Abstract
The remarkable selectivity of N-methyl mesoporphyrin IX (NMM) for G-quadruplexes (GQs) is long known, however its ability to stabilize and bind GQs has not been investigated in detail. Through the use of circular dichroism, UV-visible spectroscopy and fluorescence resonance energy transfer (FRET) melting assay we have shown that NMM stabilizes human telomeric DNA dAG3(TTAG3)3 (Tel22) and is selective for its parallel conformation to which it binds in 1:1 stoichiometry with a binding constant of ∼1.0 × 105 M−1. NMM does not interact with an antiparallel conformation of Tel22 in sodium buffer and is the second example in the literature, after TOxaPy, of a ligand with an excellent selectivity for a specific GQ structure. NMM's stabilizing ability toward predominantly parallel GQ conformation is universal: it stabilizes a variety of biologically relevant G-rich sequences including telomeres and oncogene promoters. The N-methyl group is integral for selectivity and stabilization, as the unmethylated analogue, mesoporphyrin IX, does not stabilize GQ DNA in FRET melting assays. Finally, NMM induces the isomerization of Tel22 into a structure with increased parallel component in K+ but not in Na+ buffer. The ability of NMM to cause structural rearrangement and efficient stabilization of Tel22 may bear biological significance.
INTRODUCTION
Guanine-rich regions of genomic DNA are suggested to fold into non-canonical secondary structures known as G-quadruplexes (GQ). GQs are formed by π–π stacking of G-quartets, which are composed of four planar guanines held together by Hoogsteen hydrogen-bonds. The presence of monovalent cations at the core channel of the GQ is necessary to balance charges. Sequences from telomeres, oncogene promoters (c-myc, c-myb, c-fos, c-abl) (1,2), Ribosomal DNA, minisatellites and immunoglobulin switch regions (3) form GQs in vitro. The genomic distribution of these regions makes GQs biomedically relevant (4–7). Data are being accumulated about in vivo existence of GQ structures (8–11).
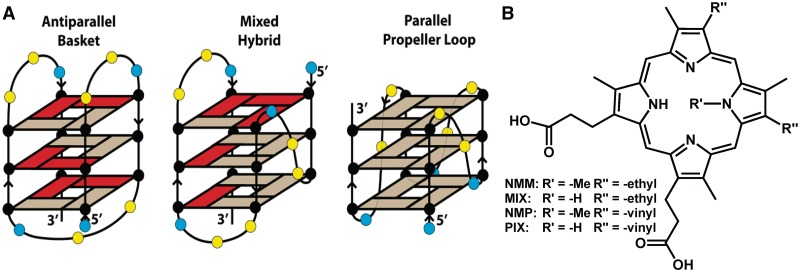
The folding topology of human telomeric repeat sequence, d(TTAGGG)n, under a variety of conditions has been researched extensively (12–18); for a recent review see Phan (19). In total, at least five different intramolecular GQ structures were reported. In a Na+-rich environment, dAG3(TTAG3)3 sequence, Tel22, forms an antiparallel basket type structure (12), whereas in a crystalline form (high K+ and high DNA) (13) and in PEG-containing solutions (20) Tel22 adopts a parallel GQ conformation (Figure 1A). Other potentially physiologically relevant structures include the mixed hybrid topologies that have both parallel and antiparallel character, named Forms 1, 2 and 3. They are found in solutions of K+ and mixtures of K+ and Na+ (16,21,22) as well as other cations (Mg2+ and Ca2+). Forms 1 and 2 are hybrid (3 + 1) GQs with one parallel and three antiparallel strands. Form 3 is an antiparallel GQ with only two G-tetrads. Tel22 in dilute K+ buffers, used in this work, is believed to fold into Forms 1 and 2, though it is likely that there is an equilibrium with Form 3 and possibly with an unfolded oligonucleotide. The folding of human telomeric DNA into a specific quadruplex or structural transition from one fold to another is believed to occur during telomere formation and function (23). In vitro it could be triggered by the presence of divalent cations (24,25), cosolutes (23,26) and a number of small molecule ligands (27–34) including porphyrins (35).
Figure 1.
Structures of Tel22 and porphyrins used. (A) Major physiologically relevant structural topologies of Tel22. Red rectangles represent guanines with glycosidic bonds in syn-conformation, while beige rectangles show those with anti-conformations. Yellow and blue circles indicate thymine and adenine bases, respectively. Shown are antiparallel basket structure (left), mixed hybrid (3 + 1) structure of Form 2 (16) (center) and all-parallel propeller loop structure (right). (B) Structures of NMM, NMP and their non-methylated analogues, MIX and PIX. Note, the central N-methyl group, in principle, could be attached to any of the four nitrogens of the porphyrin core, leading to a mixture of four isomers.
Small molecule ligands have been developed to bind GQ with high affinity, to effectively stabilize GQ structures and to be selective for quadruplex DNA versus other DNA structures, such as abundant duplex DNA (36). Selectivity for one type of quadruplex versus another also needs to be considered in ligand design. GQ structures could form in different parts of the human genome and play both positive and negative roles in human biology. For example, GQ formation at the RNA component of human telomerase was shown to play a role in telomerase assembly and stimulate telomere maintenance in vivo (37). Hence, ligands with broad quadruplex recognition that were originally designed to inhibit telomerase could increase telomerase processivity and genome instability that might lead to secondary tumors (38). With new screening methods aimed at the detection of a ligands' preference for a given GQ structure (39–41), a variety of quadruplex stabilizers were tested and only a few displayed some preference for a given quadruplex type. Recently, the acyclic oligoheteroaryle, TOxaPy, was shown to recognize human telomeric DNA in Na+, but not in K+ buffer (42). TOxaPy is the first example of a ligand with specificity for an antiparallel quadruplex fold.
N-methyl mesoporphyrin IX (NMM, Figure 1B), N-core methylated non-planar derivative of mesoporphyrin IX, was first identified as a potent inhibitor of Fe2+ insertion into protoporphyrin IX by ferrochelatase, the terminal enzyme in heme biosynthetic pathway (43). Following this work, NMM-selective DNA aptamers were developed (44). Interestingly, these aptamers had significant quadruplex forming potential, suggesting GQ as a possible binding structure for NMM. Hence, NMM was one of the first small molecules reported to bind quadruplexes. In later work, NMM was found to be highly selective for GQ DNA versus ssDNA, dsDNA, dsRNA, RNA–DNA hybrid, Z-DNA, triplex and Holliday junctions (45–47). More recently, NMM has been shown to inhibit quadruplex unwinding by the Escherichia coli RecQ helicase and by the eukaryotic helicases BLM and Sgs1p, leaving unwinding of Holliday junctions and partial-duplex substrates largely unaffected (47,48). However, NMM is regarded by some as a weak GQ binder because its negative charge at physiological pH prevents favorable electrostatic interactions with DNA. The details of NMM's interaction with GQ DNA and its stabilizing ability toward GQ structures remain largely unexplored. Yet, NMM's ability to interact with GQs is utilized in biology and chemistry, e.g. in fluorescence screening assays (46,49), in microarrays (50), in affinity chromatography (51) or in quantitative Cu(II) detection (52).
In this work, we explore the details of the interactions between NMM and human telomeric DNA, Tel22, and probe NMM's ability to discriminate between various GQ structures using circular dichroism (CD), UV-vis and fluorescence spectroscopies, and gel electrophoresis. We demonstrate that NMM induces the isomerization of Tel22 from a hybrid to a parallel structure in dilute K+ buffers; NMM effectively stabilizes parallel-stranded GQs with excellent selectivity (>480-fold compared to dsDNA), has a moderate binding constant (∼1.0 × 105 M−1) and displays 1:1 binding stoichiometry in its interactions with Tel22. NMM does not alter or stabilize antiparallel GQ structures of Tel22. Along with TOxaPy, NMM is a unique ligand with highly desired specificity for a selected GQ structure. NMM's selectivity for parallel-stranded GQs and its ability to cause a conformational change in GQ structure, make it a promising platform for further ligand development.
MATERIALS AND METHODS
Porphyrins, oligonucleotides and buffers
All porphyrins were purchased from Porphyrin Products (Logan, UT, USA). NMM and 5,10,15,20–tetrakis(N-methyl-4-pyridyl) (TMPyP4) porphyrins were dissolved in water at 5–10 mM and stored at −20°C. N-methyl protoporphyrin IX (NMP), mesoporphyrin IX dihydrochloride (MIX) and protoporphyrin IX (PIX) were dissolved in DMSO at 5–10 mM and stored at 4°C. For all experiments, porphyrin stock solutions were freshly diluted with desired buffers, and concentrations were determined using extinction coefficients reported in Supplementary Table S1.
The DNA oligonucleotides used in this work (for complete list see Supplementary Table S1) were purchased from Midland (TX, USA) or from Eurogentec (Seraing, Belgium); the fluorescently labeled oligonucleotides 5′-6-FAM-G3(TTAG3)3-Dabcyl-3′ (F21D) and 5′-6-FAM-G3(TGTG3)3-Dabcyl-3′ (FGD) were purchased from IDT (Coralville, IA, USA); calf-thymus (CT) DNA was purchased from Sigma Aldrich. DNA dAG3(TTAG3)3 (hereafter abbreviated as Tel22) was purified by ethanol precipitation, dissolved in water at 1 mM and stored at 4°C. The concentration of DNA was calculated based on absorption at 260 nm at 90°C using extinction coefficients determined from a nearest neighbor approximation (53) and reported in Supplementary Table S1. For Tel22 this coefficient is 228.5 mM−1 cm−1. Concentration of all quadruplex-forming sequences is expressed per quadruplex. To induce quadruplex formation, stocks of Tel22 were diluted with an appropriate buffer to 25 µM and annealed by heating to 90°C for 10 min, cooling to room temperature during 2–3 h and equilibrating at 4°C for >12 h. Fully equilibrated thermodynamic Tel22/NMM samples were prepared either by annealing Tel22/NMM mixtures as described earlier followed by equilibration at 30°C for 12 h, or by mere mixing of annealed Tel22 with NMM followed by equilibration at 30°C for ∼30 h. The time interval necessary for complete equilibration was determined based on preliminary kinetics experiments (data not shown).
The following buffers were used in this work: 10 mM lithium cacodylate, pH 7.2, 5 mM KCl, 95 mM LiCl (hereafter abbreviated as 5K); 10 mM lithium cacodylate, pH 7.2, 50 mM NaCl, 50 mM LiCl (50Na); 10 mM lithium cacodylate, pH 7.2, 100 mM LiCl (100Li).
UV-vis studies
Thermal difference spectra (TDS) (54) were collected on Cary300 (Varian) spectrophotometer equipped with a Peltier-thermostated cuvette holder (temperature accuracy± 0.3°C) using a 1-cm quartz cuvette. Tel22 samples annealed and fully equilibrated in 5K, 50Na or 100Li buffer with or without 2 or 4 equivalents of NMM were diluted to a concentration of ∼4 µM. UV-vis spectra were collected in the range 220–350 nm at 25°C, then the samples were heated to 90°C for 10 min and the spectra were collected again. The difference between the two spectra, TDS, was calculated and normalized to the highest peak for ease of comparison. TDS were collected also for F21D and FGD in order to confirm that the presence of dyes does not interfere with quadruplex formation. In this case, the oligonucleotides were diluted to 10 µM in the appropriate buffer in a 1-mm quartz cuvette and incubated in the UV-vis cuvette holder at 90°C for 10 min after which UV-vis spectra were collected. The heating was turned off, samples were cooled slowly during 4 h and placed at 4°C overnight. After this equilibration the second UV-vis spectrum was collected at 25°C. Data were analyzed as described earlier.
Titrations of NMM with Tel22 in 5K, 50Na and 100Li buffers were carried out in a 1-cm methacrylate or 2-mm quartz cuvettes at 25°C by the stepwise additions of 0.3–0.7 mM Tel22 into a solution of ∼2–5 µM NMM. The solution of Tel22 contained an equal amount of NMM to keep porphyrin concentration constant. Titrations were terminated when two to three consecutive additions of Tel22 yielded the same spectra or when [GQ]/[NMM] exceeded 10. Note, for the case of 5K buffer, titrations performed in this manner reflect ‘non-equilibrium’ condition due to slow rearrangement of original Tel22 structure to a new fold induced by addition of NMM. In order to obtain ‘equilibrium’ data UV-vis titrations were repeated in a batch method where NMM samples containing increasing amounts of Tel22 were incubated for 30 hours at 30°C. Accurate stoichiometry of NMM/Tel22 complex was determined using continuous variation analysis, also known as a Job plot (55), following closely the protocol published earlier (35). Concentrations of NMM and Tel22 were matched at 18 µM within 5%.
Interactions of NMM with telomeric and non-telomeric GQ forming oligonucleotides were studied by UV-vis. All oligonucleotides were annealed in 5K or 50Na buffers at ∼30 µM in the presence of 2.3 µM NMM at 90°C for 10 min, cooled slowly and incubated at 30°C for >12 h. Both the oligonucleotide concentration and high quadruplex to NMM ratio (∼13) were chosen to assure that all NMM molecules were bound to GQ DNA. A sample containing 2.3 µM NMM without DNA was treated in identical way. UV-vis spectra were collected on all samples, and red shift and hypochromicity were determined as described earlier (35).
Circular dichroism
CD experiments were performed on an AVIV 410 spectrometer equipped with a Peltier heating unit (temperature accuracy ± 0.3°C). Spectra were collected between 220 and 330 nm, using three to five scans, a 1-nm bandwidth and an averaging time of 1 s at 25°C in 1-cm or 1-mm quartz cuvettes. Some spectra were collected on JASCO J-815 spectropolarimeter with a 2-nm bandwidth, 500 nm min−1 scan speed, 1-nm step and five scans. The data were baseline corrected using the CD spectra of a buffer alone in the same cuvette, zeroed by subtracting the average absorbance between 320 and 330 nm and converted into molar ellipticity using the following equation:
where θ is the CD signal in mdeg, C is concentration in mol L−1 and l is a cuvette pathlength in cm. Data were smoothed using a Savitzky–Golay smoothing filter with a 13-point quadratic function.
CD annealing studies were performed to determine if porphyrins induce structural rearrangement of Tel22 in water, 5K, 50Na and 100Li buffers. Samples of 2.5 µM Tel22 were annealed with NMM, NMP or TMPyP4 (1:1 or 1:2 ratio), cooled slowly and placed in a 30°C incubator for 12 h. In parallel, 2.5 µM Tel22 samples were annealed alone cooled down to 30°C, after which porphyrins were added and samples were incubated at 30°C for ∼30 h. As NMM was the only porphyrin capable of changing Tel22 structure, its ability to interact in a similar manner with other GQ-forming sequences (for full list see Supplementary Table S1) was also tested.
Equilibrium CD titration of Tel22 with NMM in 5K buffer were performed in a batch format where individual samples of 2.0 µM Tel22 were annealed with increasing equivalents of NMM (from 0.5 to 12) and incubated at 30°C for >12 h to achieve thermodynamic equilibrium prior to collecting the CD spectra.
Binding constant determination
Singular value decomposition (SVD) analysis of CD and UV-vis titration data was used to verify that binding of Tel22 to NMM is a two-state process. Details of SVD analysis are provided in Supplementary Data and in a recently published protocol (56). Binding constant, Ka, for NMM—Tel22 interactions was extracted using direct fitting of the titration data to the equation:
| (1) |
where Y0 and Yf are the initial and final signal (absorbances at 379 and 399 nm in UV-vis or molar ellipticity at 264 nm in CD), Ct is the total NMM concentration and x is derived from the equilibrium expression:
| (2) |
which is based on the simple two-state 1:1 binding reaction: NMM (Ct) + Tel22 (Cs) → Complex (x). The fits yield Ka, which was averaged for all titrations. This data analysis was done in GraphPad Prism software. In addition, the value of the CD signal at 264 nm was normalized and fitted with the Hill equation:
| (3) |
Hill coefficient, n, can be interpreted as the average number of binding sites. This model assumes that all binding sites have the same affinity.
Gel electrophoresis
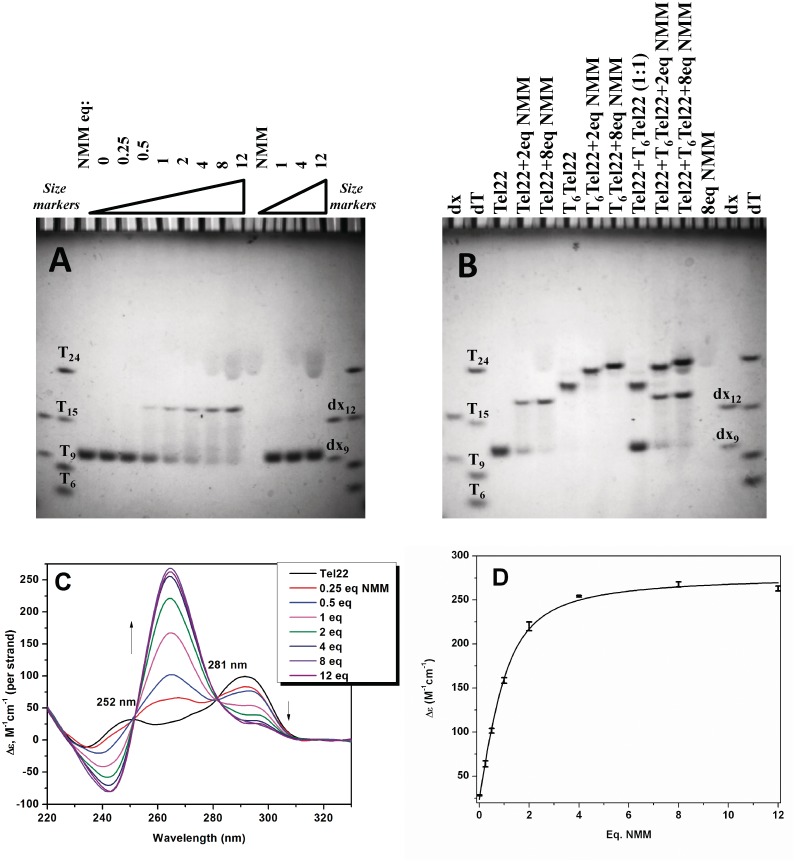
For gel electrophoresis, samples were annealed and equilibrated in 5K buffer at 40 µM Tel22 and increasing concentration of NMM from 0 to 12 equivalents. Samples were run on 20% non-denaturing polyacrylamide gel in 1 × TBE buffer (89 mM Tris–borate, 2 mM EDTA, pH 8.3) with 5 mM KCl (or 10 mM NaCl or 10 mM LiCl) at 220 V at 14–16°C for 4 h. Oligothymidylate markers 5′ dTn (where n = 6, 9, 15 and 24) were used as internal migration standards (and not necessarily as length markers) along with duplex markers dx9: 5′-GCGTATCGG + 5′-CCGATACGC and dx12: 5′-GCGTGACTTCGG + 5′-CCGAAGTCACGC. Control samples included NMM alone as well as Tel22 to which NMM was added right before loading the gel. DNA bands were visualized by UV-shadowing and using NMM's fluorescence (excitation and emission maxima at 399 and 610 nm, respectively (49); gels were excited with an LED of 470 nm and visualized utilizing a red filter). Part of each gel sample was diluted to a final concentration of 1–2 µM for CD measurements. Data were collected immediately after dilution with the assumption that CD signature observed corresponds to the Tel22 conformation at high concentration of 40 µM used for the gel, and not to the conformation present at 1–2 µM DNA (if these are indeed different).
To determine the molecularity of the quadruplex bound to NMM, Tel22 was mixed either with T6Tel22 or Tel22T6 in 1:1 ratio and annealed with 2 or 8 equivalents of NMM. T6Tel22 and Tel22T6 were chosen because they have larger size but secondary structure similar to that of Tel22 as determined by CD. Gels were prepared, loaded, run and visualized in the way described earlier.
Fluorescence resonance energy transfer melting studies
Fluorescence resonance energy transfer (FRET) was performed according to established literature procedure (57) using doubly labeled oligonucleotides F21D, 5′-6-FAM-G3(TTAG3)3-Dabcyl-3′ and FGD, 5′-6-FAM-G3(TGTG3)3-Dabcyl-3′. Specific experimental details are presented in Supplementary Data.
CD melting studies
Tel22 was prepared at 3–4 µM in 5K buffer alone and with increasing amounts of NMM. Samples were heated from 15 to 95°C, maintained at 95°C for 5 min and cooled to 15°C at the same rate of 0.27 deg min−1. CD melting was monitored at 264 and 295 nm with 0.5-min equilibration time, 1°C step, 4 s averaging time and 2-nm bandwidth. Other quadruplex forming oligonucleotides (∼4 µM, for sequences see Table 1) alone and in the presence of 2 equivalents of NMM were melted in an identical way. Melting data were analyzed assuming linear starting and final baselines. Melting temperatures and enthalpies of unfolding were adjusted to get the best fits (58). For data sets where starting or final baselines were not clearly defined (e.g. when T1/2 exceeded 80°C) the melting temperatures were determined using the first derivatives. All melting temperatures reported were obtained from melting curves. All data analysis was done in Origin 8.1.
Table 1.
Sequences, bathochromic (red-) shifts (>10-fold excess of DNA), melting temperatures (T1/2) and (2 eq.) NMM thermal stabilization values (ΔT1/2) for a variety of GQ-forming oligonucleotides in 5K buffer (unless noted otherwise)
| Name | Sequence | Red-shifta (nm) | T1/2 (°C)b | ΔT1/2 (°C)c | Conformation w/o NMMd | Conformation with NMMd |
|---|---|---|---|---|---|---|
| Tel22 | AGGGTTAGGGTTAGGGTTAGGG | 19.3 ± 0.4 | 48.0 ± 0.8 | 7 ± 1 | M | P |
| Tel22, 50Na | AGGGTTAGGGTTAGGGTTAGGG | 0.0 ± 0.2 | 51 ± 2 | 0 ± 2 | A | A |
| 26TelG4 | AGGGGTTAGGGGTTAGGGGTTAGGGG | 18 ± 1 | 73.5 ± 0.4 | 6.8 ± 0.6 | A/M | M |
| 26TelG4, 50Na | AGGGGTTAGGGGTTAGGGGTTAGGGG | 19.5 | 64.4 ± 0.9 | 0 ± 1 | A | A |
| Bcl-2 | GGGCGCGGGAGGGAATTGGCGGGG | 17.2 ± 0.3 | 48.9 ± 0.8 | 20. ± 1 | P/M | P |
| cKit1 | GGGAGGGCGCTGGGAGGAGGG | 18.6 ± 0.9 | 44.7 ± 0.6 | 11.4 ± 0.9 | P/M | P |
| cKit2 | GGGCGGGCGCGAGGGAGGGG | 19.2 ± 0.4 | 53.7 ± 0.7 | 13 ± 1 | P | P |
| cMyc | TGAGGGTGGGTAGGGTGGGTAA | 20 ± 1 | 67.5 ± 0.3 | 14.3 ± 0.3 | P/M | P |
| G4TERT | AGGGGAGGGGCTGGGAGGGC | 17.7 ± 0.3 | 55 ± 2 | 18 ± 2 | P | P |
| HIF-1α | GGGAGGGAGGGAAGGAGGGAGGGAGGG | 18.5 ± 0.7 | 65 ± 1 | 1 ± 1 | P | P |
| TBA | GGTTGGTGTGGTTGG | 19.5 ± 0.5 | 38.7 ± 0.8 | 1 ± 2 | A | A |
| THM | GGGTTGGGTTGGGTTGGG | 17.3 ± 0.9 | 55.7 ± 0.3 | 23 ± 2 | P | P |
| VEGF | GGGAGGGTTGGGGTGGG | 17.3 ± 0.8 | 57.1 ± 0.8 | 20.1 ± 0.9 | P | P |
| G8 | TGGGGGGGGT | 17.8 ± 0.4 | n/m | n/m | P | P |
| G5 | TGGGGGT | 17.3 ± 0.4 | n/m | n/m | P | P |
aRed-shift values were obtained with a >10-fold excess of DNA. bT1/2 values were calculated from CD melting experiments monitoring wavelengths at 264 or 295 nm. NMM:Tel22 ratio was 2:1. cΔT1/2 values were calculated by subtracting T1/2 of DNA alone from the NMM-stabilized value.
dP, parallel; A, antiparallel; M, mixed (either mixture of two conformations or hybrid). The first letter signifies the major conformation. n/m, not measured.
RESULTS
In this report the details of the interaction between NMM and human telomeric GQ DNA, Tel22, was investigated. NMP, the structural analogue of NMM (Figure 1B), was also included in the study. Both NMM and NMP exist in a monomeric form in the concentration range from 1 to 50 µM (Supplementary Figure S1). One needs to be aware that commercial NMM (and NMP) is a mixture of four isomers that differ by the position of the core N-methyl group. Results reported in this article are therefore average of the action produced by four NMM isomers.
Structure of Tel22 alone and in the presence of NMM and other porphyrins
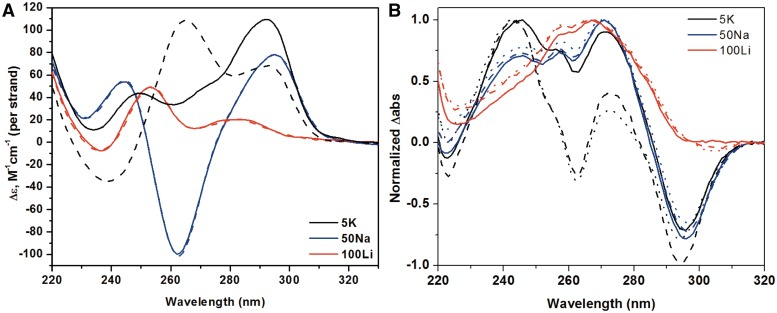
The secondary structure of Tel22 alone under various experimental conditions was determined by CD spectroscopy. In 5K buffer the CD spectrum of Tel22 has two peaks at 294 and 255 nm, and a trough at 235 nm (Supplementary Figure S2). This signal most likely corresponds to mixed (3 + 1) hybrid structure which concurs with the literature NMR evidence (14,16,18,59), but disagrees with conclusions from a recent CD study where an antiparallel structure was assigned this type of signal (60). It has been shown that in 5 mM K+ buffer at 25°C >95% of Tel22 was folded into quadruplex (61). In 50Na buffer, Tel22 displays positive peaks at 296 and 246.5 nm and a negative peak at 264 nm (Supplementary Figure S2) indicating an antiparallel structure in agreement with literature (12). Finally, in 100Li buffer no significant secondary structure is detected.
To test the effect of NMM on the structure of Tel22, this porphyrin was added to Tel22 before or after the annealing step, followed by a prolonged incubation at 30°C. In 5K buffer, NMM causes a dramatic increase in the 264 nm signal of Tel22 at the expense of the signal at 295 nm (Figure 2A), suggesting a shift from hybrid to a conformation with substantial parallel component. This NMM-induced structural conversion is slow and requires 12 h when NMM is added during the annealing step and ∼30 h at 30°C when NMM is added to Tel22 after annealing (data not shown). Previously, NMM has been shown to shift the equilibrium from an antiparallel to a parallel fold when annealed with the bimolecular telomere sequence dTAGGGUTAGGG in the presence of 100 mM KCl but not when added after annealing step (62). This could be due to inherent differences in the Tel22 and dTAGGGUTAGGGT sequences or to the fact that insufficient incubation time was allowed for the completion of rearrangement in the latter case. The isomerization process induced by NMM does not take place in 50Na or 100Li buffers as shown in Figure 2A indicating an essential role of K+ ions. Similarly, it was shown previously that NMM does not affect the antiparallel GQ formed by dG4T4G4 in 140 mM NaCl (62).
Figure 2.
(A) CD spectra of 2.5 µM Tel22 annealed alone (solid line) and in the presence of 5.0 µM NMM (dashed line) in 5K, 50Na and in 100Li buffers. Data were collected at 25°C. (B) UV-vis TDS spectra of 4 µM Tel22 annealed in 5K, 50Na and 100Li buffer in the presence of 0 (solid line), 2 (dash line) and 4 (dot line) equivalents of NMM obtained by subtracting 90°C spectra from the 25°C spectra. Data was zeroed at 320 nm and normalized to the highest peak.
There are similar but smaller changes observed upon annealing of Tel22 in the presence of NMP in 5K buffer (Supplementary Figure S2B). On the other hand, the commonly used porphyrin TMPyP4 was not able to induce the formation of parallel structure of Tel22 under identical condition (Supplementary Figure S2C). Similar to NMM, NMP did not alter Tel22 CD signal in 50Na or in 100Li buffer, while TMPyP4 somewhat decreased the Tel22 CD signal in both cases (data not shown).
TDS were used successfully to identify the presence of GQs by their characteristic signature, such as positive peaks at 243 and 273 nm and a negative peak at 295 nm (54). TDS shown in Figure 2B indicate that Tel22 forms GQ structure in 5K and 50Na buffers. In 100Li, Tel22 displays TDS signal with a peak at 267 nm and a shoulder at 258 nm suggesting a mostly unfolded structure. The addition of NMM to Tel22 in 50Na and 100Li does not change the TDS signature of either, which is consistent with the CD annealing study data. The TDS signature of Tel22 in 5K, however, is altered by the addition of 2 and 4 equivalents of NMM. The troughs at 295 and 262 nm become more prominent, and the intensity of the peak at 273 nm decreases, but the diagnostic maxima and minimum are still present, pointing toward a quadruplex structure. The differences in the TDS signature of Tel22 with and without NMM could be attributed to the different topology of GQ structures (63), corroborating CD evidence.
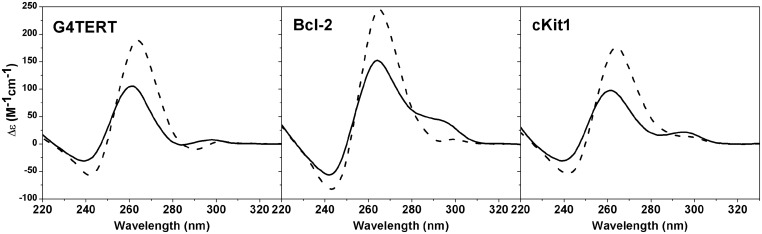
The ability of NMM to induce structural rearrangement of quadruplex DNA in potassium buffer is more universal and not restricted to Tel22 sequence. Comparable transitions were observed for other related human telomeric sequences such as dA3G3(TTAG3)3AA, dGG(ATTG3)4, T6Tel22, Tel22T6 and dAG4(TTAG4)3 (26TelG4), Supplementary Figure S3, as well as non-telomeric quadruplex DNA such as Bcl-2, cKit and G4TERT, Figure 3. These non-telomeric sequences have been shown to form parallel (or hybrid) GQs (64–66) and serve as oncogene promoters: Bcl-2 inhibits cell apoptosis mechanism in B-cell lymphoma cancer cell lines; cKit is a tyrosine kinase receptor that controls cell growth and TERT is the catalytic domain of telomerase. Quadruplexes already in a parallel conformation (THM, c-Myc, HIF-1α and VEGF) were largely unaffected by the addition of NMM (Supplementary Figure S4A), probably due to the presence of maximum possible parallel character in their structures. c-Myc oncogene is a transcriptional regulator of ∼15% of human genes and is involved in a variety of cancers (67); VEGF is a promoter gene for vascular endothelial growth factor that is upregulated in a variety of cancers (68); HIF-1α gene encodes the alpha subunit of hypoxia-inducible factor1, which is a transcription factor that regulates response to hypoxia and is essential for tumor angiogenesis (69); and THM is a telomeric sequence from Tetrahymena thermophila. The thrombin binding aptamer, TBA, which forms a chair-type antiparallel GQ in 5K buffer (70), was unaffected by NMM as well (Supplementary Figure S4A), probably due to its antiparallel nature in agreement with the data for Tel22 in 50Na buffer.
Figure 3.
Annealing study of non-telomeric GQ-forming sequences G4TERT, Bcl-2 and cKit1 in the presence of NMM in 5K buffer. Samples of 4–6 µM DNA were annealed at 90°C for 10 min without (solid lines) or with (dotted lines) 2 equivalents of NMM, cooled to 30°C slowly and stored at 30°C for >12 h before collecting CD.
It is worth noting that weak induced negative signals were observed upon NMM binding to all tested quadruplex structures but TBA in 5K buffer in the visible region of the CD spectra (Supplementary Figure S4C). No induced signal is seen for Tel22/NMM in 50Na. Presence of induced signal is an indication of interaction between NMM and GQ DNA. On the other hand, absence of induced CD signal does not necessarily signify lack of interaction as NMM was shown to interact with 10 different GQ structures but displayed no induced CD signal (49).
Potassium titration of Tel22:NMM 1:2 mixture
NMM-induced structural transition of Tel22 only occurs in the presence of K+ ion. Therefore we proceeded by testing the effect of increased potassium concentration on Tel22 structure in the presence of 2 equivalents of NMM (Supplementary Figure S5). As expected, a substantial increase in the CD signal at 264 nm was observed. Fitting the data with the Hill equation yielded a cooperativity coefficient, n, of 1.1 ± 0.1 and a binding constant of (2.9 ± 0.3) × 102 M−1 [forcing n to be exactly 1 yields a binding constant of (3.2 ± 0.2) × 102 M−1]. The 1:1 binding model is also confirmed by the fact that excellent fits were obtained with direct fitting [using Equations (1) and (2)], which yielded a similar binding constant of (3.6 ± 0.9) × 102 M−1. While the fits at 264 nm are excellent, the results should be used with care as the titration profile looks rather complex (Supplementary Figure S5A) and possibly reflects more than simple K+ binding. In control experiments Tel22 was titrated with KCl in the absence of NMM. CD signal at 264 nm did not increase appreciably (Supplementary Figure S5C). For more details see Supplementary Data.
NMM binding to Tel22 and other GQs studied by UV-vis spectroscopy
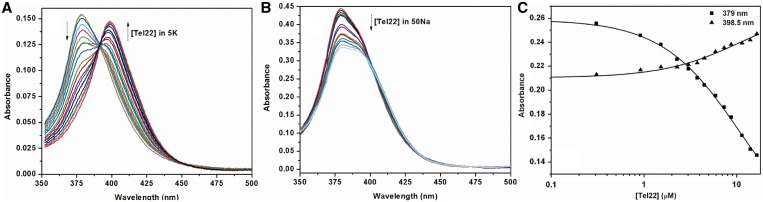
Detailed information about NMM binding to quadruplex structure of Tel22 and other sequences was obtained through UV-vis absorbance titrations by monitoring NMM's Soret band at 379 nm. In 5K buffer NMM exhibited a large bathochromic shift of 19.3 ± 0.4 nm and small hypochromicity of 4% upon binding to Tel22 (Figure 4A). While red shift was independent of incubation time, the hypochromicity changed into mild hyperchromicity (H = −3%) when samples were allowed to equilibrate for >24 h at 30°C. The Tel22–NMM complex has an extinction coefficient of (1.48 ± 0.10) × 105 M−1 cm−1 at 398.5 nm. When other GQ-forming sequences were titrated into NMM under equilibrium conditions, red shifts of 17–20 nm (Table 1) and small hypochromicities or hyperchromicities of 20 to −21% were observed (Supplementary Table S2 and Supplementary Figure S4B). The large variation in Soret band intensity could possibly indicate differences in binding modes depending on DNA sequence and quadruplex geometry. The Bolton laboratory reported ∼20 nm red shift of NMM's Soret band upon addition of 50-fold excess of TBA and d(G4T4G4) accompanied by relatively high hyperchromicity in the latter case (46), results consistent with our data. While red shift and change in Soret band intensity can provide information about the ligand's binding mode when long duplex DNA is concerned (71), no reliable numbers exist for porphyrin binding to quadruplex structures.
Figure 4.
(A) Representative UV-vis absorption spectra of 5.3 µM NMM titrated with 0.72 mM Tel22 in 5K buffer. Data were collected in 2 mm quartz cuvette. The final [GQ]/[NMM] ratio was 14.2. (B) Representative UV-vis absorption spectra of 3.0 µM NMM titrated with 0.49 mM Tel22 in 50Na buffer. Data were collected in 10 mm quartz cuvette. (C) A plot of NMM's absorbances at 379 and 398.5 nm as a function of added Tel22. Solid line represents fit to 1:1 binding model. Concentration of NMM was 1.8 µM and that of stock Tel22 was 0.154 mM in 5K buffer.
The method of continuous variation analysis was used to accurately determine the binding stoichiometry of the Tel22–NMM interaction (Supplementary Figure S6). The plot of absorbance difference versus mole fraction of NMM peaked at 0.50, indicating 1:1 binding. SVD confirmed that the 1:1 binding was a two-state process (data not shown). Direct fit of the titration data yielded a relatively weak binding constant of (1.0 ± 0.3) × 105 M−1 under equilibrium conditions (Figure 4C). Modest binding constants were also reported for NMM interactions with other quadruplex structures (44–46). Binding parameters of NMM to Tel22 were determined for short incubation times (assuming possible NMM binding to hybrid GQ structure of Tel22) and long incubation times (reflecting equilibrium condition and binding of NMM to presumably parallel GQ structure of Tel22). In both cases the data were rather similar: the same red shift, binding stoichiometry and similar binding constants [(1.0 ± 0.3) × 105 M−1 and (3 ± 2) × 105 M−1] and hypochromicities. Therefore NMM binding to hybrid and parallel GQ is rather similar with possible differences in the orientation of the porphyrin core with respect to the DNA axis, as evidenced by the slight difference in hypochromicities.
UV-vis titration experiments were repeated in 50Na (Figure 4B) and 100Li (data not shown) buffers. These titrations did not lead to any red shift and resulted in modest hypochromicity of 19 and 20% for 50Na and 100Li conditions, respectively, at [GQ]/[NMM] ratio of ≥10. Absence of red shift could be interpreted as the inability of antiparallel conformation of Tel22 that exists in 50Na or single stranded DNA that exists in 100Li buffer to interact with NMM. Note, the observed change in hypochromicity might suggest some interactions between NMM and Tel22 that does not involve any degree of porphyrin stacking. This is an important and unexpected result that demonstrates NMM's selectivity not only for quadruplex structures over other DNA structures (in this case single stranded DNA), but also for parallel GQ topology.
To further test whether binding of NMM to quadruplexes is governed by the choice of cation (Na+ versus K+) or by the nature of GQ present, we employed [dTG5T]4 (G5) which forms a parallel-stranded GQ regardless of buffer conditions. Upon addition of G5 to NMM in 5K or in 50Na buffers similar red shifts of ∼18 nm were observed, suggesting the same binding modes and underlying the importance of GQ structure rather than choice of cation.
NMM binding to and isomerization of Tel22 studied by gel electrophoresis and CD
Since CD spectroscopy could only provide indirect evidence of GQ structure but not number or size of DNA species present in solution, the details of NMM binding to Tel22 was further investigated using native PAGE gel electrophoresis. Tel22 samples at 40 µM were annealed with increasing amount of NMM (0.25–12 equivalents) and loaded on the gel as well as scanned on CD in order to get a correlation between the observed gel species and the CD spectra. Tel22 in 5K buffer appears as a single high mobility band on the gel, Figure 5A, suggesting the existence of a single conformation. Addition of NMM to Tel22 with the subsequent equilibration leads to appearance of a new slower migrating band on the PAGE gel and an increase in CD signal at 264 nm in expense of the signal at 295 nm (Figure 5). On the gel the new species seems to be structurally homogeneous. The CD spectrum of the resulting GQ is consistent with a parallel propeller-loop structure. Gel and CD data indicate that Tel22 undergoes structural rearrangement in the presence of NMM. The rearrangement is slow as the addition of NMM to Tel22 right before loading the gel (last three lanes on Figure 5A) does not lead to the formation of Tel22–NMM complex. The CD spectra of these samples resemble that of Tel22 alone (Supplementary Figure S7C).
Figure 5.
Isomerization of Tel22 in the presence of NMM. (A) Gel electrophoresis experiments. Non-denaturing gel was prepared at 20% acrylamide supplemented with 5 mM KCl. Tel22 strand concentration was 40 µM. Tel22 samples were annealed with NMM (0.25–12 equivalents), cooled to 30°C and incubated at 30°C for >12 h, except for the last three lanes (before size marker lanes) where NMM was added to Tel22 right before loading the gels. (B) Determination of the molecularity of Tel22–NMM complex. Samples of Tel22 were mixed with T6Tel22 in 1:1 ratio and annealed with 2 and 8 equivalents of NMM. Tel22 and T6Tel22 strand concentration was 40 µM. Gels were visualized using UV-shadowing. Strand mixing experiments were also repeated with Tel22 and Tel22T6 yielding similar results (data not shown). (C) CD wavelength scans for the Tel22–NMM samples from the gel in (A) at 25°C. Samples were diluted to 1.9 µM final Tel22 concentration with 5K buffer. Isoelliptic points at 252 and 281 nm are marked on the graph. (D) Molar ellipticity at 264 nm and fit to 1:1 binding model that yielded Ka of (0.7 ± 0.1) × 105 M−1.
In order to determine the distribution of NMM among observed DNA species, PAGE gels were also visualized using NMM's fluorescence (Supplementary Figure S7). NMM fluoresces weakly in the free state but its fluorescence increases in the presence of a quadruplex (46). Our results suggest that NMM acts on Tel22 in a stoichiometric fashion (single turn-over event) and remains bound to the final isomerized Tel22. On the other hand, it is not bound to the starting hybrid Tel22 structure. We cannot exclude the possibility that NMM is bound to the original Tel22 structure, but either dissociates from it during electrophoresis; or its florescence is completely quenched by the DNA.
The slow conversion of Tel22 structure in the presence of NMM as well as slower migration of resulting complex on PAGE gel could suggest that the structural transition reflects formation of bimolecular quadruplex. To test this possibility and determine unambiguously the molecularity of NMM-induced Tel22 structure we performed strand mixing experiments. T6Tel22 was chosen to have larger size but similar secondary structure to that of Tel22 and interact with NMM in a similar manner, Supplementary Figure S8. Tel22 was mixed with T6Tel22, annealed with NMM and run on the 20% native PAGE gel. Two bands are expected for monomolecular complex and three [(Tel22)2–NMM; (Tel22 × T6Tel22)–NMM and (T6Tel22)2–NMM] for bimolecular. Two bands are seen on the gel, Figure 5B, clearly indicating monomolecular nature of quadruplex core in Tel22–NMM complex. The Trent group observed similar conversion of Tel22 structure induced by 50% of acetonitrile (72) and suggested based on NMR line-width and relaxation rate constants that the newly formed GQ is most likely monomolecular supporting our conclusion.
CD data from gel samples, Figure 5C, were analyzed in order to determine binding parameters and compare them to those obtained from UV-vis titrations (see above). While in UV-vis the focus is on NMM, in CD it is on DNA. SVD analysis was employed first to determine the number of significant spectral species present in the NMM–Tel22 mixture (Supplementary Figure S9). The analysis indicates that NMM binding to Tel22 under equilibrium is a simple two-state process reflecting an NMM-induced gain in Tel22's parallel character at the expense of antiparallel structure. Direct fitting of the data with 1:1 binding model, Figure 5D, yielded binding constant of (0.7 ± 0.1) × 105 M−1 in good agreement with results from UV-vis titrations.
Gel experiments were repeated in Na+ and Li+ containing buffers (Supplementary Figure S10). In 50Na Tel22 alone ran as a single band with a slightly slower migration than in 5K buffer, reflecting the difference in quadruplex conformation. In 100Li, Tel22 alone was heterogeneous and ran as major and minor bands with slow mobility suggesting structures with large hydrodynamic radius, possibly single-stranded conformation, in agreement with CD and TDS results. Addition of NMM to Tel22 in 50Na or 100Li buffers caused no change in the mobility of Tel22 or NMM's fluorescence, confirming the UV-vis results that NMM does not interact with the antiparallel (in 50Na) or single-stranded (in 100Li) conformation of Tel22, and underscoring the importance of K+ ions.
Stabilizing ability and selectivity of NMM and its analogues determined by FRET melting assay
FRET melting assay is the benchmark technique used to test stabilizing ability and selectivity of quadruplex ligands. We employed FRET with labeled human (F21D) and yeast (FGD) telomeric DNA to test NMM and NMP in comparison to non-methylated analogues, MIX and PIX. To confirm that under experimental conditions F21D and FGD form GQ structures, CD and TDS spectra were collected (Supplementary Figure S11). Both structures displayed a quadruplex-specific TDS signature (54). The CD spectrum of F21D closely resembles that of Tel22 in 5K buffer. FGD has significantly more parallel character than F21D, displaying a major peak at 265 nm. Collectively, TDS and CD results indicate that the fluorescent probes do not interfere with the formation GQ structures of F21D and FGD.
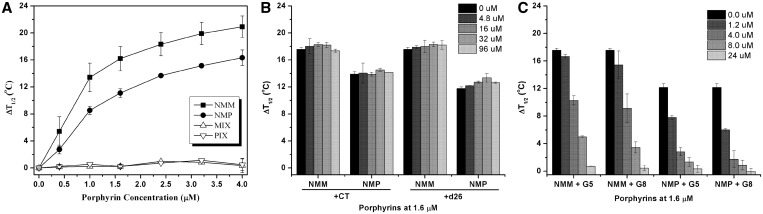
The stability of GQ in the presence of porphyrins was characterized using ΔT1/2 parameter that reflects a change in the melting temperature as compared to GQ alone. First, concentration dependence of ΔT1/2 was determined for NMM, NMP and their non-methylated analogues, MIX and PIX in 5K buffer (Figure 6A for F21D and Supplementary Figure S12A for FGD). The plots show dose-dependent behavior with stabilization temperatures of 14 ± 2°C for F21D + NMM, 12.5 ± 0.3°C for FGD + NMM, 8.5 ± 0.6°C for F21D + NMP and 9.8 ± 0.3°C for FGD + NMP at 1 µM ligand concentration in 5K buffer (Table 2). Under saturating porphyrins concentrations the values of ΔT1/2 for F21D were yet higher, >20°C for NMM and >16°C for NMP. This excellent stabilization of human telomeric DNA by NMM (and NMP) could be due to porphyrins' binding to DNA or could result from the NMM-induced structural conversion of F21D to a more stable conformation. Overall, NMM displayed stabilizing abilities superior to those of NMP and both porphyrins stabilized human telomeric DNA, F21D, to a greater extent than yeast telomeric DNA, FGD (except at concentrations <0.5 µM). MIX and PIX, on the other hand, did not stabilize either of the GQs. This finding underlines the importance of the core N-methyl group for GQ stabilization and suggests its role in porphyrins interactions with GQ DNA.
Figure 6.
FRET melting of F21D in the presence of porphyrins and competitors. (A) Increase in the stabilization temperature, ΔT1/2, of 0.2 µM F21D as a function of porphyrin concentration. (B) Stabilization of F21D by NMM or NMP in the presence of duplex DNA competitors, ds26 or CT or (C) parallel stranded GQ competitors [dTG5T]4, G5, and [dTG8T]4, G8. Concentrations of porphyrins and F21D were fixed at 1.6 and 0.2 µM, respectively. Amount of competitor is indicated in the figure legend. All experiments were performed in 5K buffer.
Table 2.
Stabilization temperature, ΔT1/2 (°C), for porphyrins under study in FRET melting assays
| DNA/porphyrin | NMM | NMP | MIX | PIX | TMPyP4 | ZnTMPyP4 |
|---|---|---|---|---|---|---|
| F21D, 5K | 14 ± 2 | 8.5 ± 0.6 | 0.2 ± 0.3 | 0.2 ± 0.3 | 34 ± 2 | 23 ± 1 |
| FGD, 5K | 12.5 ± 0.3 | 9.8 ± 0.3 | 0.8 ± 0.3 | 0.2 ± 0.3 | 31 ± 2 | 22.1 ± 0.9 |
| F21D, 50Na | 0.4 ± 0.5 | 0.3 ± 0.3 | – | – | 30.6 ± 0.3 | 16.4 ± 0.7 |
The melting temperatures were recorded at 0.2 µM DNA and 1 µM porphyrins.
F21D in 50Na buffer forms an antiparallel quadruplex according to CD and TDS (Supplementary Figure S11). In FRET assays both NMM and NMP were unable to stabilize this type of GQ structure (Supplementary Figure S13). These results clearly indicate NMM's and NMP's preference for parallel over antiparallel GQ DNA in agreement with UV-vis and CD results described earlier.
FRET melting assay allows facile determination of ligand's selectivity. The effect of two dsDNA models, CT and ds26, on T1/2 of F21D and FGD in the presence of fixed amount of NMM or NMP (1.6 µM) is shown in Figure 6 for F21D and Supplementary Figure S12 for FGD. The data demonstrate exceptional selectivity of both porphyrins as the T1/2 of F21D and FGD did not decrease upon addition of up to 480 equivalents of competitors (as compared to the labeled DNA) setting the selectivity ratio at least at 480. On the other hand, the stabilizing ability of widely used TMPyP4 toward F21D decreased substantially in the presence of competitors (Supplementary Figure S14), in agreement with literature (40). These results corroborate earlier UV-vis data for NMM and TMPyP4 binding to CT DNA, where no change in the position and intensity of NMM's Soret band was detected upon addition of 50-fold excess of CT DNA, while TMPyP4 displayed substantial hypochromicity and red shift under identical conditions (46).
FRET assays represent a convenient technique to test porphyrins' binding modes. Specifically to determine if NMM could stabilize F21D by interacting with loops or through intercalation, the non-loop containing GQ of different length [dTG5T]4 (G5) and [dTG8T]4 (G8) were used as competitors in FRET. These quadruplexes form parallel GQ structure in both Na+ and K+ (73,74). The T1/2 of F21D stabilized by NMM decreased substantially in the presence of large amounts of G5 or G8 as shown in Figure 6C. Similar results were obtained for FGD, Supplementary Figure S12C. The data indicate that both G5 and G8 compete effectively for NMM binding underlying once again exceptional selectivity of NMM toward parallel GQ. Since both G5 and G8 at equal concentration decrease the T1/2 of F21D by virtually the same amount, NMM's intercalation could be excluded, as G5 and G8 have different numbers of intercalation sites. Loop binding could also be excluded as neither G5 nor G8 have loops. However this statement is based on the assumption that NMM's binding mode is the same for Tel22 and G5/G8 quadruplexes, which might not be true. Selectivity ratios could be determined from FRET competition assays (for details see Supplementary Data) and based on values obtained, NMM's (and NMP's) preference for specific GQ follows the order FGD (5K) > F21D (5K) > G8 (5K) ≈ G5 (5K) >> F21D (50Na). This higher selectivity of NMM toward FGD and F21D as compared to G5 and G8 might suggest different modes of binding. More importantly, NMM displays clear discrimination against antiparallel GQ structure of F21D in 50Na.
Our FRET data taken together indicate that, against common belief, NMM and NMP have rather good GQ stabilizing properties, >480-fold selectivity for GQ over dsDNA, and remarkable selectivity for parallel versus antiparallel GQ structures of human telomeric DNA. The latter finding is extremely important as up to date only TOxaPy is reported to clearly favor a specific GQ structure (42) making NMM a second ligand in this class, but with opposite binding preference.
Stabilizing ability of NMM determined via CD melting
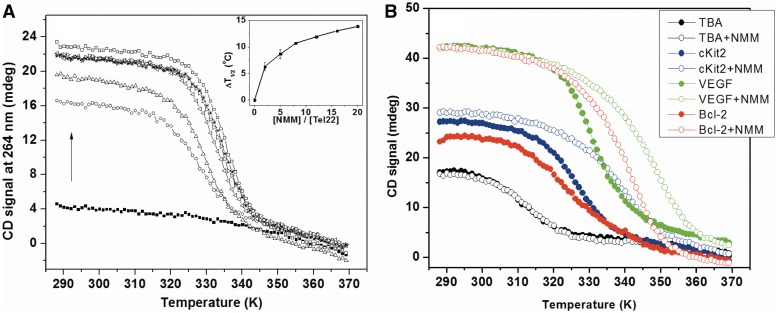
We next conducted CD melting of Tel22 alone and in the presence of NMM in order to ensure that stabilization of F21D by NMM observed in FRET was due to NMM's interaction with quadruplex and not with the fluorophores. Data were monitored at 264 and 295 nm, CD maxima for parallel and antiparallel GQ, respectively. In the absence of NMM T1/2 for Tel22 is 48.0 ± 0.8°C as determined by CD at 295 nm, while that for F21D is 44.9 ± 0.6°C as determined from FRET. Slight destabilization of a quadruplex due to fluorophore-labeling has been observed previously (75). In the presence of NMM Tel22 adopts predominantly parallel conformation whose melting temperature (determined from CD signal at 264 nm) displays dose-dependent behavior (Figure 7, inset) observed earlier in FRET (Figure 6A). At the same time, T1/2 determined from CD data at 295 nm showed no dependence on NMM concentration. In the presence of 20 equivalents of NMM, Tel22 is stabilized by 13.9°C. It is worth noting, however, that this stabilization temperature is calculated relative to T1/2 of hybrid conformation of Tel22 (monitored at 295 nm) as in the absence of NMM Tel22 does not adopt parallel GQ structure and CD signal at 264 nm does not display temperature dependence.
Figure 7.
(A) CD melting of 3.4 µM Tel22 annealed in 5K buffer alone or in the presence of 2, 5, 8, 12, 16 or 20 equivalents of NMM. All samples were equilibrated for >12 h at 30°C. Melting was monitored at 264 nm. Dependence of stabilization temperatures (ΔT1/2) on NMM concentration is shown in inset. (B) Representative CD melting curves for non-telomeric G-rich sequences (∼4.0 µM) in 5K buffer. Melting of GQs alone is show as solid circles. Melting of GQs in the presence of 2 equivalents of NMM is shown as open circles. The CD signal was monitored at 295 nm for TBA and at 264 nm for all other sequences.
In order to extract thermodynamic parameters from melting profiles, the reversibility of Tel22 folding/unfolding process was tested by recording melting and cooling curves which displayed small hysteresis of 5.2 ± 0.5°C (at 0.27°C min−1 temperature change rate). The hysteresis was independent of NMM concentration indicating that NMM most likely does not participate in the rate-determining step of Tel22 folding. The hysteresis indicates slow kinetics of Tel22 folding in the presence of NMM, also seen in annealing studies. Because hysteresis was rather small, the thermodynamic parameters were evaluated: ΔH = 261 ± 17 kJ mol−1 and ΔG(298 K) = −27 ± 1 kJ mol−1 (for heating curves). These data, while only estimates due to hysteresis, are in excellent agreement with thermodynamic parameters for unfolding of human telomeric sequences as well as other intramolecular GQs in the absence of ligands (76,77).
NMM's stabilizing abilities were also determined with respect to other quadruplex forming sequences and the results are summarized in Table 1. As in the case of Tel22 small hysteresis of 2–6°C is seen in these experiments. Two equivalents of NMM stabilized different quadruplex structures to a different degree. Specifically, structures with substantial antiparallel component were minimally stabilized: by 0 ± 2°C for Tel22 in 50Na, by 1 ± 2°C for TBA in 5K and by 0 ± 1°C for 26TelG4 in 50Na. Structures with both parallel and antiparallel components, 26TelG4 and Tel22 in 5K buffer were moderately stabilized by ∼7°C. Quadruplexes that contain predominantly parallel character (with the exception of HIF-1α) were stabilized rather effectively. VEGF, THM, G4TERT and Bcl-2 displayed the highest stabilization temperatures of ≥18°C, Table 1. Interestingly these structures also caused the largest hyperchromicity of NMM's Soret band in UV-vis titrations (Supplementary Table S2). These results yet again highlight NMM's good stabilizing ability and its preference for parallel over antiparallel GQ DNA.
Surprisingly, NMM displays no apparent stabilization of HIF-1α structure (ΔT1/2 = 1 ± 1°C) in spite of the parallel nature of this quadruplex and its ability to bind NMM as judged by a red-shift of 18.5 nm in UV-vis. HIF-1α has six GGG stretches and can form more than one stable GQ under experimental conditions. The biphasic nature of the HIF-1α melting curve (Supplementary Figure S15) indicates that at least two quadruplexes are present (a thermodynamic and a kinetic GQ). The thermodynamic GQ is more prominent under equilibrium conditions. Upon cooling a monophasic curve is observed which may be attributed to the formation of a single kinetic GQ. The melting process is accompanied by a substantial hysteresis of 17 ± 2°C. Addition of NMM leads to monophasic melting curve and small hysteresis of 1.8 ± 0.4°C, indicating that NMM favors a single (thermodynamic) GQ structure. While NMM is not required for the formation of thermodynamic product, it certainly speeds up its formation.
DISCUSSION
The main goals of this work were to provide detailed characterization of NMM's binding to human telomeric DNA, to determine its general GQ stabilizing ability, and to clarify which structural elements confer its unusual selectivity toward GQ DNA. NMM-induced structural rearrangement of Tel22 and NMM's ability to selectively interact with Tel22 in K+ buffer but not in Na+ buffer were discovered during this work, characterized and presented here as well.
NMM-induced structural rearrangement of Tel22
NMM, and to a lesser extent NMP, displayed the ability to convert Tel22 in 5K buffer to a new structure whose CD signature resembles that of a parallel GQ DNA. This transition requires potassium ions as no structural changes were observed in 100Li or 50Na buffers or in water. NMM was also able to enhance the parallel conformation of several non-telomeric GQs, including G4TERT, Bcl-2 and cKit1, highlighting the breadth of this ability. Similarly, it was reported earlier that NMM created ‘its own binding site’ on DNA aptamer possibly by rearranging the original DNA structure as the methylation protection pattern differed substantially in the presence and absence of NMM and was K+ dependent in the former case (44). As NMP caused structural rearrangement of Tel22 to a much lesser extent than NMM, the peripheral substituents on the porphyrin core might be important. There are steric and electronic differences between the ethyl groups of NMM and vinyl groups of NMP. In addition, NMP is less soluble in water than NMM.
A number of other ligands have been reported to induce structural rearrangement of human telomeric DNA (78). The majority of these ligands induced transitions to antiparallel or mixed hybrid structures and only four to a parallel conformation. The list includes a conjugate of anthracene and polyamine (79), octacationic quaternary ammonium Zn(II) phtalocyanine (80), Ni(II) salphen complex with cyclic amine head groups (33) and ZnTMPyP4 (35). In all respects, NMM acts as a very potent driver of Tel22 structural rearrangement, equal or superior to the other four molecules reported in the literature.
Details of NMM binding to and stabilization of Tel22
The NMM–Tel22 complex consists of one DNA strand and one NMM molecule binding between which is characterized by modest Ka of ∼1.0 × 105 M−1. The value of binding constant is in a good agreement with literature data: equilibrium dialysis experiments with a variety of GQ structures yielded NMM's Ka of 104–105 M−1 (40). NMM binding to G-rich DNA aptamer was an order of magnitude tighter, 1–2 × 106 M−1 (44). Finally, NMM binding constants estimated based on the NMM's inhibition of GQ unwinding by Sgs1 and BLM DNA helicases was 106 M−1 (47). Difference in GQ structure or technique of Ka determination might be responsible for this modest variation in the value of binding constants.
In spite of relatively weak binding, NMM and NMP showed excellent stabilizing ability toward fluorescently labeled human telomeric DNA in 5K buffer, >20°C for NMM and >16°C for NMP under saturating conditions. For NMM, good stabilizing ability was also confirmed in CD melting studies of Tel22 in 5K buffer with ΔT1/2 of 13.9°C at [NMM]/[Tel22] = 20. The lack of GQ stabilization by both MIX and PIX indicate that the N-methyl group of the porphyrins is integral for their association with GQ DNA. This methyl group can alter the interactions between Tel22 and NMM in at least two ways. It can interact directly with the quadruplex or it could lead to the distortion of NMM's macrocyle (81), which, in turn, can confer NMM's remarkable selectivity.
When compared to other porphyrins, NMM's and NMP's GQ stabilizing abilities are somewhat weaker (Table 2). This trend was expected as both NMM and NMP are negatively charged at physiological pH and could not engage in electrostatic interactions with the DNA backbone. However, with the added advantage of exceptional selectivity [(45,46) and here], the stabilizing abilities of NMM and NMP are rather good. In fact, at the highest concentration of the duplex competitor ds26 used (480-fold excess), the stabilization of F21D caused by TMPyP4 (ΔT1/2 = 10.0°C) is lower than that caused by both NMM (ΔT1/2 = 18.2°C) and NMP (ΔT1/2 = 15.9°C). The same is true in the case of FGD. This means that under physiological conditions NMM and NMP will perform far better than TMPyP4.
The unique structural specificity of NMM–GQ interaction raises questions about NMM’s binding mode. No structural data is available for NMM binding to GQ and the information about the binding modes is derived solely based on the spectroscopic data. UV-vis data and FRET competition experiments with dsDNA and parallel quadruplexes of different length ([dTG5T]4 and [dTG8T]4) indicate that, as expected but rarely tested, true intercalation is not likely. Groove binding could be excluded as well, because parallel stranded propeller GQ, preferred by NMM, has three of its grooves blocked by loops.
Other possible binding modes that require consideration are end-stacking and loop binding. End-stacking was observed in the NMR structure of TMPyP4-bound c-myc promoter (82) but loop binding was reported for the only crystal structure of TMPyP4-bound human telomeric DNA (83). As for NMM, the end-stacking binding mode is compatible with the fact that parallel GQ of Tel22 has easily accessible G-tetrads on both ends of its core (13), whereas the end G-tetrads of antiparallel Tel22 are crowded with loops (12). In addition, NMM binds to an antiparallel structure of TBA (although NMM does not stabilize it) possibly because TBA forms chair type antiparallel GQ whose end G-tetrads are not obstructed by loops (70). In view of NMM's remarkable selectivity, it is very possible that the interaction is more specific than simple end-stacking of conjugated systems. Parallel and antiparallel structures of Tel22 have no loop type overlap, therefore NMM's selectivity could be explained by combination of end-stacking and specific loop recognition. Loops could be considered promising targets for development of GQ-structure specific ligands. At the same time, FRET experiments suggest that NMM is well capable of binding to parallel stranded quadruplexes [dTG5T]4 and [dTG8T]4 that have no loops, therefore loop binding, while possible, might not be essential for NMM interactions with GQ DNA. Based on spectroscopic data alone, binding of NMM to quadruplex structures probably involves both end-stacking and loop-binding to a different extent for different quadruplexes. Structural data therefore are highly desirable as they can provide detailed molecular view of NMM's binding to quadruplex DNA.
Selectivity of NMM toward GQ versus dsDNA and toward different GQ structures
Greater than 10-fold selectivity of NMM toward variety of GQ structures versus a total of 46 single, double and triple stranded DNA, RNA and DNA–RNA hybrids has been demonstrated by the Chaires and Bolton groups (40,45,46). Biological studies of helicase inhibition by NMM suggested selectivity for GQ DNA versus Holliday junctions or partial duplex substrate to be >30-fold (47). Our work supports these findings and demonstrates exceptional selectivity of NMM and NMP (∼480-fold) for quadruplex versus dsDNA in FRET assays. It is important to note, that a variety GQ ligands have good to excellent selectivity for GQ DNA versus dsDNA (36,78). This selectivity, while absolutely necessary, is probably not sufficient for a promising drug candidate whose mode of action involves GQ stabilization. Selectivity of a ligand for a specific GQ target must be considered. This is a challenging task due to heterogeneity of quadruplex structures on one hand and to the presence of the same tetrad structure in each quadruplex on the other hand.
NMM displays unexpected preference for a parallel GQ structure of Tel22 in K+ buffer and no interaction with antiparallel Tel22 in Na+ buffer. Three experimental results support this conclusion. First, no red shift was observed in UV-vis titrations of NMM with Tel22 annealed in 50Na buffer. Second, NMM failed to stabilize F21D in FRET melting assays performed under the same buffer condition. Lastly, NMM was not able to alter the antiparallel structure of Tel22 in 50Na (shown by CD and gel electrophoresis) because binding of NMM to the original structure is necessary for the rearrangement. Similarly, no binding of NMM to human telomeric DNA in Na+-rich buffer was detected in competition dialysis experiments by Chaires group (40). In addition, NMM failed to alter or stabilize antiparallel structure of TBA in 5K buffer. However, red shift of 19.5 nm was observed in UV-vis titrations suggesting possible interaction. Antiparallel structure of TBA lacks diagonal loop (all loops are lateral) (70) and has an open G-tetrad available for NMM binding via end-stacking.
Contrary to our observation of NMM's selectivity for a specific GQ conformation, equilibrium dialysis experiments in a Na+-rich buffer using 14 quadruplex and i-motif structures demonstrated only 2-fold selectivity. In these experiments NMM showed the highest preference for the parallel G-wire formed by 5′-dG10T4G10 and, surprisingly, for the antiparallel quadruplex 5′-d(G4T4)3 (40). To explain the latter case we conducted preliminary annealing experiments and observed NMM-induced structural conversion of 5′-d(G4T4)3 in 50Na buffer to quadruplex with substantially increased parallel component (data not shown). The time used for equilibrium dialysis experiment is sufficient for such a transition to take place.
The first reported quadruplex ligand capable of clear discrimination between parallel and antiparallel quadruplex of human telomeric repeat is TOxaPy (42). Interestingly, with respect to Tel22, TOxaPy behaves in a manner opposite to NMM. It stabilizes F21T (where TAMRA has been used instead of Dabcyl) in Na+-rich buffer (ΔTm = 10.8°C), but not in K+ rich buffer (ΔTm < 1°C, at 1 µM ligand) (42). However, both ligands bind the parallel structure of [d(TG5T)]4 regardless of buffer (K+ or Na+). When other properties are compared, TOxaPy displays more efficient binding to quadruplex DNA (Ka = 5 × 106 M versus Ka = ∼1 × 105 M for NMM), but lower stabilizing ability and selectivity than NMM. This comparison is only qualitative as experiment type and exact conditions were similar but not identical.
NMM thus represents a second complementary example of GQ stabilizer with clear preference for a specific GQ structure or structure type. It is, therefore, a promising platform for development of efficient cancer therapeutics or probes for detection and isolation of specific GQ structures. NMM's structural elements present convenient points for synthetic modifications. Side chains and core nitrogens could be chemically modified with the hope of preparing compounds that will retain NMM's exceptional selectivity, but will have improved quadruplex binding and stabilization properties.
CONCLUSION
The all parallel propeller-loop structure of the human telomeric repeat, which has previously been shown to form only in high potassium buffer, under crowding conditions, or in the crystalline state, becomes kinetically and thermodynamically favorable in dilute, low potassium environments when Tel22 is complexed with NMM. At the same time NMM is completely insensitive to Na+ form of Tel22 and displays low binding to TBA and 26TelG4, both of which have substantial antiparallel component. NMM therefore is a second example, after TOxaPy, of GQ-ligand that has clear preference for a unique GQ structure, a property necessary for being a successful drug candidate. These two compounds have opposite preference: NMM favors parallel structures, whereas TOxaPy favors antiparallel GQs. NMM displays an excellent stabilizing ability toward human, yeast and T. thermophila telomeric sequences and a variety of oncogene promoters in dilute K+ buffer. All of these sequences contain substantial parallel component either present already or induced by NMM. While the stabilizing ability of NMM is weaker than that of TMPyP4, its selectivity is far superior making it a better GQ-stabilizer under the physiological excess of duplex DNA. The question remains about the structural details of the NMM–Tel22 complex and the kinetics and mechanism of NMM-induced structural rearrangement of Tel22 in K+ buffer. Our group is currently pursuing this work. Results, taken together, indicate that NMM presents an interesting platform toward developing a promising cancer drug that acts via structural transformation and subsequent stabilization of specific GQ targets.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figures 1–15, Supplementary Methods and Supplementary References [84–87].
FUNDING
To L.A.Y.: Camille and Henry Dreyfus Faculty Start-up Award; Cottrell College Science Award from Research Corporation (grant number 7843); Swarthmore funds. To J.L.M.: Association pour la Recherche sur le Cancer (programme libre ARC); Région Aquitaine; Fondation pour la Recherche Médicale (FRM); INCa and ANR grants (F-DNA and G4-TOOLBOX). Funding for open access charge: Swarthmore College and INSERM.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr. Johnson from University of Pennsylvania for inspiring us to work with NMM, Dr. Chaires from University of Louisville for his help with data fitting for Ka determination and Dr. Pasternack from Swarthmore College for helpful discussions.
REFERENCES
- 1.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rankin S, Reszka AP, Huppert J, Zloh M, Parkinson GN, Todd AK, Ladame S, Balasubramanian S, Neidle S. Putative DNA quadruplex formation within the human c-kit oncogene. J. Am. Chem. Soc. 2005;127:10584–10589. doi: 10.1021/ja050823u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 4.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ribeyre C, Lopes J, Boulé J-B, Piazza A, Guédin A, Zakian VA, Mergny J-L, Nicolas A. The yeast Pif1 helicase prevents genomic instability caused by G-quadruplex-forming CEB1 sequences in vivo. PLoS Genet. 2009;5:e1000475. doi: 10.1371/journal.pgen.1000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paeschke K, Capra JA, Zakian VA. DNA replication through G-quadruplex motifs Is promoted by the saccharomyces cerevisiae Pif1 DNA helicase. Cell. 2011;145:678–691. doi: 10.1016/j.cell.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopes J, Piazza A, Bermejo R, Kriegsman B, Colosio A, Teulade-Fichou M-P, Foiani M, Nicolas A. G-quadruplex-induced instability during leading-strand replication. EMBO J. 2011;30:4033–4046. doi: 10.1038/emboj.2011.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat. Struct. Mol. Biol. 2006;13:1055–1059. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- 11.Lipps HJ, Rhodes D. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol. 2009;19:414–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, Patel DJ. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 13.Parkinson GN, Lee MPH, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 14.Luu KN, Phan A, Kuryavyi V, Lacroix L, Patel DJ. Structure of the human telomere in K+ solution: an intramolecular (3 + 1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phan AT, Luu KN, Patel DJ. Different loop arrangements of intramolecular human telomeric (3 + 1) G-quadruplexes in K+ solution. Nucleic Acids Res. 2006;34:5715–5719. doi: 10.1093/nar/gkl726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Z, Dai J, Veliath E, Jones RA, Yang D. Structure of a two-G-tetrad intramolecular G-quadruplex formed by a variant human telomeric sequence in K+ solution: insights into the interconversion of human telomeric G-quadruplex structures. Nucleic Acids Res. 2010;38:1009–1021. doi: 10.1093/nar/gkp1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Y, Noguchi Y, Sugiyama H. The new models of the human telomere d[AGGG(TTAGGG)3] in K+ solution. Bioorg. Med. Chem. 2006;14:5584–5591. doi: 10.1016/j.bmc.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 19.Phan AT. Human telomeric G-quadruplex: structures of DNA and RNA sequences. FEBS J. 2010;277:1107–1117. doi: 10.1111/j.1742-4658.2009.07464.x. [DOI] [PubMed] [Google Scholar]
- 20.Heddi B, Phan AT. Structure of human telomeric DNA in crowded solution. J. Am. Chem. Soc. 2011;133:9824–9833. doi: 10.1021/ja200786q. [DOI] [PubMed] [Google Scholar]
- 21.Li J, Correia JJ, Wang L, Trent JO, Chaires JB. Not so crystal clear: the structure of the human telomere G-quadruplex in solution differs from that present in a crystal. Nucleic Acids Res. 2005;33:4649–4659. doi: 10.1093/nar/gki782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dai J, Carver M, Yang D. Polymorphism of human telomeric quadruplex structures. Biochimie. 2008;90:1172–1183. doi: 10.1016/j.biochi.2008.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyoshi D, Nakao A, Sugimoto N. Molecular crowding regulates the structural switch of the DNA G-quadruplex. Biochemistry. 2002;41:15017–15024. doi: 10.1021/bi020412f. [DOI] [PubMed] [Google Scholar]
- 24.Miyoshi D, Nakao A, Toda T, Sugimoto N. Effect of divalent cations on antiparallel G-quartet structure of d(G4T4G4) FEBS Lett. 2001;496:128–133. doi: 10.1016/s0014-5793(01)02416-4. [DOI] [PubMed] [Google Scholar]
- 25.Miyoshi D, Nakao A, Sugimoto N. Structural transition from antiparallel to parallel G-quadruplex of d(G4T4G4) induced by Ca2+ Nucleic Acids Res. 2003;31:1156–1163. doi: 10.1093/nar/gkg211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hänsel R, Löhr F, Foldynová-Trantírková S, Bamberg E, Trantírek L, Dötsch V. The parallel G-quadruplex structure of vertebrate telomeric repeat sequences is not the preferred folding topology under physiological conditions. Nucleic Acids Res. 2011;39:5768–5775. doi: 10.1093/nar/gkr174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Cian A, Mergny J-L. Quadruplex ligands may act as molecular chaperones for tetramolecular quadruplex formation. Nucleic Acids Res. 2007;35:2483–2493. doi: 10.1093/nar/gkm098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han H, Cliff CL, Hurley LH. Accelerated assembly of G-quadruplex structures by a small molecule. Biochemistry. 1999;38:6981–6986. doi: 10.1021/bi9905922. [DOI] [PubMed] [Google Scholar]
- 29.Pivetta C, Lucatello L, Paul Krapcho A, Gatto B, Palumbo M, Sissi C. Perylene side chains modulate G-quadruplex conformation in biologically relevant DNA sequences. Bioorg. Med. Chem. 2008;16:9331–9339. doi: 10.1016/j.bmc.2008.08.068. [DOI] [PubMed] [Google Scholar]
- 30.Gonçalves DPN, Rodriguez R, Balasubramanian S, Sanders JKM. Tetramethylpyridiniumporphyrazines - a new class of G-quadruplex inducing and stabilizing ligands. Chem. Commun. 2006:4685–4687. doi: 10.1039/b611731g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H, Xiao X, Wang P, Pang S, Qu F, Ai X, Zhang J. Conformational conversion of DNA G-quadruplex induced by a cationic porphyrin. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2009;74:243–247. doi: 10.1016/j.saa.2009.06.018. [DOI] [PubMed] [Google Scholar]
- 32.Zhou J, Yuan G. Specific recognition of human telomeric G-quadruplex DNA with small molecules and the conformational analysis by ESI mass spectrometry and circular dichroism spectropolarimetry. Chem. Eur. J. 2007;13:5018–5023. doi: 10.1002/chem.200601605. [DOI] [PubMed] [Google Scholar]
- 33.Arola-Arnal A, Benet-Buchholz J, Neidle S, Vilar R. Effects of metal coordination geometry on stabilization of human telomeric quadruplex DNA by square-planar and square-pyramidal metal complexes. Inorg. Chem. 2008;47:11910–11919. doi: 10.1021/ic8016547. [DOI] [PubMed] [Google Scholar]
- 34.Gonçalves DPN, Ladame S, Balasubramanian S, Sanders JKM. Synthesis and G-quadruplex binding studies of new 4-N-methylpyridinium porphyrins. Org. Biomol. Chem. 2006;4:3337–3342. doi: 10.1039/b608494j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhattacharjee AJ, Ahluwalia K, Taylor S, Jin O, Nicoludis JM, Buscaglia R, Chaires JB, Kornfilt DJP, Yatsunyk LA. Induction of G-quadruplex DNA structure by Zn(II) 5,10,15,20-tetrakis(N-methyl-4-pyridyl)porphyrin. Biochimie. 2011;93:1297–1309. doi: 10.1016/j.biochi.2011.05.038. [DOI] [PubMed] [Google Scholar]
- 36.Monchaud D, Teulade-Fichou MP. A hitchhiker's guide to G-quadruplex lilgands. Org. Biomol. Chem. 2008;6:627–636. doi: 10.1039/b714772b. [DOI] [PubMed] [Google Scholar]
- 37.Sexton AN, Collins K. The 5’ guanosine tracts of human telomerase RNA are recognized by the G-quadruplex binding domain of the RNA helicase DHX36 and function to increase RNA accumulation. Mol. Cell. Biol. 2011;31:736–743. doi: 10.1128/MCB.01033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Cian A, Cristofari G, Reichenbach P, De Lemos E, Monchaud D, Teulade-Fichou M-P, Shin-ya K, Lacroix L, Lingner J, Mergny J-L. Reevaluation of telomerase inhibition by quadruplex ligands and their mechanisms of action. Proc. Natl Acad. Sci. USA. 2007;104:17347–17352. doi: 10.1073/pnas.0707365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lacroix L, Séosse A, Mergny J-L. Fluorescence-based duplex–quadruplex competition test to screen for telomerase RNA quadruplex ligands. Nucleic Acids Res. 2011;39:e21. doi: 10.1093/nar/gkq1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ragazzon P, Chaires JB. Use of competition dialysis in the discovery of G-quadruplex selective ligands. Methods. 2007;43:313–323. doi: 10.1016/j.ymeth.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tran PLT, Largy E, Hamon F, Teulade-Fichou M-P, Mergny J-L. Fluorescence intercalator displacement assay for screening G4 ligands towards a variety of G-quadruplex structures. Biochimie. 2011;93:1288–1296. doi: 10.1016/j.biochi.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 42.Hamon F, Largy E, Guédin-Beaurepaire A, Rouchon-Dagois M, Sidibe A, Monchaud D, Mergny J-L, Riou J-F, Nguyen C-H, Teulade-Fichou M-P. An acyclic oligoheteroaryle that discriminates strongly between diverse G-quadruplex topologies. Angew. Chem. Int. Ed. 2011;50:8745–8749. doi: 10.1002/anie.201103422. [DOI] [PubMed] [Google Scholar]
- 43.De Matteis F, Gibbs AH, Smith AG. Inhibition of protohaem ferro-lyase by N-substituted porphyrins. Biochem. J. 1980;189:645–648. doi: 10.1042/bj1890645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y, Geyer CR, Sen D. Recognition of anionic porphyrins by DNA aptamers. Biochemistry. 1996;35:6911–6922. doi: 10.1021/bi960038h. [DOI] [PubMed] [Google Scholar]
- 45.Ren J, Chaires JB. Sequence and structural selectivity of nucleic acid binding ligands. Biochemistry. 1999;38:16067–16075. doi: 10.1021/bi992070s. [DOI] [PubMed] [Google Scholar]
- 46.Arthanari H, Basu S, Kawano TL, Bolton PH. Fluorescent dyes specific for quadruplex DNA. Nucleic Acids Res. 1998;26:3724–3728. doi: 10.1093/nar/26.16.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huber MD, Lee DC, Maizels N. G4 DNA unwinding by BLM and Sgs1p: substrate specificity and substrate-specific inhibition. Nucleic Acids Res. 2002;30:3954–3961. doi: 10.1093/nar/gkf530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu X, Maizels N. Substrate-specific inhibition of RecQ helicase. Nucleic Acids Res. 2001;29:1765–1771. doi: 10.1093/nar/29.8.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paramasivan S, Bolton PH. Mix and measure fluorescence screening for selective quadruplex binders. Nucleic Acids Res. 2008;36:e106. doi: 10.1093/nar/gkn487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hershman SG, Chen Q, Lee JY, Kozak ML, Yue P, Wang L-S, Johnson FB. Genomic distribution and functional analyses of potential G-quadruplex-forming sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 2008;36:144–156. doi: 10.1093/nar/gkm986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith JS, Johnson FB. In: Baumann,P. (ed), Methods in Molecular Biology, Vol. 608. Humana Press, NYC; 2010. G-quadruplex DNA: methods and protocols; pp. 207–221. [DOI] [PubMed] [Google Scholar]
- 52.Qin H, Ren J, Wang J, Wang E. G-quadruplex facilitated turn-off fluorescent chemosensor for selective detection of cupric ion. Chem. Commun. 2010;46:7385–7387. doi: 10.1039/c0cc01695k. [DOI] [PubMed] [Google Scholar]
- 53.Cantor CR, Warshaw MM, Shapiro H. Oligonucleotide interactions. 3. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers. 1970;9:1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- 54.Mergny J-L, Li J, Lacroix L, Amrane S, Chaires JB. Thermal difference spectra: a specific signature for nucleic acid structures. Nucleic Acids Res. 2005;33:1182–1192. doi: 10.1093/nar/gni134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Job P. Formation and stability of inorganic complexes in solution. Ann. Chim. 1928;9:113–203. [Google Scholar]
- 56.Gray RD, Chaires JB. Current Protocols in Nucleic Acid Chemistry. John Wiley & Sons, Inc., Hoboken NJ; 2011. pp. 17.14.11–17.14.16. [Google Scholar]
- 57.De Cian A, Guittat L, Kaiser M, Saccà B, Amrane S, Bourdoncle A, Alberti P, Teulade-Fichou M-P, Lacroix L, Mergny J-L. Fluorescence-based melting assays for studying quadruplex ligands. Methods. 2007;42:183–195. doi: 10.1016/j.ymeth.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 58.Ramsay GD, Eftink MR. Analysis of multidimensional spectroscopic data to monitor unfolding of proteins. Methods Enzymol. 1994;240:615–645. doi: 10.1016/s0076-6879(94)40066-0. [DOI] [PubMed] [Google Scholar]
- 59.Gray RD, Li J, Chaires JB. Energetics and kinetics of a conformational switch in G-quadruplex DNA. J. Phys. Chem. B. 2009;113:2676–2683. doi: 10.1021/jp809578f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Renciuk D, Kejnovska I, Skolakova P, Bednarova K, Motlova J, Vorlickova M. Arrangement of human telomere DNA quadruplex in physiologically relevant K+ solutions. Nucleic Acids Res. 2009;37:6625–6634. doi: 10.1093/nar/gkp701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gray RD, Chaires JB. Linkage of cation binding and folding in human telomeric quadruplex DNA. Biophys. Chem. 2011;159:205–209. doi: 10.1016/j.bpc.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paramasivan S, Rujan I, Bolton PH. Circular dichroism of quadruplex DNAs: applications to structure, cation effects and ligand binding. Methods. 2007;43:324–331. doi: 10.1016/j.ymeth.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 63.Karsisiotis AI, Hessari NMA, Novellino E, Spada GP, Randazzo A, Webba da Silva M. Topological characterization of nucleic acid G-quadruplexes by UV absorption and circular dichroism. Angew. Chem. Int. Ed. 2011;50:10645–10648. doi: 10.1002/anie.201105193. [DOI] [PubMed] [Google Scholar]
- 64.Dai J, Dexheimer TS, Chen D, Carver M, Ambrus A, Jones RA, Yang D. An intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands formed in the human BCL-2 promoter region in solution. J. Am. Chem. Soc. 2006;128:1096–1098. doi: 10.1021/ja055636a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Palumbo SL, Ebbinghaus SW, Hurley LH. Formation of a unique end-to-end stacked pair of G-quadruplexes in the hTERT core promoter with implications for inhibition of telomerase by G-quadruplex-interactive ligands. J. Am. Chem. Soc. 2009;131:10878–10891. doi: 10.1021/ja902281d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Phan AT, Kuryavyi V, Burge S, Neidle S, Patel DJ. Structure of an unprecedented G-quadruplex scaffold in the human c-kit promoter. J. Am. Chem. Soc. 2007;129:4386–4392. doi: 10.1021/ja068739h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.González V, Hurley LH. The c-MYC NHE III1: function and regulation. Annu. Rev. Pharmacol. Toxicol. 2010;50:111–129. doi: 10.1146/annurev.pharmtox.48.113006.094649. [DOI] [PubMed] [Google Scholar]
- 68.Sun D, Guo K, Shin Y-J. Evidence of the formation of G-quadruplex structures in the promoter region of the human vascular endothelial growth factor gene. Nucleic Acids Res. 2011;39:1256–1265. doi: 10.1093/nar/gkq926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.De Armond R, Wood S, Sun D, Hurley LH, Ebbinghaus SW. Evidence for the presence of a guanine quadruplex forming region within a polypurine tract of the hypoxia inducible factor 1α promoter. Biochemistry. 2005;44:16341–16350. doi: 10.1021/bi051618u. [DOI] [PubMed] [Google Scholar]
- 70.Macaya RF, Schultze P, Smith FW, Roe JA, Feigon J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl Acad. Sci. USA. 1993;90:3745–3749. doi: 10.1073/pnas.90.8.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pasternack RF, Gibbs EJ, Villafranca JJ. Interactions of porphyrins with nucleic acids. Biochemistry. 1983;22:2406–2414. doi: 10.1021/bi00279a016. [DOI] [PubMed] [Google Scholar]
- 72.Miller MC, Buscaglia R, Chaires JB, Lane AN, Trent JO. Hydration is a major determinant of the G-quadruplex stability and conformation of the human telomere 3′ sequence of d(AG3(TTAG3)3) J. Am. Chem. Soc. 2010;132:17105–17107. doi: 10.1021/ja105259m. [DOI] [PubMed] [Google Scholar]
- 73.De Cian A, DeLemos E, Mergny J-L, Teulade-Fichou M-P, Monchaud D. Highly efficient G-quadruplex recognition by bisquinolinium compounds. J. Am. Chem. Soc. 2007;129:1856–1857. doi: 10.1021/ja067352b. [DOI] [PubMed] [Google Scholar]
- 74.Lu M, Gou Q, Kallenbach NR. Structure and stability of sodium and potassium complexes of dT4G4 and dT4G4T. Biochemistry. 1992;31:2455–2459. doi: 10.1021/bi00124a003. [DOI] [PubMed] [Google Scholar]
- 75.Alberti P, Mergny J-L. DNA duplex–quadruplex exchange as the basis for a nanomolecular machine. Proc. Natl Acad. Sci. USA. 2003;100:1569–1573. doi: 10.1073/pnas.0335459100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chaires JB. Human telomeric G-quadruplex: thermodynamic and kinetic studies of telomeric quadruplex stability. FEBS J. 2010;277:1098–1106. doi: 10.1111/j.1742-4658.2009.07462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mergny J-L, Phan A-T, Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Letters. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 78.Georgiades SN, Abd Karim NH, Suntharalingam K, Vilar R. Interaction of metal complexes with G-quadruplex DNA. Angew. Chem. Int. Ed. 2010;49:4020–4034. doi: 10.1002/anie.200906363. [DOI] [PubMed] [Google Scholar]
- 79.Rodriguez R, Pantoş GD, Gonçalves DPN, Sanders JKM, Balasubramanian S. Ligand-driven G-quadruplex conformational switching by using an unusual mode of interaction. Angew. Chem. Int. Ed. 2007;46:5405–5407. doi: 10.1002/anie.200605075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ren L, Zhang A, Huang J, Wang P, Weng X, Zhang L, Liang F, Tan Z, Zhou X. Quaternary ammonium zinc phthalocyanine: inhibiting telomerase by stabilizing G-quadruplexes and inducing G-quadruplex structure transition and formation. ChemBioChem. 2007;8:775–780. doi: 10.1002/cbic.200600554. [DOI] [PubMed] [Google Scholar]
- 81.Senge MO, Kalisch WW, Runge S. N-Methyl derivatives of highly substituted porphyrins – the combined influence of both core and peripheral substitution on the porphyrin conformation. Liebigs Annalen. 1997;1997:1345–1352. [Google Scholar]
- 82.Phan AT, Kuryavyi V, Gaw HY, Patel DJ. Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter. Nat. Chem. Biol. 2005;1:167–173. doi: 10.1038/nchembio723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Parkinson GN, Ghosh R, Neidle S. Structural basis for binding of porphyrin to human telomeres. Biochemistry. 2007;46:2390–2397. doi: 10.1021/bi062244n. [DOI] [PubMed] [Google Scholar]
- 84.Haq I, Chowdhry BZ, Chaires JB. Singular value decomposition of 3-D DNA melting curves reveals complexity in the melting process. Eur. Biophys. J. 1997;26:419–426. doi: 10.1007/s002490050096. [DOI] [PubMed] [Google Scholar]
- 85.Henry RW, Hofrichter J. Singular Value Decomposition: Application to the analysis of experimental data. In: Brand L, Johnson ML, editors. Methods in Enzymology. Vol. 210. Academic Press, Waltham, MA; 1992. pp. 129–191. [Google Scholar]
- 86.Gray RD, Chaires JB. Kinetics and mechanism of K+- and Na+-induced folding of models of human telomeric DNA into G-quadruplex structures. Nucleic Acids Res. 2008;26:4191–4203. doi: 10.1093/nar/gkn379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Scolaro LM, Castriciano M, Romeo A, Patane S, Cefali E, Allegrini M. Aggregation behavior of Protoporphyrin IX in aqueous solutions: clear evidence of vesicle formation. J. Phys. Chem. B. 2002;106:2453–2459. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.