, heart |
| Zengcheng City | 1 | SC0116 | Lung |
| Dongguan City | 2 | SC0022, SC0023 | Lung, brain |
| Hunan | |||
| Changsha City | 3 | SC0024–SC0026 | Lung, synovium, brain, heart, nose |
| Hengyang City | 4 | SC0037–SC0040 | Lung, synovium, heart |
| Chenzhou City | 7 | SC0054–SC0057, SC0034–SC0036 | Lung, synovium, heart, nose |
| Zhuzhou City | 4 | SC0058–SC0061 | Lung, synovium, heart |
| Jiangxi | |||
| Nanchang City | 4 | SC0041–SC0044 | Lung, synovium, heart |
| Jiujiang City | 3 | SC0046–SC0048 | Lung, synovium, brain |
| Yingtan City | 3 | SC0064–SC0066 | Lung, synovium |
| Longyan City | 2 | SC0062, SC0063 | Lung |
| Guangxi | |||
| Nanning City | 4 | SC0087–SC0090 | Lung, synovium, brain, heart |
| Baise City | 3 | SC0093–SC0095 | Lung, synovium, brain, nose |
| Fujian | |||
| Fuzhou City | 5 | SC00101–SC0105 | Lung, synovium, heart, nose |
| Longyan City | 4 | SC0108, SC0110, SC0111, SC0114 | Lung, synovium |
For serotyping, hyperimmune antiserum reactive to H. parasuis was generated in rabbits as previously described (8), with use of H. parasuis serovar strains 1 to 15. Adsorbed antiserum was then used, in combination with indirect hemagglutination (IHA) testing, to serotype the field isolates (29). Autoclaved antigen for the gel diffusion (GD) test was prepared as described by Turni and Blackall (30) except that the agar used was TSA. For the IHA test, saline extracts were used as the antigen source (29). The GD and IHA tests were conducted as previously described (30), the GD test being done first; if a definitive serovar could not be determined for an isolate, the IHA test was then done. Isolates that could not be identified by either test were defined as nontypeable (NT).
For genotyping, the PFGE method (25) was used. Briefly, agarose blocks containing H. parasuis DNA were digested with 40 units of CpoI (TaKaRa, Dalian, Liaoning Province, China) for 2 h at 30°C. Digested slices were then electrophoresed in a CHEF-Mapper system (Bio-Rad Laboratories, Hercules, California, USA) in 1% SeaKem Gold Agarose (FMC, Bioproducts, Rockland, ME, USA) in 0.5 × Tris-borate and ethylene diamine tetraacetic acid as the running buffer at 14°C and 6 V/cm for 21 h, the pulse time ramping up from 2.16 to 63.8 s. Salmonella enterica serovar Braenderup H9812 standards served as size standards after digestion with Xba I (TaKaRa).
The banding patterns were analyzed with Molecular Analyst Fingerprinting Plus software (Bio-Rad) with 1.5% tolerance for fragment shifts. Similarities between banding patterns corresponding to respective strains were determined by means of the Dice coefficient (Dice coefficient according to the manufacturer’s instructions.). All digestion patterns were evaluated visually by generation of a dendrogram. The discriminatory ability of the 2 typing methods was determined with the use of Simpson’s index of diversity (31).
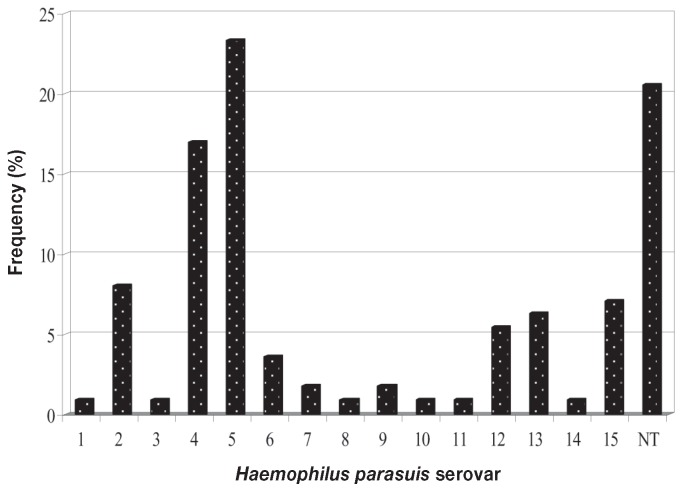
The serovar distribution of the 112 H. parasuis isolates, determined by combining the GD and IHA data, is shown in Figure 1; 23 (20%) of the isolates could not be assigned to a serovar. All 15 serovars were identified. Serovars 5 and 4 were the most prevalent, accounting for 23% and 17%, respectively. Serovars 2, 15, 13, and 12 were next most common, with frequencies of 8%, 7%, 6%, and 5%, respectively. Serovars 1, 3, 6, 7, 8, 9, 10, 11, and 14 were also identified but in low numbers.
Figure 1.
Percentage of each serovar among 112 Haemophilus parasuis isolates from clinical samples collected from pigs with Glässer’s disease in southern China.
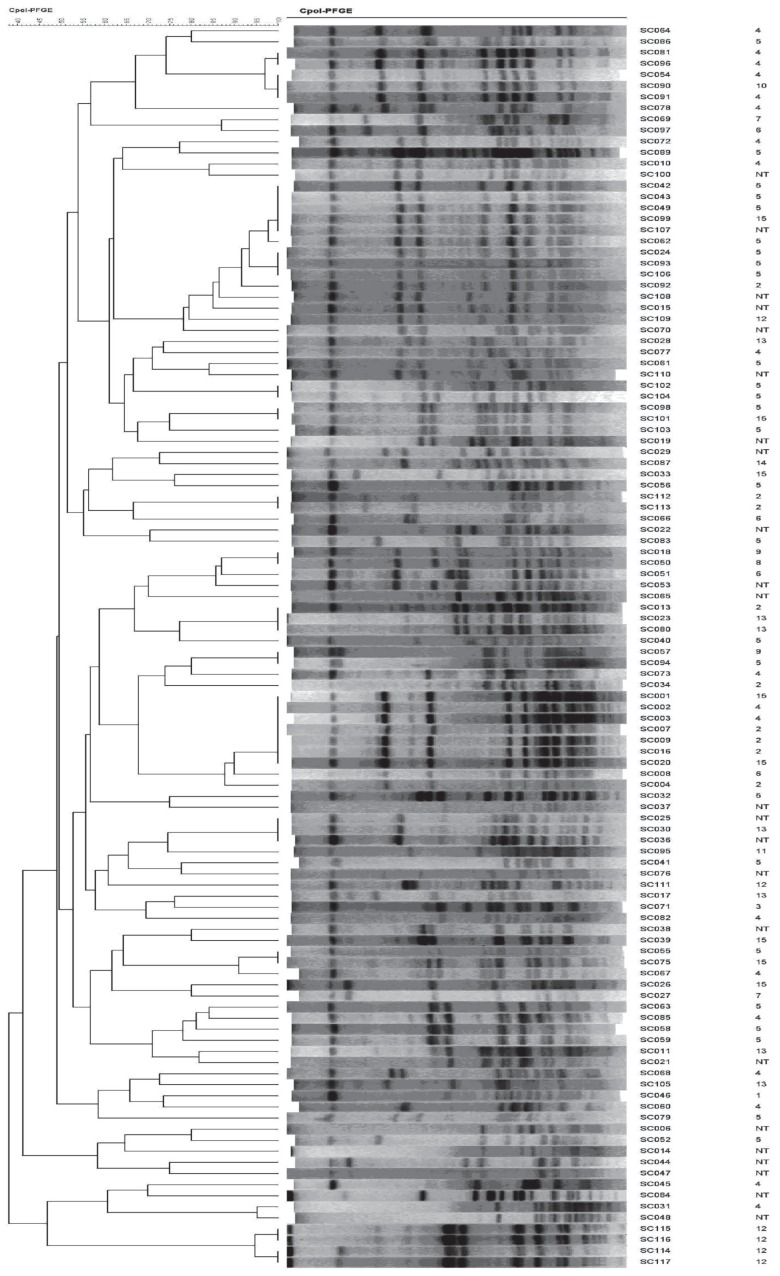
All 112 isolates were typeable by the PFGE method previously established by our laboratory, and great genetic diversity was observed: 85 different PFGE patterns were obtained (Figure 2). The discriminatory ability of PFGE, calculated with Simpson’s index of diversity, was 0.992. The 89 isolates typeable by GD and IHA testing and belonging to the 15 H. parasuis serovars displayed 63 different PFGE profiles. The 23 H. parasuis isolates that could not be assigned to a serovar displayed 22 different PFGE profiles.
Figure 2.
Dendrogram analysis of banding patterns obtained by pulsed-field gel electrophoresis of digested DNA from the isolates. The serovars are indicated on the right.
Analysis of the correlation between serovar and PFGE profile showed a relatively high diversity index for all 112 H. parasuis field isolates, including the nontypeable group (Table II): strains belonging to the same serovar showed various distinctive PFGE patterns. Comparison of the genomic DNA fingerprints of all the isolates yielded no defined correlations between serovar and PFGE profile.
Table II.
Variations in pulsed-field gel electrophoresis patterns between the isolates
| Serovar number | Number of isolates | Number of patterns | Diversity index |
|---|---|---|---|
| 1 | 1 | 1 | 1.00 |
| 2 | 9 | 6 | 0.67 |
| 3 | 1 | 1 | 1.00 |
| 4 | 19 | 15 | 0.79 |
| 5 | 26 | 21 | 0.81 |
| 6 | 4 | 4 | 1.00 |
| 7 | 2 | 2 | 1.00 |
| 8 | 1 | 1 | 1.00 |
| 9 | 2 | 2 | 1.00 |
| 10 | 1 | 1 | 1.00 |
| 11 | 1 | 1 | 1.00 |
| 12 | 6 | 4 | 0.67 |
| 13 | 7 | 6 | 0.86 |
| 14 | 1 | 1 | 1.00 |
| 15 | 8 | 7 | 0.88 |
| Nontypeable | 23 | 22 | 0.96 |
Some field strains that belonged to different serovars had similar PFGE patterns.
Distinctive restriction patterns were identified: 21 among the 26 H. parasuis isolates in serovar 5, 15 among the 19 isolates in serovar 4, and 22 among the 23 nontypeable isolates. Some field strains that belonged to different serovars had similar PFGE patterns. The variations in PFGE patterns between the serovars are illustrated in Figure 2.
It is essential to know the prevalent H. parasuis serovars in a given area to control Glässer’s disease since vaccination confers only limited protection across certain serovars (32). However, the distribution and prevalence of serovars and genotypes can vary considerably from region to region and over time within a region. Thus, local epidemiologic study can determine if 1 or more strains are causing an outbreak or, in the case of persistent infection, if treatment has failed or a new virulent strain has been introduced.
This study of 112 H. parasuis isolates obtained from pigs with clinical signs of Glässer’s disease between 2008 and 2010 has demonstrated that H. parasuis strains were widely prevalent in pig farms in southern China and that all 15 serovars could be identified. The distribution of serovars was similar to the distributions described by Cai et al (33) and Zhou et al (34) in China and by investigators in various countries (4–7,29,35,36). However, there was a slight difference. We found the H. parasuis serovars identified to be diverse, including all 15 known H. parasuis serovars, and we therefore hypothesized that this was a potential reason for the elevated mortality rates associated with H. parasuis infection in this geographic location and the difficulty in controlling the infections. Serovars 5 and 4 were the most prevalent in our study, with the number of serovar 5 isolates somewhat higher than the number of serovar 4 isolates. In contrast, the studies by Cai et al (33) and Zhou et al (34) found serovar 5 strains to account for a great proportion of all H. parasuis isolates identified and to be highly virulent, which made this serovar the most dangerous to the swine industry in southern China. We found nontypeable isolates to represent a large proportion of all field isolates, which suggests that significant genetic changes are taking place among the H. parasuis strains in this geographic location. Therefore, it is of great importance to study the effect of vaccines used in the prevention of infections caused by typeable strains in the context of infections with nontypeable H. parasuis.
This study also confirmed that serotyping alone does not provide sufficient discrimination between isolates for epidemiologic studies: 20% of the isolates could not be serotyped. Molecular techniques represent a major advance for epidemiologic studies since they allow unambiguous identification of every isolate in a timely manner. Widely used and highly discriminatory, PFGE is a molecular typing technique based on comparing patterns of chromosomal DNA fragments obtained by digestion with restriction enzymes. In recent years, PFGE has become a useful tool for typing, differentiating, and classifying bacterial strains in epidemiologic studies (37–40). In the current study, all 112 H. parasuis isolates, including those nontypeable by serotyping, were genotyped by PFGE, which confirmed that PFGE analysis is useful for conducting epidemiologic studies of Glässer’s disease. The present study also showed that isolates of 2 different serovars could be classified into the same group by genotyping, indicating a lack of correlation between serotyping and PFGE typing. The high diversity of the H. parasuis field isolates as well as the poor correlation between serotyping and PFGE profiling is similar to previous findings (41).
Our previous research demonstrated that the PFGE method is rapid and easy to do for molecular subtyping of H. parasuis (25). Furthermore, this study not only provided prevalence data and characterized the genotypic diversity of H. parasuis in southern China but also validated PFGE as an excellent approach to studying the molecular epidemiology of H. parasuis, all practical information that could be of great value in implementing effective prevention and control programs. Also, the PFGE method established by our laboratory provides a high level of discrimination, which has allowed investigation of the spread of mechanisms of antibiotic resistance between strains of H. parasuis. We observed great genetic diversity among 20 antimicrobial-resistant and 6 PMQR-positive isolates; as well, we found 3 isolates from different farms to be clonally related (26).
Our study has shown that H. parasuis infection is widespread in southern China. Therefore, more attention should be paid to diagnosis and infection control. In addition, since infection in this region is caused by numerous serovars and genotypes, further studies need to be done to investigate use of the PFGE technique in finding correlations between virulence, cross-immunity, and genotype.
Acknowledgments
We thank Professor Huanchun Chen College of Veterinary Medicine, Huazhong Agricultural University, Wuhan, Hubei Province, China, for providing the 15 H. parasuis strains. This work was supported by grants from the Program for New Century Excellent Talents in University (NCET-06-0752) and the Guangdong Technology Planning Committee (2006B0152, 2009A0201006, and 2009B03083050).
References
- 1.Biberstein EL, White DC. A proposal for the establishment of two new Haemophilus species. J Med Microbiol. 1969;2:75–78. doi: 10.1099/00222615-2-1-75. [DOI] [PubMed] [Google Scholar]
- 2.Little TWA. Haemophilus parasuis infection in pigs. Vet Rec. 1970;87:399–402. doi: 10.1136/vr.87.14.399. [DOI] [PubMed] [Google Scholar]
- 3.Rapp-Gabrielson VJ, Oliveira SR, Pijoan C. Haemophilus parasuis. In: Straw B, Zimmerman JJ, D’Allaire S, Taylor DJ, editors. Diseases of Swine. Williston, Florida: Blackwell Publishing; 2006. pp. 681–690. [Google Scholar]
- 4.Kielstein P, Rapp-Gabrielson VJ. Designation of 15 serovars of Haemophilus parasuis on the basis of immunodiffusion using heat-stable antigen extracts. J Clin Microbiol. 1992;30:862–865. doi: 10.1128/jcm.30.4.862-865.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rapp-Gabrielson VJ, Gabrielson DA. Prevalence of Haemophilus parasuis serovars among isolates from swine. Am J Vet Res. 1992;53:659–664. [PubMed] [Google Scholar]
- 6.Blackall PJ, Rapp-Gabrielson VJ, Hampson DJ. Serological characterization of Haemophilus parasuis isolates from Australian pigs. Aust Vet J. 1996;73:93–95. doi: 10.1111/j.1751-0813.1996.tb09984.x. [DOI] [PubMed] [Google Scholar]
- 7.Rubies X, Kielstein P, Costs L, Riera P, Artigas C, Espuna E. Prevalence of Haemophilus parasuis serovars isolated in Spain from 1993 to 1997. Vet Microbiol. 1999;66:245–248. doi: 10.1016/s0378-1135(99)00007-3. [DOI] [PubMed] [Google Scholar]
- 8.Rafiee M, Blackall PJ. Establishment, validation and use of Kielstein–Rapp-Gabrielson serotyping scheme for Haemophilus parasuis. Aust Vet J. 2000;78:172–174. doi: 10.1111/j.1751-0813.2000.tb10586.x. [DOI] [PubMed] [Google Scholar]
- 9.Nielsen R. Pathogenicity and immunity of Haemophilus parasuis serotypes. Acta Vet Scand. 1993;34:193–198. doi: 10.1186/BF03548209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rapp-Gabrielson VJ, Kocus GJ, Clark JT, Stephen KM. Haemophilus parasuis: Immunity in swine after vaccination. Vet Med. 1997;92:83–90. [Google Scholar]
- 11.Bak H, Riising HJ. Protection of vaccinated pigs against experimental infections with homologous and heterologous Haemophilus parasuis. Vet Rec. 2002;151:502–505. doi: 10.1136/vr.151.17.502. [DOI] [PubMed] [Google Scholar]
- 12.Oliveira S, Pijoan C. Haemophilus parasuis: New trends on diagnosis, epidemiology and control. Vet Microbiol. 2004;99:1–12. doi: 10.1016/j.vetmic.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 13.Oliveira S, Blackall PJ, Pijoan C. Characterization of the diversity of Haemophilus parasuis field isolates by serotyping and genotyping. Am J Vet Res. 2003;64:435–442. doi: 10.2460/ajvr.2003.64.435. [DOI] [PubMed] [Google Scholar]
- 14.Rafiee M, Bara M, Stephens CP, Blackall PJ. Application of ERIC-PCR for the comparison of isolates of Haemophilus parasuis. Aust Vet J. 2000;78:846–849. doi: 10.1111/j.1751-0813.2000.tb10507.x. [DOI] [PubMed] [Google Scholar]
- 15.Ruiz A, Oliveira S, Torremorell M, Pijoan C. Outer membrane proteins and DNA profiles in strains of Haemophilus parasuis recovered from systemic and respiratory sites. J Clin Microbiol. 2001;39:1757–1762. doi: 10.1128/JCM.39.5.1757-1762.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olvera A, Calsamiglia M, Aragon V. Genotypic diversity of Haemophilus parasuis field strains. Appl Environ Microbiol. 2006;72:3984–3992. doi: 10.1128/AEM.02834-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.del Rio ML, Martin CB, Navas J, Gutierrez-Muniz B, Rodriguez-Barbosa JI, Rodriguez Ferri EF. aroA gene PCR-RFLP diversity patterns in Haemophilus parasuis and Actinobacillus species. Res Vet Sci. 2006;80:55–61. doi: 10.1016/j.rvsc.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 18.Basim E, Basim H. Pulsed-field gel electrophoresis (PFGE) technique and its use in molecular biology. Turk J Biology. 2001;25:405–418. [Google Scholar]
- 19.Seifert H, Dolzani L, Bressan R, et al. Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis-generated fingerprints of Acinetobacter baumannii. J Clin Microbiol. 2005;43:4328–4335. doi: 10.1128/JCM.43.9.4328-4335.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bagattini M, Crivaro V, Di Popolo A, et al. Molecular epidemiology of extended-spectrum β-lactamase-producing Klebsiella pneumoniae in a neonatal intensive care unit. J Antimicrob Chemother. 2006;57:979–982. doi: 10.1093/jac/dkl077. Epub 2006 Mar 10. [DOI] [PubMed] [Google Scholar]
- 21.Pitout JD, Church DL, Gregson DB, et al. Molecular epidemiology of CTX-M-producing Escherichia coli in the Calgary Health Region: Emergence of CTX-M-15-producing isolates. Antimicrob Agents Chemother. 2007;51:1281–1286. doi: 10.1128/AAC.01377-06. Epub 2007 Feb 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mammina C, Noto AM, Ricci A, Nastasi A. Phenogenotyping of Salmonella enterica serotype Enteritidis isolates identified in Sicily during a reemergence period. Foodborne Pathog Dis. 2004;1:195–199. doi: 10.1089/fpd.2004.1.195. [DOI] [PubMed] [Google Scholar]
- 23.Tassios PT, Chadjichristodoulou C, Lambiri M, et al. Molecular typing of multidrug-resistant Salmonella Blockley outbreak isolates from Greece. Emerg Infect Dis. 2000;6:60–64. doi: 10.3201/eid0601.000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woo YK, Lee SH. Genetic diversity of multi-resistant Salmonella enterica serotype Typhimurium isolates from animals and humans. J Microbiol. 2006;44:106–112. [PubMed] [Google Scholar]
- 25.Zhang JM, Xu CG, Guo LL, et al. A rapid pulsed-field gel electrophoresis method of genotyping Haemophilus parasuis isolates. Lett Appl Microbiol. 2011;52:589–595. doi: 10.1111/j.1472-765X.2011.03048.x. [DOI] [PubMed] [Google Scholar]
- 26.Guo LL, Zhang JM, Xu CG, et al. Molecular characterization of fluoroquinolone resistance in Haemophilus parasuis isolated from pigs in South China. J Antimicrob Chemother. 2011;66:539–542. doi: 10.1093/jac/dkq497. [DOI] [PubMed] [Google Scholar]
- 27.Kielstein P, Wuthe H, Angen O, Mutters R, Ahrens P. Phenotypic and genetic characterization of NAD-dependent Pasteurellaceae from the respiratory tract of pigs and their possible pathogenic importance. Vet Microbiol. 2001;81:243–255. doi: 10.1016/s0378-1135(01)00351-0. [DOI] [PubMed] [Google Scholar]
- 28.Oliveira S, Galina L, Pijoan C. Development of a PCR test to diagnose Haemophilus parasuis infections. J Vet Diagn Invest. 2001;13:495–501. doi: 10.1177/104063870101300607. [DOI] [PubMed] [Google Scholar]
- 29.del Rio ML, Gutierrez CB, Rodriguez Ferri EF. Value of indirect hemagglutination and coagglutination tests for serotyping Haemophilus parasuis. J Clin Microbiol. 2003;41:880–882. doi: 10.1128/JCM.41.2.880-882.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turni C, Blackall PJ. Comparison of the indirect haemagglutination and gel diffusion test for serotyping Haemophilus parasuis. Vet Microbiol. 2005;106:145–151. doi: 10.1016/j.vetmic.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 31.Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: An application of Simpson’s index of diversity. J Clin Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miniats OP, Smart NL, Rosendal S. Cross-protection among Haemophilus parasuis strains in immunized gnotobiotic pigs. Can J Vet Res. 1991;55:37–41. [PMC free article] [PubMed] [Google Scholar]
- 33.Cai X, Chen H, Blackall PJ, Yin Z, Wang L, Liu Z, Jin M. Serological characterization of Haemophilus parasuis isolates from China. Vet Micro. 2005;111:231–236. doi: 10.1016/j.vetmic.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 34.Zhou XL, Xu XJ, Zhao YX, et al. Distribution of antimicrobial resistance among different serovars of Haemophilus parasuis isolates. Vet Microbiol. 2010;141:168–173. doi: 10.1016/j.vetmic.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 35.Tadjine M, Mittal KR, Bourdon S, Gottschalk M. Development of a new serological test for serotyping Haemophilus parasuis isolates and determination of their prevalence in North America. J Clin Microbiol. 2004;42:839–840. doi: 10.1128/JCM.42.2.839-840.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Angen Ø, Svensmark B, Mittal KR. Serological characterization of Danish Haemophilus parasuis isolates. Vet Microbiol. 2004;103:255–258. doi: 10.1016/j.vetmic.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 37.Ichiyama S, Ohta M, Shimokata K, Kato N, Takeuchi J. Genomic DNA fingerprinting by pulsed-field gel electrophoresis as an epidemiological marker for study of nosocomial infections caused by methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 1991;29:2690–2695. doi: 10.1128/jcm.29.12.2690-2695.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kristjansson M, Samore MH, Gerding DN, et al. Comparison of restriction endonuclease analysis, ribotyping, and pulsed-field gel electrophoresis for molecular differentiation of Clostridium difficile strains. J Clin Microbiol. 1994;32:1963–1969. doi: 10.1128/jcm.32.8.1963-1969.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fujita M, Fujimoto S, Morooka T, Amako K. Analysis of strains of Campylobacter fetus by pulsed-field gel electrophoresis. J Clin Microbiol. 1995;33:1676–1678. doi: 10.1128/jcm.33.6.1676-1678.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thong KL, Ngeow YF, Altwegg M, Navaratnam P, Pang T. Molecular analysis of Salmonella enteritidis by pulsed-field gel electrophoresis and ribotyping. J Clin Microbiol. 1995;33:1070–1074. doi: 10.1128/jcm.33.5.1070-1074.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de la Puente-Redondo VA, Navas Mendez J, Garcia del Blanco N, Ladron Boronat N, Gutierrez Martin CB, Rodriguez Ferri EF. Typing of Haemophilus parasuis strains by PCR-RFLP analysis of the tbpA gene. Vet Microbiol. 2003;92:253–262. doi: 10.1016/s0378-1135(02)00362-0. [DOI] [PubMed] [Google Scholar]