Motion and force in biological systems are produced by motor proteins, which reach out from one set of filaments or organelles and translate these relative to a second set of filaments. These motor proteins take the chemical energy of ATP and convert it into mechanical energy. The laws of physics dictate that the production of mechanical energy must involve the exertion of a force through a displacement. Recent work has shown that forces are of the order of 5–10 pN and displacements are between 5–10 nm (1, 2). Basically, there are two simple ways in which a motor protein could produce a force through a distance. In one model the motor protein could produce relative translation by attaching to one of the filaments followed by a shortening of some element, most likely the element tethering it to the first set of filaments, thereby effecting a translation. In the second model the motor protein could attach to the second set of filaments and then undergo a rotation, thus acting as a “rowing oar” propelling one set of filaments past the other. Studies of both the structure and function of myosin have suggested that it functions by the second of the two above hypotheses. Electron microscopy showed that myosin could assume different orientations in the filament lattice of the muscle, a conclusion that was supported by x-ray diffraction patterns (3, 4). In addition, it was shown that the head region of myosin interacting with actin could generate force and displacement, eliminating any structural element that could reasonably shorten (5). Thus the current paradigm for explaining the action of myosin is that at least a portion of the myosin head must rotate during the power stroke. The goal of the field for many years has been to identify which portions rotate and to accurately measure this rotation. The results of Baker et al., reported in this issue of the Proceedings, make a major step forward toward this goal (6).
Crystallographic structures of both actin and the head region of myosin have been determined (7, 8), and these, along with previous data from spectroscopic probes, have suggested which portions of myosin might rotate. The myosin head has a large, globular region known as the catalytic domain, which contains the site for interaction with actin and ATP. A second region of the myosin head, known as the light chain domain, consists of a single α-helix, stabilized by two light chains that surround it. This helix connects the catalytic domain to the α-helical myosin tail, which both forms the thick filament and tethers the myosin head to the thick filament. Myosin heads bind tightly to actin in the “rigor complex,” which is thought to exist at the end of the power stroke, and models of this complex show that the catalytic domain attaches strongly and apparently in a rigid fashion to the actin, whereas the light chain domain extends away from the actin filament, forming an approximately 45° angle with the axis of the filament (Fig. 1; refs. 9 and 10). Early work with paramagnetic and fluorescent spectroscopic probes, along with electron micrographs of myosin heads bound to actin in the presence of ATP, had all suggested that the catalytic domain attaches rigidly to actin and does not rotate during the power stroke (11–14). Together with the structures, this suggested that the light chain domain might form the rowing oar that generated tension and displacement. This hypothesis has been supported by the observation that the velocity generated by myosin fragments depends on the length of the light chain domain (15). In addition, the orientation of the light chain domain was observed to move upon release of ADP, but this did not occur in skeletal muscle and may not be associated with a power stroke (16–18).
Figure 1.
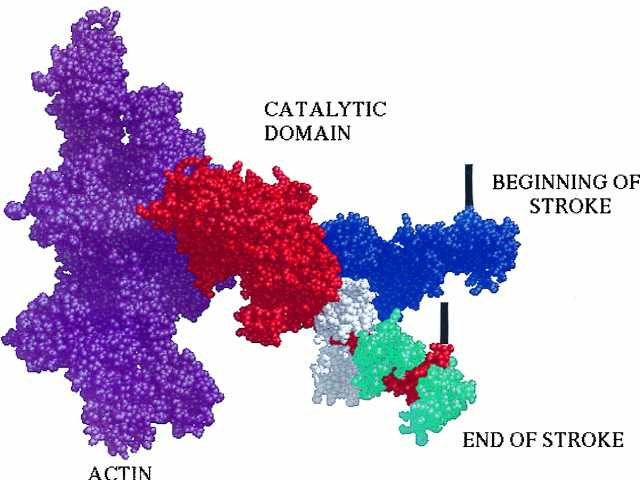
A model of the actomyosin complex illustrates how the spectroscopic results described by Baker et al. (6) might translate into the domain movements of myosin. The actin filament is shown in purple on the left. The catalytic domain of the myosin head, shown in red, attaches rigidly to the actin filament, with its light chain domain extending down at about a 45° angle. The two light chains that help form this domain are shown in white and cyan, with the spectroscopic label of Baker et al. (6) attached to the latter. The light chain domain is also shown in a second orientation in blue, rotated upward by approximately 36° to simulate the beginning of the power stroke suggested by the results of Baker et al. (6). The power stroke would consist of a rotation of this domain from the upper position to the lower. This rotation would pull the tether that connects the myosin head to the thick filament, down by about 5 nm. The position of the tether is depicted schematically by the thick black vertical lines. The model is derived from the coordinates of the crystal structures and displayed in Midas (7–10).
The light chain domain of myosin has been the object of a number of spectroscopic studies using extrinsic probes to determine whether it indeed does rotate. Fluorescent probes attached to one of the light chains, the LC2, have suggested that it undergoes small rotations, through angles of about 3°, when active muscle is shortened or lengthened (19). The interpretation of this result depends on the fraction of the myosin heads that were actively attached to actin. This fraction is not known, but if it is small the angular change of the active heads could be larger than the 3° observed for the whole ensemble of heads. However, fluorescence does not have the resolution to resolve several species of probes with different angles. Electron paramagnetic resonance spectroscopy of nitroxide spin labels has such angular resolution, and Baker et al. (6) use it to show that large angular changes of the light chain domain do occur and that changes in these angles are associated with the generation of force.
Muscle fibers are commonly observed in three states. (i) In rigor—i.e., rigor mortis—the state obtained in the absence of ATP, all myosin heads are bound rigidly to actin in a configuration thought to resemble that found at the end of the power stroke. (ii) In relaxation, myosin heads are largely detached from actin and may be bound in a helical array around the thick filament or may be disordered and not bound to either filament. (iii) In active muscle, the myosin heads are undergoing a cyclic interaction with actin producing force. Baker et al. (6) observe their spin probe in these three states, finding results that are radically different from those found by previous investigators.
Like other investigators, they find that the light chain domain is relatively well ordered with respect to the fiber axis in rigor fibers. This has been a surprising result, because one might expect that the steric restraints of the filament array would create some disorder in this part of the myosin. Recent unpublished work from the same lab, in which the spin probe is placed on isolated myosin heads that are not attached to a thick filament, shows a spectrum similar to that seen for rigor fibers. Thus the lattice restraints produce little distortion of the light chain domain, suggesting that distortions occur in the element that tethers the heads to the thick filament.
The big surprise comes from the spectra of relaxed fibers, where previous spin probes and fluorescent probes have reported a fairly high degree of disorder in both the catalytic and light chain domains. Here Baker et al. find two distinct orientations, one centered at 74°, the same as found in rigor fibers, and a second, centered at 38°, showing that the angles of the probes themselves differ by at least 36° (Fig. 1). This is the first time that a spectroscopic technique has seen two distinct angles for any portion of the myosin molecule, and the angles differ by an amount that is appropriate for driving a power stroke of some 5 nm or more. The exact correspondence of these two orientations with distinct biochemical states remains to be determined, but one orientation, that found for rigor muscle, must represent myosin heads that are oriented via an interaction with the actin filament. The second orientation must represent myosin heads oriented by a weak interaction either with the actin filament or the myosin filament.
In active fibers Baker et al. (6) find the same two orientations, but their relative populations have shifted with a slightly greater fraction of myosin heads associated with the rigor orientation. Thus in active fibers some fraction of myosin heads has altered the angle of the light chain domain by some 36°. Baker et al. (6) present a model in which the orientation at 38° arises from myosin heads bound to actin at the beginning of the power stroke whereas that at 74° represents myosin heads bound at the end of the power stroke. A cyclic interaction drives attached heads between these two orientations, resulting in force and relative filament translation. Because the spectra of active fibers can be deconvoluted into two orientations, with no evidence of a distribution of heads between these, the data suggest that myosin heads are oriented either at the beginning or at the end of their power stroke, but not distributed to any extent in between. This puts quite a constraint on possible models of force generation.
The results of Baker et al. (6) mark a distinct advance in the field of motor proteins with the finding of two distinct angles for the light chain region of myosin. Why have they succeeded where others have failed? First, they use EPR spectroscopy, which allows the resolution of different angular distributions. Second, they attain a greater degree of exchange under milder conditions, made possible by the use of scallop muscle fibers. Scallop, unlike mammalian striated fibers, is regulated by binding of calcium to the light chain domain. Are their results peculiar to scallop fibers, or are they general? Baker et al. find similar angles when their light chain is exchanged in skeletal fibers, but there is a greater disordered component, and the spectral changes occurring on activation are almost absent. Further work will be required to understand species differences and to fit the results of Baker et al. into a larger biochemical scheme. However, they have achieved one of the holy grails in the field of motility—the observation of two distinct and substantially different angles for a portion of the myosin head.
These results help at last to establish a model that was first proposed more than 30 years ago. A portion of myosin, the light chain domain, rotates during the power stroke, acting as a lever arm, which produces both force and displacement. Now that we appear to understand at least superficially how myosin works as a motor protein, along comes the kinesin family of motors. The motor domain of these motors has strong structural homology with the motor domain of myosin, suggesting that they work by a similar mechanism (20). However, these motors have no apparent structural element that could act as a lever arm. Nature is full of surprises.
References
- 1.Finer J T, Simmons R M, Spudich J A. Nature (London) 1994;368:113–119. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- 2.Svoboda K, Schmidt C F, Schnapp B J, Block S M. Nature (London) 1993;365:721–727. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- 3.Reedy R T, Holmes K C, Tregear R T. Nature (London) 1965;207:1276–1280. doi: 10.1038/2071276a0. [DOI] [PubMed] [Google Scholar]
- 4.Huxley H E. Science. 1969;164:1356–1366. [PubMed] [Google Scholar]
- 5.Toyoshima Y Y, Kron E M, McNally K R, Neibling C, Toyoshima C, Spudich J A. Nature (London) 1987;328:536–539. doi: 10.1038/328536a0. [DOI] [PubMed] [Google Scholar]
- 6.Baker J E, Brust-Mascher I, Ramachandran S, LaConte L E W, Thomas D D. Proc Natl Acad Sci USA. 1998;95:2944–2949. doi: 10.1073/pnas.95.6.2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rayment I, Rypniewski W R, Schmidt-Base K, Smith R, Thomchick D R, Benning M M, Winkelman D A, Wesenberg G, Holden H M. Science. 1993;261:50–57. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 8.Kabsch W, Mannherz H G, Suck D, Pai E F, Holmes K C. Nature (London) 1990;347:37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- 9.Rayment I, Holden H M, Whittaker M, Yohn C B, Lorenz M, Holmes K C, Milligan R A. Science. 1993;261:58–65. doi: 10.1126/science.8316858. [DOI] [PubMed] [Google Scholar]
- 10.Schroder R R, Manstein D J, Jahn W, Holden H, Rayment I, Helmong K, Spudich J A. Nature (London) 1993;364:171–174. doi: 10.1038/364171a0. [DOI] [PubMed] [Google Scholar]
- 11.Cooke R, Crowder M S, Thomas D. Nature (London) 1982;30:776–778. doi: 10.1038/300776a0. [DOI] [PubMed] [Google Scholar]
- 12.Yanagida T. J Mus Res Cell Motility. 1985;6:43–52. doi: 10.1007/BF00712310. [DOI] [PubMed] [Google Scholar]
- 13.Cooke R. Crit Rev Biochem. 1986;21:53–118. doi: 10.3109/10409238609113609. [DOI] [PubMed] [Google Scholar]
- 14.Pollard T D, Bhandari D, Maupin P, Wachsstock D, Weeds A G, Zot H. Biophys J. 1993;64:454–471. doi: 10.1016/S0006-3495(93)81387-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uyeda T, Abramson P, Spudich J. Proc Natl Acad Sci USA. 1996;93:4459–4464. doi: 10.1073/pnas.93.9.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jontes J D, Wilson-Kubalek E M, Milligan R A. Nature (London) 1995;378:751–753. doi: 10.1038/378751a0. [DOI] [PubMed] [Google Scholar]
- 17.Whittaker M, Wilson-Kubalek E M, Smith J E, Faust L, Milligan R A, Sweeney H L. Nature (London) 1995;378:748–751. doi: 10.1038/378748a0. [DOI] [PubMed] [Google Scholar]
- 18.Gollub J, Cremo C R, Cooke R. Nat Struct Biol. 1996;3:796–802. doi: 10.1038/nsb0996-796. [DOI] [PubMed] [Google Scholar]
- 19.Irving M, Allen T S, Sabidodavid C, Craik J S, Brandmeier B, Kendrickjones J, Corrie J E T, Trentham D R, Goldman Y E. Nature (London) 1995;375:688–691. doi: 10.1038/375688a0. [DOI] [PubMed] [Google Scholar]
- 20.Kull F J, Sablin E P, Lau R, Fletterick R J, Vale R D. Nature (London) 1996;380:550–555. doi: 10.1038/380550a0. [DOI] [PMC free article] [PubMed] [Google Scholar]


