Abstract
We have investigated the structural, biochemical and cellular roles of the two single-stranded (ss) DNA-binding proteins from Bacillus subtilis, SsbA and SsbB. During transformation, SsbB localizes at the DNA entry pole where it binds and protects internalized ssDNA. The 2.8-Å resolution structure of SsbB bound to ssDNA reveals a similar overall protein architecture and ssDNA-binding surface to that of Escherichia coli SSB. SsbA, which binds ssDNA with higher affinity than SsbB, co-assembles onto SsbB-coated ssDNA and the two proteins inhibit ssDNA binding by the recombinase RecA. During chromosomal transformation, the RecA mediators RecO and DprA provide RecA access to ssDNA. Interestingly, RecO interaction with ssDNA-bound SsbA helps to dislodge both SsbA and SsbB from the DNA more efficiently than if the DNA is coated only with SsbA. Once RecA is nucleated onto the ssDNA, RecA filament elongation displaces SsbA and SsbB and enables RecA-mediated DNA strand exchange. During plasmid transformation, RecO localizes to the entry pole and catalyzes annealing of SsbA- or SsbA/SsbB-coated complementary ssDNAs to form duplex DNA with ssDNA tails. Our results provide a mechanistic framework for rationalizing the coordinated events modulated by SsbA, SsbB and RecO that are crucial for RecA-dependent chromosomal transformation and RecA-independent plasmid transformation.
INTRODUCTION
Genetically programmed natural transformation is a widely distributed mechanism for genetic recombination in many bacterial genera (1,2,3). In the Firmicutes Phylum, little is known about the fate of internalized DNA of any source and even less is known about the transformation process in Gram-negative bacteria, with some species only taking up DNA from their own clade (1,3–5). In Bacillus subtilis, only a small fraction of cells differentiate and become naturally competent. These cells have distinct physiological characteristics that include an inability to synthesize DNA or undergo cell division as well as the transient expression of at least 20 polar-localized membrane-associated and cytosolic proteins that are collectively dedicated to the internalization, processing and chromosomal integration of foreign DNA or plasmid establishment (1,3,4,6). The membrane-associated competence proteins bind environmental double-stranded (ds) DNA, degrade one of the strands to produce single-stranded (ss) DNA, and internalize the linear ssDNA into the cytosol (3,6–8). The cytosolic proteins protect the internalized ssDNA and facilitate RecA nucleation and RecA·ssDNA filament formation (7–11).
Previous genetic, cytological and biochemical studies suggest that SsbB (also termed YwpH), DprA (Smf or CilB), CoiA (YjbF), RecA and RecU, which localize at the entry pole, are required for chromosomal and/or plasmid transformation (6–10,12). However, the exact roles of these proteins as well as other functions (e.g. CoiA) remain largely unknown. In addition, RecN and/or SbcE are known to localize at the entry pole whenever foreign DNA is internalized, and RecO localizes when DNA with self-annealing potential (e.g. plasmid or viral DNA) is present (6,10,12–14).
SsbA, RecA, RecU, RecN, SbcE and RecO are expressed during exponential growth with SsbA and RecA also being transiently induced during competence, whereas SsbB, DprA and CoiA are specifically induced during competence development (15–17). Additionally, the expression of SbcE and AddAB proteins are indirectly affected by the ComA regulator (18).
With few exceptions (e.g. Helicobacter pylori, Deinococcus radiodurans and Campylobacter jejuni), naturally transformable bacteria contain SsbA- and SsbB-like proteins (based on sequence), whereas the non-naturally transformable contain a single SsbA-like protein (19). Bacillus subtilis SsbA (counterpart of Escherichia coli SSB [SSBEco]) is a 172-residue polypeptide that shares strong sequence similarity with the DNA-binding N-terminal domain and protein-binding C-terminus of SSBEco. SsbA is an essential homotetrameric protein involved in genome maintenance (5). Unlike B. subtilis SsbA, Streptococcus pneumoniae SsbA (SsbASpn) is not induced during competence development (20). During recombinational repair, SsbA physically interacts with RecO as well as with many other proteins (21–23). Such RecO·SsbA interaction is important for growth as shown by the partial compensation upon over-expression of RecO of the thermosensitivity of B. subtilis cells expressing an SsbA variant lacking the last C-terminal 35 residues (SsbAΔ35) (23). In recent studies, an interaction platform for SsbA and RecO has been discovered in both E. coli and Thermus thermophilus where the C-terminal tail of SSBEco or SSBTth interacts with the hydrophobic pocket on the C-terminal domain of RecOEco or RecOTth, respectively (24,25). While little is known about SsbA localization and relative protein concentration in competent cells, it is possible that the protein is induced to levels similar to those of SsbB in a small subset, likely at amounts equal to or greater than levels of SsbA found during exponential growth (>750 tetramers per cell) (15,17,19).
In contrast to SsbA, SsbB is a 113-residue polypeptide that is specialized for activity in transformational recombination, namely protection of internalized ssDNA. SsbB shares 63% identity with the N-terminal DNA-binding domain of SsbA (amino acids 1–106), but lacks the characteristic C-terminal tail that mediates protein interactions in SsbA. In vivo analyses in B. subtilis reveal that SsbB is located at the DNA entry poles in competent cells and is in contact or close proximity with RecA, CoiA and DprA (7,8). Unlike SsbA, no interactions have been shown between SsbB and RecO (our unpublished results). The absence of SsbB only moderately reduces chromosomal transformation (3- to 10-fold) in both B. subtilis and S. pneumoniae cells (15,17,26), suggesting other protein(s) might protect the internalized ssDNA. While B. subtilis SsbB lacks the prototypical C-terminal domain for protein interactions, some naturally competent bacteria, e.g. S. pneumoniae, have SsbB proteins with an acidic C-terminal tail that might serve for protein interactions (20). Unlike B. subtilis SsbB, SsbBSpn is ∼20-fold more abundant than SsbASpn (20).
To gain insight into the early events of the genetic recombination process in B. subtilis competent cells and to compare it with the recruitment of RecA onto SsbA-coated ssDNA during double strand break (DSB) repair, the roles of SsbA and SsbB are explored in this report. Although SsbA and SsbB are both thought to cover and protect ssDNA from nucleolytic attacks and to exert negative effects on RecA nucleation onto ssDNA, we present evidence that there is a division of labor between SsbA and SsbB. SsbA stimulates RecO-mediated strand annealing required for plasmid transformation, overcoming interference exerted by SsbB. SsbA also facilitates RecO-mediated RecA recruitment onto ssDNA for chromosomal transformation along with the help of SsbB. Conversely, SsbB protects the internalized ssDNA and enhances RecA nucleation when co-assembled with SsbA on ssDNA.
MATERIALS AND METHODS
Bacterial strains and plasmids
Bacillus subtilis isogenic rec-deficient strains were described in Supplementary Table S1. pCB722-borne ssbA or pCB669-borne recO gene, under the control of a phage T7 promoter, were used to over-express SsbA and RecO proteins, respectively, in E. coli BL21(DE3)[pLysS] cells as described (22,27). pBT61-borne recA gene, under the control of its own promoter, was used to over-express RecA in B. subtilis BG214 cells (28). The ssbB gene was fused at the 3′-end with the terminal 5′-end 30 nt of the ssbA gene, to generate a ssbB variant termed ssbB* gene. pCB777-borne ssbB or pCB892-borne ssbB* gene, under the control of a phage T7 promoter, were used to over-express SsbB or SsbB* in E. coli BL21(DE3)[pLysS] cells.
Enzymes, reagents and protein purification
DNA modification enzymes were supplied by Roche, BioLabs or Fermentas and DTT was from Sigma. The cross-linking agent glutaraldehyde was from Sigma. SsbA (18.7 kDa) and RecO (29.3 kDa) proteins were purified as described (22) and free from corresponding E. coli proteins. RecA (38.0 kDa) was purified as previously described (28). SsbB (12.4 kDa) and SsbB* (13.5 kDa) proteins were purified as described in Supplemental Material.
All proteins were purified to homogeneity >98%. The NH2 terminus of the purified proteins was sequenced by automatic Edman degradation. The corresponding molar extinction coefficients for SsbA, SsbB, SsbB*, RecA and RecO were calculated as 11 400, 13 000, 12 950, 15 200 and 19 600 M−1 cm−1, respectively, at 280 nm, as previously described (28). The protein concentrations were determined using the above molar extinction coefficients. RecA is expressed as mol of protein as monomers, RecO as dimers, and SsbA, SsbB and SsbB* as tetramers.
For determination of the oligomeric state of SsbA or SsbB protein cross-linking experiments were performed as described in Supplemental Material.
Limiting Trypsin (0.25 µg/ml) degradation of SsbB or SsbB* proteins was performed as described elsewhere (29,30) and in the Supplemental Material.
Protein and DNA interactions
The formation of SsbA-, SsbB- or SsbB*-ssDNA complexes were measured by EMSA or filter binding assays. The 30-, 40-, 50-, 60-, 70- and 80-nt long [γ-32P]-poly[dT] ssDNA (dT30 – dT80, 0.2 nM in ssDNA molecules) was incubated with various amounts of SsbA or SsbB proteins for 15 min at 37°C in buffer C (50 mM Tris–HCl, pH 7.5, 1 mM DTT, 50 mM NaCl, 50 µg/ml BSA, 5% glycerol) containing or not 10 mM magnesium acetate (MgOAc) in a final volume of 20 µl. The reaction was stopped and separated either using a 10% PAGE or filtered trough KOH-treated filters as described in Supplemental Material. The PAGEs were run with Tris–borate at 45 V at 4°C and dried prior to autoradiography.
The rate of dissociation of the SsbA· or SsbB·ssDNA complexes was measured by using alkali-treated filters as described elsewhere (31,32) and in the Supplemental Material.
Crystallization, SAD phasing and refinement of dT35-bound SsbB
Crystals of the SsbB·dT35 complex were obtained as described in Supplemental Material.
The structure of the SsbB·dT35 complex was solved to 2.8-Å resolution using a combination of SAD phasing and molecular replacement (Table 2). Data were indexed and scaled using HKL2000 (33). A single mercury site was identified and refined using Phenix (34) and solvent flattening resulted in interpretable experimental electron density maps for model building. A partial model of the SsbB·dT35 complex was built into the SAD maps from the 3.55-Å resolution derivative data using Coot (35) and then used as a molecular replacement model against the 2.8-Å resolution native data set using Phaser (36). The final complex, which included two SsbB monomers and extensive stretches of dT35, was refined through iterative rounds of manual building and refinement using Coot (35) and Refmac (37), respectively. The full SsbB tetramer is generated by applying symmetrical constraints. Coordinate and structure factor files have been deposited in the Protein Data Bank (PDB ID 3VDY).
Table 2.
Diffraction data and crystal structure solution
| Native | HgCl2- derivative | |
|---|---|---|
| Data collection | ||
| Space group | P41212 | P41212 |
| Unit cell [a, b, c (Å)] | 101.75, 101.75, 118.57 | 103.65, 103.65, 113.65 |
| Wavelength, Å | 1.53 | 1.008 |
| Resolution (last shell), Å | 20–2.8 (2.85–2.8) | 50–3.55 (3.61–3.55) |
| Reflection measured/unique | 117 177 (15 468) | 86 280 (7916) |
| Multiplicity (last shell) | 7.6 (3.2) | 10.9 (11.5) |
| Completeness (last shell), % | 97.4 (71.3) | 99.8 (100) |
| Rsyma (last shell), % | 8.1 (52.5) | 11.0 (46.2) |
| I/σ (last shell) | 31.2 (1.7) | 36.2 (7.1) |
| Phasing statistics | ||
| Resolution, Å | 50–3.55 | |
| Figure of merit | 0.398 | |
| Refinement | ||
| Resolution, Å | 20–2.80 | |
| Rwork/Rfreeb, % | 23.0/26.2 | |
| rms deviation bond lengths, Å | 0.010 | |
| rms deviation bond angles, ° | 1.4 | |
| Ramachandran statistics (% most favored/allowed/ generously allowed/disallowed) | 92.1/6.8/1.1/0 |
aRsym = ΣΣj|Ij − <I>|ΣIj, where Ij is the intensity measurement for reflection j and <I> is the mean intensity for multiply recorded reflections.
bRwork/Rfree = Σ||Fobs| − |Fcalc||/|Fobs|, where the working and free R factors are calculated by using the working and free reflection sets, respectively. The free R reflections (5% of the total) were held aside throughout refinement.
RecA dATP hydrolysis assays
The ssDNA-dependent dATP hydrolysis activity of RecA protein was observed via a coupled spectrophotometric enzyme assay (38,39). Absorbance measurements were taken with a Shimadzu CPS-240A dual-beam spectrophotometer equipped with a temperature controller and 6-position cell chamber. The cell path length and band pass were 1 cm and 2 nm, respectively. The regeneration of dATP from dADP and phosphoenolpyruvate driven by the oxidation of NADH can be followed by a decrease in absorbance at 340 nm. Rates of ssDNA-dependent RecA-mediated dATP hydrolysis and the lag times were measured in buffer D (50 mM Tris–HCl, pH 7.5, 1 mM DTT, 90 mM NaCl, 10 mM MgOAc, 50 µg/ml BSA, 5% glycerol) containing 5 mM dATP for variable time at 37°C in a 100-µl reaction mixture. A dATP regeneration system (0.5 mM phosphoenolpyruvate, 10 U/ml pyruvate kinase) and a coupling system (0.25 mM NADH, 10 U/ml lactate dehydrogenase, 3 mM potassium glutamate) were also included. The orders of addition of 3199-nt pGEM ssDNA (10 µM in nt), the proteins and their concentrations were indicated in the text. The amount of dADP was calculated as describe (40).
RecA-mediated dATP-dependent DNA strand exchange
The 3199-bp KpnI-cleaved pGEM dsDNA (20 µM in nt) and homologous circular 3199-nt ssDNA (10 µM in nt) were incubated with the indicated concentrations of protein or protein combination in buffer D containing 2 mM dATP for variable periods up to 60 min at 37°C in a final volume of 20 µl. A dATP regeneration system (8 U/ml creatine phosphokinase and 8 mM phosphocreatine) was included when indicated. The samples were deproteinized as described (41,42), and fractionated through 0.8% agarose gel electrophoresis (AGE) with ethidium bromide. The signal was quantified using a Geldoc (BioRad) system as described (22).
RecO-mediated DNA strand annealing
Linear 440-bp [γ-32P]-dsDNA was heat denatured during 10 min at 100°C and shifted to water-ice for 2 min. Heat-denatured linear 440-nt [γ-32P]-ssDNA (7 µM in nt) was pre-incubated with SsbA, SsbB, SsbB* or both SsbA and SsbB (100 nM) for 10 min at 30°C in buffer E (50 mM Tris–HCl, pH 7.5, 1 mM DTT, 2 mM EDTA, 110 mM NaCl, 50 µg/ml BSA, 5% glycerol) as described (11). Variable amounts of RecO (1–3 µM) were then added and reactions incubated for 60 min. The complexes formed were deproteinized as described (41), and fractionated through 6% PAGE. The signal was quantified as described earlier.
RESULTS
SsbA and SsbB DNA binding
SSBEco is a homotetramer that exhibits multiple binding modes differing in the number of monomers that interact with the ssDNA (43,44). In general, under modest Mg2+ concentrations and low protein to ssDNA ratios, SSBEco uses all four subunits of the tetramer to bind ssDNA in the so-called (SSBEco)65 binding mode, where 65-nt of ssDNA are occluded per SSBEco tetramer. However, in the absence of Mg2+ and with higher protein to ssDNA ratios, SSBEco uses only two of its four subunits to interact with the ssDNA in the (SSBEco)35 binding mode (43,44). To gain insight into the ssDNA binding and the type of nucleoprotein complexes formed by SsbA and SsbB proteins, binding assays with homopolymeric or heteropolymeric ssDNA were performed and the effects of Mg2+ were examined.
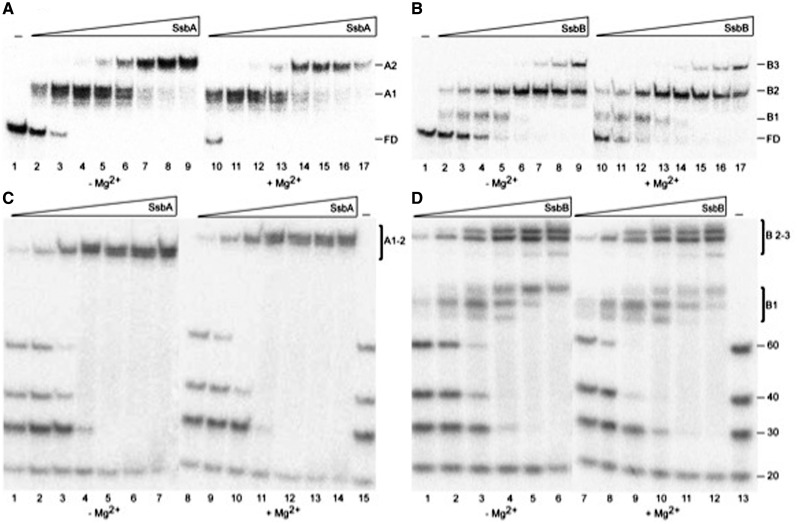
SsbA bound homopolymeric dT80 in a concentration-dependent manner with an apparent dissociation constant (KDapp) of <0.2 nM in the absence or presence of Mg2+ (Figure 1A and Table 1). In low protein to dT80 ratios, an initial complex (A1) was formed with gel mobility lower than that of free dT80 (FD), likely corresponding to ssDNA interacting with all four subunits of the tetramer (Figure 1A, lanes 2–4 and 10–13). However, in the presence of higher protein to dT80 ratios (1 SsbA tetramer per 32-nt or lower), the A1 product was no longer present and an A2 complex accumulated (Figure 1A, lanes 5–9 and 14–17).
Figure 1.
Binding of SsbA or SsbB to poly dT ssDNA. (A and B) An 80-nt long [γ-32P]-dT ssDNA (0.1 nM in ssDNA molecules) was incubated with increasing concentrations of SsbA (0.03, 0.06, 0.12, 0.25, 0.5, 1, 2 and 4 nM) (A) or SsbB (0.8, 1.5, 3, 6, 12, 25, 50 and 100 nM) (B) in buffer C containing 5 mM EDTA (−Mg2+) or 10 mM MgOAc (+Mg2+) for 15 min at 37°C. (C and D) [γ-32P]-poly(dT) ssDNA of different length (20-, 30-, 40- and 60-nt) (0.1 nM in ssDNA molecules) was incubated with increasing concentrations of SsbA (0.015, 0.03, 0.06, 0.12, 0.25 and 1 nM) (C) or SsbB (3, 6, 12, 25, 50 and 100 nM) (D) in buffer C −Mg2+ or +Mg2+ for 15 min at 37°C. The reactions were analyzed by 10% PAGE using a gel running buffer consisting of Tris–borate (pH 7.5) and the same concentration of Mg2+ or EDTA as in the reaction solutions at 45 V at 4°C and dried. The bands corresponding to unbound poly(dT)(FD) and the various protein·ssDNA complexes (A1–A2 and B1–B3) were visualized by autoradiography. Each experiment was carried out a minimum of three times with similar results.
Table 1.
SsbA binds ssDNA with higher affinity than SsbB
| DNA substrate | DNA binding affinity (in nM) |
|||||
|---|---|---|---|---|---|---|
| SsbA |
SsbB |
SsbB* |
||||
| −Mg2+ | +Mg2+ | −Mg2+ | +Mg2+ | −Mg2+ | +Mg2+ | |
| dT80a | <0.2 | <0.2 | 1.5 ± 0.5 | 1.2 ± 0.2 | 0.9 ± 0.1 | 0.8 ± 0.2 |
| ssDNA80a | <0.2 | 0.20 ± 0.1 | 10 ± 5 | 30 ± 4 | ND | ND |
| dT80b | 1.5 ± 0.5 | ND | >200 | ND | ND | ND |
The KDapp values (in nM) are the average of at least three independent experiments and are within a 10% SE.
aProteins were incubated with the indicated substrate for 15 min at 37°C in buffer C containing or not 10 mM MgCl2. Samples were separated by 10% PAGE, and the formation of protein–DNA complexes was quantified as described in ‘Materials and Methods’ section.
bProteins were incubated with the indicated substrate for 15 min at 37°C in buffer C lacking MgCl2. The mixture was filtered through KOH-treated filters (millipore, type HAWP 45 mm), the filters dried and the amount of radioactivity bound to the filter was determined by scintillation counting.
ND, not done.
SsbB bound homopolymeric dT80 with a KDapp of ∼1.0 nM in the absence or presence of Mg2+, a >5-fold lower affinity than SsbA (Figure 1B and Table 1). At low SsbB to dT80 ratios, two complexes were formed with gel mobilities lower than that of FD (B1 and B2; Figure 1B, lanes 2–5 and 10–13). Higher SsbB to dT80 ratios resulted in the disappearance of B1 and accumulation of B2, while saturating SsbB concentrations resulted in the appearance of a third, higher molecular-weight complex (B3; Figure 1B, lanes 7–9 and 15–17). Unfortunately, nucleotide ratios could not be calculated due to indistinct formation of B1, B2 and B3. In general, a higher protein to dT80 ratio produced SsbB·ssDNA complexes with lower gel mobility. These data, along those of SsbA, support a model in which both SsbA and SsbB can bind homopolymeric dT80 in two binding modes, similar to those observed with SSBEco.
SsbA and SsbB binding to a mixed-sequence ssDNA (heteropolymeric) of 80-nt in length with self-annealing potential was distinct from that observed with dT80. SsbA bound this ssDNA with a KDapp of <0.2 nM and ∼0.2 nM in the absence and presence of Mg2+, respectively, while SsbB bound with a KDapp of 10 and 30 nM in the absence and presence of Mg2+ (Table 1). In terms of the number of complexes formed, SsbA binding of ssDNA led to the formation of the slow mobility complex (C3) (Supplementary Figure S1A and S1B, lanes 2–9 and 11–18) regardless of Mg2+. SsbB in the absence of Mg2+ formed both C2 and C3 complexes, while only C3 was observed in the presence of Mg2+ (Supplementary Figure S1C and S1D, lanes 6–9 and 15–18). In both SsbA and SsbB interactions, a higher ratio of protein to heteropolymeric ssDNA was necessary for the complete binding of free ssDNA independent of the presence or absence of Mg2+ (Supplementary Figure S1A–D). This is possibly the result of SsbA and SsbB binding ssDNA with secondary structure potential with lower affinity than homopolymeric ssDNA as shown by the KDapp. In contrast to the dT80 results where one or two SsbA tetramers appeared to bind homopolymeric dT80 in the presence or absence of Mg2+, respectively, only one tetramer appeared to bind heteropolymeric ssDNA regardless of Mg2+ status. SsbB showed similar results but with dependence on Mg2+.
To gain insight in the length of ssDNA needed for stable interactions with SsbA or SsbB, binding assays using dT20, dT30, dT40 and dT60 were performed in the presence or the absence of Mg2+. Both proteins failed to bind the dT20 but could bind the remaining ssDNAs regardless of Mg2+ status, (Figure 1C and D). In addition, both SsbA and SsbB appeared to have higher affinities for longer ssDNA segments (60 > 40 > 30), as lower protein concentrations were required to gel shift longer DNA (Figure 1C and D). Paralleling the dT80 results, SsbA·dTn complexes migrated as a single species regardless of SsbA concentration (A1–2), whereas the SsbB·dTn complexes migrated as multiple species depending on the SsbB to ssDNA ratio (B1–2 and B3) (Figure 1C and D). Binding experiments done with individual dTn oligos confirmed these overall observations (data not shown).
To understand the origin of the above differences we measured the apparent thermodynamic stability (binding affinity) and kinetic stability (half-life) of the protein·ssDNA complexes by filter binding assays at low NaCl concentrations (∼100 mM) in the absence of Mg2+. Both SsbA and SsbB form complexes with dT80 with KDapp ∼ 1.5 and >200 nM, respectively (Supplementary Figure S2A and S2B). The SsbA·ssDNA complexes were short lived when the ssDNA was 50-nt or shorter and the half-life increased significantly with dT60 or longer oligos (Supplementary Figure S2C). A similar pattern was observed for SsbB·ssDNA, except that SsbB also formed short-lived complexes with dT60 ssDNA (Supplementary Figure S2D). These data indicate that formation of SsbA·ssDNA and SsbB·ssDNA complexes was reduced ∼7- and >200-fold, respectively, when comparing EMSA (Figure 1A and B) and filter binding assays (Supplementary Figure S2A and S2B). In general, SsbA appears to bind ssDNA with greater kinetic stability than SsbB in the absence of Mg2+, corresponding to the homopolymeric and heteropolymeric results.
Crystal structure of the SsbB·ssDNA complex
The tetrameric SSB proteins involved in DNA replication and repair have similar structures (44). To better understand the structure and function of an SSB protein specifically involved in genetic recombination, we crystallized full-length SsbB bound to dT35 (in a molar ratio of two dT35 oligos per one SsbB tetramer) and determined its 2.8-Å resolution X-ray crystal structure (Table 2 and Figure 2A). The SsbB·dT35 complex crystallized with two protein monomers per asymmetric unit; the full SsbB tetramer being comprised of four monomers or two symmetric SsbB pairs. The structure was refined with good bond geometry and crystallographic quality statistics (Table 2). The electron density maps revealed the presence of significant segments of dT35 bound to the surface of SsbB (Figure 2B). In total, 48 nt were fit to electron density, wrapping around the surface of the tetramer (24-nt in each crystallographic asymmetric unit). Gaps between the observed dT segments could be estimated to account for the remaining nucleotides, consistent with the apparent site size of ∼60-nt for SsbB.
Figure 2.
Structure of the SsbB·ssDNA complex. (A) Structure of SsbB tetramer bound to dT35. Ribbon (left) and surface (right) diagrams show the SsbB tetramer (green and blue) with resolved dT35 (stick form). (B) 2Fo-Fc electron density contoured to 1.8 σ showing an example of the dT35 bound to SsbB through stacking and electrostatic interactions. (C) Comparison of the SsbB·ssDNA (left) and SSBEco·ssDNA (right) complexes. The protein subunits and ssDNA binding surfaces are strikingly similar between the two proteins.
In terms of the overall arrangement of monomers within the SSB tetramer and the path of the ssDNA bound to the surface of the protein, the SsbB·ssDNA complex structure strongly resembles that of SSBEco (Figure 2C) and H. pylori SSB (SSBHpy, which plays an active role during vegetative growth and natural transformation) bound to ssDNA (45,46). One interesting difference is that for 8–10 bases of the ssDNA in the SsbB·ssDNA, the bases face the protein whereas the corresponding bases point away from the protein core (toward solvent) in the SSBEco·ssDNA complex. In one tract of ∼4 of these bases, apparent stacking between the ssDNA bases and SsbB residues Trp54 and Phe102 appear to promote the protein-facing bias of the ssDNA. SSBEco residues at equivalent positions are not aromatic, which could explain the difference. The first base of a second tract of ∼4 bases appears to stack against Tyr82 from one of the SsbB monomers, which could be important for establishing the differential ssDNA packing. Interestingly, the corresponding residue in SSBEco, Trp88, also stacks with a base in ssDNA, but this stacking is not propagated to adjacent bases in the SSBEco·ssDNA complex. These differences in ssDNA binding could possibly be related to the distinct functions of the two proteins, sequestering of the ssDNA by SsbB and protection of ssDNA in an active complex by SSBEco. Since SsbB appears similar to the ternary structure of other SSB proteins regardless of primary function (replication versus natural competence), the structure of SsbA will likely resemble SsbB.
SsbA and SsbB constrain RecA nucleation onto ssDNA to different extents
In both natural transformation and recombinational repair, RecA is required for binding to ssDNA in the first of the multi-step recombination processes. Yet, ssDNA is rarely present in the cell without SsbA; therefore, the dynamics of RecA and SsbA binding to the same ssDNA play an important role in understanding these processes. RecA nucleation onto ssDNA and subsequent extension of RecA filaments can be monitored by measuring the rate of dATP hydrolysis under RecA-limiting conditions (27,47,48). The rate of hydrolysis of dATP also provides an indirect measure of the displacement of SsbA and/or SsbB from ssDNA by RecA. We used this approach to examine RecA nucleation and filament extension onto SsbA-, SsbB- or SsbA- and SsbB-coated ssDNA (Figure 3).
Figure 3.
SsbA or SsbB plays a role in the rate-limiting nucleation of RecA and RecO activation. (A) The 3199-nt ssDNA (10 µM in nt) was pre-incubated with SsbA or SsbB (300 nM) and then incubated or not with RecO (100 nM) in buffer D containing 5 mM dATP. Then RecA (800 nM) was added and the absorption measured for 30 min. (B) The ssDNA was pre-incubated with SsbA, SsbB or with SsbA and then with SsbB (SsbA → SsbB) or vice versa (SsbB → SsbA) in buffer D containing 5 mM dATP. RecO was added and incubated for 5 min. RecA was then added and the absorption measured for 30 min. All reactions were repeated three or more times with similar results.
The rate of RecA nucleation onto naked 3199-nt long ssDNA was not significantly affected by the concentration of free RecA protein; however, the rate of dATP hydrolysis correlated with the amount of RecA bound to ssDNA within the experimental uncertainty (Supplementary Figure S3A). The rate of nucleation (one RecA per 12-nt) onto naked ssDNA and subsequent filament formation was biphasic, with a <5-min lag phase preceding establishment of the maximal hydrolysis rate (Figure 3A). Pre-binding of SsbA or SsbB (one SsbA or SsbB tetramer per 33-nt) to ssDNA extended the RecA lag phase to ∼11 or ∼7 min, respectively (Figure 3A). This is consistent with competitive binding between RecA and SsbA or SsbB for the ssDNA, limiting RecA nucleation. Since the half-lives for both SsbA·ssDNA and SsbB·ssDNA complexes with dT80 or longer ssDNAs were longer than the time of reaction (Supplementary Figure S2), nucleated RecA is likely displacing SsbA and SsbB during filament extension, albeit at a low rate. Similar results are seen with SSBEco and RecAEco in that SSBEco delays nucleation of RecAEco onto SSBEco-coated ssDNA (∼20 min lag time) (39).
To determine the effect of both SsbA and SsbB on RecA nucleation onto 3199-nt long ssDNA, both proteins were co-assembled onto ssDNA (creating an SsbA·ssDNA·SsbB complex) and RecA-mediated dATP hydrolysis analyzed. When ssDNA was pre-incubated with SsbB (one SsbB tetramer per 33-nt) followed by addition of excess of SsbA, the RecA nucleation time onto ssDNA was increased to levels comparable to SsbA alone (Supplementary Figure S4B). However, the same was not true for the addition of excess amount of SsbB to saturating amounts of SsbA (one tetramer per 33-nt) pre-bound to ssDNA; there was no decrease in RecA nucleation time (Supplementary Figure S4A). It is likely that in mixed SsbA·ssDNA·SsbB complexes, SsbA exerts a dominant negative effect on RecA nucleation over SsbB (Supplementary Figure S4A).
Alternative pathways for genetic recombination
The ultimate outcome of natural transformation is chromosomal transformation or plasmid establishment. Interestingly, ΔrecA strains block chromosomal transformation yet only marginally (<3-fold) affect plasmid transformation, suggesting that the formation of a RecA nucleoprotein filament is the basis of only chromosomal transformation (49,50,51; Table 3). RecA and its eukaryotic homologs often rely on ‘mediator’ proteins (RecO, RecOREco, Rad52, BRCA2) to assist loading RecA onto SSB-coated ssDNA during recombinational repair (5,52,53). DprASpn is known to recruit RecAEco onto SSBEco-coated ssDNA and DrpASpn accelerates single-stranded annealing (SSA) of naked complementary ssDNAs >5-fold relative to protein-free reactions (54). In the absence of DprA, chromosomal transformation is reduced 10- to 100-fold (12,15,17), but neither the pattern of RecA localization nor RecA thread formation after the addition of DNA is altered (12), indicating a second RecA-mediator is present.
Table 3.
Effect of the absence of both RecO and DprA on genetic recombination
| Relevant genotype | Normalized chromosomal transformationa | Normalized plasmid transformationb |
|---|---|---|
| rec+ | 100 | 100 |
| ΔrecA | <0.01 (<0.01)c | 97 (95)c |
| ΔrecO | 48 (45)c | 3.0 (2.7)c |
| ΔdprA | 1.7 (1–10)c | 2.5 (1.6)c |
| ΔrecO ΔdprA | <0.1 | <0.1 |
| ΔrecO ΔrecA | <0.01 | 48 |
aThe yield of met+ transformants (chromosomal transformation).
bpUB110 kanamycin-resistant transformants (plasmid transformation) was corrected for DNA uptake and cell viability and the values obtained normalized relative to that of the rec+ strain, taken as 100. The results are the average of at least five independent experiments and are within a 10% SE.
RecO, which physically interacts with SsbA, could be an alternate RecA-mediator during chromosomal transformation. Even though RecO is only 29% identical to the first 164 amino acids of the 255-residue RecOEco protein, it loads RecA onto SsbA-coated ssDNA during recombinational repair (22,27) and catalyzes SSA during plasmid transformation (10,11). In addition, the absence of RecO reduces overall plasmid transformation ∼30-fold (55) as well as the formation of RecA threads needed for chromosomal transformation (10).
To determine whether RecO helps RecA overcome the interference imposed by SSB proteins to stimulate nucleation onto ssDNA, a null recO dprA (ΔrecO ΔdprA) double mutant strain was constructed. When compared to the ΔrecA strain, chromosomal transformation were drastically impaired in ΔrecO ΔdprA cells, but not abolished (Table 3), suggesting that RecO, in the absence of DprA, likely works as a RecA mediator, contributing to RecA-mediated chromosomal transformation. Plasmid transformation was drastically impaired in ΔrecO ΔdprA cells when compared to the single mutant strains (Table 3). In agreement with previous studies, ΔdprA and ΔrecO single mutant strains both impaired plasmid transformation whereas only ΔdprA impaired chromosomal transformation (Table 3). Moreover, DprA plays an essential, but unknown role in plasmid transformation, even though DprA fails to catalyze DNA strand annealing (described in 3). Interestingly, in the absence of both RecO and DprA, RecA seems to overcome the inference imposed by the SSB proteins to bind and nucleate onto SSB-coated ssDNA, albeit with low efficiency. Although, the absence of RecA partially suppressed the RecO requirement for plasmid transformation (Table 3).
Interaction between RecO and SsbA facilitates RecA nucleation onto ssDNA
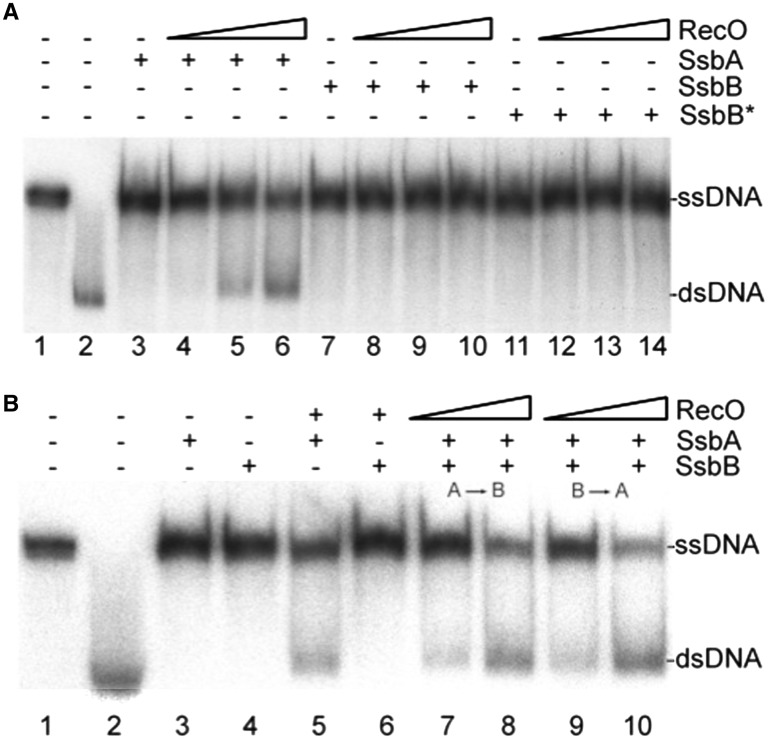
Since the genetic data suggest that RecO can act as an alternate RecA-mediator, we determined the effect of adding RecO to the RecA ATPase assays, providing evidence of functional interaction. While the addition of RecO did not significantly affect RecA nucleation on naked or SsbB-coated ssDNA (Supplementary Figure S3B), RecO profoundly accelerated RecA nucleation onto SsbA·ssDNA, reducing the RecA lag phase to ∼6 min (∼2-fold), and markedly stimulated RecA filament formation (Figure 3A).
RecO (RecOEco) physically interacts with SsbA (SSBEco) both in vivo (23) and in vitro (11,22,25,39,57) through the C-terminal residues of SsbA (SSBEco) (23,44). Conversely, interaction between RecO and SsbB, which lacks the C-terminal acidic tail, has not been detected (data not shown). To determine whether these C-terminal residues of SsbA play a significant role in RecO activation of RecA nucleation onto ssDNA, a hybrid ssbB-ssbA gene was constructed. A DNA segment encoding the last nine codons of ssbA, including the hexapeptide protein-binding motif DDDI/LPF (58), was fused to the 3′-end of the ssbB gene. This 122 codon-long ssbB* gene expressed SsbB*, the full-length SsbB fused to the nine C-terminal residues of SsbA. Purified SsbB* decreased RecA nucleation onto SsbB*-coated ssDNA compared to SsbB·ssDNA alone. Also, addition of RecO prior to RecA moderately assisted RecA loading onto SsbB*·ssDNA (Supplementary Figure S5A). Interestingly the addition of the C-terminal residues on SsbB* did not show the same response to RecO as SsbA even though SsbB* bound ssDNA with an ∼1.7-fold higher affinity than SsbB (Supplementary Figure S5B and Table 1) and the C-terminal end of SsbB* was solvent exposed as shown by sensitivity to trypsin proteolysis (Supplementary Figure S5C and S5D). It is likely that SsbA does not solely interact with RecO through the nine C-terminal-most residues; this is consistent with the observation that SSBTth interacts with RecOTth through more than just its C-terminal region (24).
Since SsbA has a significant effect on RecA nucleation mitigated by interacting with RecO, we predicted that RecO could dislodge both SsbA and SsbB bound to ssDNA at a different rate than either SsbA or SsbB alone. To test this hypothesis, SsbA was pre-incubated with ssDNA and SsbB added (or vice versa) followed by addition of RecO (one RecO per 100-nt). RecA-mediated dATP hydrolysis was then measured for the heterologous SSB-coated ssDNA (Figure 3B). Since the second SSB protein was added after the first was already in complex with ssDNA, formation of heterotetrameric proteins was unlikely. A co-assembled SsbA·ssDNA·SsbB complex markedly reduced the rate-limiting RecA nucleation to <2 min (Figure 3B). This co-assembly of SsbA and SsbB might enable RecO to recognize SsbA and carry out the limited release of SsbA or both SsbA and SsbB from ssDNA, subsequently loading RecA more efficiently. In addition, RecA displaced the SSB proteins from the heterologous complex more effectively than SsbA or SsbB alone, suggesting that the functional interaction between SsbB and RecA might be facilitated by the presence of SsbA, RecO or both.
RecO facilitates RecA-mediated DNA strand exchange in the presence of both SsbA and SsbB
SsbA or SSBEco pre-bound to ssDNA inhibits RecA nucleoprotein filament formation and dATP hydrolysis, but when added after RecA, SSBs generally aid RecA-mediated DNA strand exchange by melting inhibitory secondary structure in the ssDNA substrate and coating the displaced strand (22,52,59). To better understand the effects of the co-assembled SsbA·ssDNA·SsbB complex on RecA function, we next examined the effects of adding either SSB protein to RecA-catalyzed DNA strand exchange reactions. In the absence of SsbA or SsbB, RecA catalyzed dATP-dependent strand exchange between circular ssDNA (css) and a linear dsDNA (lds), converting ∼10% of the homologous ldsDNA into joint molecules (jm) and the final nicked-circular (nc) product during a 60-min reaction (Figure 4A, lanes 2 and 9). The addition of half-saturating to saturating SsbA or SsbB (one tetramer per 66-, 40- and 33-nt), added prior to RecA significantly stimulated RecA strand exchange (∼3- and 2-fold, respectively) as judged by the accumulation of dATP-dependent jm intermediates and nc products (Figure 4A, lanes 3–5 and 6–8). The presence of SsbA, even in limited quantities, along with SsbB also enhanced strand exchange, ∼30% of the ldsDNA substrate was converted to jm intermediates and nc products (Figure 4A, lanes 10–17). This result suggests that SsbA plays a larger role in facilitating RecA-mediated strand exchange than does SsbB, though both could be enhancing strand exchange by removing ssDNA secondary structure and sequestering the newly displaced ssDNA.
Figure 4.
RecO facilitates RecA loading onto SsbA·ssDNA or SsbA·ssDNA·SsbB. (A) Circular ssDNA (10 µM in nt) and homologous KpnI-linearized dsDNA (20 µM in nt) were pre-incubated with increasing concentrations of SsbA or SsbB (150, 250, 300 nM; lanes 3–5, 6–8) or decreasing concentrations (450, 300, 250 and 150 nM) of SsbB and then increasing concentrations of SsbA (lanes 10–14) or vice versa (lanes 14–17) for 5 min at 37°C in buffer D containing 2 mM dATP. Then a constant amount of RecA (700 nM, lanes 2–17) was added and the reaction incubated for 60 min at 37°C. (B) Circular ssDNA and homologous linear dsDNA were pre-incubated with increasing concentrations of SsbA or SsbB (lanes 2–4, 5–7) or decreasing concentrations of SsbB and then increasing concentrations of SsbA (lanes 10–14) or vice versa (lanes 14–17) for 5 min at 37°C in buffer D containing 2 mM dATP. The complex was incubated with a constant amount of RecO (100 nM, lanes 2–17) for 5 min at 37°C, followed by addition of a constant amount of RecA (700 nM, lanes 2–17) and incubated for 60 min at 37°C. The products of the reactions were deproteinized, separated and monitored by 0.8% AGE with ethidium bromide. The position of the bands corresponding to css, lds, nc, jm and ccc are indicated. ± denote the presence or absence of the indicated protein.
As previously reported, the accumulation of jm intermediates increases with the presence of RecO, suggesting that RecO modulates the extent of RecA-mediated DNA strand exchange (22). To test whether the RecO acts by targeting RecA using SsbA or SsbB, RecA-mediated strand exchange in the presence of RecO and SsbA, SsbB or both was measured. RecO significantly increased the accumulation of jm intermediates and nc product with SsbA (Figure 4B, lanes 2–4) as compared to the absence of RecO (Figure 4A, lanes 3–5). The addition of RecO to SsbB·ssDNA did not stimulate RecA-mediated accumulation of nc products (Figure 4B, lanes 5–7), and only increased the accumulation of jm to a similar extent compared to RecO alone (see 22). The hybrid protein SsbB* showed similar results to SsbB (data not shown). In the presence of both SsbA and SsbB, in the form of the SsbA·ssDNA·SsbB complex, addition of RecO increased RecA-mediated DNA strand exchange when SsbA was in excess compared to SsbB, independent of the order of addition (Figure 4B, lanes 10–17). RecO interaction with SsbA likely enables RecA utilization of SsbA·ssDNA and SsbA·ssDNA·SsbB and promotes RecA re-invasion of the displaced ssDNA as deduced by the accumulation of jm intermediates, but re-invasion cannot take place on SsbB-coated ssDNA due to the lack of RecO interaction. Similarly, RecO is unable to overcome the inhibitory effect of SSBSPP1 or SSBEco when added before RecA (22,27).
SsbA reverses the negative effect exerted by SsbB on RecO-mediated DNA strand annealing
Plasmid transformation in B. subtilis requires RecO (55 and Table 3). RecO localizes to the entry pole when oligomeric plasmid DNA, which can self-anneal, enters the cell (10). In addition, SsbA-coated ssDNA facilitates RecO-mediated annealing of complementary ssDNA strands (11). To study the contribution of SsbA and SsbB on RecO-dependent plasmid transformation, the effects of SsbA, SsbB, or both on the rate of RecO-mediated SSA were measured. When compared with the absence of SSBs (11), the addition of SsbA, SsbB or SsbB* blocked spontaneous strand annealing of complementary homologous 440-nt ssDNA (Figure 5A, lanes 3, 7 and 11). Only SsbA facilitated RecO-mediated annealing of the complementary ssDNA molecules (10,11; Figure 5A, lanes 5 and 6). SsbB did not stimulate RecO-mediated strand annealing nor was the C-terminal end of SsbA, in the context of SsbB*, sufficient to contribute in the interaction with RecO and stimulate activity (Figure 5A, lanes 8–10 and 12–14). SsbA as part of the heterologous SsbA·ssDNA·SsbB complex facilitated RecO-mediated DNA strand annealing (Figures 5B, 8 and 10), again suggesting the significance of the functional interaction between SsbA and RecO.
Figure 5.
RecO anneals complementary strands complexed with SsbA protein. (A) Heat-denatured 440-nt long [α-32P]-ssDNA (7 µM in nt) was quickly cooled and pre-incubated with a fix amount of SsbA, SsbB or SsbB* (100 nM) for 10 min at 30°C in buffer E, and then incubated with increasing concentrations of RecO (1, 2 and 3 µM) for 60 min at 30°C. (B) The heat-denatured ssDNA was pre-incubated with a fix amount of SsbA, SsbB, SsbA followed by SsbB (lanes 7 and 8) or vice versa (lanes 9 and 10) (100 nM) for 10 min at 30°C in buffer E, and then incubated with a fix amount (2 µM, lanes 5 and 6) or increasing concentrations of RecO (1 and 2 µM, 7–10) for 60 min at 30°C in buffer E. Lane 2, heat-denatured ssDNA was slowly cold down (spontaneous annealing). The products of the reactions were deproteinized, separated by 6% PAGE and monitored by using a Geldoc (BioRad) system.
DISCUSSION
Our data reveal a division of labor between the SsbA and SsbB proteins in modulating RecA nucleation and filament extension and in reactions with the RecA-mediator protein RecO. SsbA facilitates RecO-mediated RecA nucleation and filament extension onto SsbA·ssDNA or SsbA·ssDNA·SsbB. In addition, SsbA helps RecO mediate DNA strand annealing between two complementary ssDNA molecules coated by SsbA or both SsbA and SsbB during plasmid transformation.
Distinct functions for SsbA and SsbB
The N-terminal domains of SsbA and SsbB share 63% identity in the ssDNA binding and subunit tetramerization domains (residues 1–106). Both SsbA and SsbB appear to have two modes for binding ssDNA correlating to the (SSBEco)65 and (SSBEco)35 binding modes (Figure 1). However, in B. subtilis, as well as S. pneumoniae, SsbB needs longer segments of ssDNA than SsbA in order to form two complexes in the presence of Mg2+ (Figure 1 and Supplemental Figure S2) (60,61). Consistent with its ssDNA binding properties, the crystal structure of SsbB in complex with ssDNA (Figure 2) shows commonalities with other SSB proteins in its ssDNA binding surfaces. However, SsbB appears to interact with ssDNA in a manner that buries the DNA bases toward the protein surface (Figure 2), unlike the SSBEco·ssDNA complex where the bases point toward the solvent (45). The biochemical and biological impact of this difference is not yet clear but it could account for some of the effects described herein.
Although the DNA-binding domains are similar, SsbA binds ssDNA with a much greater affinity (>5-fold) over that of SsbB (Figure 1 and Supplementary Figure S2). Interestingly, this contrasts the roles of SsbASpn and SsbBSpn, where the secondary SSB binds ssDNA with greater affinity (60,61). This difference in binding affinities for the two B. subtilis proteins could be partially due to the difference in the C-terminal domains. The C-terminus of SsbB (amino acids 107–113) lacks the acidic tail that serves as the interaction platform between SsbA and other proteins involved various DNA interactions. When the nine-most C-terminal residues of SsbA are attached to SsbB, in the chimeric protein SsbB*, SsbB* shows increased ssDNA binding affinity (Supplementary Figure S5). Unlike SsbBSpn which has an acidic C-terminal tail crucial for interaction with other proteins (20), the B. subtilis SsbB domain(s) for interaction with RecA, CoiA or DprA is not yet identified (8).
Taken together, these data show that the primary role of SsbB is to protect incoming ssDNA from nucleases, remove DNA secondary structures and inhibit non-productive RecA or RecO binding to ssDNA (Figures 4 and 5). Though the mechanism of SsbB interaction with RecA is poorly understood, we have shown here that SsbB has complex effects on RecA function. SsbB inhibits RecA nucleation onto SsbB-coated ssDNA (Figure 3A) but still marginally stimulates RecA-mediated DNA recombination (Figure 4A), suggesting that SsbB might be facilitating RecA activities through the removal of DNA secondary structures. Due to the lack of interaction between RecO and SsbB, RecO fails to recruit RecA onto SsbB-coated ssDNA or to catalyze strand annealing between two complementary ssDNA strands coated by SsbB (Figure 5).
SsbA has three seemingly opposing roles: prevention of RecA nucleation onto SsbA-coated ssDNA, facilitation of RecA·ssDNA filament extension via recruitment of RecO onto SsbA-coated ssDNA, and facilitation of RecO-mediated SSA. The recruitment of RecO by SsbA is mediated by the C-terminal end of SsbA (SSBEco) and the hydrophobic pocket on the C-terminal of RecO (24,25). However, the presence of the flexible nine C-terminal residues of SsbA in the context of SsbB (SsbB*) was not sufficient to promote SsbA·RecO interactions (Figure 5), suggesting that SsbA interacts with RecO through more than its C-terminal region (24). After RecO recruitment, the specific RecO·ssDNA interaction leads to limited displacement of SsbA from ssDNA and RecA nucleation onto SsbA·RecO·ssDNA or the annealing of two complementary ssDNA strands coated by SsbA (10,11).
SsbA and accessory SsbB modulates RecA and RecO activities
A model begins to emerge where SsbB plays an accessory role when co-assembled with SsbA on ssDNA. SsbB binds and protects the internalized ssDNA (20), but in its absence SsbA might also protect the foreign ssDNA. However, it has been suggested that SsbASpn cannot substitute for SsbBSpn (20). Importantly, SsbB cannot substitute for SsbA in the modulation of RecA activities and in the facilitation of RecO-mediated SSA. This is consistent with the observation that a heterologous SSB (e.g. SSBEco, SsbSPP1) cannot replace SsbA during RecO-mediated loading of RecA onto SSBEco-coated ssDNA (22,27). The specific interaction between RecO and SsbA, within the co-assembled SsbA·ssDNA·SsbB complex, enhances RecA nucleation and filament extension (Figure 3) and might alter the dissociation of both SsbA and SsbB proteins, resulting in faster net disassembly of both SsbB and SsbA. Indeed, the absence of SsbB decreases chromosomal transformantion at high donor DNA concentrations (20).
Chromosomal transformation of cells lacking SsbB decreases 3- to 10-fold in both B. subtilis and S. pneumoniae cells (15,17,26), suggesting that both SSB proteins act redundantly and SsbB might play a specific role elsewhere during natural transformation, perhaps in DprA-mediated RecA nucleation and filament extension. However, DrpASpn has poor selectivity for which SSB protein coats ssDNA because it can recruit the heterologous RecAEco onto SSBEco-coated ssDNA (54). It is possible that each mediator in B. subtilis, RecO or DprA, might preferentially work with specific SSB proteins.
RecO mediates RecA nucleation and filament extension during DSB repair and genetic recombination specifically using SsbA
DSB-initiated recombination, a multistage process that must occur between homologous sister strands or chromosomes, and the early stages of genetic transformation, between internalized ssDNA and a homologous recipient chromosome, each have two pathways for loading RecA onto ssDNA. In both processes, RecO is the RecA-mediator for one of the pathways. In DSB recombination, RecO and RecR are the most common RecA-mediator proteins in bacteria (52,62). During DNA repair and after end recognition and processing (5), RecO alone (or in association with RecN and/or RecR) recruits RecA onto SsbA·ssDNA at the RecN-mediated repair center (10,13). Similarly, RecOEco and RecREco facilitate the nucleation and the formation of RecAEco filaments onto SSBEco-coated ssDNA (39,57,63). In the other pathway, which is specific for DSB repair, end-processing and RecA recruitment are coupled (64).
During genetic recombination using competent cells, RecO facilitates limited displacement of SsbA from ssDNA or from the co-assembled SsbA·ssDNA·SsbB and promotes RecA nucleation and filament extension. No other accessory proteins beyond RecO (e.g. RecR or RecF) have yet been implicated using biochemical or genetic studies (22,55), meaning RecA nucleation and complete displacement of SsbA, SsbB and RecO from ssDNA appears to be achieved without additional factors. The alternate pathway comprises DprA mediating RecA nucleation onto SsbA- and SsbB-coated ssDNA. Regardless of the pathway for RecA nucleation and filament extension, the resulting RecA·ssDNA filament then searches for homology and catalyzes DNA strand invasion with subsequent integration of the internalized ssDNA (chromosomal transformation).
RecO interaction with SsbA is essential for plasmid transformation
Plasmid transformation, which is a RecA-independent event, proceeds via RecO and perhaps DprA, albeit the latter avenue is poorly understood. The SsbB·ssDNA complex inhibits RecO-mediated annealing of complementary strands (Figure 5), whereas SsbA·ssDNA recruits RecO to form a ternary SsbA·RecO·ssDNA (11). In the co-assembled SsbA·ssDNA·SsbB complex, RecO interaction with SsbA leads to the formation of bridged structures and strand annealing, rather than mutually exclusive interactions. These structures either decrease the half-life of SsbA- and SsbB-coated ssDNA or alter the structure of ssDNA to facilitate the dissociation of both SsbA and SsbB from ssDNA, possibly resulting in faster net disassembly of both SsbA and SsbB. The SsbA-mediated assembly of RecO then promotes DNA strand annealing.
How might an intact dsDNA circular replicon be reconstituted (plasmid establishment) at the molecular level? In B. subtilis plasmid transformation exhibits a linear dependence on the concentration of oligomeric donor plasmid DNA and the monomers are inactive (50,65,66). In one model, SsbA or both SsbA and SsbB limit RecA filament formation by coating segments of the improperly hybridized complementary strands of the internalized linear oligomeric ssDNA (66). If no homology is found with recipient DNA, RecU dislodges any RecA nucleoprotein filament (10,67,68) and SsbA or both SsbA and SsbB bind to the internalized ssDNA. SsbA would then facilitate RecO-mediated annealing of the internalized strands and subsequent re-circularization of the tailed duplex molecule (10,11,50,65,66). In another model, the internalized strand of oligomeric plasmid DNA coated by SsbA or SsbA and SsbB is synthesized at its lagging strand replication origin to generate a tailed duplex molecule (50,69) that is subsequently re-circularized by RecO-mediated DNA strand annealing (11). This is consistent with the observation that the absence of SsbBSpn decreases plasmid transformation ∼10-fold, but in the presence of high plasmid DNA concentrations increases plasmid transformation ∼10-fold (20), suggesting that SsbB covers and protects ssDNA from nucleolytic attacks and might also antagonize RecO-mediated strand annealing. In this report, we have presented evidence in support of a division of labor between SsbA and SsbB, with SsbA stimulating RecO-mediated strand annealing required for plasmid transformation, overcoming the interference exerted by SsbB.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figures 1–5 and Supplementary Methods.
FUNDING
Ministerio de Ciencia e Innovación, Dirección General de Investigación (grants BFU2009-07167 and CSD2007-00010 to J.C.A.); Comunidad de Madrid (grant S2009MAT-1507 to J.C.A.); NIH (grant GM068061 to J.L.K.). T.Y. is a PhD fellow of the International Fellowship Programme of La Caixa Foundation (La Caixa/CNB). Funding for open access charge: Ministerio de Ciencia e Innovación, Dirección General de Investigación (BFU2009-07167 to J.C.A.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank S. Ayora for comments on the manuscript.
REFERENCES
- 1.Dubnau D. DNA uptake in bacteria. Annu. Rev. Microbiol. 1999;53:217–244. doi: 10.1146/annurev.micro.53.1.217. [DOI] [PubMed] [Google Scholar]
- 2.Sanchez H, Carrasco B, Ayora S, Alonso JC. Homologous Recombination in Low dC + dG Gram-Positive Bacteria. Berlin, Heidelberg: Springer Berlin/Heidelberg; 2007. [Google Scholar]
- 3.Claverys JP, Martin B, Polard P. The genetic transformation machinery: composition, localization, and mechanism. FEMS Microbiol. Rev. 2009;33:643–656. doi: 10.1111/j.1574-6976.2009.00164.x. [DOI] [PubMed] [Google Scholar]
- 4.Chen I, Christie PJ, Dubnau D. The ins and outs of DNA transfer in bacteria. Science. 2005;310:1456–1460. doi: 10.1126/science.1114021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ayora S, Carrasco B, Cárdenas PP, César CE, Cañas C, Yadav T, Marchisone C, Alonso JC. Double-strand break repair in bacteria: a view from Bacillus subtilis. FEMS Microbiol. Rev. 2011;35:1055–1081. doi: 10.1111/j.1574-6976.2011.00272.x. [DOI] [PubMed] [Google Scholar]
- 6.Burton B, Dubnau D. Membrane-associated DNA transport machines. Cold Spring Harb. Perspect. Biol. 2010;2:a000406. doi: 10.1101/cshperspect.a000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hahn J, Maier B, Haijema BJ, Sheetz M, Dubnau D. Transformation proteins and DNA uptake localize to the cell poles in Bacillus subtilis. Cell. 2005;122:59–71. doi: 10.1016/j.cell.2005.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kramer N, Hahn J, Dubnau D. Multiple interactions among the competence proteins of Bacillus subtilis. Mol. Microbiol. 2007;65:454–464. doi: 10.1111/j.1365-2958.2007.05799.x. [DOI] [PubMed] [Google Scholar]
- 9.Kidane D, Graumann PL. Intracellular protein and DNA dynamics in competent Bacillus subtilis cells. Cell. 2005;122:73–84. doi: 10.1016/j.cell.2005.04.036. [DOI] [PubMed] [Google Scholar]
- 10.Kidane D, Carrasco B, Manfredi C, Rothmaier K, Ayora S, Tadesse S, Alonso JC, Graumann PL. Evidence for different pathways during horizontal gene transfer in competent Bacillus subtilis cells. PLoS Genet. 2009;5:e1000630. doi: 10.1371/journal.pgen.1000630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manfredi C, Suzuki Y, Yadav T, Takeyasu K, Alonso JC. RecO-mediated DNA homology search and annealing is facilitated by SsbA. Nucleic Acids Res. 2010;38:6920–6929. doi: 10.1093/nar/gkq533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tadesse S, Graumann PL. DprA/Smf protein localizes at the DNA uptake machinery in competent Bacillus subtilis cells. BMC Microbiol. 2007;7:105. doi: 10.1186/1471-2180-7-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kidane D, Sanchez H, Alonso JC, Graumann PL. Visualization of DNA double-strand break repair in live bacteria reveals dynamic recruitment of Bacillus subtilis RecF, RecO and RecN proteins to distinct sites on the nucleoids. Mol. Microbiol. 2004;52:1627–1639. doi: 10.1111/j.1365-2958.2004.04102.x. [DOI] [PubMed] [Google Scholar]
- 14.Krishnamurthy M, Tadesse S, Rothmaier K, Graumann PL. A novel SMC-like protein, SbcE (YhaN), is involved in DNA double-strand break repair and competence in Bacillus subtilis. Nucleic Acids Res. 2010;38:455–466. doi: 10.1093/nar/gkp909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berka RM, Hahn J, Albano M, Draskovic I, Persuh M, Cui X, Sloma A, Widner W, Dubnau D. Microarray analysis of the Bacillus subtilis K-state: genome-wide expression changes dependent on ComK. Mol. Microbiol. 2002;43:1331–1345. doi: 10.1046/j.1365-2958.2002.02833.x. [DOI] [PubMed] [Google Scholar]
- 16.Hamoen LW, Smits WK, de Jong A, Holsappel S, Kuipers OP. Improving the predictive value of the competence transcription factor (ComK) binding site in Bacillus subtilis using a genomic approach. Nucleic Acids Res. 2002;30:5517–5528. doi: 10.1093/nar/gkf698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ogura M, Yamaguchi H, Kobayashi K, Ogasawara N, Fujita Y, Tanaka T. Whole-genome analysis of genes regulated by the Bacillus subtilis competence transcription factor ComK. J. Bacteriol. 2002;184:2344–2351. doi: 10.1128/JB.184.9.2344-2351.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Comella N, Grossman AD. Conservation of genes and processes controlled by the quorum response in bacteria: characterization of genes controlled by the quorum-sensing transcription factor ComA in Bacillus subtilis. Mol. Microbiol. 2005;57:1159–1174. doi: 10.1111/j.1365-2958.2005.04749.x. [DOI] [PubMed] [Google Scholar]
- 19.Lindner C, Nijland R, van Hartskamp M, Bron S, Hamoen LW, Kuipers OP. Differential expression of two paralogous genes of Bacillus subtilis encoding single-stranded DNA binding protein. J. Bacteriol. 2004;186:1097–1105. doi: 10.1128/JB.186.4.1097-1105.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Attaiech L, Olivier A, Mortier-Barriére I, Soulet AL, Granadel C, Martin B, Polard P, Claverys JP. Role of the single-stranded DNA-binding protein SsbB in pneumococcal transformation: maintenance of a reservoir for genetic plasticity. PLoS Genet. 2011;7:e1002156. doi: 10.1371/journal.pgen.1002156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lecointe F, Séréna C, Velten M, Costes A, McGovern S, Meile JC, Errington J, Ehrlich SD, Noirot P, Polard P. Anticipating chromosomal replication fork arrest: SSB targets repair DNA helicases to active forks. EMBO J. 2007;26:4239–4251. doi: 10.1038/sj.emboj.7601848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manfredi C, Carrasco B, Ayora S, Alonso JC. Bacillus subtilis RecO nucleates RecA onto SsbA-coated single-stranded DNA. J. Biol. Chem. 2008;283:24837–24847. doi: 10.1074/jbc.M802002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Costes A, Lecointe F, McGovern S, Quevillon-Cheruel S, Polard P. The C-terminal domain of the bacterial SSB protein acts as a DNA maintenance hub at active chromosome replication forks. PLoS Genet. 2010;6:e1001238. doi: 10.1371/journal.pgen.1001238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Inoue J, Nagae T, Mishima M, Ito Y, Shibata T, Mikawa T. A mechanism for single-stranded DNA-binding protein (SSB) displacement from single-stranded DNA upon SSB-RecO interaction. J. Biol. Chem. 2011;286:6720–6732. doi: 10.1074/jbc.M110.164210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryzhikov M, Koroleva O, Postnov D, Tran A, Korolev S. Mechanism of RecO recruitment to DNA by single-stranded DNA binding protein. Nucleic Acids Res. 2011;39:6305–6314. doi: 10.1093/nar/gkr199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bergé M, Mortier-Barriére I, Martin B, Claverys JP. Transformation of Streptococcus pneumoniae relies on DprA- and RecA-dependent protection of incoming DNA single strands. Mol. Microbiol. 2003;50:527–536. doi: 10.1046/j.1365-2958.2003.03702.x. [DOI] [PubMed] [Google Scholar]
- 27.Carrasco B, Manfredi C, Ayora S, Alonso JC. Bacillus subtilis SsbA and dATP regulate RecA nucleation onto single-stranded DNA. DNA Repair. 2008;7:990–996. doi: 10.1016/j.dnarep.2008.03.019. [DOI] [PubMed] [Google Scholar]
- 28.Carrasco B, Ayora S, Lurz R, Alonso JC. Bacillus subtilis RecU Holliday-junction resolvase modulates RecA activities. Nucleic Acids Res. 2005;33:3942–3952. doi: 10.1093/nar/gki713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lioy VS, Martín MT, Camacho AG, Lurz R, Antelmann H, Hecker M, Hitchin E, Ridge Y, Wells JM, Alonso JC. pSM19035-encoded ζ toxin induces stasis followed by death in a subpopulation of cells. Microbiology. 2006;152:2365–2379. doi: 10.1099/mic.0.28950-0. [DOI] [PubMed] [Google Scholar]
- 30.Soberón NE, Lioy VS, Pratto F, Volante A, Alonso JC. Molecular anatomy of the Streptococcus pyogenes pSM19035 partition and segrosome complexes. Nucleic Acids Res. 2011;39:2624–2637. doi: 10.1093/nar/gkq1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alonso JC, Stiege AC, Dobrinski B, Lurz R. Purification and properties of the RecR protein from Bacillus subtilis 168. J. Biol. Chem. 1993;268:1424–1429. [PubMed] [Google Scholar]
- 32.Riggs AD, Bourgeois S, Cohn M. The lac repressor-operator interaction. 3. Kinetic studies. J. Mol. Biol. 1970;53:401–417. doi: 10.1016/0022-2836(70)90074-4. [DOI] [PubMed] [Google Scholar]
- 33.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1977;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 34.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 36.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–253. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 38.Morrical SW, Lee J, Cox MM. Continuous association of Escherichia coli single-stranded DNA binding protein with stable complexes of RecA protein and single-stranded DNA. Biochemistry. 1986;25:1482–1494. doi: 10.1021/bi00355a003. [DOI] [PubMed] [Google Scholar]
- 39.Hobbs MD, Sakai A, Cox MM. SSB protein limits RecOR binding onto single-stranded DNA. J. Biol. Chem. 2007;282:11058–11067. doi: 10.1074/jbc.M611007200. [DOI] [PubMed] [Google Scholar]
- 40.Arenson TA, Tsodikov OV, Cox MM. Quantitative analysis of the kinetics of end-dependent disassembly of RecA filaments from ssDNA. J. Mol. Biol. 1999;288:391–401. doi: 10.1006/jmbi.1999.2705. [DOI] [PubMed] [Google Scholar]
- 41.Ayora S, Missich R, Mesa P, Lurz R, Yang S, Egelman EH, Alonso JC. Homologous-pairing activity of the Bacillus subtilis bacteriophage SPP1 replication protein G35P. J. Biol. Chem. 2002;277:35969–35979. doi: 10.1074/jbc.M204467200. [DOI] [PubMed] [Google Scholar]
- 42.Ayora S, Piruat JI, Luna R, Reiss B, Russo VE, Aguilera A, Alonso JC. Characterization of two highly similar Rad51 homologs of Physcomitrella patens. J. Mol. Biol. 2002;316:35–49. doi: 10.1006/jmbi.2001.5336. [DOI] [PubMed] [Google Scholar]
- 43.Lohman TM, Ferrari ME. Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu. Rev. Biochem. 1994;63:527–570. doi: 10.1146/annurev.bi.63.070194.002523. [DOI] [PubMed] [Google Scholar]
- 44.Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL. SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 2008;43:289–318. doi: 10.1080/10409230802341296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raghunathan S, Kozlov AG, Lohman TM, Waksman G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat. Struct. Biol. 2000;7:648–652. doi: 10.1038/77943. [DOI] [PubMed] [Google Scholar]
- 46.Chan KW, Lee YJ, Wang CH, Huang H, Sun YJ. Single-stranded DNA-binding protein complex from Helicobacter pylori suggests an ssDNA-binding surface. J. Mol. Biol. 2009;388:508–519. doi: 10.1016/j.jmb.2009.03.022. [DOI] [PubMed] [Google Scholar]
- 47.Kowalczykowski SC, Clow J, Somani R, Varghese A. Effects of the Escherichia coli SSB protein on the binding of Escherichia coli RecA protein to single-stranded DNA. Demonstration of competitive binding and the lack of a specific protein-protein interaction. J. Mol. Biol. 1987;193:81–95. doi: 10.1016/0022-2836(87)90629-2. [DOI] [PubMed] [Google Scholar]
- 48.Lindsley JE, Cox MM. Assembly and disassembly of RecA protein filaments occur at opposite filament ends. Relationship to DNA strand exchange. J. Biol. Chem. 1990;265:9043–9054. [PubMed] [Google Scholar]
- 49.Dubnau D, Davidoff-Abelson R, Scher B, Cirigliano C. Fate of transforming deoxyribonucleic acid after uptake by competent Bacillus subtilis: phenotypic characterization of radiation-sensitive recombination-deficient mutants. J. Bacteriol. 1973;114:273–286. doi: 10.1128/jb.114.1.273-286.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Canosi U, Morelli G, Trautner TA. The relationship between molecular structure and transformation efficiency of some S. aureus plasmids isolated from B. subtilis. Mol. Gen. Genet. 1978;166:259–267. doi: 10.1007/BF00267617. [DOI] [PubMed] [Google Scholar]
- 51.Alonso JC, Lüder G, Tailor RH. Characterization of Bacillus subtilis recombinational pathways. J. Bacteriol. 1991;173:3977–3980. doi: 10.1128/jb.173.13.3977-3980.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cox MM. Regulation of bacterial RecA protein function. Crit. Rev. Biochem. Mol. Biol. 2007;42:41–63. doi: 10.1080/10409230701260258. [DOI] [PubMed] [Google Scholar]
- 53.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 54.Mortier-Barriére I, Velten M, Dupaigne P, Mirouze N, Piétrement O, McGovern S, Fichant G, Martin B, Noirot P, Le Cam E, et al. A key presynaptic role in transformation for a widespread bacterial protein: DprA conveys incoming ssDNA to RecA. Cell. 2007;130:824–836. doi: 10.1016/j.cell.2007.07.038. [DOI] [PubMed] [Google Scholar]
- 55.Fernández S, Kobayashi Y, Ogasawara N, Alonso JC. Analysis of the Bacillus subtilis recO gene: RecO forms part of the RecFLOR function. Mol. Gen. Genet. 1999;261:567–573. doi: 10.1007/s004380051002. [DOI] [PubMed] [Google Scholar]
- 56.Ceglowski P, Lüder G, Alonso JC. Genetic analysis of recE activities in Bacillus subtilis. Mol. Gen. Genet. 1990;222:441–445. doi: 10.1007/BF00633853. [DOI] [PubMed] [Google Scholar]
- 57.Umezu K, Kolodner RD. Protein interactions in genetic recombination in Escherichia coli. Interactions involving RecO and RecR overcome the inhibition of RecA by single-stranded DNA-binding protein. J. Biol. Chem. 1994;269:30005–30013. [PubMed] [Google Scholar]
- 58.Lu D, Keck JL. Structural basis of Escherichia coli single-stranded DNA-binding protein stimulation of exonuclease I. Proc. Natl Acad. Sci. USA. 2008;105:9169–9174. doi: 10.1073/pnas.0800741105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Grove DE, Bryant FR. Effect of Mg2+ on the DNA binding modes of the Streptococcus pneumoniae SsbA and SsbB proteins. J. Biol. Chem. 2006;281:2087–2094. doi: 10.1074/jbc.M510884200. [DOI] [PubMed] [Google Scholar]
- 61.Salerno B, Anne G, Bryant FR. DNA Binding Compatibility of the Streptococcus pneumoniae SsbA and SsbB Proteins. PLoS One. 2011;6:e24305. doi: 10.1371/journal.pone.0024305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beernink HT, Morrical SW. RMPs: recombination/replication mediator proteins. Trends Biochem. Sci. 1999;24:385–389. doi: 10.1016/s0968-0004(99)01451-6. [DOI] [PubMed] [Google Scholar]
- 63.Umezu K, Chi NW, Kolodner RD. Biochemical interaction of the Escherichia coli RecF, RecO, and RecR proteins with RecA protein and single-stranded DNA binding protein. Proc. Natl Acad. Sci. USA. 1993;90:3875–3879. doi: 10.1073/pnas.90.9.3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dillingham MS, Kowalczykowski SC. RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiol. Mol. Biol. Rev. 2008;72:642–671. doi: 10.1128/MMBR.00020-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dubnau D, Contente S, Gryzan TJ. DNA – Recombination Interactions and Repair. Oxford: Pergamon Press; 1980. pp. 365–386. [Google Scholar]
- 66.de Vos WM, Venema G, Canosi U, Trautner TA. Plasmid transformation in Bacillus subtilis: fate of plasmid DNA. Mol. Gen. Genet. 1981;181:424–433. doi: 10.1007/BF00428731. [DOI] [PubMed] [Google Scholar]
- 67.Cañas C, Carrasco B, Ayora S, Alonso JC. The RecU Holliday junction resolvase acts at early stages of homologous recombination. Nucleic Acids Res. 2008;36:5242–5249. doi: 10.1093/nar/gkn500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cañas C, Carrasco B, García-Tirado E, Rafferty JB, Alonso JC, Ayora S. The stalk region of the RecU resolvase is essential for holliday junction recognition and distortion. J. Mol. Biol. 2011;410:39–49. doi: 10.1016/j.jmb.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 69.Fernández S, Ayora S, Alonso JC. Bacillus subtilis homologous recombination: genes and products. Res. Microbiol. 2000;151:481–486. doi: 10.1016/s0923-2508(00)00165-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.