Abstract
Non-coding RNAs (ncRNAs) are versatile regulators in cellular networks. While most trans-acting ncRNAs possess well-defined mechanisms that can regulate transcription or translation, they generally lack the ability to directly sense cellular signals. In this work, we describe a set of design principles for fusing ncRNAs to RNA aptamers to engineer allosteric RNA fusion molecules that modulate the activity of ncRNAs in a ligand-inducible way in Escherichia coli. We apply these principles to ncRNA regulators that can regulate translation (IS10 ncRNA) and transcription (pT181 ncRNA), and demonstrate that our design strategy exhibits high modularity between the aptamer ligand-sensing motif and the ncRNA target-recognition motif, which allows us to reconfigure these two motifs to engineer orthogonally acting fusion molecules that respond to different ligands and regulate different targets in the same cell. Finally, we show that the same ncRNA fused with different sensing domains results in a sensory-level NOR gate that integrates multiple input signals to perform genetic logic. These ligand-sensing ncRNA regulators provide useful tools to modulate the activity of structurally related families of ncRNAs, and building upon the growing body of RNA synthetic biology, our ability to design aptamer–ncRNA fusion molecules offers new ways to engineer ligand-sensing regulatory circuits.
INTRODUCTION
Small, non-coding RNAs (ncRNAs) are key regulators and defenders of the genome in all organisms (1). In prokaryotes, these ncRNAs are being discovered at an accelerating rate, and have been shown to coordinate with proteins to form complex networks that govern important cellular responses to stress and other environmental cues (2). This ubiquity suggests their utility in the control and tuning of cellular regulatory networks. In synthetic biology, trans-acting ncRNAs have been shown to have advantages in circuit engineering (3), and orthogonally acting ncRNA regulators that work independently in the same cell have been used to engineer higher-order regulatory functions (4–8).
A large number of trans-acting ncRNA molecules, however, function solely as wires of gene circuits, linking the activity of one genetic element to another (9). They therefore possess a well-defined and often highly tunable mechanism to regulate transcription or translation of target genes, but lack the ability to directly sense cellular signals. To expand the versatility of these ncRNAs, we propose to engineer ligand-inducible switching capability into trans-acting ncRNA molecules by fusing them with molecule-sensing RNA aptamers. We expect the strategy of adding ligand-sensing ability to ncRNA regulators will add an extra sensory layer to the increasingly sophisticated designs of circuits that utilize ncRNA regulations.
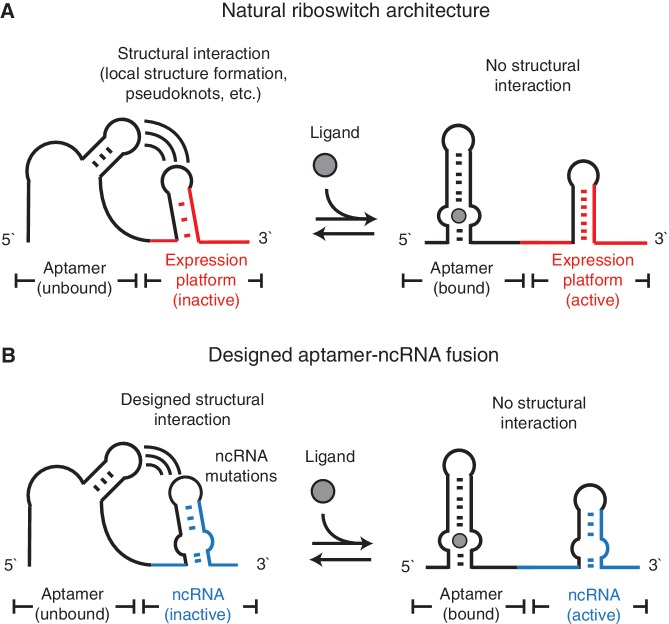
Bacterial trans-acting ncRNAs usually possess highly structured conformations (9). Examples of allosteric interactions between structured RNA elements and aptamers exist in natural cis-acting riboswitches (10), which usually consist of two structural motifs fused together—an aptamer, and an expression platform that converts ligand binding at the aptamer into a structural rearrangement that can block or allow the formation of functional hairpins (11). Our aptamer–ncRNA fusions are designed to recapitulate this ligand-dependent structural rearrangement, and we hypothesize that disruptions on ncRNA structure could be actuated through the designed fusion of RNA aptamers that would interfere with the ncRNA structure until bound to its cognate ligand (Figure 1).
Figure 1.
The proposed riboswitch-like design for the aptamer–ncRNA fusion. (A) A riboswitch usually consists of an aptamer (black color) coupled to a structured expression platform (red color). The structural interaction between the aptamer and the expression platform could inactivate (left) or activate (right) the expression platform depending on the presence of ligand. (B) We propose to fuse the aptamer and the ncRNA (blue color) in a similar architecture. The designed structural interaction between the aptamer and the ncRNA mutation regions (black color) would inactivate ncRNA without the ligand (left). Ligand binding could eliminate such structural interactions and activate the ncRNA (right).
Two primary types of natural ncRNA regulators acting on the 5′-untranslated regions (UTRs) are engineered for designing custom regulations (Supplementary Figure S1). The first are regulators of transcriptional elongation, and recent studies have shown a family of orthogonal ncRNA regulators derived from the pT181 antisense RNA could attenuate transcription independently (4). The second are regulators of translational initiation, and recent work have shown a family of orthogonal ncRNA regulators derived from the IS10 antisense RNA could inhibit translation with high specificity (12). Both pT181-derived and IS10-derived ncRNA systems have been shown as versatile regulatory platforms for constructing synthetic gene circuits in vivo. In this work, we describe design principles that allow fusion of these two types of trans-acting ncRNAs to RNA aptamers for allosteric modulation of ncRNA functions by ligands in Escherichia coli. We begin by fusing the IS10 ncRNA to the well-studied theophylline aptamer using a designed ‘pseudoknot’ interaction to create a theophylline-responsive translational regulator. We show that a similar design leads to a theophylline-responsive pT181-derived transcriptional regulator. We then demonstrate that an alternative design using ‘strand exchange’ exhibits similar allosteric switching functions. Using this design, we can modularly fuse different aptamers sensing either theophylline or the MS2 coat protein to the pT181 ncRNA. We show that this design strategy is robust to modifications of the ncRNA target recognition motif, which have been engineered to create orthogonal ncRNA-attenuator pairs. Since these orthogonal pairs serve as a toolbox for engineering RNA-based gene circuits, the sensing capability adds an extra ‘sensory’ layer of these RNA-based circuits. We finish by showing that this is indeed the case by creating a two-input NOR gate using two designed ncRNA fusions that integrate a small molecule and a protein signal to perform genetic logic.
MATERIALS AND METHODS
Plasmid construction
Plasmids used in this study are described in Supplementary Table S1. Their sequences are listed in Supplementary Table S2. The sequences of functional aptamer–ncRNA fusions are summarized in Supplementary Table S3. Supplementary Tables S4–S7 list the sequences of all fusion designs used for screening. Supplementary Figure S1C outlines the plasmids that harbor IS10 and pT181 systems used in the study. All restriction enzymes were purchased from New England Biolabs. Oligonucleotides were ordered from Integrated DNA Technologies. All chemicals were purchased from Sigma–Aldrich unless otherwise indicated. Plasmids were sequencing verified by QuintaraBio. The IS10 reporter plasmid with the pSC101 replication origin and a chloramphenicol resistance marker, contained 40-nt IS10 target sequences with an embedded Shine–Dalgarno sequence and an AUG start codon that were translationally fused to the second amino acid of the reporter gene sfGFP (13). The pT181 reporter plasmid with the p15A replication origin and a chloramphenicol resistance marker, contained the 287-nt pT181 target sequences that were transcriptionally fused to the downstream reporter gene sfGFP. Different aptamer–ncRNA fusion mutants were cloned onto a vector with the ColE1 replication origin and an ampicillin resistance marker. Pairs of ultramers (i.e. oligonucleotides >60 nt) containing 30-nt complementary regions were synthesized and PCR amplified to generate aptamer–ncRNA mutant cassettes, which were then digested and ligated to the vector. The E. coli promoter J23119 (http://partsregistry.org/Part:BBa_J23119), modified to include a SpeI site right before the start of transcription, was used for expression of the reporter gene and aptamer–ncRNA fusions. The MS2 aptamer–ncRNA fusion plasmids also contained cassettes expressing either MS2 coat protein or MS2–RFP fusion protein under the inducible PLlacO-1 promoter (14). This was done by cloning the whole cassettes from the plasmid pCT119 for the MS2 coat protein (a gift from Dr D. Peabody) or from the plasmid K133 for the MS2–RFP fusion protein (a gift from Dr I. Golding) onto the MS2 aptamer–ncRNA fusion plasmids.
Strains, growth media and in vivo fluorescence assays using plate reader
All in vivo fluorescence experiments were performed in E. coli Top10 strain (Invitrogen). Plasmid combinations were transformed into competent cells, plated on Difco LB+Agar plates containing 100 µg/ml carbenicillin and 34 µg/ml chloramphenicol, and incubated overnight at 37°C. In all experiments, three colonies were picked into 300 µl of Difco LB containing 100 µg/ml carbenicillin and 34 µg/ml chloramphenicol in a 2-ml, 96-well block (Costar 3960) and grown overnight in a Labnet Vortemp 56 bench top shaker at 37°C with 1000 rpm shaking speed. Ten microliters of this overnight culture was then added to 240 µl (1:25 dilution) of supplemented M9 Minimal Media (pH 7.4) with 0.2% casamino acids (Fisher Scientific), selective antibiotics, and proper concentrations of theophylline or IPTG (Fisher Scientific), and incubated overnight at 37°C and 1000 rpm. A second fresh 96-well plate containing 240 µl of the same supplemented M9 minimal media with same concentrations of theophylline or IPTG was inoculated by overnight minimal media culture with 1:25 dilution factor. Once the cells grew to mid-log phase, fluorescence was assayed. Cell cultures with volume of 100 µl mixed with 100 µl of phosphate-buffered saline (PBS) were transferred to 96-well plates, and fluorescence [excitation at 485 nm, emission at 510 nm for sfGFP; excitation at 587 nm, emission at 610 nm for RFP (15)] and ODs (optical densities, measured at 600 nm, were similar among experiments and fell in the linear range of our instruments) were then measured using a fluorescence plate reader (Tecan Safire2). The ratio of fluorescence to optical density (RFU/OD) was calculated and the background RFU/OD corresponding to Top10 cells without fluorescent proteins was subtracted.
In vivo fluorescence using flow cytometry
Cell culture with volume of 5 µl mixed with 250 µl PBS containing 2 mg/ml kanamycin were transferred to 96-well plates, and assayed using the flow cytometer (Partec Cyflow Space) in the five parameters of time, forward scatter (FSC), side scatter (SSC), GFP fluorescence (excitation at 500 nm, emission at 527 nm) and RFP fluorescence (excitation at 590 nm, emission at 610 nm). Data for at least 50 000 cellular counts (triggered by SSC) were collected for each sample. The mean autofluorescence distribution of Top10 cells without fluorescent proteins was also measured. Counts were gated by FSC and SSC, and the arithmetic mean of each distribution is calculated using FCS Express Version 3.0 (De Novo Software) with mean autofluorescence subtracted.
Single mutation assays
Single mutation plasmids were constructed by inverse PCR amplification of the wild-type ncRNA plasmid using two phosphorylated primers containing the desired mutations, and self-ligated. These single mutations plasmids were then transformed together with pT181 reporter plasmid into E. coli Tg1 cells (Zymo Research). Three colonies were picked and inoculated as described earlier. Fluorescence and OD were assayed using the plate reader (Tecan Safire2).
In vitro RNA synthesis
A DNA template for transcription of the theo–P–IS10 ncRNA fusion molecules, inserted in the context of the 5′ and 3′ flanking selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) structure cassette (16), was generated by PCR [1 ml; containing 20 mM Tris (pH 8.4), 50 mM KCl, 2.5 mM MgCl2, 200 µM each dNTP, 500 nM each forward and reverse primer, 5 pM template and 0.025 U/µl Taq polymerase; denaturation at 94°C, 45 s; annealing 55°C, 30 s; and elongation 72°C, 1 min; 34 cycles]. The PCR product was recovered by ethanol precipitation and resuspended in 150 µl of TE [10 mM Tris (pH 8.0), 1 mM EDTA]. Transcription reactions (1.0 ml, 37°C, 12–14 h) contained 40 mM Tris (pH 8.0), 20 mM MgCl2, 10 mM DTT, 2 mM spermidine, 0.01% (v/v) Triton X-100, 5 mM each NTP, 50 µl of PCR-generated template, 0.12 U/µl RNase Inhibitor (Promega) and 0.1 mg/ml of T7 RNA polymerase. The RNA products was purified by denaturing polyacrylamide gel electrophoresis (8% polyacrylamide, 7 M urea, 29:1 acrylamide:bisacrylamide, 32 W, 2 h), excised from the gel and recovered by passive elution and ethanol precipitation. The purified RNA (∼3 nmol) was resuspended in 200 µl TE.
SHAPE experiment
Structure-selective RNA modification with and without theophylline was performed following the experimental design of (17). For the without theophylline reaction, 10 pmol of RNA was suspended in 12 µl of nuclease free water in a PCR tube. Using a thermocycler, the RNA was heated to 95°C for 2 min, then placed on ice for 1 min. Six microliters of 3.3 × folding buffer [333 mM HEPES (pH 8.0), 333 mM NaCl, 33 mM MgCl2] was added, followed by incubation at 37°C in the thermocycler for 20 min. Nine microliters of this folded RNA solution was added to either 1 µl 10 × 1M7 (65 mM) (+ reaction), or 1 µl neat DMSO (− reaction), and further incubated at 37°C. Modified or control RNAs were then ethanol precipitated following (18). For the with theophylline reaction, 10 pmol of RNA was suspended in 10 µl of water in a PCR tube. After identical heat denaturation steps, 2 µl of 9 × theophylline (4.5 mM dissolved in water) was added, and 6 µl of 3.3 × folding buffer was added followed by identical steps as above. The general procedure of primer extension and data analysis followed (18), using 5 pmol of RNA for ddA and ddT sequencing reactions.
Secondary structure prediction using SHAPE-Seq reactivity constraints
SHAPE intensities were converted into a pseudo-free energy change term in the RNAstructure program (19):
ΔGSHAPE = m × ln[SHAPE-Seq reactivity + 1.0] + b
The intercept, b, is the free-energy bonus for formation of a base pair with zero or low SHAPE reactivity, whereas m, the slope, drives an increasing penalty for base pairing as the SHAPE reactivity increases. These parameters dictate the strength of the experimental contribution to the energy function. The b and m parameters used were −0.5 and 3.4 kcal/mol, respectively.
RESULTS
Engineering a translational ncRNA to sense a small molecule with a designed pseudoknot
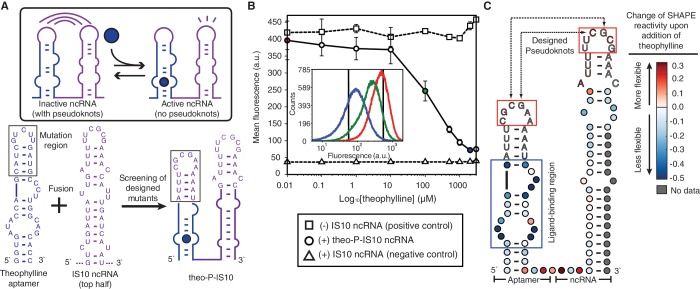
The wild-type E. coli insertion sequence IS10 utilizes antisense RNA-mediated translational inhibition to regulate the expression of transposase (20). Following previous work on engineering orthogonally acting variants of the IS10 system (12), we used a single hairpin of the IS10 ncRNA. In-depth biochemical studies have shown that the loop of IS10 ncRNA binds to the 5′ most nucleotides of its target transcript, which nucleates further hybridization into the ribosome binding site of the target (21). Furthermore, these studies have shown that single nucleotide mutation of the loop nucleotides resulted in almost complete loss of repression. We therefore sought to design a pseudoknot interaction between the loop regions of the aptamer and the ncRNA, such that the aptamer loop nucleotides could interact with the ncRNA loop nucleotides to disrupt its regulatory function (Figure 2A).
Figure 2.
Designed theophylline aptamer–IS10 ncRNA fusions. (A) The theophylline aptamer sequences (blue) were fused to the 5′ end of IS10 ncRNA (purple), and screening of seven designed pseudoknot mutants (Supplementary Figure S2) resulted in one functional fusion, theo–P–IS10 (P for pseudoknot). The proposed mechanism for allosteric switching of the fusion molecule is shown in the box. (B) Fluorescence assay of theo–P–IS10 using flow cytometry. The induction curves were plotted from the average values of three biological replicates at each theophylline concentration. The inset shows the cytometry histograms of three ligand concentrations (red—0.01 µM, green—100 µM, blue—2 mM), with the two black vertical lines showing the mean values of the positive and negative controls. The repression percentage between 2 mM and 0.01 µM theophylline is 83.0%, compared to 91.0% between the positive and negative controls. (C) SHAPE data of theo–P–IS10. The difference in nucleotide reactivity with and without the ligand is overlaid on a hypothesized secondary structure model of theo–P–IS10 based on Refs (21) and (24). Colors represent the changes of SHAPE reactivity upon addition of the ligand, with red colors showing positive changes (more flexible) and blues colors showing negative changes (more stable). The blue box is the known ligand-binding pocket. The red boxes are designed pseudoknot interactions. Original SHAPE data is available in Supplementary Figure S3A.
To start, we utilized the well-known theophylline aptamer obtained by the Systematic Evolution of Ligands by EXponential enrichment method (SELEX) (22). The aptamer is a single RNA hairpin that binds theophylline in an inner loop region with high affinity (Kd = ∼300 nM) (23,24). Previous studies have shown mutations in the loop region were tolerated as long as the loop structure was preserved (24). This allowed us to mutate the loop of the theophylline aptamer to create a hypothesized pseudoknot interaction between the aptamer and the IS10 ncRNA. When fused to IS10 ncRNA, we hypothesized that this pseudoknot would inhibit its regulatory activity in the absence of theophylline. In the presence of theophylline, we expected that the aptamer structure would be stabilized by the ligand, eliminating the pseudoknot interaction between the loops and restoring IS10 ncRNA function (Figure 2A).
We rationally designed seven different aptamer–IS10 ncRNA fusion mutants bearing different aptamer loop mutations and tested them in vivo (Supplementary Text S1 for design procedures). Each variant was transformed into E. coli with a reporter plasmid containing the IS10 ncRNA UTR target controlling the downstream fluorescent protein sfGFP. Colonies were picked and grown with and without theophylline, and fluorescence was measured using the plate reader with two controls: a positive control that lacked an aptamer–ncRNA sequence (high GFP), and a negative control that expressed the IS10 ncRNA not fused to any aptamer (low GFP). Four variants (#3–#6) showed different levels of switching ability in the presence of theophylline (Supplementary Figure S2). From this set, mutant #6 was distinguished by both obtaining nearly the same maximal GFP expression as the positive control and exhibiting the largest dynamic range of repression between the two theophylline conditions. To further characterize this mutant, called theo–P–IS10, we assayed GFP expression for varying amounts of theophylline using flow cytometry (Figure 2B). All fluorescence histograms showed a single peak. A large dynamic range (83.0%) was observed, which was close to the difference between positive and negative controls (91.0%). This data implied that, as designed, the fused aptamer interfered with the function of ncRNA only in the absence of theophylline, and that interaction of theophylline with the aptamer released the ncRNA hairpin to regulate its target.
To support this proposed mechanism, we carried out SHAPE experiments on the theo–P–IS10 fusion molecule to characterize its structure with and without theophylline. The SHAPE experiment uses a structure-dependent chemical probe, here 1-methyl-7-nitroisatoic anhydride (1M7), to modify RNA molecules preferentially at positions of high nucleotide flexibility (25). After modification, RNAs are converted to cDNAs by a reverse transcriptase primer extension reaction, which is blocked by adducts. cDNA products are then analyzed by capillary electrophoresis to infer the location of adduct formation via cDNA length, and the intensity of adduct formation at each nucleotide via the amount of cDNA of a given length. These intensities are converted into reactivities, with higher reactivity interpreted as positions of lower or no RNA structure, and lower reactivity interpreted as positions of strong RNA structure or nucleotide constraint (25).
We transcribed the theo–P–IS10 fusion molecule in vitro, which was then folded with and without theophylline, followed by SHAPE probing and analysis (Supplementary Figure S3). Figure 2C plots the difference in nucleotide reactivity between the two folding conditions overlaid on a secondary structure model of the theo–P–IS10 fusion. From this data, two reactivity changes can be discerned. First, as expected, the ligand-binding pocket of the aptamer shows a large drop in reactivity upon addition of theophylline, indicating that these nucleotides become constrained as they directly interact with theophylline. Second, both aptamer and IS10 loops show consistent but small increases in reactivity, suggesting that these nucleotides probably become unconstrained in the presence of theophylline. While the SHAPE analysis data is consistent with our hypothesis and in vivo fluorescence assay data that a designed pseudoknot interaction forms only in the absence of theophylline to prevent the ncRNA loop region from interacting with its target, we cannot rule out the possibility that multiple aptamer-ncRNA fusions dimerize with each other to affect their structures and functions in a synergetic way.
Engineering a transcriptional ncRNA to sense a small molecule with designed strand exchange
The Staphylococcus aureus plasmid pT181 uses antisense RNA-mediated transcription attenuation to control copy number (26). Similar to many trans-acting ncRNAs, the pT181 ncRNA is highly structured (27). The pT181 ncRNA makes initial contact with the sense attenuator through a loop–loop kissing interaction, and mutating the loop nucleotides can decrease its attenuation function (28). We therefore followed our pseudoknot design strategy to fuse the theophylline aptamer to the pT181 ncRNA.
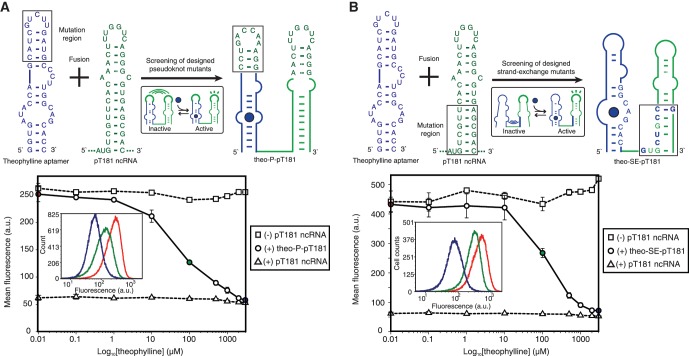
Previous work has shown that mutations in only one hairpin of pT181 ncRNA were sufficient to change its specificity of interaction with the sense attenuator (4), and we confirmed here that this single hairpin was sufficient to repress the target (Supplementary Figure S4A). The theophylline aptamer was fused to this single hairpin, with the aptamer loop nucleotides mutated to form pseudoknot base-pairing with the pT181 ncRNA loop nucleotides (Figure 3A). Similar to the above screening procedure of IS10 fusion mutants, we tested eight different fusion mutants with varying loop–loop pseudoknot interactions in E. coli along with a reporter plasmid containing the pT181 sense attenuator transcriptionally fused to sfGFP. The mutant with the best performance (#4) under screening (Supplementary Figure S5), called theo–P–pT181, was further assayed with various concentrations of theophylline using flow cytometry. Our results showed that the pseudoknot design prevented the ncRNA from acting in the absence of theophylline, and allowed the ncRNA to repress the sense target when theophylline was present (Figure 3A).
Figure 3.
Alternative designs of theophylline-sensing pT181 ncRNA fusions. (A) Screening of eight designed pseudoknot mutants (Supplementary Figure S5) results in theo–P–pT181, which displays strong theophylline-inducible repressive effects on its target. (B) An alternative design of theophylline-sensing pT181 ncRNA fusions based on the strand-exchange strategy by mutating the lower bottom stem region of the ncRNAs. The sequences of the best mutant screened from 15 mutants (Supplementary Figure S6) is shown as theo–SE–pT181 (SE for strand exchange). In vivo fluorescence data from flow cytometry is shown on the bottom. The repression percentage between 3 mM and 0.01 µM theophylline is 83.4%, compared to 86.6% between positive and negative controls. The insets show the cytometry histograms corresponding to three ligand concentrations in both (A) and (B).
While the pseudoknot design strategy was effective, it was not inherently modular as the aptamer loop sequences must be adjusted to the ncRNA loop sequences. This was particularly relevant to the pT181 ncRNA system, where the loop region of the pT181 ncRNA has been exclusively mutated to create orthogonally acting ncRNA-attenuator systems (4). We hypothesized that we could engineer a more modular aptamer–ncRNA fusion by designing an interaction between the aptamer and the ncRNA stem region, while leaving the ncRNA loop nucleotides for modifying target specificity (Figure 3B). In this design, named ‘strand exchange’, one strand on the ncRNA stem is mutated to exhibit exchange between two possible conformations of the aptamer–ncRNA fusion molecule: one where the strand is base paired to the aptamer causing the ncRNA to be non-functional (ncRNA inactivated), and one where the strand is base paired to the other strand of ncRNA to restore ncRNA function (ncRNA activated). We hypothesized that theophylline binding would cause strand exchange between the conformations and bias the population of fusion molecules toward the activated conformation.
To verify that the mutation strategy would not disrupt the function of pT181 ncRNA, we first carried out single mutations to determine the regulatory importance of each nucleotide (Supplementary Figure S4B and C). Each nucleotide was mutated to its complementary nucleotide (such as G to C, A to T, etc.), giving 58 mutant ncRNAs each bearing a single mutation. The regulatory activities of all 58 mutant ncRNAs were then assayed with the pT181 reporter plasmid. By comparing with the wild-type pT181 ncRNA, we found that nucleotides in the lower stem can tolerate moderate modifications without affecting the regulatory function.
We therefore mutated this lower stem region of the pT181 ncRNA to base pair with the ligand-binding pocket of the theophylline aptamer. We designed 15 aptamer–ncRNA fusion variants following the strand exchange strategy, and measured their repression on the pT181 sense target with and without theophylline (Supplementary Figure S6). One mutant (#14) with the highest dynamic range, called theo–SE–pT181, was selected for assaying with varying amounts of theophylline using flow cytometry. Figure 3B showed that repression by the theo–SE–pT181 fusion molecule (83.4%) almost covered the full dynamic range (86.6%) of the wild-type ncRNA regulator. The data implied that the strand exchange design was acting like an allosteric switch, with theophylline triggering a change in the conformations to allow the pT181 ncRNA to repress its target. To confirm this switching behavior, we measured the temporal expression of the pT181 reporter gene under the control of theo–SE–pT181, which showed that its expression was gradually repressed to the level of the negative control (Supplementary Figure S7). The theophylline aptamer used in the study does not bind caffeine that differs from theophylline by a single methyl group (23). We found that caffeine did not, as expected, repress gene expression in this system, implying that these aptamer–ncRNA fusions retained a high level of ligand specificity (Supplementary Figure S8).
To support our mechanistic hypothesis, we attempted to perform SHAPE on the theo–SE–pT181 fusion. However, the ncRNA hairpin prevented reverse transcriptase from transcribing the full-length molecule (Supplementary Figure S3B), thus precluding direct measurement of the proposed structural transitions. As an alternative approach, we computed the thermodynamic free energies of substructures of four closely related aptamer–pT181 fusion variants using the RNA secondary structure prediction algorithm Mfold (29) (Supplementary Text S2 and Supplementary Figure S9). The data suggested that the thermodynamic free energy of the two folds of the aptamer–pT181 fusion were balanced in such a way that allowed theophylline binding to bias the fold toward the active conformation. The computation results also suggested several principles in the design of switchable aptamer–ncRNA fusions as was detailed in the Discussion section.
The strand exchange design is modular with respect to specificity-changing ncRNA mutations
Recent work has demonstrated that the pT181 system served as a platform for engineering cellular gene networks (4). In particular, multiple orthogonally acting pT181 attenuators could be composed on the same transcript to logically control gene expression, and one sense attenuator could control the transcription of another orthogonal pT181 ncRNA that propagated the signal in RNA-mediated transcriptional cascades. Underlying these two features was the ability to mutate the loop region of the pT181 ncRNA hairpin to change its target specificity, which led to the creation of ncRNA-attenuator pairs that regulated multiple targets independently in the same cell. Here we show that the strand exchange strategy allows modularly fusing orthogonal pT181 ncRNAs with RNA aptamers.
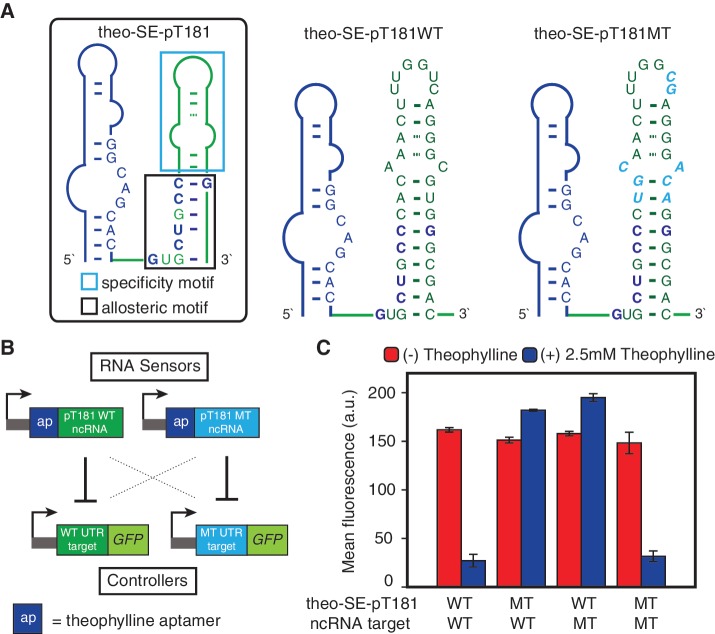
Since the specificity mutations located outside of the lower stem region of the ncRNA that base-paired with the aptamer, we used the same strand exchange design to produce fusions with the mutant pT181 ncRNA, called theo–SE–pT181MT (Figure 4A). We tested the orthogonality of theo–SE–pT181WT and theo–SE–pT181MT against their cognate targets with and without theophylline (Figure 4B). Measured GFP expression for all four possible combinations of ncRNAs and their targets under different theophylline conditions showed that the theo–SE–pT181MT fusion responded to theophylline in the same way as the theo–SE–pT181WT fusion did (Figure 4C). Furthermore, both fusions were orthogonal relative to each other's target. These results demonstrated the modularity of the strand exchange design, and implied that these ligand-sensing fusion molecules could perform similarly as the pT181 antisense RNAs in sophisticated genetic networks, but with added functionality to fine-tune their activity with small molecules.
Figure 4.
Orthogonal theophylline-sensing pT181 fusions based on the strand-exchange design. (A) The region that determines the target-specificity of pT181 ncRNA is in the cyan box (4), and allosteric region is in the black box. The sequences of theophylline-sensing WT pT181 and MT pT181 fusions are shown. (B) Experimental setup to test theophylline-induced target specificity of theo–SE–pT181WT and theo–SE–pT181MT. All four possible combinations between aptamer–ncRNA fusions and reporters were assayed with and without theophylline. (C) Fluorescence assay data with red bars showing (−) theophylline and blue bars showing (+) 2.5 mM theophylline.
The strand exchange design can be used to create protein-sensing ncRNAs
Equally important to sensing small molecules would be the ability of ncRNAs to sense protein concentrations and alter their regulatory functions accordingly. Here, we used the MS2 coat protein-sensing aptamer for several reasons. First, the natural MS2 coat protein aptamer is well studied and binds to MS2 coat protein with high affinity (Kd = 20 pM) (30). Second, it has been shown that the MS2 coat protein could be fused to other proteins (31), serving as an adapter to sense other intracellular proteins in vivo (32). Third, it has recently been used to demonstrate a class of RNA control devices that can couple the abundance of desired proteins to targeted gene expression through alternative RNA splicing (33).
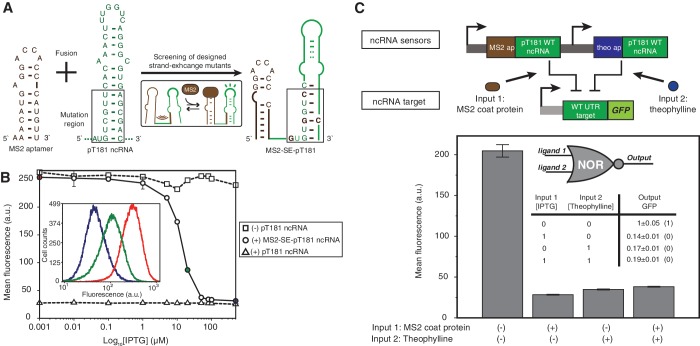
Following the strand exchange design strategy, we mutated the lower stem of the pT181 ncRNA hairpin to base pair with the MS2 aptamer nucleotides that were critical for MS2 coat protein binding (Figure 5A) (34). We expected that without the MS2 coat protein, the aptamer and ncRNA would fold together to disrupt the ncRNA structure and inhibit its function; when the MS2 coat protein was present and bound to the aptamer, the protein ligand would restore the pT181 ncRNA structure and function. We designed five fusion mutants with different levels of base pairing between the ncRNA and the aptamer, and tested them in E. coli with the pT181 reporter plasmid. We used an IPTG-inducible promoter PLlacO-1 to induce the expression of MS2 coat protein from the same ColE1 plasmids that harbored the fusion variants (Supplementary Figure S1C). GFP expression was measured with and without IPTG for each variant. All designs were able to switch into an active state upon induction of the MS2 coat protein (Supplementary Figure S10), and one variant (#2) with the best performance, called MS2–SE–pT181, was selected for further study using flow cytometry (Figure 5B). The repression of MS2–SE–pT181 fusion molecule almost covered the full dynamic range of the wild-type ncRNA regulator. Once again, the data suggested that strand exchange design was acting like a switch, with the presence of MS2 coat protein triggering a change in the conformation of the fusion molecule to that allowed pT181 ncRNA to repress its target. Further, using the fusion protein of the MS2 coat protein and RFP, we observed a near-identical induction curve, indicating that MS2 protein could indeed serve as an adapter for sensing other intracellular proteins (Supplementary Figure S11).
Figure 5.
Designed MS2 coat protein-sensing pT181 ncRNA fusions. (A) Sequences of the MS2–SE–pT181 aptamer–ncRNA fusion screened from five designed mutants (Supplementary Figure S10). (B) Fluorescence assay of MS2–SE–pT181 ncRNA fusions with intracellular MS2 coat protein induced by IPTG. The repression percentage between 500 µM and 1 nM IPTG is 87.5% compared to 89.1% between positive and negative controls. The inset shows the cytometry histogram of three IPTG concentrations. (C) A NOR logic engineered from theo–SE–pT181WT and MS2–SE–pT181WT aptamer–ncRNA fusions. The two aptamer–ncRNA fusions control the same target and integrate ligand signals in a way that presence of any ligand represses the target gene expression. A theoretical NOR truth table is shown in the plot.
To confirm that the design of MS2–SE–pT181 was modular with respect to pT181 mutants with different target specificities, we constructed the MS2–SE–pT181MT variant by directly swapping the target specificity motif of the ncRNA, and tested the orthogonality with and without MS2 coat protein expression (Supplementary Figure S12). Similar to the theo–SE–pT181WT and theo–SE–pT181MT designs, the MS2–SE–pT181WT and MS2–SE–pT181MT fusions acted orthogonally and only showed repression on their cognate targets in the presence of MS2 coat proteins.
Orthogonal ncRNA sensors can integrate cellular signals in logic circuits
To demonstrate the flexibility of using these engineered ncRNA sensors to regulate gene expression, we used the theo–SE–pT181WT and MS2–SE–pT181WT ncRNA fusion molecules to regulate the same gene via the wild-type pT181 sense attenuator. We expected that in the presence of either theophylline or MS2 coat protein, one of the aptamer–ncRNAs would be functional and repress the expression of the target gene. In this way, this system should be able to integrate two cellular signals and act like a NOR logic. Indeed, In vivo fluorescence data indicated that GFP expression was high only when there was no ligand present, and presence of either theophylline or MS2 coat protein induced by IPTG or both consistently repressed the reporter gene expression to a low expression (Figure 5C). This demonstrated the use of designed orthogonal aptamer–ncRNA fusions in logically integrating cellular signals on the sensory-level. Compared to the NOR logic using two orthogonal pT181 ncRNA-attenuator systems (4), here we showed that only one pT181 ncRNA-attenuator pair was required for the same regulatory function, implying sensory-level engineering using orthogonal ncRNA sensors could further increase the scales and complexities of synthetic circuits.
DISCUSSION
Design principles for switchable aptamer–ncRNA fusions
In this work, we have demonstrated two alternative designs to rationally engineer natural ncRNAs to sense ligands by fusing them with RNA aptamers. The IS10 and pT181 ncRNAs are both highly structured and likely to represent a class of ncRNA regulators whose function highly depends on the structure (9). In our designs, we fused RNA aptamer sequences to the 5′ end of the ncRNA molecules in the similar architecture as natural riboswitches. We introduced mutations into different locations in different designs to disrupt the ncRNA structure: the mutations were on the aptamer loop region to form pseudoknots with the ncRNA loop (‘pseudoknot’ design); or the mutations were on the ncRNA lower stem region to make this region exchangeable between alternative conformations (‘strand exchange’ design). In both designs, ligand binding eliminated disruptions on the ncRNA hairpin structures and activated their functions (Supplementary Text S1).
In all designs, we have utilized the RNA structure prediction algorithm, Mfold, to compute and verify that the disrupted conformation had the lowest thermodynamic free energy. Thus, without ligands, the structure of ncRNA was disrupted. To explain the requirements for engineered allosteric switching, we dissected the architecture of the designed fusion molecule of the theo–SE–pT181 design, and calculated the hybridization free energy of possible local RNA structures (Supplementary Text S2 and Supplementary Figure S9). Comparing three closely related variants (MT-1, MT-2, MT-3) with the functional design (WT), we discovered that allosteric switching was not only determined by the interaction between the ncRNA and the aptamer, but also by the ncRNA folding itself. If there was over-disruption on ncRNA (MT-1), the fusion molecule was always inactivated; if the disruption was much weaker than the ncRNA folding energy (MT-3), the ncRNA was always activated. Only mutants with similar free energy between ncRNA disruption and formation showed allosteric switching (WT and MT-2). Surprisingly, one base-pair formation or elimination in the ncRNA stem could completely change the allosteric switching, indicating the allosteric properties of designed aptamer–ncRNA fusions were sensitive to nucleotide compositions.
Furthermore, we calculated free energy differences for all 15 theophylline–SE–pT181 fusion mutants between alternative conformations (Supplementary Figure S13). We fitted the measured fluorescence data to the calculated free energy differences, and observed almost linear correlation between the two quantities for both with and without the ligand. These experiments also suggest that the hybridization free energy between adjacent RNA strands can serve as a guide for designing allosteric properties of other aptamer–ncRNA fusions.
Strengths and limitations of switchable aptamer–ncRNA fusions
We have demonstrated a modular strategy for engineering ligand-responsive ncRNAs. This builds from a growing body of work on engineering gene circuits using RNA mechanisms that respond to ligands. Previous work has demonstrated that cis-acting RNA elements residing in the 5′ (35,36) or 3′- (37,38) UTR sequences can respond to small molecules, and there was pioneering work to engineer riboregulators in yeast to control translation in response to small molecules (6). Our work here further expands the category of RNA molecules that can control gene expression in a ligand-inducible way to the naturally occurring trans-acting ncRNAs that modulate 5′-UTR functions. This is particularly valuable for transcriptional ncRNA regulators, as transcriptional regulators have been shown as useful genetic parts that could be systematically tethered together to form logics and other higher-order regulatory systems.
For the demonstration purpose, we fused theophylline- or MS2 coat protein-sensing aptamers to ncRNAs. So far, many RNA aptamers have been reported by in vitro selection (39,40) or through the discovery of natural riboswitches (10). These aptamers can bind specifically to different ligands, including nucleotide-like molecules such as flavin mononucleotide (FMN) (41) and dopamine (42), complex compounds such as tetracycline (43) and vitamin B12 (44), and proteins such as HIV-1 rev peptide (45) or nicotine acetylcholine receptor (46). While our design strategy worked well for theophylline and MS2 coat protein aptamers that possess relatively simple structures, to design functional fusions between complex aptamers and ncRNAs, the detailed information about their structures and mechanisms is needed. We foresee the advent of high throughput RNA structure characterization methods such as SHAPE-Seq (47) will facilitate our design process. Such methods will identify the regions of aptamers that exhibit the appropriate flexibility for intramolecular interaction with the ncRNA following our design strategy. When connected to environmentally or medically relevant ligands, we expect these ligand-responsive RNA regulators offer novel capabilities to engineer cellular behaviors in response to different stimuli.
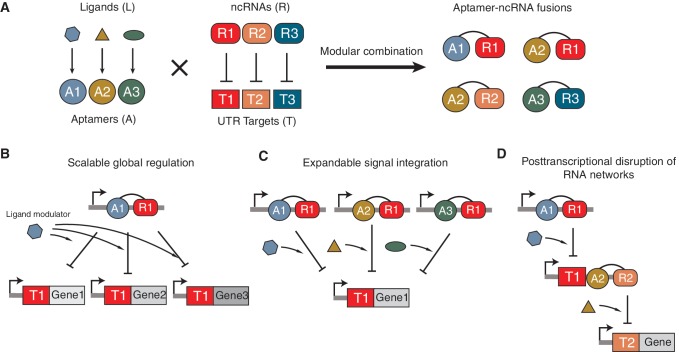
By focusing on switchable trans-acting aptamer–ncRNA fusions, we are extending this capability by introducing modularity on both molecular and network levels. First, at the molecular level, the modular design of aptamer–ncRNA fusions allows the ligand sensed to be switched with no further adjustment (Figure 6A). Figures 3B and 5A show transcriptional regulation by the same pT181 ncRNA molecule but modulated by two different ligands—theophylline or MS2 coat protein. The only difference between the two experiments is the particular aptamer–ncRNA fusion molecule expressed, and the target of these aptamer–ncRNA fusions remains the same. To achieve this with cis-acting elements, the genetic context of the regulatory target would have to be changed by manipulating the 5′- or 3′-UTRs since cis-acting elements are inherently tightly coupled with their regulatory target. However, unlike cis-acting elements, the in trans nature of our regulatory elements allows the possibility of regulation of off-target genes. Nonetheless, many ncRNAs are highly specific or can be engineered to be so (4,12). An assumption of our study is that the fusion of an aptamer to the ncRNA does not strongly affect the properties of components. This is supported in the present case by the maintenance of the quantitative behavior of the fused ncRNA compared to the wild-type ncRNA (Figure 3A and B). However, it is possible that the fusion may interfere with the function and specificity of both components—an eventuality that would need to be diagnosed by structural and functional studies. Although we only demonstrated the NOR logic by having two different aptamer–ncRNA fusions to control a single target, it is conceivable that other types of sensory-level logic such as AND and OR can be theoretically implemented by placing the complex logic into the fusion molecules as suggested by other researchers (37). For example, previous studies have demonstrated that AND and OR functions could be engineered by fusing two different aptamers to the same ribozyme (37), and we expect a similar design could also work for the aptamer–ncRNA fusions. We plan to investigate this topic in our future study.
Figure 6.
Modularity of aptamer–ncRNAs at the molecular and network levels. (A) Combining orthogonal ncRNAs with different aptamers could provide a toolbox of orthogonally acting aptamer–ncRNA fusions that sense multiple ligands and control their cognate targets with high specificity. (B) The scalable global regulation allows regulation of multiple gene targets in the same cell by responding to a global ligand modulator. (C) The expandable signal integration allows regulation of a single gene target by integrating multiple signal inputs in a logic way. (D) The post-transcriptional disruption of ncRNA networks allows us to debug and fine-tune the performance of individual regulators in complicated networks such as transcriptional cascades.
Second, at the gene network level, trans-acting aptamer–ncRNA fusions have the potential to globally change the regulation of many targets in response to customizable cellular signals (Figure 6B). Many natural ncRNAs have multiple targets in the cell (2), and by engineering the capability to fine-tune the post-transcriptional function of these ncRNAs in response to ligands, this work creates the opportunity to globally switch on or off many targets in the cell simultaneously. In addition to adding flexibility in an engineering context, this could offer powerful capabilities to the scientific toolkit to study ncRNA biology through dynamic switching of global ncRNA regulators (48). Since we have shown that our aptamer-pT181 designs are modular with respect to both the aptamer and orthogonal pT181 variants, there are numerous places that we can integrate these fusion molecules into existing capabilities to create more sophisticated RNA/protein or RNA-only circuits with unique regulatory properties. Generally, transcriptional aptamer–ncRNAs could be used as alternatives to inducible promoters by having ncRNA responding directly to the ligands after being expressed from constitutive promoters; multiple signal integration at the sensor level can compute logics on external stimuli whose outputs could be fed into transcriptional logics for creation of advanced logic gates (Figure 6C); and aptamer–ncRNA fusion molecules themselves could serve as signal-transmitting molecules in RNA/protein or RNA-only networks to provide ligand-inducible control over network connection (Figure 6D). The ability to sense diverse signals (ncRNAs, proteins and metabolites) and integrate them to execute gene regulatory programs is key to the detection or implementation of specific cellular responses in complex environments (49). The modularity of aptamer–ncRNAs might offer powerful extensions to our capabilities for rationally engineering trans-acting RNA regulators-mediated genetic circuits, and the ubiquity of such mechanisms makes them potentially useful across a diverse range of organisms, from prokaryotes to humans (6). Coupled with the rapid increase in our understanding of the regulatory roles of trans-acting ncRNAs, this work opens the door for engineering sensory-level gene regulations that are modular, flexible and versatile based on naturally occurring trans-acting ncRNAs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Texts 1 and 2, Supplementary Tables 1–7 and Supplementary Figures 1–13.
FUNDING
U.S. Department of Energy, Lawrence Berkeley National Laboratory (Contract No. DEAC02-05CH11231); National Science Foundation, Synthetic Biology Engineering Research Center (grant number 0540879); Miller Institute for Basic Research in Science (to J.B.L. and C.C.L.). Funding for open access charge: Synthetic Biology Engineering Research Center (SynBERC) under National Science Foundation (grant number 0540879).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank D. Peabody for the gracious gift of pCT119 plasmid. We thank I. Golding for the gracious gift of K133 plasmid. We thank Shana Topp and Yohei Yokobayashi for helpful advice.
REFERENCES
- 1.Sharp PA. The centrality of RNA. Cell. 2009;136:577–580. doi: 10.1016/j.cell.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Gottesman S, Storz G. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb. Perspect Biol. 2010;3:1–16. doi: 10.1101/cshperspect.a003798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Callura JM, Dwyer DJ, Isaacs FJ, Cantor CR, Collins JJ. Tracking, tuning, and terminating microbial physiology using synthetic riboregulators. Proc. Natl Acad. Sci. USA. 2010;107:15898–15903. doi: 10.1073/pnas.1009747107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lucks JB, Qi L, Mutalik VK, Wang D, Arkin AP. Versatile RNA-sensing transcriptional regulators for engineering genetic networks. Proc. Natl Acad. Sci. USA. 2011;108:8617–8622. doi: 10.1073/pnas.1015741108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rinaudo K, Bleris L, Maddamsetti R, Subramanian S, Weiss R, Benenson Y. A universal RNAi-based logic evaluator that operates in mammalian cells. Nat. Biotechnol. 2007;25:795–801. doi: 10.1038/nbt1307. [DOI] [PubMed] [Google Scholar]
- 6.Bayer TS, Smolke CD. Programmable ligand-controlled riboregulators of eukaryotic gene expression. Nat. Biotechnol. 2005;23:337–343. doi: 10.1038/nbt1069. [DOI] [PubMed] [Google Scholar]
- 7.Isaacs FJ, Dwyer DJ, Ding C, Pervouchine DD, Cantor CR, Collins JJ. Engineered riboregulators enable post-transcriptional control of gene expression. Nat. Biotechnol. 2004;22:841–847. doi: 10.1038/nbt986. [DOI] [PubMed] [Google Scholar]
- 8.Friedland AE, Lu TK, Wang X, Shi D, Church G, Collins JJ. Synthetic gene networks that count. Science. 2009;324:1199–1202. doi: 10.1126/science.1172005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottesman S. The small RNA regulators of Escherichia coli: roles and mechanisms*. Annu. Rev. Microbiol. 2004;58:303–328. doi: 10.1146/annurev.micro.58.030603.123841. [DOI] [PubMed] [Google Scholar]
- 10.Roth A, Breaker RR. The structural and functional diversity of metabolite-binding riboswitches. Annu. Rev. Biochem. 2009;78:305–334. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 12.Mutalik V, Qi L, Guimaraes J, Lucks J, Arkin A. Rationally designed families of orthogonal RNA regulators of translation. Nat. Chem. Biol. 2012;8:447–454. doi: 10.1038/nchembio.919. [DOI] [PubMed] [Google Scholar]
- 13.Pédelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 14.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. A monomeric red fluorescent protein. Proc. Natl Acad. Sci. USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 17.Steen KA, Malhotra A, Weeks KM. Selective 2′-hydroxyl acylation analyzed by protection from exoribonuclease. J. Am. Chem. Soc. 2010;132:9940–9943. doi: 10.1021/ja103781u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mortimer SA, Weeks KM. Time-resolved RNA SHAPE chemistry: quantitative RNA structure analysis in one-second snapshots and at single-nucleotide resolution. Nat. Protoc. 2009;4:1413–1421. doi: 10.1038/nprot.2009.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Low JT, Weeks KM. SHAPE-directed RNA secondary structure prediction. Methods. 2010;52:150–158. doi: 10.1016/j.ymeth.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simons RW, Kleckner N. Translational control of IS10 transposition. Cell. 1983;34:683–691. doi: 10.1016/0092-8674(83)90401-4. [DOI] [PubMed] [Google Scholar]
- 21.Kittle JD, Simons RW, Lee J, Kleckner N. Insertion sequence IS10 anti-sense pairing initiates by an interaction between the 5′ end of the target RNA and a loop in the anti-sense RNA. J. Mol. Biol. 1989;210:561–572. doi: 10.1016/0022-2836(89)90132-0. [DOI] [PubMed] [Google Scholar]
- 22.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 23.Jenison RD, Gill SC, Pardi A, Polisky B. High-resolution molecular discrimination by RNA. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 24.Zimmermann GR, Jenison RD, Wick CL, Simorre JP, Pardi A. Interlocking structural motifs mediate molecular discrimination by a theophylline-binding RNA. Nat. Struct. Biol. 1997;4:644–649. doi: 10.1038/nsb0897-644. [DOI] [PubMed] [Google Scholar]
- 25.Mortimer SA, Weeks KM. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J. Am. Chem. Soc. 2007;129:4144–4145. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- 26.Novick RP, Iordanescu S, Projan SJ, Kornblum J, Edelman I. pT181 plasmid replication is regulated by a countertranscript-driven transcriptional attenuator. Cell. 1989;59:395–404. doi: 10.1016/0092-8674(89)90300-0. [DOI] [PubMed] [Google Scholar]
- 27.Brantl S, Wagner EGH. An antisense RNA-mediated transcriptional attenuation mechanism functions in Escherichia coli. J. Bacteriol. 2002;184:2740–2747. doi: 10.1128/JB.184.10.2740-2747.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brantl S, Wagner EG. Antisense RNA-mediated transcriptional attenuation: an in vitro study of plasmid pT181. Mol. Microbiol. 2000;35:1469–1482. doi: 10.1046/j.1365-2958.2000.01813.x. [DOI] [PubMed] [Google Scholar]
- 29.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romaniuk PJ, Lowary P, Wu HN, Stormo G, Uhlenbeck OC. RNA binding site of R17 coat protein. Biochemistry. 1987;26:1563–1568. doi: 10.1021/bi00380a011. [DOI] [PubMed] [Google Scholar]
- 31.Gesnel MC, Del Gatto-Konczak F, Breathnach R. Combined use of MS2 and PP7 coat fusions shows that TIA-1 dominates hnRNP A1 for K-SAM exon splicing control. J. Biomed. Biotechnol. 2009;2009:104853. doi: 10.1155/2009/104853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peabody DS, Lim F. Complementation of RNA binding site mutations in MS2 coat protein heterodimers. Nucleic Acids Res. 1996;24:2352–2359. doi: 10.1093/nar/24.12.2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Culler SJ, Hoff KG, Smolke CD. Reprogramming cellular behavior with RNA controllers responsive to endogenous proteins. Science. 2010;330:1251–1255. doi: 10.1126/science.1192128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Convery MA, Rowsell S, Stonehouse NJ, Ellington AD, Hirao I, Murray JB, Peabody DS, Phillips SE, Stockley PG. Crystal structure of an RNA aptamer-protein complex at 2.8 A resolution. Nat. Struct. Biol. 1998;5:133–139. doi: 10.1038/nsb0298-133. [DOI] [PubMed] [Google Scholar]
- 35.Sinha J, Reyes SJ, Gallivan JP. Reprogramming bacteria to seek and destroy an herbicide. Nat. Chem. Biol. 2010;6:464–470. doi: 10.1038/nchembio.369. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Mandal M, Breaker RR. Gene regulation by riboswitches. Nat. Rev. Mol. Cell. Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- 37.Win MN, Smolke CD. Higher-order cellular information processing with synthetic RNA devices. Science. 2008;322:456–460. doi: 10.1126/science.1160311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Win MN, Smolke CD. A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proc. Natl Acad. Sci. USA. 2007;104:14283–14288. doi: 10.1073/pnas.0703961104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee JF, Hesselberth JR, Meyers LA, Ellington AD. Aptamer database. Nucleic Acids Res. 2004;32:D95–D100. doi: 10.1093/nar/gkh094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hermann T, Patel DJ. Adaptive recognition by nucleic acid aptamers. Science. 2000;287:820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- 41.Vicens Q, Mondragón E, Batey RT. Molecular sensing by the aptamer domain of the FMN riboswitch: a general model for ligand binding by conformational selection. Nucleic Acids Res. 2011;39:8586–8598. doi: 10.1093/nar/gkr565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mannironi C, Di Nardo A, Fruscoloni P, Tocchini-Valentini GP. In vitro selection of dopamine RNA ligands. Biochemistry. 1997;36:9726–9734. doi: 10.1021/bi9700633. [DOI] [PubMed] [Google Scholar]
- 43.Steber M, Arora A, Hofmann J, Brutschy B, Suess B. Mechanistic basis for RNA aptamer-based induction of TetR. ChemBioChem. 2011;12:2608–2614. doi: 10.1002/cbic.201100503. [DOI] [PubMed] [Google Scholar]
- 44.Sussman D, Nix JC, Wilson C. The structural basis for molecular recognition by the vitamin B 12 RNA aptamer. Nat. Struct. Biol. 2000;7:53–57. doi: 10.1038/71253. [DOI] [PubMed] [Google Scholar]
- 45.Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Swanstrom R, Burch CL, Weeks KM. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–716. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sivaprakasam K, Pagán OR, Hess GP. Minimal RNA aptamer sequences that can inhibit or alleviate noncompetitive inhibition of the muscle-type nicotinic acetylcholine receptor. J. Membr. Biol. 2010;233:1–12. doi: 10.1007/s00232-009-9215-z. [DOI] [PubMed] [Google Scholar]
- 47.Lucks JB, Mortimer SA, Trapnell C, Luo S, Aviran S, Schroth GP, Pachter L, Doudna JA, Arkin AP. Multiplexed RNA structure characterization with selective 2′-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq) Proc. Natl Acad. Sci. USA. 2011;108:11063–11068. doi: 10.1073/pnas.1106501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alper H, Stephanopoulos G. Global transcription machinery engineering: a new approach for improving cellular phenotype. Metab. Eng. 2007;9:258–267. doi: 10.1016/j.ymben.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 49.Xie Z, Wroblewska L, Prochazka L, Weiss R, Benenson Y. Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science. 2011;333:1307–1311. doi: 10.1126/science.1205527. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.