Abstract
Transposable elements play a fundamental role in genome evolution. It is proposed that their mobility, activated under stress, induces mutations that could confer advantages to the host organism. Transcription of the Ty1 LTR-retrotransposon of Saccharomyces cerevisiae is activated in response to a severe deficiency in adenylic nucleotides. Here, we show that Ty2 and Ty3 are also stimulated under these stress conditions, revealing the simultaneous activation of three active Ty retrotransposon families. We demonstrate that Ty1 activation in response to adenylic nucleotide depletion requires the DNA-binding transcription factor Tye7. Ty1 is transcribed in both sense and antisense directions. We identify three Tye7 potential binding sites in the region of Ty1 DNA sequence where antisense transcription starts. We show that Tye7 binds to Ty1 DNA and regulates Ty1 antisense transcription. Altogether, our data suggest that, in response to adenylic nucleotide reduction, TYE7 is induced and activates Ty1 mRNA transcription, possibly by controlling Ty1 antisense transcription. We also provide the first evidence that Ty1 antisense transcription can be regulated by environmental stress conditions, pointing to a new level of control of Ty1 activity by stress, as Ty1 antisense RNAs play an important role in regulating Ty1 mobility at both the transcriptional and post-transcriptional stages.
INTRODUCTION
Transposable elements constitute a large fraction of eukaryotic genomes (nearly half of the human genome, up to 85% of plant genomes and 3% of the compact genome of the yeast Saccharomyces cerevisiae as examples). Once seen as simple genomic parasites with potential mutagenic effects, they are currently believed to play a fundamental role in shaping genomes and triggering genetic innovations (1,2). Activation of transposable elements in response to stress conditions has been reported in a wide range of organisms (3–5) and has been proposed to promote genetic variability that could help the cell to adapt to environmental changes (6). Stress conditions generally stimulate transcription of the element, which is the first step of the transposition cycle, as shown with Tnt1A and TLC-1 in Solanaceae (3,7), Mutator in Maize (8,9), Tf2 in Schizosaccharomyces Pombe (10) and Ty1 in S. cerevisiae (11–18). Generally, this process involves regulatory sequences located in the promoter region of transposable elements, which are similar to the well-characterized motifs required for the induction of stress-responsive genes (3,4). Five Long Terminal Repeat (LTR)-retrotransposon families (Ty1–Ty5) reside in the genome of S. cerevisiae (19). They share the same basic structure, which consists of two direct LTRs and two open reading frames (ORFs), TYA and TYB, analogs of the retroviral gag and pol genes. They transpose through an RNA intermediate that is reverse-transcribed following encapsidation into a virus-like particle (VLP). Thereafter, the resulting cDNA copy is integrated into the yeast genome. With ∼30 full-length copies per haploid genome, the Ty1 family is responsible for most of the mutagenic events associated with Ty elements (4). Different environmental stresses such as ionizing radiation, DNA damage and nutrient starvation activate Ty1 transcription and retrotransposition (11–18).
The full Ty1 promoter extends over 1 kb, both upstream and downstream of two TATA boxes, and includes the 5′ LTR and part of the TYA ORF (Figure 1A). Several transcription factors bind to the Ty1 promoter to regulate Ty1 transcription (4). The DNA-binding transcription factor Tye7, was originally identified as a multicopy activator of Ty1-adjacent gene transcription (20). More recently, TYE7 was shown to be necessary for the up-regulation of Ty1 transcription in yeast cells lacking the adenylate kinase Adk1 (21). TYE7 also contributes to the activation of several glycolytic genes (22–24).
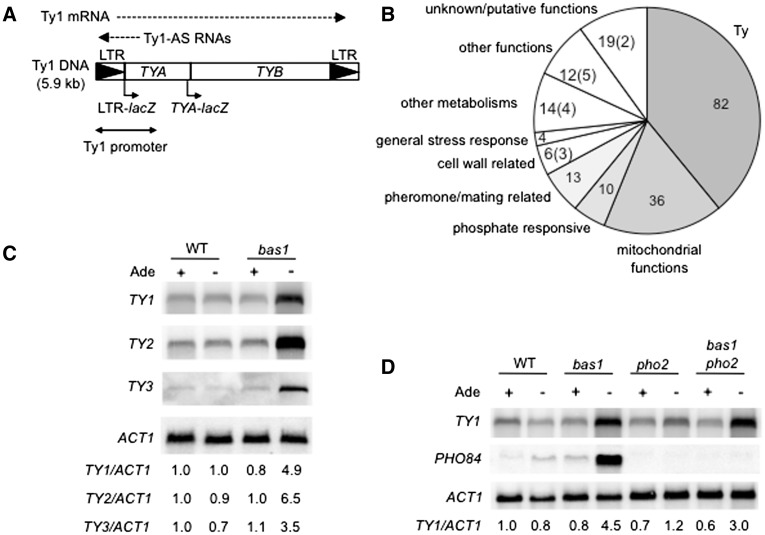
Figure 1.
(A) Ty1 structure and transcription. Ty1 structure consists of two direct long terminal repeats (LTR, symbolized by black triangles) and two open reading frames, TYA and TYB, analogs of the retroviral gag and pol genes. Ty1 transcription regulatory sequences are located within the first kilobase of the retrotransposon. Dotted arrows indicate Ty1 mRNA and Ty1-AS RNA. LTR-lacZ and TYA-lacZ fusions are indicated by bent arrows under the box. (B) Functional classes of genes differentially expressed in adenine-deprived bas1Δ cells relative to adenine-supplied bas1Δ cells. The number of genes whose expression was up- or down-regulated (in parentheses) by at least a factor of 2.5 is indicated for each class. (C) Northern-blot analysis of Ty1, Ty2 and Ty3 mRNA levels in wild-type (FYBL1-23D) and bas1Δ (LV426) cells grown in SDc minimum medium supplemented or not with adenine. For each sample, ∼15 µg of RNA were loaded onto the gel. The sizes of the mRNA molecules are 5.6 kb (Ty1 and Ty2), 5.1 kb (Ty3) and 1.3 kb (ACT1). Ratios were determined on a Molecular Dynamics Phosphorimager with ImageQuant software and set as 1 for wild-type (WT) cells grown with adenine. (D) Northern-blot analysis of Ty1 mRNA in WT (FYBL1-23D), bas1Δ (LV426) pho2Δ (LV1010) and bas1Δ pho2Δ (LV1012) cells grown with and without adenine. PHO84, which is activated by PHO2 served as a positive control (21). Growth conditions, the northern-blot experimental procedure and mRNA quantifications are described in the legend of Figure 1(C).
In addition to Ty1 mRNA, Ty1 transcription produces antisense non-coding RNAs (25,26) whose transcription starts in TYA and encompasses Ty1 promoter sequences (Figure 1A: Ty1-AS RNAs). Transcription of non-coding sequences plays an important role in the regulation of gene expression (27). In S. cerevisiae, there are several examples of non-coding RNAs, or of their transcription, regulating the expression of genes in response to nutrient deprivation, such as the IMD2 and URA2 genes of the GMP and UMP biosynthesis pathways, respectively (28–30), the phosphate responsive PHO84 and PHO5 genes (31,32), the serine biosynthesis SER3 gene (33) and the galactose-inducible GAL1-GAL10 locus (34,35). Non-coding RNAs can either be stable (36) or be rapidly degraded by the nuclear exosome or the cytoplasmic Xrn1 5′–3′ exoribonuclease (referred as Cryptic Unstable Transcripts, CUTs and Xrn1-sensitive Unstable Transcripts, XUTs, respectively) (37–40). Ty1-AS RNAs (also named Ty1-RTL), which are stabilized in the absence of Xrn1 (25,41,42), repress Ty1 transcription by a trans-silencing mechanism requiring Set1-dependent histone H3 methylation (25). Ty1-AS RNAs are also detected in VLPs, where they interfere with the accumulation of Ty1-encoded integrase and reverse-transcriptase proteins, and inhibit post-transcriptional steps of Ty1 lifecycle (26). Ty1-AS RNAs could participate to transcriptional and post-transcriptional silencing of Ty1 (43,44), as S. cerevisiae lacks the classical RNAi machinery that usually silences transposable elements in other organisms (45).
We have previously shown that Ty1 transcription is activated in adenine-deprived cells defective in de novo AMP biosynthesis (referred as conditions of severe adenine starvation), the consequence of this activation being an increase in retrotransposition (18). This activation overcomes the absence of Ste12, a transcription factor which is required for basal levels of Ty1 transcription, is independent of the Bas1 transcription activator of the de novo AMP biosynthesis pathway and involves the Swi/Snf chromatin-remodeling complex (18,46). In this report, we used a global approach to characterize the transcriptome of adenine-deprived bas1Δ cells in order to get insights into the mechanism of activation of Ty1 transcription by severe adenine starvation. We found that Ty2 and Ty3 are also activated under these stress conditions. Genes involved in ATP regeneration are also up-regulated. Their stimulation is consistent with low ATP and ADP levels measured in adenine-deprived bas1Δ cells, which suggests that a decrease in adenylic nucleotide content might be a signal of activation of Ty1 transcription. Consistently, Ty1 transcription also increases in adk1Δ cells, which have low ATP and ADP levels (21). We found that expression of the Tye7 transcription factor is induced in response to adenylic nucleotide reduction and that Tye7 contributes to activation of Ty1 expression. Importantly, Ty1-AS RNA levels decrease in adenine-deprived bas1Δ cells. We provide evidence that Tye7 is implicated in the control of Ty1-AS RNA synthesis and that its action requires sequences located in TYA, where Ty1 antisense transcription starts. Based on these data, we propose a model in which the activation of TYE7 in response to adenylic nucleotide depletion may contribute to the increase in Ty1 transcription by controlling Ty1-AS RNA synthesis.
MATERIALS AND METHODS
Yeast strains and plasmids
All strains used in this study are S288C derivatives, contain the same number of Ty retrotransposons in their genome and are described in Supplementary Table S1. Strains containing TYA-lacZ or LTR-lacZ fusions at the chromosomal locus of a native Ty1 element have already been described (14,46). All deletions were created in strains carrying TYA-lacZ or LTR-lacZ fusions, FYBL1-23D and BY4742, by one-step gene replacement, using polymerase chain reaction (PCR) fragment of HIS3, hphMX or kanMX cassettes, flanked with 5′ and 3′ sequences of the deleted gene. The 3′ end of TYE7 was tagged with 13Myc sequences by cloning, as described (47).
To construct the pPL297 plasmid that expresses a MYC-TYE7 allele, from the tetracyclin repressible promoter, TYE7 coding sequence was amplified by PCR and cloned into a derivative of pCM189 (48) containing an MYC sequence at the BamHI site of the polylinker (p2717, TETop-MYC, URA3, centromeric). Details of constructions can be obtained upon request.
Growth conditions
Yeast strains were grown in rich YPD, Hartwell's synthetic complete (HC) and synthetic minimum (SDc: SD minimal medium containing arginine, isoleucine, tryptophan, leucine and valine) media, all supplemented with 2% glucose (49). Adenine, hypoxanthine or guanine was added to a final concentration of 0.3 mM.
Microarray analyses
Precultures of wild-type (BY4742) and bas1Δ mutant (Y2951) cells were grown in SDc medium containing adenine, at 30°C, and diluted 1:200 in 10 ml of the same medium either containing or not containing adenine to reach a concentration of 107 cells/ml the next day (A600 = 1). The cultures were then diluted again in the same media to grow for 3–4 generations and harvested at a concentration of 107 cells/ml. RNA extraction, purification and labeling and cDNA probing were performed as described at http://www.transcriptome.ens.fr/sgdb/protocols/. The arrays were read with a Genepix 4000 scanner. Two hybridizations were performed for each comparison using a dye-swap procedure. Normalization was done with the lowest global method (50). The complete datasets have been deposited at the GEO database (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=xpsxlqicaqyusrk&acc=GSE28579).
Northern blots and β-galactosidase assays
Growth conditions, β-galactosidase assays, RNA extraction and northern-blot assays were performed as described in ref. 46. Probes against Ty1 [coordinates 3137 to 3682 in the TYB sequence of Ty1-H3 (51)], Ty2 [coordinates 3194 to 3747 in the TYB sequence of YLRWTy2-1 (4), Ty3 (coordinates 3213 to 4216 in the TYB sequence of YGRWTy3-1 (4)] and ACT1 mRNA were generated by random-priming (Roche). Ty1 and Ty2 probes were chosen with low homology to avoid cross-hybridization. A Ty1 antisense RNA probe [coordinates 686 to 340 in Ty1-H3 (51)] was synthesized by T7 RNA polymerase (Promega), following a procedure described in ref. 46. Northern blot assays were reproduced at least twice from independent cultures. Results were quantified on a Molecular Dynamics PhosphorImager with Image-Quant software.
Intracellular adenine derivatives content determination
Cellular extracts were prepared by an ethanol extraction method adapted from the one described by ref. (52), and metabolites were separated by high-performance liquid chromatography (HPLC), detected by UV-diode array detector and quantified as described (53). Cellular volume was determined by using a Multisizer 4 (Beckman Coulter).
Chromatin immunoprecipitation
To analyze Tye7 occupancy at the Ty1 promoter, the Tye7 protein was tagged at its C-terminus with a 13Myc-Tag, by cloning (54). The TYE7-MYC allele was expressed from TYE7 native promoter at the chromosomal locus. Chromatin immunoprecipitations (ChIPs) and real-time PCR reactions were performed essentially as described in ref. 55. Yeast strains were grown to A600 = 0.8–1 in SDc medium either supplemented or not with adenine at 22°C, and cross-linked for 10 min by the addition of formaldehyde (1.2%). The cross-linking reaction was quenched by adding glycine (0.4 M). Chromatin was sonicated to yield average DNA size fragments of 400 bp (range 100–700 bp). The chromatin solution (1:3 of total chromatin) was immunoprecipitated with 1.4 mg of antibody against the Myc epitope (9E10, Santa Cruz Biotechnology) coupled to 3 mg of Dynabeads anti-mouse IgG (Invitrogen). Immunoprecipitated DNA was quantified by real-time PCR (Platinum SYBR green qPCR supermix-UDG, Invitrogen) using a Master Cycler Realplex (Eppendorf). Primers were designed to amplify Ty1 [coordinates 561–675 of Ty1-H3 (51), forward primer O-PL499 5′ATGATGACCCAAAACCAAGC3′, reverse primer O-PL500 5′TGGATACTGCGGAAACTGTG3′], Ty2 [coordinates 559–687 of YLRWTy2-1 (4) forward primer O-PL505 5′ATGATGACCCCAAACAAAGC3′, reverse primer O-PL506 5′CTGTGGCAACGGATAGTGTG3′] and ENO1 sequences [coordinates −497, −405, relative to ATG start codon (24), forward primer O-PL503 5′TCTACTGATCCGAGCTTCCA3′, reverse primer O-PL504 5'GAGAGGCGAAAGTGGTTTTT3′] and an intergenic sequence on chromosome II (coordinates 408360–418469, O-GS55 5′GTCCCGAAGTAAGATGAGGTT3′, O-GS56 5′AGGTCTCGCAAATCAGAGG3′). For each pair of primers, a 10-fold dilution series of input DNA was used to calibrate the quantification. Real-time PCR reactions were done in triplicate in two independent experiments, using the following conditions of amplification: one cycle 2 min 95°C, 40 cycles 15 s 95°C, 15 s 55°C and 15 s 68°C.
RESULTS
The yeast transcriptome is strongly affected in adenine-deprived bas1Δ mutant
To examine the regulation of Ty1 transcription under conditions of severe adenine starvation, we compared the transcriptome of a bas1Δ strain grown with or without adenine. Expression of 197 genes was up-regulated, while that of 14 genes was down-regulated, in the absence of adenine (Supplementary Figure S1: activation/repression threshold of 2.5-fold). As a control, we compared the transcriptome of a wild-type strain grown with or without adenine and found that only 16 genes were up- or down-regulated (Supplementary Figure S1), most belonging to the AMP biosynthesis pathway (56). Strikingly, 82 out of the 197 (42%) up-regulated genes in bas1Δ cells grown in the absence of adenine matched with the TYA and TYB ORFs of Ty retrotransposons (Figure 1B): of these 82 genes, 51 corresponded to Ty1, 21 to Ty2 and five to Ty3 (Supplementary Figure S1), and the five remaining genes corresponded to Ty truncated sequences. The numbers of up-regulated Ty genes did not correspond to the numbers of Ty1, Ty2 and Ty3 elements present in the strain [31 Ty1, 12 Ty2 and 2 Ty3 (19)] because the probes that were designed to identify TYA and TYB ORFs are not specific within a Ty family, due to the strong sequence homology between the elements of each Ty family. Nevertheless, the up-regulation of Ty1, Ty2 and Ty3 ORFs in adenine-deprived bas1Δ cells indicated that the three families of Ty retrotransposons might be activated in these cells. We confirmed by northern-blot assay that steady-state levels of Ty2 and Ty3 mRNA increased in adenine-deprived bas1Δ cells (Figure 1C). Even though we already described the activation of Ty1 transcription by severe adenine starvation (18,46), these results establish that the mRNA level of Ty1, Ty2 and Ty3 elements increases simultaneously in cells defective in de novo AMP biosynthesis.
Besides Ty retrotransposons, 36 genes that were stimulated in adenine-deprived bas1Δ cells encoded proteins with mitochondrial functions (Figure 1B and Supplementary Figure S1). Among these proteins, 24 belong to oxidative phosphorylation chain complexes, such as ATP synthesis coupled proton transport, cytochrome C oxidase, cytochrome C reductase, cytochrome C and succinate dehydrogenase. Ten out of 14 genes of the PHO regulon (57), involved in phosphate uptake and storage, were also activated (Figure 1B and Supplementary Figure S1). Finally, several genes with functions in conjugation (13 genes), general stress response (four genes), and cell wall (nine genes) were also deregulated in adenine-deprived bas1Δ cells. We conclude from this analysis that conditions impairing de novo AMP biosynthesis activate the expression of Ty1, Ty2 and Ty3 retrotransposons, stress-related genes and a large number of genes related to energy production, such as the PHO and respiratory genes.
The PHO genes are activated in response to adenylic nucleotide variations by Pho2 (also known as Bas2), which is involved with Bas1 in the activation of the de novo AMP biosynthetic genes in adenine-depleted cells (21,58). This suggests that Pho2 could also be responsible for the activation of Ty1 transcription in adenine-deprived bas1Δ cells. However, Ty1 mRNA levels increased in adenine-deprived bas1Δ pho2Δ cells, indicating that Pho2 was dispensable for Ty1 activation (Figure 1D). As a control, PHO84 was activated in adenine-deprived bas1Δ cells and the activation was dependent on PHO2. Additionally, the absence of adenine did not significantly activate Ty1 expression in pho2Δ cells (Figure 1D). Altogether, these observations rule out a role of Pho2 in the activation of Ty1 by severe adenine starvation.
Activation of Ty1 transcription correlates with a decrease in intracellular ATP and ADP levels
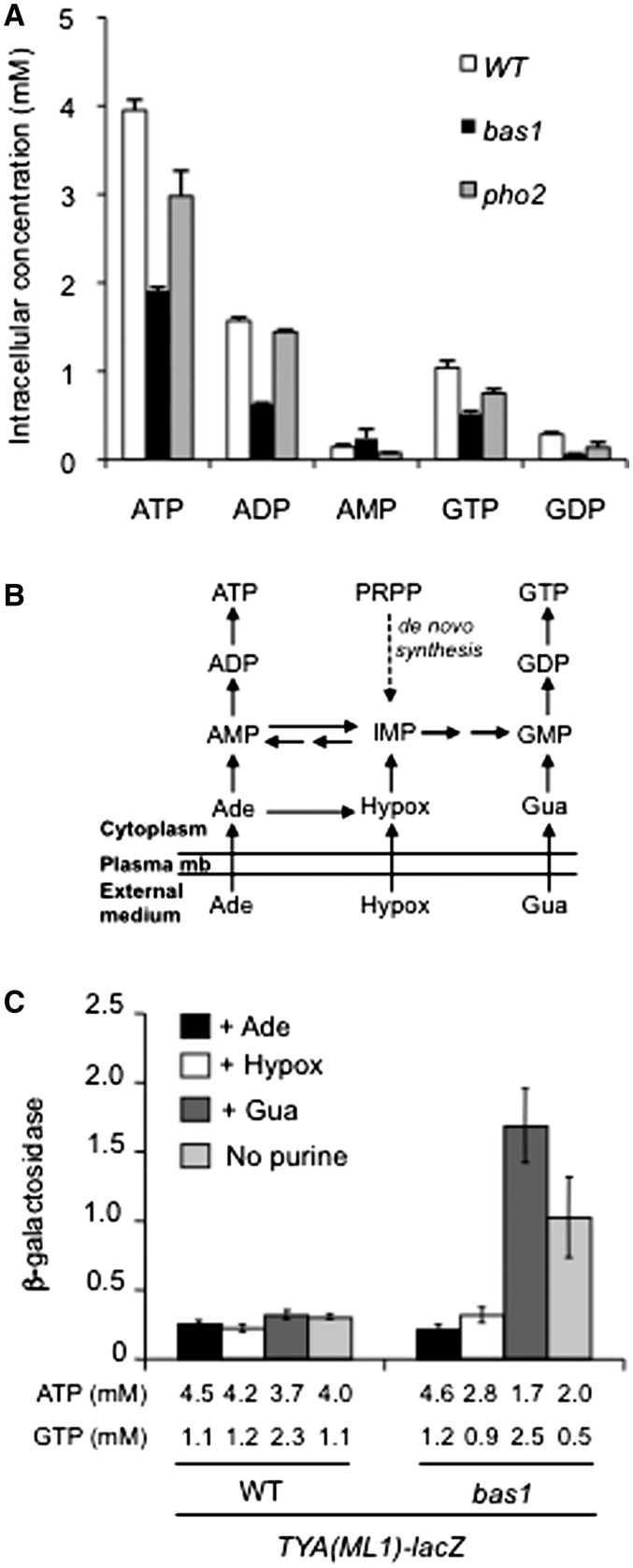
In yeast, the activation of mitochondrial and PHO genes is correlated with decrease in ATP and/or ADP levels (21,59,60). Thus, the up-regulation of mitochondrial and PHO genes in adenine-deprived bas1Δ cells indicated that intracellular adenylic nucleotide pools could be affected in these cells, leading to ATP deficiency. To search for depleted or accumulated metabolites in adenine-deprived bas1Δ cells that could account for Ty1 activation, we analyzed the intracellular concentration of purine nucleotides by HPLC in wild-type, pho2Δ and bas1Δ cells grown without adenine. We used pho2Δ cells as control, since these cells did not significantly activate Ty1 transcription in the absence of adenine (Figure 1D). Thus, we speculated that the comparison of adenine-deprived bas1Δ and pho2Δ cells would identify nucleotide variations specific to bas1Δ cells. Amounts of guanine nucleotides were also characterized, since they can be altered under certain conditions that affect de novo AMP biosynthesis (61). Quantification of the peaks indicated a significant reduction in intracellular ATP, ADP, GTP and GDP levels in adenine-deprived bas1Δ cells compared to wild-type and pho2Δ cells (Figure 2A).
Figure 2.
(A) Intracellular purine derivative contents from the WT (FYBL1-23D), bas1Δ (LV426) and pho2Δ (LV1010) strains. Cells were grown in SDc medium to mid-log phase, metabolites were extracted and intracellular nucleotide concentrations were determined by HPLC. (B) Schematic representation of de novo purine pathway in S. cerevisiae. Ade, adenine; Hypox, hypoxanthine; Gua, guanine ; IMP, inosine 5′-monophosphate; PRPP, 5-phosphoribosyl-1-pyrophosphate. (C) β-Galactosidase activity of a TYA-lacZ fusion at Ty1-ML1 in WT (LV33 Ty1(ML1)-lacZ) and bas1Δ (LV436) cells grown in SDc minimum medium supplemented or not with adenine, hypoxanthine or guanine. β-Galactosidase specific activities are expressed in nanomoles of 2-nitrophenyl β-d-galactopyranoside hydrolyzed per minute per milligram of protein. Data represent the average and standard error of three independent cultures. Intracellular concentrations of ATP and GTP were determined by HPLC as in Figure 2A and are indicated for each culture below the bars.
We previously constructed a set of strains, each expressing lacZ from the full promoter sequence of a different chromosomal Ty1 copy (i.e. the 5′LTR and part of the TYA ORF, Figure 1A, TYA-lacZ), such that the β-galactosidase activity of these strains reflects the expression of each Ty1 element (14). To establish whether the variations in adenine or guanine derivatives could account for Ty1 activation, we compared the expression of a TYA-lacZ fusion expressed from the complete Ty1 promoter sequence of the Ty1-ML1 endogenous element, in wild-type and bas1Δ cells grown in the presence or absence of guanine, adenine or hypoxanthine, which is a precursor of adenine nucleotides (Figure 2B). The fusion was expressed at high levels when bas1Δ cells were grown in the absence of both adenine and guanine. The addition of adenine or hypoxanthine but not of guanine strongly decreased β-galactosidase activity (Figure 2C). HPLC determination of intracellular ATP and GTP concentrations in these cells indicated that the addition of adenine or hypoxanthine in adenine-deprived bas1Δ cells increased intracellular ATP and GTP levels, while guanine addition restored only high intracellular GTP levels (Figure 2C). Thus, there is a strong correlation between the decrease in adenylic nucleotides, but not guanylic nucleotides in the activation of Ty1 transcription. Of note, the levels of these nucleotides were much less affected in adenine-deprived pho2Δ cells (Figure 2A), which may explain why Ty1 transcription was less activated in these cells. From these experiments, we conclude that Ty1 transcription is activated under conditions that decrease intracellular ATP and ADP levels.
TYE7 contributes to the activation of Ty1 mRNA transcription in adenine-deprived bas1Δ mutant
The DNA-binding protein Tye7 is required for the activation of Ty1 transcription in yeast cells lacking the adenylate kinase (Adk1p) responsible for the conversion of AMP into ADP (21). Interestingly, as for adenine-deprived bas1Δ cells, adk1Δ mutants have low ATP and ADP content and their transcriptome is characterized by an increase in the expression of Ty1 elements and genes of the energetic and PHO pathways (21). However, BAS1-activated genes are up-regulated in adk1Δ cells. We reproduced the activation of Ty1 transcription in adk1Δ cells using our conditions of culture and the expression of TYA-lacZ fusions introduced at three Ty1 elements (Supplementary Figure S2). Cells lacking Adk1 activated Ty1 transcription with the same preference as adenine-deprived bas1Δ cells for weakly expressed Ty1 elements (18). Our transcriptome analysis indicated that TYE7 was up-regulated 3-fold in adenine-deprived bas1Δ cells (Supplementary Figure S1). Therefore, Ty1 transcription might also be activated by a mechanism involving TYE7 in bas1Δ cells.
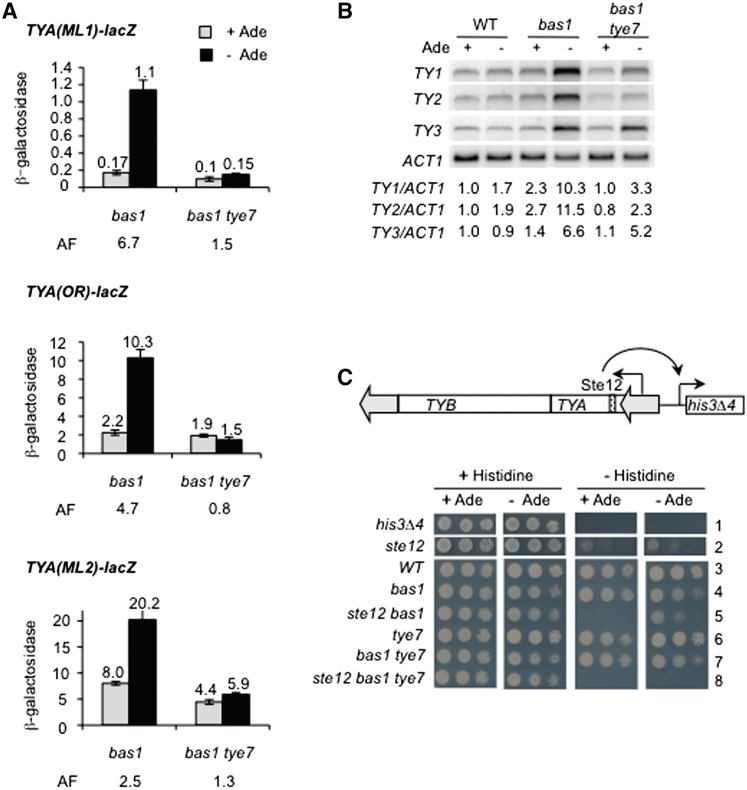
To test this hypothesis, we compared the activation of TYA-lacZ fusions introduced at three Ty1 elements, Ty1-ML1, Ty1-OR and Ty1-ML2, in bas1Δ and bas1Δ tye7Δ cells grown with or without adenine. Adenine deprivation in bas1Δ cells stimulated lacZ transcription from the three fusions 6.7-, 4.7- and 2.5-fold respectively, but activation was abolished for the three elements when TYE7 was deleted (Figure 3A). This indicates that TYE7 is essential for TYA-lacZ activation in adenine-deprived bas1Δ cells. Northern-blot analysis showed that the increase of Ty1 mRNA levels was also 3-fold lower in tye7Δ bas1Δ cells than in bas1Δ cells, under adenine deprivation (Figure 3B). Note that TYE7 deletion alone had no effect on Ty1 mRNA levels (Figure 5A) and a weak effect on expression of TYA-lacZ fusion at Ty1-DR3 and Ty1-ML2 (Supplementary Figure S3). Together, these findings indicate that TYE7 is required for the activation of Ty1 transcription under conditions of severe adenine starvation. Interestingly, TYE7 was also necessary for the activation of Ty2, but not of Ty3, in adenine-deprived bas1Δ cells, as shown in Figure 3B.
Figure 3.
(A) β-Galactosidase activity of TYA-lacZ fusions at Ty1-ML1, Ty1-OR and Ty1-ML2 in bas1Δ cells (LV436, LV658 and LV500, respectively) and bas1Δ tye7Δ cells (LV1213, LV1287 and LV1285, respectively). Growth conditions and data representations are described in the legend of Figure 2C. Exact averages of β-galactosidase specific activities are given above the bars. AF, activation factor (No adenine versus + adenine). (B) Northern-blot analysis of Ty1, Ty2 and Ty3 mRNA levels in WT (FYBL1-23D), bas1Δ (LV426) and bas1Δ tye7Δ (LV1234) cells. Growth conditions, northern-blot experimental procedure and mRNA quantifications are described in the legend of Figure 1C. (C) Growth assay of WT cells carrying a his3Δ4 allele (LV69, row 1) and WT (LV150, row 3), ste12Δ (LV993, row 2), bas1Δ (LV922, row 4), ste12Δ bas1Δ (LV926, row 5), tye7Δ (LV1370, row 6), bas1Δ tye7Δ (LV1372, row 7) and ste12Δ bas1Δ tye7Δ (LV1374, row 8) cells, carrying a Ty1-his3Δ4 allele. Cells were spotted onto plates in a series of 10-fold dilutions of 1 A600 of overnight precultures. All plates were incubated at 30°C for four days. Rows are numbered for clarity.
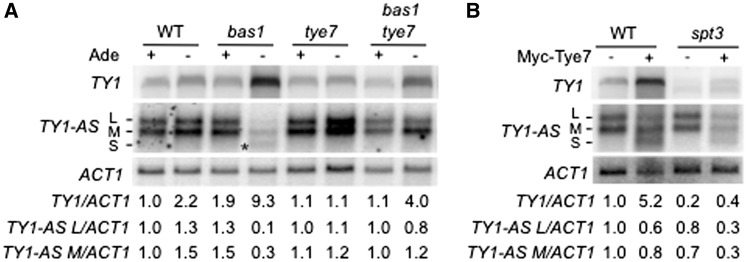
Figure 5.
(A) Northern-blot analysis of RNA extracted from WT (FYBL1-23D), bas1Δ (LV426), tye7Δ (LV1232) and bas1Δ tye7Δ (LV1234) cells. L, M and S stand for large, medium and short Ty1-AS RNA species, respectively. (B) Northern-blot analysis of Ty1 mRNA and Ty1-AS RNA levels in WT (FYBL1-23D) and spt3Δ (LV940) cells transformed with pPL297 (TETop-Myc-TYE7, URA3, CEN) or the empty vector p2717. Growth conditions, northern-blot experimental procedure and mRNA quantifications are described in the legend of Figure 1C.
We have previously shown that transcription of a gene adjacent to a full-length Ty1 element is activated in adenine-deprived bas1Δ cells (46). Since TYE7 has been identified as a gene involved in Ty1-mediated gene expression (20), we asked whether it could be responsible for activating expression of a Ty1-adjacent gene under conditions of severe adenine starvation. To address this point, we used yeast cells, which are unable to grow in the absence of histidine, unless a Ty1 element is present upstream of the promoterless his3Δ4 allele, such that his3Δ4 expression is driven from Ty1 promoter sequences (Figure 3C, rows 1 and 3). The growth of Ty1-his3Δ4 cells depends on the Ste12 transcriptional activator, which binds to the Ty1 promoter and is required for Ty1 transcription (Figure 3C, row 2 and ref. 46). We have previously shown that the histidine prototrophy of Ty1-his3Δ4 ste12Δ cells is recovered in adenine-deprived Ty1-his3Δ4 ste12Δ bas1Δ cells (Figure 3C, row 5 and ref. 46). As shown in Figure 3C row 8, this activation was dependent on TYE7. As expected, TYE7 deletion alone had no impact on the growth of Ty1-his3Δ4 in wild-type and bas1Δ cells (Figure 3C rows 6 and 7).
Altogether, these results indicate that the increase in TYE7 expression in adenine-deprived bas1Δ cells could be part of the mechanism of activation of Ty1 and Ty1-adjacent gene transcription.
TYE7 requires sequences located in TYA to activate Ty1 expression
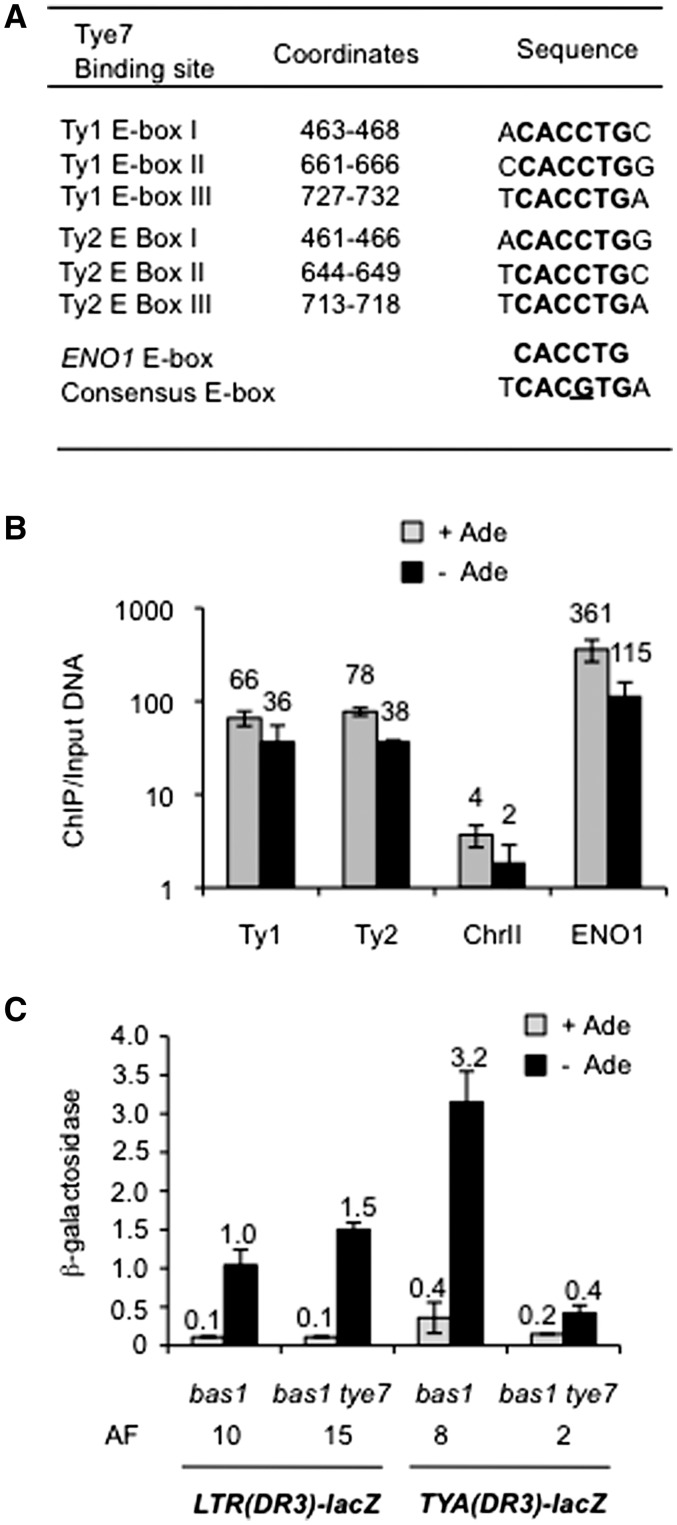
In vivo, the Tye7 protein binds to the CACCTG E-box of the ENO1 gene promoter and activates its transcription (22–24). Genome-wide analyses of Tye7-binding sites have established an extended but slightly different TCACGTGA consensus sequence (62–65). Three conserved DNA motifs identical to the ENO1 E-box are present in the TYA portion of Ty1 promoter and are conserved in the promoter of Ty2 elements (Figure 4A). By ChIP experiments, we confirmed the presence of the Tye7 protein at the Ty1 and Ty2 promoter in bas1Δ cells carrying a chromosomal Myc-tagged TYE7 allele, expressed from TYE7 promoter sequences (Figure 4B). There was no difference in the relative Tye7–Myc occupancy whether cells were grown with or without adenine. However, we could not establish whether Tye7–Myc bound to a subset of sites under normal growth conditions or to the three sites simultaneously in the absence of adenine. Indeed, the three potential E-boxes are located within a window of 269 bp, which is smaller than the average size of DNA fragments obtained upon chromatin sonication and required for the experiment. Moreover, ChIP experiment gives an average of Tye7-binding to all TYA sequences in the genome (from Ty1 or Ty2), but provides no information on Tye7-binding to individual elements, within a family.
Figure 4.
(A) Coordinates and sequences of potential Tye7 binding sites in Ty1 and Ty2 (E-box I to III). Coordinates are given relative to the numbering of Ty1-H3 (51) and YLRWTy2-1 (4). ENO1 E-box (24) and Tye7p consensus binding site (62) are indicated. (B) Chromatin immunoprecipitation (ChIP) analysis of Tye7 occupancy at the Ty1 promoter in bas1Δ (LV1058) and bas1Δ TYE7-MYC (LV1368) strain. Signals are expressed as ratios of ChIP/Total DNA and are set as 1 for untagged TYE7 bas1Δ cells grown with adenine. Data represent the average and standard error of two independent real-time PCR amplifications. (C) β-Galactosidase activity of LTR-lacZ and TYA-lacZ fusions at Ty1-DR3 in bas1Δ cells (LV1013 and LV722, respectively) and bas1Δ tye7Δ cells (LV1320 and LV1341, respectively). Growth conditions and data representations are described in the legend of Figure 2C. Exact averages of β-galactosidase specific activities are given above the bars. AF, activation factor (versus adenine).
To further analyze whether TYE7 activates Ty1 transcription through TYA sequences where the three potential E-boxes are located, we compared the expression of LTR-lacZ and TYA-lacZ fusions (Figure 1A), at the Ty1-DR3 element, in bas1Δ and bas1Δ tye7Δ cells. We previously showed that the 5′LTR of several endogenous Ty1 elements is sufficient to activate transcription of lacZ fusions in adenine-deprived bas1Δ cells, although additional sequences in TYA are necessary to optimize such activation (18,46). As expected, expression of both fusions increased significantly in bas1Δ cells grown without adenine (Figure 4C). However, in the double mutant tye7Δ bas1Δ and in the absence of adenine, the activation of the TYA-lacZ fusion was abolished, whereas the LTR-lacZ fusion was still fully activated. This result indicates that TYE7 does not stimulate Ty1 transcription through promoter sequences located in the 5′LTR but rather through sequences located in TYA ORF, and is consistent with the binding of Tye7 to potential E-boxes located in TYA.
TYE7 regulates Ty1 antisense transcription in adenine-deprived bas1Δ mutant
Ty1-AS RNAs have been reported to repress Ty1 at both transcriptional and post-transcriptional levels (25,26). Since the accumulation of Ty1-AS RNAs in an xrn1 mutant represses Ty1 transcription (25), we asked whether, conversely, adenine-deprived bas1Δ cells could prevent Ty1-AS RNA synthesis as part of the mechanism of activation of Ty1 transcription. Northern-blot analysis showed two different species of Ty1-AS RNA in wild-type cells [Figure 5A, lanes 1 and 2, Large (L) and Medium (M) species], with an estimated size comprised between 0.5 and 1-kb, as described in ref. (26). Remarkably, this pattern was altered in adenine-deprived bas1Δ cells, since these two species decreased in intensity, whereas a species of lower molecular weight appeared [Figure 5A, lane 4, Short (S) species]. Interestingly, the alteration of Ty1-AS RNA pattern correlated with an increase in Ty1 mRNA levels (Figure 5A). This suggests that the mechanism of activation of Ty1 transcription is linked to a change in Ty1-AS RNA expression in adenine-deprived bas1Δ cells. Strikingly, the Ty1-AS RNA profile was not modified in adenine-deprived tye7Δ bas1Δ cells (Figure 5A, lane 8). On the other hand, TYE7 overexpression from the tetracycline-repressed operator in wild-type and adenine-supplied bas1Δ cells reproduced the increase in Ty1 mRNA levels concomitantly to the decrease in the L and M Ty1-AS RNA and the increase in the S species, observed in adenine-deprived bas1Δ cells (Figure 5B lane 2 and Supplementary Figure S4). In adenine-deprived bas1Δ cells, TYE7 overexpression enhanced this phenotype (Supplementary Figure S4). These observations reveal the implication of TYE7 in the alteration of Ty1-AS RNA expression in adenine-deprived bas1Δ cells.
TYE7 could repress transcription of Ty1-AS RNA directly or indirectly, by activating Ty1 mRNA transcription. If Tye7 directly affects Ty1-AS RNA synthesis, down-regulation of Ty1-AS RNAs should be observed even in the absence of Ty1 mRNA transcription. To address this point, TYE7 was overexpressed from the tetracycline-repressed operator in an spt3Δ mutant, which is defective in Ty1 mRNA transcription, but has elevated levels of Ty1-AS RNAs (41,42). In the spt3Δ mutant, a similar change in the Ty1-AS RNA profile to that observed in wild-type cells was detected upon TYE7 overexpression, although Ty1 mRNA transcription did not increase significantly. These findings strongly suggest that Tye7 directly controls Ty1-AS RNA synthesis. In the spt3Δ mutant, a smaller transcript running below the full-length Ty1 transcript was detected and corresponds to an already described 5′-end truncated Ty1 transcript (66). Altogether, these results indicate that the synthesis of Ty1-AS RNAs is modified in adenine-deprived bas1Δ cells and that TYE7 is involved in the control of Ty1-AS transcription.
DISCUSSION
This work shows that a decrease in intracellular ATP and ADP levels correlates with an increase in Ty1, Ty2 and Ty3 mRNA transcription. Activation of Ty1 requires the Tye7 transcription factor, which binds to the TYA portion of Ty1 promoter, where Ty1-AS transcription occurs. We provide evidence that Tye7 also regulates Ty1-AS transcription. These data support a model in which activation of TYE7 in response to adenylic nucleotide depletion contributes to the increase in Ty1 transcription by controlling Ty1-AS RNA synthesis.
The data presented here establish that the transcription of three of the five Ty families in the yeast genome, i.e. Ty1, Ty2 and Ty3, are activated in adenine-deprived bas1Δ cells. Activation of Ty4 and Ty5 was not detected; however, Ty4 expression is extremely low and gives rise to truncated transcripts, and no functional Ty5 elements are present in S. cerevisiae laboratory strains (67,68). Although Ty1 and Ty2 are both copia-like elements sharing a high level of sequence similarity (19), the regulation of their transcription presents some differences. For instance, the transcription of Ty1 but not of Ty2 strongly depends on transcription factors Ste12 and Tec1 (13,69,70). Ty3 is a gypsy-like element with a weak homology with Ty1 and its transcription is differently regulated (71,72). Our results indicate that activation of transcription of Ty1 and Ty2 but not Ty3 depends on the Tye7 transcription factor in adenine-deprived bas1Δ cells (see below), suggesting that the transcription of Ty1 and Ty2 is regulated by a similar mechanism. Simultaneous activation of several active mobile elements, which are structurally different, has already been described in Drosophila virilis upon hybrid dysgenesis and in Maize in response to chromosome breakage (6,73). However, simultaneous transcriptional activation of different families of Ty elements by nutrient starvation is a novel finding in S. cerevisiae, although Ty1 and Ty3 share many host factors that control their transposition, mostly at post-transcriptional levels (74).
Several lines of evidence indicate that suboptimal ATP and ADP intracellular concentrations could be a signal for the activation of Ty1, Ty2 and Ty3 transcription. First, adenine-deprived bas1Δ cells contain abnormally low ATP and ADP levels and activate mitochondrial and PHO genes, which is consistent with a deficit in ATP (21,59). Second, cells lacking the major adenylate kinase Adk1 also display low ATP and ADP levels and activate Ty1 transcription. Third, although the amounts of other purine metabolites are affected in adenine-deprived bas1Δ cells (i.e. GTP and GDP), only the deficiency in ATP and ADP is consistently associated with the induction of Ty1 transcription. In the case of Ty1, we have shown that the increase in transcription is accompanied by an increase in retrotransposition (18) and activation of the expression of genes adjacent to Ty1 insertions (46). We could not identify laboratory conditions that would decrease ATP and ADP concentrations in wild-type cells, to the same extent as in adenine deprived bas1Δ cells. However, limited nutrient availability is a common situation in nature, and microorganisms are able to decrease their rate of metabolism and to survive using rare nutrient sources (75). Thus, adenine-deprived bas1Δ cells might reproduce a metabolic state of yeasts in their natural environment. Noteworthy, ‘domestication’ of yeast cells isolated from nature to grow on rich medium in the laboratory is accompanied by a decrease in Ty1 mRNA levels (76).
We provide strong evidence that the Tye7 transcription factor is involved in the mechanism of activation of Ty1 transcription. First, TYE7 transcription is activated in adenine-deprived bas1Δ cells as in adk1Δ cells (21). Second, TYE7 deletion abolishes the activation of several TYA-lacZ fusions containing the full Ty1 promoter and reduces Ty1 mRNA levels in adenine-deprived bas1Δ cells. Third, TYE7 overexpression activates Ty1 mRNA transcription in wild-type cells. We have previously shown that transcription of genes adjacent to Ty1 insertions is stimulated in adenine-deprived ste12Δ bas1Δ cells (46). Here, we demonstrate that TYE7 is necessary for this activation to occur. Fourth, Tye7 protein is present at the Ty1 promoter. We have identified three consecutive potential Tye7 binding sites (E-boxes), downstream of Ty1 transcription start site. Their location is consistent with our result indicating that TYE7 does not stimulate Ty1 transcription through promoter sequences located in the 5′LTR but rather through sequences located in TYA ORF. However, there is no evidence that all or a subset of the E-boxes are involved in Tye7-dependent regulation and we cannot exclude that other sequences in TYA could be involved in this regulation. Nevertheless, it is noteworthy that Ty1 and Ty2 contain three E-boxes and are regulated by Tye7, while Ty3 does not contain E-boxes and is not regulated by Tye7. Importantly, TYE7 deletion does not alter basal Ty1 transcription, indicating that the Tye7 protein might be essential for the full activation of Ty1 transcription under certain environmental stress conditions, only. Since the LTR-lacZ fusion at Ty1-DR3 is activated and there is a residual activation of Ty1 mRNA transcription, in adenine-deprived bas1Δ cells independently of Tye7, additional transcription factor(s) could participate in the activation of Ty1 transcription by interacting with the 5′LTR.
TYE7 alters Ty1-AS RNA synthesis in adenine-deprived bas1Δ cells or in response to Tye7 overexpression in wild-type cells. The alteration is characterized by a decrease in the levels of two Ty1-AS RNA species present under normal growth conditions and already described in ref. 26 and the appearance of a new species of lower molecular weight, whose synthesis might interfere with the synthesis of the two other Ty1-AS species. One possible model is that the alteration of Ty1 antisense transcription is the consequence of the activation of Ty1 transcription by Tye7. Supporting this hypothesis, the activation of Ty1 and Ty2 expression from sequences located downstream of the transcription start has already been described (13,69,77). However, Tye7 also down-regulates the already described Ty1 antisense transcription in a spt3Δ mutant, which is defective in Ty1 mRNA transcription. Thus, it is likely that Tye7 directly controls Ty1 antisense transcription. An attractive hypothesis is that the Tye7 protein represses Ty1 antisense transcription by binding to one or more of the three E-boxes. In support of a potential repressive role of TYE7, a previous study has reported that it could repress the transcription of the E-box-containing CIT2 gene (78). In a first model, Ty1 sense and antisense transcription could interfere in cis. A reduction in Ty1-AS RNA synthesis would therefore increase Ty1 mRNA transcription. Such a mechanism has already been described for the control of the stress-responsive SER3 gene by the SRG1 non-coding gene although, in contrast to the Ty1 situation, these two genes are adjacent and transcribed in the same direction (33). It is noteworthy that Tye7 is required for the activation of adjacent gene transcription under adenine starvation, while it down-regulates Ty1 antisense transcription, since both transcription occur in the same direction. This discrepancy could be explained by the fact that the transcription of genes adjacent to Ty1 starts from cryptic sites located in the 5′LTR (46). Thus, reducing Ty1 antisense transcription would stimulate RNA synthesis from the 5′LTR, bi-directionally. In a second model, the decrease in Ty1-AS RNA levels could relieve the basal level of Ty1 trans-silencing, since antisense Ty1-RTL RNA, which accumulate in xrn1Δ cells, has been reported to inhibit Ty1 transcription in trans, by helping to install repressive chromatin over the Ty1 promoter (25). Although we cannot discriminate between the cis and trans models, the reduction of Ty1-AS RNA levels in cells severely depleted in ATP and ADP provides evidence that Ty1-AS RNA can be regulated by environmental stress conditions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figures 1–4 and Supplementary References [14,18,79].
FUNDING
The Centre National de la Recherche Scientifique, CNRS (Institut de Biologie Physico-Chimique UPR9073, Institut Universitaire d'Hématologie UMR7212, Institut de Biochimie et Génétique Cellulaires UMR5095, Institut de Biologie de l'ENS, UMR8197); The Institut national de la santé et de la recherche médicale, Inserm (Institut Universitaire d'Hématologie U944, Institut de Biologie de l'ENS, U1024); University Paris Diderot Sorbonne Paris Cité and University Bordeaux 2; PhD fellowship from the CNRS and from the Association pour la Recherche sur le Cancer (ARC) to G.S.; PhD fellowship from the French Government (Ministère de l'Enseignement Supérieur et de la Recherche, MESR to A.T.-C. and A.L.T.; PhD fellowship from the CNRS (to A.B.-N.). Funding for open access charge: Inserm.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are very grateful to M. Springer and H. de Thé for supporting this study, A. Bensoussan, C. Condon and S. Marcand for their hospitality to perform radioactive (CC and AB) and ChIP (SM) experiments in their laboratories, A. Maes for technical help with riboprobe synthesis and members of A. Saïb laboratory for stimulating discussions. Special thanks go to S. Marcand for helpful discussion, experimental input and comments on the manuscript. We also thank JC Gluckman, A. El Hage, A. Saïb and A. Zamborlini for critical reading of the manuscript.
REFERENCES
- 1.Biemont C. A brief history of the status of transposable elements: from junk DNA to major players in evolution. Genetics. 2010;186:1085–1093. doi: 10.1534/genetics.110.124180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gogvadze E, Buzdin A. Retroelements and their impact on genome evolution and functioning. Cell. Mol. Life Sci. 2009;66:3727–3742. doi: 10.1007/s00018-009-0107-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grandbastien MA, Audeon C, Bonnivard E, Casacuberta JM, Chalhoub B, Costa AP, Le QH, Melayah D, Petit M, Poncet C, et al. Stress activation and genomic impact of Tnt1 retrotransposons in Solanaceae. Cytogenet. Genome Res. 2005;110:229–241. doi: 10.1159/000084957. [DOI] [PubMed] [Google Scholar]
- 4.Lesage P, Todeschini AL. Happy together: the life and times of Ty retrotransposons and their hosts. Cytogenet. Genome Res. 2005;110:70–90. doi: 10.1159/000084940. [DOI] [PubMed] [Google Scholar]
- 5.Oliver KR, Greene WK. Transposable elements: powerful facilitators of evolution. Bioessays. 2009;31:703–714. doi: 10.1002/bies.200800219. [DOI] [PubMed] [Google Scholar]
- 6.McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- 7.Salazar M, González E, Casaretto JA, Casacuberta JM, Ruiz-Lara S. The promoter of the TLC1.1 retrotransposon from Solanum chilense is activated by multiple stress-related signaling molecules. Plant Cell Rep. 2007;26:1861–1868. doi: 10.1007/s00299-007-0375-y. [DOI] [PubMed] [Google Scholar]
- 8.Qian Y, Cheng X, Liu Y, Jiang H, Zhu S, Cheng B. Reactivation of a silenced minimal Mutator transposable element system following low-energy nitrogen ion implantation in maize. Plant Cell Rep. 2010;29:1365–1376. doi: 10.1007/s00299-010-0922-9. [DOI] [PubMed] [Google Scholar]
- 9.Qüesta JI, Walbot V, Casati P. Mutator transposon activation after UV-B involves chromatin remodeling. Epigenetics. 2010;5:352–363. doi: 10.4161/epi.5.4.11751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sehgal A, Lee CY, Espenshade PJ. SREBP controls oxygen-dependent mobilization of retrotransposons in fission yeast. PLoS Genet. 2007;3:e131. doi: 10.1371/journal.pgen.0030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bradshaw VA, McEntee K. DNA damage activates transcription and transposition of yeast Ty retrotransposons. Mol. Gen. Genet. 1989;218:465–474. doi: 10.1007/BF00332411. [DOI] [PubMed] [Google Scholar]
- 12.McClanahan T, Mc Entee K. Specific transcripts are elevated in Saccharomyces cerevisiae in response to DNA damage. Mol. Cell. Biol. 1984;4:2356–2363. doi: 10.1128/mcb.4.11.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morillon A, Springer M, Lesage P. Activation of the Kss1 invasive-filamentous growth pathway induces Ty1 transcription and retrotransposition in Saccharomyces cerevisiae. Mol. Cell. Biol. 2000;20:5766–5776. doi: 10.1128/mcb.20.15.5766-5776.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morillon A, Bénard L, Springer M, Lesage P. Differential effects of chromatin and Gcn4 on the 50-fold range of expression among individual yeast Ty1 retrotransposons. Mol. Cell. Biol. 2002;22:2078–2088. doi: 10.1128/MCB.22.7.2078-2088.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rolfe M, Spanos A, Banks G. Induction of yeast Ty element transcription by ultraviolet. Nature. 1986;319:339–340. [Google Scholar]
- 16.Sacerdot C, Mercier G, Todeschini AL, Dutreix M, Springer M, Lesage P. Impact of ionizing radiation on the life cycle of Saccharomyces cerevisiae Ty1 retrotransposon. Yeast. 2005;22:441–455. doi: 10.1002/yea.1222. [DOI] [PubMed] [Google Scholar]
- 17.Staleva Staleva L, Venkov P. Activation of Ty transposition by mutagens. Mutat. Res. 2001;474:93–103. doi: 10.1016/s0027-5107(00)00165-2. [DOI] [PubMed] [Google Scholar]
- 18.Todeschini AL, Morillon A, Springer M, Lesage P. Severe adenine starvation activates Ty1 transcription and retrotransposition in Saccharomyces cerevisiae. Mol. Cell. Biol. 2005;25:7459–7472. doi: 10.1128/MCB.25.17.7459-7472.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim JM, Vanguri S, Boeke JD, Gabriel A, Voytas DF. Transposable elements and genome organization: a comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 1998;8:464–478. doi: 10.1101/gr.8.5.464. [DOI] [PubMed] [Google Scholar]
- 20.Löhning C, Ciriacy M. The TYE7 gene of Saccharomyces cerevisiae encodes a putative bHLH-LZ transcription factor required for Ty1-mediated gene expression. Yeast. 1994;10:1329–1339. doi: 10.1002/yea.320101010. [DOI] [PubMed] [Google Scholar]
- 21.Gauthier S, Coulpier F, Jourdren L, Merle M, Beck S, Konrad M, Daignan-Fornier B, Pinson B. Co-regulation of yeast purine and phosphate pathways in response to adenylic nucleotide variations. Mol. Microbiol. 2008;68:1583–1594. doi: 10.1111/j.1365-2958.2008.06261.x. [DOI] [PubMed] [Google Scholar]
- 22.Nishi K, Park CS, Pepper AE, Eichinger G, Innis MA, Holland MJ. The GCR1 requirement for yeast glycolytic gene expression is suppressed by dominant mutations in the SGC1 gene, which encodes a novel basic-helix-loop-helix protein. Mol. Cell. Biol. 1995;15:2646–2653. doi: 10.1128/mcb.15.5.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sato T, Lopez MC, Sugioka S, Jigami Y, Baker HV, Uemura H. The E-box DNA binding protein Sgc1p suppresses the gcr2 mutation, which is involved in transcriptional activation of glycolytic genes in Saccharomyces cerevisiae. FEBS Lett. 1999;463:307–311. doi: 10.1016/s0014-5793(99)01654-3. [DOI] [PubMed] [Google Scholar]
- 24.Chen M, Lopes JM. Multiple basic helix-loop-helix proteins regulate expression of the ENO1 gene of Saccharomyces cerevisiae. Eukaryot. Cell. 2007;6:786–796. doi: 10.1128/EC.00383-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berretta J, Pinskaya M, Morillon A. A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S. cerevisiae. Genes Dev. 2008;22:615–626. doi: 10.1101/gad.458008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuda E, Garfinkel DJ. Posttranslational interference of Ty1 retrotransposition by antisense RNAs. Proc. Natl Acad. Sci. USA. 2009;106:15657–15662. doi: 10.1073/pnas.0908305106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wery M, Kwapisz M, Morillon A. Noncoding RNAs in gene regulation. Wiley Interdiscip. Rev. Syst. Biol. Med. 2011;3:728–738. doi: 10.1002/wsbm.148. [DOI] [PubMed] [Google Scholar]
- 28.Kuehner JN, Brow DA. Regulation of a eukaryotic gene by GTP-dependent start site selection and transcription attenuation. Mol. Cell. 2008;31:201–211. doi: 10.1016/j.molcel.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 29.Kwapisz M, Wery M, Després D, Ghavi-Helm Y, Soutourina J, Thuriaux P, Lacroute F. Mutations of RNA polymerase II activate key genes of the nucleoside triphosphate biosynthetic pathways. EMBO J. 2008;27:2411–2421. doi: 10.1038/emboj.2008.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thiebaut M, Colin J, Neil H, Jacquier A, Séraphin B, Lacroute F, Libri D. Futile cycle of transcription initiation and termination modulates the response to nucleotide shortage in S. cerevisiae. Mol. Cell. 2008;31:671–682. doi: 10.1016/j.molcel.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 31.Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell. 2007;131:706–717. doi: 10.1016/j.cell.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 32.Uhler JP, Hertel C, Svejstrup JQ. A role for noncoding transcription in activation of the yeast PHO5 gene. Proc. Natl Acad. Sci. USA. 2007;104:8011–8016. doi: 10.1073/pnas.0702431104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martens JA, Wu PY, Winston F. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 2005;19:2695–2704. doi: 10.1101/gad.1367605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Houseley J, Rubbi L, Grunstein M, Tollervey D, Vogelauer M. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol. Cell. 2008;32:685–695. doi: 10.1016/j.molcel.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinskaya M, Gourvennec S, Morillon A. H3 lysine 4 di- and tri-methylation deposited by cryptic transcription attenuates promoter activation. EMBO J. 2009;28:1697–1707. doi: 10.1038/emboj.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Münster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Régnault B, Devaux F, Namane A, Séraphin B, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 38.Thompson DM, Parker R. Cytoplasmic decay of intergenic transcripts in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007;27:92–101. doi: 10.1128/MCB.01023-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee A, Hansen KD, Bullard J, Dudoit S, Sherlock G. Novel low abundance and transient RNAs in yeast revealed by tiling microarrays and ultra high-throughput sequencing are not conserved across closely related yeast species. PLoS Genet. 2008;4:e1000299. doi: 10.1371/journal.pgen.1000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Dijk EL, Chen CL, d'Aubenton-Carafa Y, Gourvennec S, Kwapisz M, Roche V, Bertrand C, Silvain M, Legoix-Né P, Loeillet S, et al. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature. 2011;475:114–117. doi: 10.1038/nature10118. [DOI] [PubMed] [Google Scholar]
- 41.Checkley MA, Nagashima K, Lockett SJ, Nyswaner KM, Garfinkel DJ. P-body components are required for Ty1 retrotransposition during assembly of retrotransposition-competent virus-like particles. Mol. Cell. Biol. 2010;30:382–398. doi: 10.1128/MCB.00251-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dutko JA, Kenny AE, Gamache ER, Curcio MJ. 5' to 3′ mRNA decay factors colocalize with Ty1 gag and human APOBEC3G and promote Ty1 retrotransposition. J. Virol. 2010;84:5052–5066. doi: 10.1128/JVI.02477-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang YW. Transcriptional cosuppression of yeast Ty1 retrotransposons. Genes Dev. 2002;16:467–478. doi: 10.1101/gad.923502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garfinkel DJ, Nyswaner K, Wang J, Cho JY. Post-transcriptional cosuppression of Ty1 retrotransposition. Genetics. 2003;165:83–99. doi: 10.1093/genetics/165.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Girard A, Hannon GJ. Conserved themes in small-RNA-mediated transposon control. Trends Cell. Biol. 2008;18:136–148. doi: 10.1016/j.tcb.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Servant G, Pennetier C, Lesage P. Remodeling yeast gene transcription by activating the Ty1 long terminal repeat retrotransposon under severe adenine deficiency. Mol. Cell. Biol. 2008;28:5543–5554. doi: 10.1128/MCB.00416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 48.Gari E, Piedrafita L, Aldea M, Herrero E. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast. 1997;13:837–848. doi: 10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 49.Adams A, Gottschling DE, Kaiser CA, Stearns T. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- 50.Lemoine S, Combes F, Servant N, Le Crom S. Goulphar: rapid access and expertise for standard two-color microarray normalization methods. BMC Bioinform. 2006;7:467. doi: 10.1186/1471-2105-7-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boeke JD, Eichinger D, Castrillon D, Fink GR. The Saccharomyces cerevisiae genome contains functional and nonfunctional copies of transposon Ty1. Mol. Cell. Biol. 1988;8:1432–1442. doi: 10.1128/mcb.8.4.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loret MO, Pedersen L, Francois J. Revised procedures for yeast metabolites extraction: application to a glucose pulse to carbon-limited yeast cultures, which reveals a transient activation of the purine salvage pathway. Yeast. 2007;24:47–60. doi: 10.1002/yea.1435. [DOI] [PubMed] [Google Scholar]
- 53.Laporte D, Lebaudy A, Sahin A, Pinson B, Ceschin J, Daignan-Fornier B, Sagot I. Metabolic status rather than cell cycle signals control quiescence entry and exit. J. Cell. Biol. 2011;192:949–957. doi: 10.1083/jcb.201009028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 55.Takahashi T, Burguiere-Slezak G, Van der Kemp PA, Boiteux S. Topoisomerase 1 provokes the formation of short deletions in repeated sequences upon high transcription in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2010;108:692–697. doi: 10.1073/pnas.1012582108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pinson B, Vaur S, Sagot I, Coulpier F, Lemoine S, Daignan-Fornier B. Metabolic intermediates selectively stimulate transcription factor interaction and modulate phosphate and purine pathways. Genes Dev. 2009;23:1399–1407. doi: 10.1101/gad.521809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ogawa N, DeRisi J, Brown PO. New components of a system for phosphate accumulation and polyphosphate metabolism in Saccharomyces cerevisiae revealed by genomic expression analysis. Mol. Biol. Cell. 2000;11:4309–4321. doi: 10.1091/mbc.11.12.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Daignan-Fornier B, Fink GR. Coregulation of purine and histidine biosynthesis by the transcriptional activators BAS1 and BAS2. Proc. Natl Acad. Sci. USA. 1992;89:6746–6750. doi: 10.1073/pnas.89.15.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Amiott EA, Jaehning JA. Mitochondrial transcription is regulated via an ATP “sensing” mechanism that couples RNA abundance to respiration. Mol. Cell. 2006;22:329–338. doi: 10.1016/j.molcel.2006.03.031. [DOI] [PubMed] [Google Scholar]
- 60.Zhang F, Kirouac M, Zhu N, Hinnebusch AG, Rolfes RJ. Evidence that complex formation by Bas1p and Bas2p (Pho2p) unmasks the activation function of Bas1p in an adenine-repressible step of ADE gene transcription. Mol. Cell. Biol. 1997;17:3272–3283. doi: 10.1128/mcb.17.6.3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saint-Marc C, Pinson B, Coulpier F, Jourdren L, Lisova O, Daignan-Fornier B. Phenotypic consequences of purine nucleotide imbalance in Saccharomyces cerevisiae. Genetics. 2009;183:529–538. doi: 10.1534/genetics.109.105858. 521SI–527SI. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol. Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhu C, Byers KJ, McCord RP, Shi Z, Berger MF, Newburger DE, Saulrieta K, Smith Z, Shah MV, Radhakrishnan M, et al. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 2009;19:556–566. doi: 10.1101/gr.090233.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gordân R, Hartemink AJ, Bulyk ML. Distinguishing direct versus indirect transcription factor–DNA interactions. Genome Res. 2009;19:2090–2100. doi: 10.1101/gr.094144.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Winston F, Durbin KJ, Fink GR. The SPT3 gene is required for normal transcription of Ty elements in S. cerevisiae. Cell. 1984;39:675–682. doi: 10.1016/0092-8674(84)90474-4. [DOI] [PubMed] [Google Scholar]
- 67.Hug AM, Feldmann H. Yeast retrotransposon Ty4 – the majority of the rare transcripts lack a U3-R sequence. Nucleic Acids Res. 1996;24:2338–2346. doi: 10.1093/nar/24.12.2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Voytas DF, Boeke JD. Yeast retrotransposon revealed. Nature. 1992;358:717. doi: 10.1038/358717a0. [DOI] [PubMed] [Google Scholar]
- 69.Laloux I, Jacobs E, Dubois E. Involvement of SRE element of Ty1 transposon in TEC1-dependent transcriptional activation. Nucleic Acids Res. 1994;22:999–1005. doi: 10.1093/nar/22.6.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Türkel S, Farabaugh PJ. Interspersion of an unusual GCN4 activation site with a complex transcriptional repression site in Ty2 elements of Saccharomyces cerevisiae. Mol. Cell. Biol. 1993;13:2091–2103. doi: 10.1128/mcb.13.4.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bilanchone VW, Claypool JA, Kinsey PT, Sandmeyer SB. Positive and negative regulatory elements control expression of the yeast retrotransposon Ty3. Genetics. 1993;134:685–700. doi: 10.1093/genetics/134.3.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kinsey PT, Sandmeyer SB. Ty3 transposes in mating populations of yeast: a novel transposition assay for Ty3. Genetics. 1995;139:81–94. doi: 10.1093/genetics/139.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Petrov DA, Schutzman JL, Hartl DL, Lozovskaya ER. Diverse transposable elements are mobilized in hybrid dysgenesis in Drosophila virilis. Proc. Natl Acad. Sci. USA. 1995;92:8050–8054. doi: 10.1073/pnas.92.17.8050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maxwell PH, Curcio MJ. Host factors that control long terminal repeat retrotransposons in Saccharomyces cerevisiae: implications for regulation of mammalian retroviruses. Eukaryot. Cell. 2007;6:1069–1080. doi: 10.1128/EC.00092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Palkova Z. Multicellular microorganisms: laboratory versus nature. EMBO Rep. 2004;5:470–476. doi: 10.1038/sj.embor.7400145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kuthan M, Devaux F, Janderová B, Slaninová I, Jacq C, Palkova Z. Domestication of wild Saccharomyces cerevisiae is accompanied by changes in gene expression and colony morphology. Mol. Microbiol. 2003;47:745–754. doi: 10.1046/j.1365-2958.2003.03332.x. [DOI] [PubMed] [Google Scholar]
- 77.Farabaugh PJ, Vimaladithan A, Türkel S, Johnson R, Zhao H. Three downstream sites repress transcription of a Ty2 retrotransposon in Saccharomyces cerevisiae. Mol. Cell. Biol. 1993;13:2081–2090. doi: 10.1128/mcb.13.4.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen L, Lopes JM. Multiple bHLH proteins regulate CIT2 expression in Saccharomyces cerevisiae. Yeast. 2010;27:345–359. doi: 10.1002/yea.1757. [DOI] [PubMed] [Google Scholar]
- 79.Fairhead C, Llorente B, Denis F, Soler M, Dujon B. New vectors for combinatorial deletions in yeast chromosomes and for gap-repair cloning using ‘split-marker’ recombination. Yeast. 1996;12:1439–1457. doi: 10.1002/(SICI)1097-0061(199611)12:14%3C1439::AID-YEA37%3E3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.