Abstract
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive hematological malignancy. The understanding of its gene expression regulation and molecular mechanisms still remains elusive. Started from experimentally verified T-ALL-related miRNAs and genes, we obtained 120 feed-forward loops (FFLs) among T-ALL-related genes, miRNAs and TFs through combining target prediction. Afterwards, a T-ALL miRNA and TF co-regulatory network was constructed, and its significance was tested by statistical methods. Four miRNAs in the miR-17–92 cluster and four important genes (CYLD, HOXA9, BCL2L11 and RUNX1) were found as hubs in the network. Particularly, we found that miR-19 was highly expressed in T-ALL patients and cell lines. Ectopic expression of miR-19 represses CYLD expression, while miR-19 inhibitor treatment induces CYLD protein expression and decreases NF-κB expression in the downstream signaling pathway. Thus, miR-19, CYLD and NF-κB form a regulatory FFL, which provides new clues for sustained activation of NF-κB in T-ALL. Taken together, we provided the first miRNA-TF co-regulatory network in T-ALL and proposed a model to demonstrate the roles of miR-19 and CYLD in the T-cell leukemogenesis. This study may provide potential therapeutic targets for T-ALL and shed light on combining bioinformatics with experiments in the research of complex diseases.
INTRODUCTION
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive hematological malignancy accounting for about 15 and 25% of pediatric and adult acute lymphoblastic leukemia (ALL), respectively (1). T-ALL is usually characterized by proliferation of thymocytes at various stages of development with high-white blood cell counts, mediastinal lymph nodes enlargement and central nervous system involvement (2). Although this neoplastic disorder originates from the thymus, it will spread throughout all organs and will be fatal rapidly without therapy. Compared to the common B-cell lineage ALL, T-ALL has a worse prognosis in patients historically. Current multi-agent combination chemotherapy provides an overall survival rate of 60–70% in children and only 30–40% in adults (3,4). Securing further advances in treatment is dependent on our increasing knowledge on the factors and mechanism contributing to the malignant behaviour of transformed thymocytes. Currently, understanding of the etiology of T-ALL has largely come from the studies of chromosomal abnormalities. Many chromosomal translocations and gene-specific alterations have been identified, which include rearrangements of T-cell receptor genes, ectopic expression of TLX1, TLX3, LMO2, LMO1, TAL1 and HOXA, mutations of NOTCH1, PTEN and FBXW7, deletion of CDKN2A and fusion of NUP214 to ABL1 [review in (5–7)]. Although the oncogenicity of these genes is well established, understanding of the transformational programs and multi-step pathogenesis of T-ALL remains limited. Especially, the regulatory networks of T-ALL genes expression are still elusive.
MicroRNAs (miRNAs) are small noncoding RNAs of 19–24 nt in length that regulate gene expression at the post-transcriptional level. Long primary miRNAs are first transcribed by RNA polymerase II in the nucleus and modified by an enzyme complex containing DROSHA and DGCR8 to form pre-miRNA. Subsequent cleavage of pre-miRNA by an RNase III, DICER1, results in mature miRNA. The mature miRNA may suppress translation and enhances degradation of target mRNA by binding to its target site on mRNA 3′-UTR regions (8). MiRNAs play crucial roles in various physiological processes and are involved in the initiation and progression of human cancers including T-ALL (9–11). It had been reported that over-expression of miR-125b would induce leukemia independently in a mouse model (12). High expression of miR-196b was found in leukemia with aberrant activation of HOXA genes (13). MiRNA expression profiles in ALL have been identified by several groups (14,15). Human miR-17–92 cluster is sufficient to promote leukemogenesis in Notch1-induced T-ALL in vivo (16), and over-expression of pri-miR-17–92 in T-ALL cell lines will reduce E2F1 protein level to enhance the survival of leukemic T-cells (17). Recently, miR-451 and miR-709 were demonstrated as potent suppressors of oncogenesis in Notch1-induced mouse T-ALL (18). Although a few studies reported the aberrant expression and function of miRNAs in T-ALL, the miRNA regulatory network in T-ALL is a key question to be addressed urgently.
Transcription factors (TFs) are key regulators controlling the transcription of target genes by binding to specific DNA sequences on the promoter of target genes. Both the TFs and miRNAs are regulators of gene expression, and they may mutual regulate each other to form feedback loops, or they regulate the same target gene to form a feed-forward loop (FFL). It has been reported that hundreds of potential miRNA-mediated feedback and FFLs are available at the genome level (19–21). Several feedback loops and FFLs have been experimentally verified, such as PITX3 and miR-133b in midbrain dopamine neurons, cyclin D1 and miRNA-17/20 feedback loop in breast cancer and TP53/miR-106b/E2F FFL in cell proliferation (22–24). Moreover, several databases about miRNA-TF feed-forward regulatory circuits have been developed (25,26). Recently, we have identified 32 FFLs and constructed the miRNA-TF co-regulatory network in schizophrenia, which is the first study investigating the miRNA-TF regulatory network for a human complex disease (27).
In this study, we aim to identify important miRNAs and regulatory modules in T-ALL. Starting from collecting T-ALL-related miRNAs and predicting the miRNA and TF targets based on a combined strategy, then, we constructed a miRNA-TF co-regulatory network specifically for T-ALL and found some hub regulators, genes and their regulation in the network. To verify the reliability of regulatory modules and network, we experimentally confirmed that the hub miR-19 indeed inhibited hub gene CYLD in T-ALL, and CYLD, miR-19 and NF-κB participated in a FFL. Consequently, this study enhanced the understanding of regulatory mechanism of T-ALL and also provided a new approach with bioinformatics guidance in studying of cancers and other complex diseases.
MATERIALS AND METHODS
T-ALL candidate genes and miRNAs
To analyze the miRNA-TF regulation in T-ALL, we collected and curated the T-ALL-related miRNAs and genes in the following two criteria: (i) Those have been verified the functions and aberrant expression in T-ALL patients, cell lines or animal models by experiments. (ii) Those have been clearly described with pivotal roles in T-ALL review literatures. For comparison, we compiled two control miRNA data sets: miRNAs expressed in thymus (thymus miRNAs) and non-thymus tissues (non-thymus miRNAs). Thymus miRNAs were collected from miRNA microarray expression studies and a miRNA survey study (15,28). Non-thymus miRNAs were those reported not expressed in thymus from a miRNA expression profile (29). To avoid the redundancy, we removed T-ALL-related miRNAs (T-ALLmiRNAs) from thymus miRNAs and thymus miRNAs from non-thymus miRNAs. We obtained information of genomic locations, host genes and conservations of these miRNAs from miRBase (30) (Release 16: Sept 2010, genome assembly: NCBI 37.1).
Target prediction of miRNAs and TFs
MiRNA targets were predicated mainly by combinatorial utilization of four different web-based databases. We chose the overlapped predicted targets from TargetScan (TargetScan 5.1, April 2009) and miRanda (Release Date: September 2010) based on the evolutionary conservation among mammalias. Then, we merged the results with the reported miRNA targets from miR2Disease (Release Date: December 2010) and TarBase (TarBase_V5, Jun 2008) (31–34). The relationships between them can be described as (TargetScan∩miRanda)∪miR2Disease∪TarBase. We also collected the experimentally confirmed targets for the T-ALL miRNAs by literature curation and combined them with the above results. MiRNAs clustered in a genomic region are preferentially co-expressed and miRNAs in gene region are usually co-expressed with their host genes, as a part of the same transcription unit. Used the similar strategy used in our previous work, we chose a 5 kb maximum inter-miRNA distance as the miRNA cluster criteria and 5 kb upstream of the host gene, miRNA precursor or miRNA cluster as the putative promoter region for miRNA in a genic region, intergenic region or miRNA cluster, respectively (27). We retrieved predicted transcription factor binding sites (TFBSs) from the UCSC genome browser (35) and required them to be conserved among humans, mice and rats. To further reduce the false-positive prediction, we used Z score of 2.33 as a cutoff for high-quality TFBSs. A TFBS was considered associated with a target gene when it was in the gene’s 5 k promoter region and its Z score was >2.33.
FFLs and statistics tests
FFLs were constructed among TFs, miRNAs and T-ALL genes (T-ALLGenes) according to the procedure in Figure 1. Two methods were used to evaluate the significance of FFLs among T-ALLmiRNAs, T-ALLGenes and TFs as our previous study (27). Here, we described them simply. First, we used Fisher’s exact test to compare the observed FFLs from T-ALLmiRNAs with those from thymus miRNAs or non-thymus miRNAs. Second, we ran randomisation processes to extract the same number of random miRNA target pairs out of all predicted target pairs of the T-ALLmiRNAs and then calculated the number of FFL among miRNAs, random targets and TFs. We repeated this 10 000 times and set the P-value as the proportion of the random results that had no less than the number of FFLs observed in the set of T-ALLmiRNAs and T-ALLGenes.
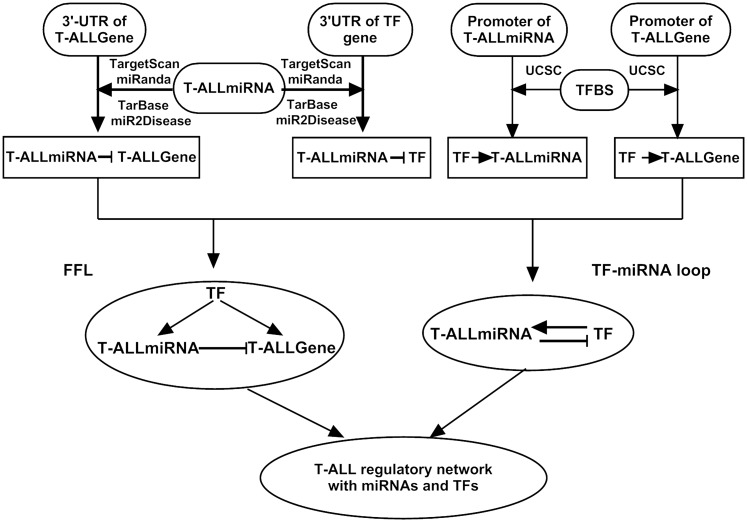
Figure 1.
Workflow of the miRNA-TF regulatory network construction in T-ALL. First, using sequence analysis strategy, we built two independent pipelines for the construction of a transcription factor and miRNA regulatory network. Then, merged them into a miRNA-TF regulatory network.
Network and hub analysis
Networks were presented with Cytoscape software (36) (version 2.6.1). In the miRNA-TF-mediated network, we defined node as a hub gene or miRNA when it directly linked to more than eight total nodes in the network. We used the Functional Annotation Tool of DAVID (37) (DAVID 6.7, http://david.abcc.ncifcrf.gov) to analyze networks and pathways for a set of genes. We set P-value < 0.01 as the cutoff for enriched GO terms or pathways identified in Gene Ontology and KEGG.
Cell lines and culture
Human tumor cell line Jurkat (acute T-cell leukemia), Molt-4 (human T-cell leukemia), HepG2 (hepatocarcinoma) and embryonic kidney cell line HEK-293 T were purchased from the China Center for Type Culture Collection (CCTCC, China) and maintained in RPMI-1640 Medium (Hyclone, USA) containing 10% fetal bovine serum (FBS), with 100 U/ml penicillin and 100 U/ml streptomycin. All the cells were grown in an atmosphere of 5% CO2 at 37°C.
RNA extraction and qRT–PCR
Whole RNAs were isolated from cell lines and tissue samples using TRIzol (Invitrogen, USA), and the protocol was done according to the manual of TRIzol. cDNAs were synthesized from 2 μg of total RNA with RevertAid™ First-strand cDNA Synthesis Kit (Fermentas, USA) in a 50 μl volume {2 μg total RNA, 5 nM reverse transcription primer [random primers for U6 srRNA and miRNA specific primers (Bulge-Loop™ miRNA qPCR primers from RiboBio, China) for miRNA], 0.8 U/μl reverse transcriptase, 4 U/μl inhibitors, 0.2 mM dNTP mix}. Real time-PCR was carried out with the reagents of SYBR Green Master (Roche, Germany) in 20 μl reaction volume (10 μl SYBR Green Master, 500 nM forward primers, 500 nM reverse primers and 2 μl cDNA template) by using StepOnePlus™ Systems (ABI, USA) with the following protocol: 95°C for 20 s; 40 cycles of 95°C for 10 s, 60°C for 20 s, 70°C for 5 s. Data analysis was performed using the 2ΔΔCt method (38).
Transfection assay
The mimics and inhibitors of miR-19, negative control miRNA were obtained from Ribobio (Guangzhou, China); Transfection was performed using X-tremeGENE siRNA Transfection Reagent (Roche, Germany) according to the manufacturer’s instructions. The cells were seeded in six-well plates (1 × 105 cells/well) with 1.5 ml RPMI-1640 medium (Hyclone, USA) containing 6% FBS, without penicillin/streptomycin. The miR-19 mimics/inhibitors/negative control groups were prediluted in 250 μl Opti-MEM® I Reduced Serum Medium (Gibco, USA). Subsequently, 5–10 μl X-tremeGENE siRNA Transfection Reagent was diluted in 250 μl Opti-MEM® I Reduced Serum Medium and combined with the miR-19 dilution solution by Opti-MEM® I Reduced Serum Medium. The 500 μl mixture was incubated for 20 min at room temperature and added to the cells.
Cell survival assay
The cells were seeded into the 96-well plates and transfected with the miR-19 mimics/inhibitors/negative control, which was performed using X-tremeGENE siRNA Transfection Reagent (Roche, Germany) according to the manufacturer’s instructions. After 24 or 48 h of transfection, WST-1 solution (Roche, Germany) was added, and the cell number was detected at 450 nm using the Tecan Sunrise microplate reader (Mannedorf, Switzerland). Relative cell survival = Asample/Auntreat control.
Western blot
The cells cytoplasm or nucleus lysates after transfection with miR-19 mimics/inhibitors/negative were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE), followed by the transfer onto nitrocellulose membranes. The membranes were blocked in TBST (Tris-buffered saline with 0.1% Triton X-100) containing 5% non-fat milk and probed with rabbit anti-human CYLD (Abcam, UK) or NF-κB P65 (SantaCruz, USA) or Histone 2A.X (Abcam, UK) antibodies or mouse anti-human β-actin antibody (SantaCruz, USA). After incubation with the secondary antibody (Li-Cor, USA), conjugated with horseradish peroxidase, membranes were extensively washed with TBST, and positive signals were visualized by Odyssey® Two-Color Infrared Imaging System (Li-Cor, USA). After indicated, the level of CYLD and P65 protein was quantitated by densitometric analysis using Quantity One® software (Bio-Rad, USA). The results were expressed as the ratio of the band density of CYLD to that of corresponding β-actin or P65 to that of corresponding Histone 2A.X.
Luciferase reporter assay
A fragment of 3′-UTR of CYLD DNA containing the target sequence (TTTGCAC) of miR-19 was amplified by PCR (sense 5′-GCCTCGAGGGCAAAGAAACTGAAGGC-3′, anti-sense 5′-CAGCGGCCGCCTGCTAAATACCCTAATC-3′; Bioladder, China). The PCR products were gel purified, digested and inserted into pMD®18-T Vector (Takara) and then confirmed by PCR. The plasmids were extracted and digested by XhoI and NotI. Then, the produces were inserted into the digested psiCHECK-2 vectors (Promega, USA) between the XhoI and NotI sites. The inserted sequences were confirmed by sequencing.
The HEK293T cells were seeded in 12-well plates with 800 μl Opti-MEM® I Reduced Serum Medium without penicillin/streptomycin. Plasmids and miR-19 mimics/inhibitors/negative control were cotransfected into HEK293T cells with Lipo 2000 Transfection Reagent (Invitrogen, USA) according to the manufacturer’s protocol. The mimics of miR-19 and luciferase reporter vectors were prediluted in 100 μl Opti-MEM® I Reduced Serum Medium. Afterwards, 2 μl Lipo2000 Transfection Reagent was diluted in 100 μl Opti-MEM® I Reduced Serum Medium and combined with the miR-19 dilution solution by Opti-MEM® I Reduced Serum Medium. The 200 μl mixture was incubated for 20 min at room temperature and added to cells. The final miR-19 concentration was 50 nM. The medium was adjusted to 10% FBS after incubation for 6 h.
The luciferase reporter assay was performed according to the manufacturer’s instructions of Dual-Luciferase Report Assay Kit (Promega, USA). The cells were collected after transfection for 48 h, and 75 μl (5 × 104) cells were lysed with 75 -μl luciferase reagent for 10 min in the white plate. The plate was read by Tecan luminescence plate reader (Mannedorf, Switzerland) as the activity of firefly luciferase. Then, 75 -μl dual luciferase reagent was added into the plate and mixed for 10 min. The plate was read as the activity of renilla luciferase. Result = Arenilla luciferase/Afirefly luciferase.
RESULTS
T-ALL-related miRNAs, genes and TFs
Aiming to explore the miRNA and TF co-regulatory network in T-ALL, it is indispensable to construct the feedback loop between miRNA and TF, and FFL among miRNA, TF and T-ALL genes (Figure 1). On one hand, we compiled 67 T-ALL-related genes (T-ALLGenes) from literatures, which play critical roles with aberrant expression in T-ALL patients, cell lines or animal models. On the other hand, after exhausted searching for miRNA expression profiles in T-ALL, we collected and curated 36 T-ALL-related miRNAs (T-ALLmiRNAs), which are experimentally verified to be crucial and aberrant expression in T-ALL (14,39,40). Furthermore, for comparison, we also collected 109 miRNAs expressed in thymus (thymus miRNAs) and 113 miRNAs, which were not expressed in thymus from published miRNA expression profiles (non-thymus miRNAs).
According to unified names in miRBase (30) (http://mirbase.org), the 36 T-ALL-related mature miRNAs, we collected match to 42 precursor miRNAs. Among the 42 T-ALL miRNAs, 31 are conserved in Vertebrata, 4 are conserved in Chordata, 1 is even conserved in Drosophila and 6 are mammal-specific (Table 1). Ten miRNAs are in host genes and others are intergenic miRNAs. Thirteen T-ALLmiRNAs (31%) are found downregulated in T-ALL, while others are upregulated. It is notable that all members of the two notorious tumor-related miRNA clusters (miR-17–92 and miR-106–363) are included in our T-ALLmiRNA list.
Table 1.
Basic information of T-ALL-related miRNAs and their aberrant expression in T-ALL
| T-ALLmiRNA | Location (Chr:start-end[strand]) | Host gene | Taxonomy conservation | Expression in T-ALL | |
|---|---|---|---|---|---|
| let-7e | chr19:52196039-52196117[+] | Intergenic | Chordata | ↓ | |
| miR-100 | chr11:122022937-122023016[−] | LOC399959 | Metazoa | ↓ | |
| miR-106a | chrX:133304228-133304308[−] | Intergenic | Mammalia | ↑ | |
| miR-125b | miR-125b-1 | chr11:121970465-121970552[−] | LOC399959 | Vertebrata | ↓ |
| miR-125b-2 | chr21:17962557-17962645[+] | C21orf34 | Vertebrata | ↓ | |
| miR-130b | chr22:22007593-22007674[+] | Intergenic | Vertebrata | ↑ | |
| miR-142 | chr17:56408593-56408679[−] | Intergenic | Vertebrata | ↑ | |
| miR-148a | chr7: 25989539- 25989606[−] | Intergenic | Vertebrata | ↑ | |
| miR-151 | chr8:141742663-141742752[−] | PTK2 | Mammalia | ↓ | |
| miR-17 | chr13:92002859-92002942[+] | Intergenic | Vertebrata | ↑ | |
| miR-181a | miR-181a-1 | chr1:198828173-198828282[−] | Intergenic | Vertebrata | ↓ |
| miR-181a-2 | chr9:127454721-127454830[+] | Intergenic | Vertebrata | ↓ | |
| miR-181b-1 | chr1:198828002-198828111[−] | Intergenic | Vertebrata | ↑ | |
| miR-181c | chr19:13985513-13985622[+] | Intergenic | Vertebrata | ↑ | |
| miR-18a | chr13:92003005-92003075[+] | Intergenic | Vertebrata | ↑ | |
| miR-18b | chrX:133304071-133304141[−] | Intergenic | Vertebrata | ↑ | |
| miR-193a | chr17:29887015-29887102[+] | Intergenic | Vertebrata | ↑ | |
| miR-196a | miR-196a-1 | chr17:46709852-46709921[−] | Intergenic | Vertebrata | ↓ |
| miR-196a-2 | chr12:54385522-54385631[+] | Intergenic | Vertebrata | ↓ | |
| miR-196b | chr7:27209099-27209182[−] | Intergenic | Vertebrata | ↓ | |
| miR-19a | chr13:92003145-92003226[+] | Intergenic | Vertebrata | ↑ | |
| miR-19b | miR-19b-1 | chr13:92003446-92003532[+] | Intergenic | Vertebrata | ↑ |
| miR-19b-2 | chrX:133303701-133303796[−] | Intergenic | Vertebrata | ↑ | |
| miR-204 | chr9:73424891-73425000[−] | TRPM3 | Vertebrata | ↑ | |
| miR-20a | chr13:92003319-92003389[+] | Intergenic | Vertebrata | ↑ | |
| miR-20b | chrX:133303839-133303907[−] | Intergenic | Vertebrata | ↑ | |
| miR-223 | chrX:65238712-65238821[+] | Intergenic | Vertebrata | ↑ | |
| miR-29a | chr7:130561506-130561569[−] | Intergenic | Vertebrata | ↓ | |
| miR-29b-1 | chr7:130562218-130562298[−] | Intergenic | Vertebrata | ↓ | |
| miR-30e | chr1:41220027-41220118[+] | NFYC | Vertebrata | ↑ | |
| miR-331 | chr12:95702196-95702289[+] | Intergenic | Mammalia | ↑ | |
| miR-34b | chr11:111383663-111383746[+] | LOC728196 | Chordata | ↑ | |
| miR-363 | chrX:133303408-133303482[−] | Intergenic | Vertebrata | ↑ | |
| miR-365 | miR-365-1 | chr16:14403142-14403228[+] | Intergenic | Vertebrata | ↑ |
| miR-365-2 | chr17:29902430-29902540[+] | Intergenic | Vertebrata | ↑ | |
| miR-424 | chrX: 133680644-133680741[−] | Intergenic | Mammalia | ↑ | |
| miR-582 | chr5:58999432-58999529[−] | PDE4D | Mammalia | ↑ | |
| miR-708 | chr11:79113066-79113153[−] | ODZ4 | Mammalia | ↑ | |
| miR-92a | miR-92a-1 | chr13:92003568-92003645[+] | Intergenic | Chordata | ↑ |
| miR-92a-2 | chrX:133303568-133303642[−] | Intergenic | Chordata | ↑ | |
| miR-93 | chr7:99691391-99691470[−] | MCM7 | Vertebrata | ↑ | |
| miR-99a | chr21:17911409-17911489[+] | C21orf34 | Vertebrata | ↓ | |
Taxonomy conservation: mammals: (human, mouse, rat, dog); vertebrates: (human, mouse, rat, dog, chicken, frog, fish); chordata: (human, mouse, chicken, fish, Ciona); metazoa: (human, mouse, chicken, fish, Ciona, fly).
After compiling the T-ALL gene and miRNA lists, we tried to predict the targets of T-ALLmiRNAs in T-ALLGenes. We mainly obtained the miRNA targets by combinatorial utilization of four different databases (see ‘Materials and Methods’ section), including predicted results from TargetScan (http://www.targetscan.org), miRanda (http://www.microrna.org) and experimental verified targets from miR2Disease (http://www.mir2disease.org./) and TarBase (http://diana.cslab.ece.ntua.gr/tarbase) (31–34). We also collected the published experimentally reported targets of the 42 collected T-ALL miRNAs by literature curation. Among the 67 T-ALLGenes, 47 are predicted targets of 39 T-ALLmiRNAs (Supplementary Figure S1). Seven of the 47 targeted T-ALLGenes (i.e. PTEN, CYLD, BCL2L11, MCL1, RUNX1, CDKN1A and JAK1) are targeted by more than 8 T-ALLmiRNAs. Among the T-ALLmiRNAs, miR-582 targets the most (21) genes in the T-ALLGene list, followed by miR-30e and miR-34b, each targets 13 T-ALLGenes. The two notorious tumor-related miRNA clusters (miR-17–92 and miR-106–363), which target 17 and 18 T-ALLGenes, respectively (Supplementary Table S1).
To obtain the TF regulatory relationship for our miRNA-TF co-regulatory network, we further predicted the TF targets in our compiled T-ALLGenes and T-ALLmiRNAs. Under a stringent criteria and conservation among the human, mouse and rat genomes for the predicted TFBS data in UCSC Genome Browser, we obtained 313 TFBSs on the promoter regions of 56 of the 67 T-ALLGenes and 242 TFBSs on the predicted promoters of 33 of the 42 T-ALLmiRNAs. These TFBSs established a complex relationship between T-ALL-related genes, TFs and miRNAs. Referring to MSigDB database (41), we extracted all the TFs corresponding to TFBSs. Thus, we obtained the miRNA targets and TF targets in T-ALLGenes.
FFLs in T-ALL
Since the targets of miRNAs and TFs were predicted in T-ALLGenes, we obtained 120 FFLs among T-ALLGenes, T-ALLmiRNAs and TFs by integrating the joint regulatory relationships of T-ALLmiRNAs and TFs to T-ALLGenes (Supplementary Table S2), which contain 24 T-ALLGenes, 26 T-ALLmiRNAs and 33 TFs. To evaluate the enrichment of these observed FFLs in the T-ALL network, we performed statistical tests with two approaches. First, we compared the FFLs obtained from T-ALLmiRNAs with those from thymus miRNAs or non-thymus miRNAs and evaluated the significance by Fisher’s exact test (Table 2). Both of the P-values of the two comparisons are highly significant (<0.01), which indicate that there were significantly more FFLs in T-ALLmiRNAs with T-ALLGenes than in thymus or non-thymus miRNAs with T-ALLGenes. We noticed that the P-value in the comparison between T-ALLmiRNAs and thymus miRNAs was larger than the comparison between T-ALLmiRNAs and non-thymus miRNAs, which likely represent that some thymus miRNAs in our data set may be highly associated with T-ALL but not yet been reported.
Table 2.
Statistics of FFLs identified by miRNAs in T-ALLmiRNAs and T-ALLGenes
| miRNA data set | No. of miRNAs | FFLs | P-value |
|---|---|---|---|
| T-ALL miRNA | 42 | 120 | |
| Thymus miRNA | 109 | 160 | 0.002 |
| Non-thymus miRNA | 113 | 33 | <2.2 E-16 |
Second, we ran 10 000 random simulations (see ‘Materials and Methods’ section) to test the significance of the observed FFLs in T-ALL. In each run, since there were 270 miRNA-target pairs between T-ALLmiRNAs and T-ALLGenes, we randomly selected 270 miRNA-target pairs out of all target pairs of the 42 T-ALLmiRNAs and calculated the number of FFLs among TFs, T-ALLmiRNAs and those randomly selected target genes. We found that the numbers of FFLs in all the simulations were less than our observed number of FFLs in T-ALLmiRNAs and T-ALLGenes, indicating that our observed FFLs significantly differ from that by chance.
MiRNA and TF co-regulatory network in T-ALL
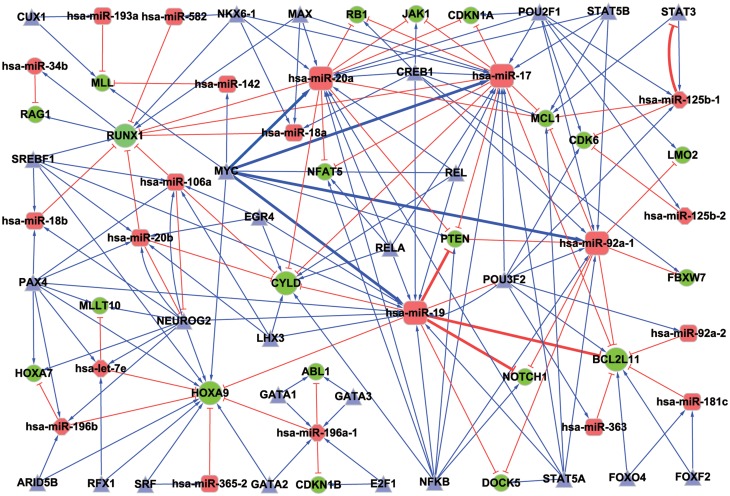
Besides the miRNA-TF co-regulate one target as in a FFL module, miRNA and TF may also regulate each other to form a feedback loop. In the compiled T-ALLmiRNAs, we identified three T-ALLmiRNA-TF mutual regulatory loops, which all components are included in our above FFLs (Supplementary Table S3). We merged the three T-ALLmiRNA-TF loops into FFLs and constructed a miRNA-TF co-regulatory network for T-ALL. To simplify the graph, we merged the miR-19a/b to miR-19 and NFKB1/2 to NFKB. After removing a few isolated nodes, the network contains 21 T-ALLmiRNAs, 21 T-ALLGenes, 28 TFs and 176 links (edges) between these molecules (nodes) (Figure 2). Among the 21 T-ALLmiRNAs in the network, most of (16/21) them are upregulated in T-ALL. Moreover, there were three pairs of regulatory relationships (miR-19 represses NOTCH1, PTEN and BCL2L11) that had been experimentally confirmed in T-ALL (16). MYC was verified as a potent and direct transcriptional activator of miR-17–92, which is consistent with the same regulatory relationship in our network (42).
Figure 2.
miRNA and TF co-regulatory network in T-ALL. Round rectangle nodes: upregulated T-ALLmiRNAs; hexagon nodes: downregulated T-ALLmiRNAs; ellipse nodes: T-ALL candidate genes; triangle nodes: TFs. Bigger nodes represent hubs in network. T shape edge: miRNA repression; arrow shape edge: transcriptional activation/repression. Thick lines denote regulatory relationships that are supported by experimental reports.
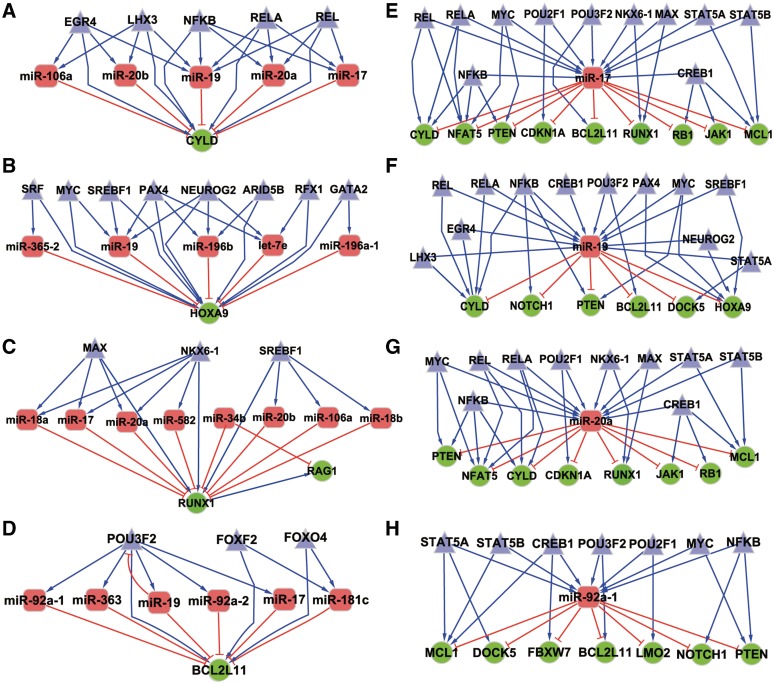
Subnetworks for hubs in the miRNA-TF regulatory network
According to the criteria described in the ‘Materials and Methods’ section, we selected four hub genes (HOXA9, CYLD, BCL2L11 and RUNX1) and four hub miRNAs (miR-19, miR-20a, miR-92a-1 and miR-17) in Figure 2 T-ALL miRNA-TF regulatory network. All the four hub genes are related to apoptosis, inflammation, proliferation and differentiation of T cell. It is interesting that the four hub miRNAs are all from the notorious cancer-related miRNA cluster miR-17–92, and they are all upregulated in T-ALL (Supplementary Figure S2A). To further investigate the regulation of these hub genes and miRNAs, we extracted their subnetworks that include all their directly linked molecules in the miRNA-TF regulatory network (Figure 3). CYLD stood out as a promising tumor suppress gene that involved in NF-κB pathway, which is regulated by five TFs and five T-ALLmiRNAs, all from the two cancer-related miRNA clusters (Supplementary Figure S2B). MiR-19 is potentially regulated by 12 TFs (e.g. NFKB, MYC, REL, CREB1 and STAT5A) and represses the expression of six important cancer genes, such as NOTCH1, PTEN, BCL2L11, HOXA9 and CYLD. We extracted the predicted targets of these four miRNAs on human genome and found 794, 856, 854 and 556 target genes for them, respectively. The enriched pathways of these predicted targets of the four hub miRNAs were examined by the DAVID Functional Annotation Tool (see ‘Materials And Methods’ section). Interestingly, we found many pathways related to proliferation, differentiation, metabolism, cancer and intracellular signaling cascade (Supplementary Table S4), which suggested that the four hub miRNAs were important in cancer development including T-ALL. We also examined the enriched GO terms of these predicted targets. Among the enriched GO terms, several terms were related to the occurrence and development of T-ALL, such as regulation of cellular protein metabolic process, intracellular signaling cascade, cell apoptosis and proliferation (Supplementary Table S5).
Figure 3.
Subnetworks of the eight hubs in our miRNA-TF co-regulatory network. The subnetworks were drawn by all the direct linked nodes for the hubs. (A) CYLD; (B) HOXA9; (C) RUNX1; (D) BCL2L11; (E) miR-17; (F) miR-19; (G) miR-20a; (H) miR-92a-1.
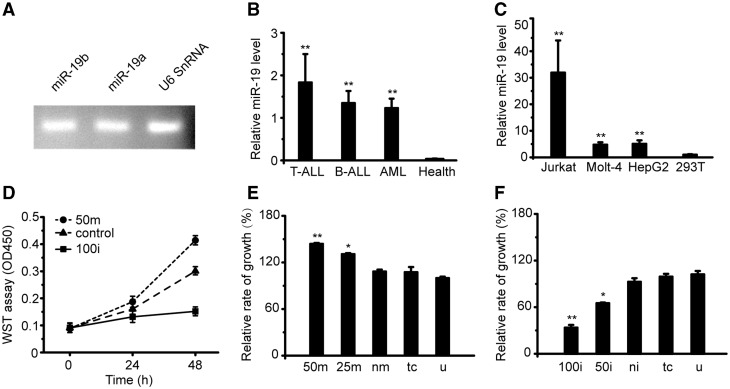
High expression of miR-19 in T-ALL patients and T-leukemia cell lines
As predicted several hub miRNAs, genes and modules among them in the T-ALL miRNA-TF co-regulatory network, we further validated the different expression and regulation for a specific hub miRNA and gene in T-ALL to partially verify our network by experiments. The cancer-related miRNA cluster miR-17–92 contains six miRNAs: miR-17-5p, miR-18a, miR-19a, miR-19b, miR-20a and miR-92a. Among them, miR-19 is one of the most important and well-studied miRNAs in different cancers including T-ALL. MiR-19a and miR-19b have the same seed sequence, and there is only one nucleotide difference in their mature sequences. We detected the expression of miR-19a and miR-19b in clinical samples and found very similar expression level of them (Figure 4A). Thus, miR-19 was applied to represent both miR-19a and miR-19b in the following context.
Figure 4.
MiR-19 is high expressed in leukemia patients and tumor cell lines. (A) The similar expression of miR-19a and miR-19b in peripheral blood mononuclear cells (PBMCs) of ALL patients. U6 snRNA was used as an internal control. (B) Expression of miR-19 in T-ALL/B-ALL/AML patients and health donors. (C) Expression of miR-19 in human tumor cell lines: HepG2 (Hepatocarcinoma), Jurkat (T cell acute leukemia), Molt-4 (Human T cell leukemia) and normal cell line HEK-293T (Human Embryo Kidney cell). (D) Changing of the relative absorbance by WST assay in the Jurkat cells treated with miR-19mimics/inhibitors for 0/24/48 h. (E) Relative cell survival level of Jurkat cells after transfected with miR-19 mimics for 48 h in different concentration. (F) Relative growth rate of Jurkat cells after transfected with miR-19 inhibitors for 48 h in different concentration. Data in histograms are represented as mean ± SD. T test, *P < 0.05. **P < 0.01 compared with the health (B), 293 T (C), untreated group (E and F). 50 m: 50 nM miR-19 mimics; 25 m: 25 nM miR-19 mimics; nm: negative mimics; tc: transfect reagent control; u: untreated group; 100i: 100 nM miR-19 inhibitors; 50i: 50 nM miR-19 inhibitors; ni: negative inhibitors.
To test the expression level of miR-19 in leukemic patients, we chose 23 diagnosed patients, which contain 11 T-ALL, 3 B-ALL and 9 acute myeloid leukemia (AML) cases. The results showed that the expressions of miR-19 in T-ALL/B-ALL/AML patients were all significantly higher than health donors, and T-ALL patients had the highest expression (Figure 4B). Furthermore, we examined the miR-19 expression in several cell lines. Compared to the low-miR-19 expression in non-tumor HEK-293T cells, miR-19 was found highly expressed in tumor cell lines, such as Jurkat (T-cell acute leukemia), Molt-4 (Human T cell leukemia) and HepG2 (Hepatocarcinoma; Figure 4C). The highest expression of miR-19 was found in Jurkat cell line, which was consistent with the investigation in patient samples.
To test the oncogenic role of miR-19, the cell survival level was detected in Jurkat cells. The cells were incubated with miR-19 mimics and inhibitors for 0, 24 and 48 h (Figure 4D). Our results showed that transfection of miR-19 mimics increased the survival level of Jurkat cells, while the inhibitors of miR-19 transfection decreased the survival level of the Jurkat cells. The effect was more significantly stronger after transfection 48 h than 24 h. In addition, we used different concentrations of miR-19 mimics/inhibitors and found that the relative cell survival level of Jurkat cells was increased when the concentration of the miR-19 mimics was enhanced (Figure 4E). The opposite phenomenon was found when dosages of the miR-19 inhibitors were increased (Figure 4F).
MiR-19 inhibits the expression of CYLD and induces downstream NF-κB
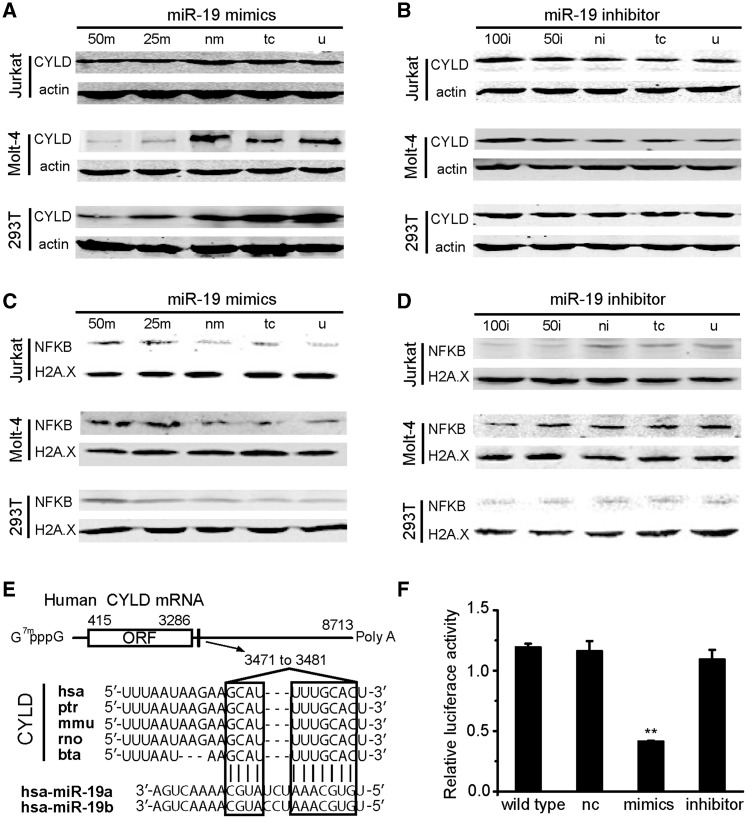
To find out the oncogenic role of miR-19, we investigated the expression level of CYLD protein through transfection with miR-19 mimics/inhibitors in Jurkat, Molt-4 and HEK293T cells. In all these three kinds of cells, overexpression of the miR-19 decreased the expression of CYLD protein when transfection with miR-19 mimics in a dosage-dependent manner, and the expression level of CYLD protein was significantly lower transfected with 50 nM than 25 nM (Figure 5A).
Figure 5.
MiR-19 inhibits CYLD and affects downstream NF-κB. (A) MiR-19 mimics decreased the expression level of CYLD in Jurkat, Molt-4 and HEK293T cells by western blot. (B) Expression level of CYLD was enhanced by miR-19 inhibitors in Jurkat, Molt-4, but no obvious effect in HEK-293T cells. (C) The miR-19 mimics increased the downstream NF-κB P65 protein level in cell line nucleus. (D) NF-κB P65 expression level decreased by miR-19 inhibitors in the tumor cell lines. (E) Sequence alignment of the miR-19a/b base-paring site in the 3' UTR of CYLD mRNAs. The region complementary to the miR-19 is highly conserved among five species (hsa: human, ptr: chimpanzee, mmu: mouse, rno: rat, and bta: cow. The predicted binding site of miR-19 with CYLD target sequences are shown in boxes. (F) The miR-19 targeted the 3′-UTR of CYLD mRNA by luciferase reporter assay in HEK293T cells by transfected with miR-19 mimics/inhibitors/negative and reporter vector. Data are represented as mean ± SD. nc: negative control. T test, **P < 0.01 compared with the negative control. The means of 50 m, 25 m, nm, tc, u, 100i, 50i and ni are the same as described in Figure 4.
The expression level of CYLD protein was increased in Jurkat and Molt-4 cells while blocking of miR-19 compared with the negative control, X-tremeGENE siRNA Transfection Reagent and untreated group, but no obvious change in HEK-293 T cells (Figure 5B). The expression level of CYLD protein was significantly higher when transfected with 100 nM than 50 nM’s miR-19 inhibitors. We have repeated these experiments three times, and all the quantified western blot data of CYLD were presented in Supplementary Figure S3 (A, B, E, F, I and J).
Since CYLD plays a predominant role in the negative regulation of NF-κB (43), we further examined the impact of CYLD on NF-κB activation by detecting the level of nucleus NF-κB P65 protein in the three cell lines transfected with miR-19 mimics/inhibitors. After transfected with miR-19 mimics, the results indicated that the level of nucleus NF-κB P65 protein was clearly increased compared with negative control, and the effect of 50 nM mimics was stronger than that of 25 nM (Figure 5C). While the expression level of P65 was clearly decreased when transfected with miR-19 inhibitors compared with the control (Figure 5D). These experiments were also repeated at least three times, and all the quantified western blot data of NF-κB were presented in Supplementary Figure S3 (C, D, G, H, J and K). Since the expression of cytokine IL-6 mRNA relied on NF-κB (44); here, we also found that the expression of IL-6 was decreased after transfecting miR-19 inhibitor (Supplementary Figure S4).
MiR-19 inhibits CYLD expression by binding to the 3′UTR conservative sites of its mRNA (Figure 5E), to further confirm that CYLD is directly regulated by miR-19, we performed miRNA luciferase assay in four groups (Figure 5F). The first group only contains CYLD 3′UTR firefly luciferase vector, and no obvious fluorescence changing was found. The second group transfected with negative miRNA also showed no significant change in fluorescence quenching. The third group transfected with miR-19 mimics, its fluorescence intensity extremely significantly decreased compared with the first two groups. The fourth group transfected with miR-19 inhibitors, we could not find significant fluorescence intensity changes.
DISCUSSION
In this study, we constructed a miRNA and TF co-regulatory network specifically for T-ALL, which provided important insights into the etiology of T-ALL. Our analysis indicated that this regulatory network is significantly enriched in T-ALL, and some published results also confirmed the reliability of the network. Furthermore, we discovered miR-19 inhibits CYLD in T-ALL for the first time. We tentatively proposed that miR-19 and CYLD play critical regulatory roles in the NF-κB pathway, while in turn NF-κB may bind to their upstream promoter regions and regulate their transcriptions. Our results identified gene regulatory network modules and extended the understanding of gene regulation mechanism in T-ALL.
In the miRNA-TF co-regulatory network, we identified four hub genes (CYLD, HOXA9, BCL2L11 and RUNX1) and four hub miRNAs (miR-19, miR-17, miR-92a-1 and miR-20a) in the miR-17–92 cluster, all of these hubs likely play important regulatory roles in T-ALL. The hub genes are fragile points in the regulatory network with many nodes linked to them and mutation of them may deadly disrupt the normal function. Meanwhile, we also confirmed some well-studied T-ALL candidate genes (e.g. NOTCH1, PTEN, FBXW7 and LMO2) and nuclear TFs (e.g. CREB1, MYC, STAT and NF-κB) in the miRNA-TF network. Many of them involved in important pathways, such as PI3K/AKT pathway, NOTCH pathway, NF-κB pathway and JAK pathway (45–49). Some targets of the miRNAs in our network participated in multiple signal pathways, which may establish cross-talks among these pathways. Therefore, our network offered more details of why some genes mutated lead to T-ALL and provided potential targets for treatment.
As detected by previous studies and our results, members of the miR-17–92 cluster were found aberrantly upregulated in T-ALL patients and most of hematological malignant cell lines (50–52). In our network, miR-19 is a hub miRNA regulate 6 T-ALLGenes and targeted by 12 TFs. The predicted targets of miR-19 were enriched in many important signaling pathways, such as MAPK signaling pathway, Wnt signaling pathway and TGF-β signaling pathway (Supplementary Table S4). Several groups have identified that miR-19 was significantly upregulated in Notch-induced T-ALL, and its target genes like PTEN, BCL2L11 (Bim) and NOTCH1 have been identified in mouse models (16). It had been reported that deletion of the complete miR-17–92 cluster repressed the MYC-induced oncogenesis (53,54). This effect was rescued by reintroduction of the full cluster, but not by the cluster lacking miR-19. Recovery of miR-19 largely rescued the tumorigenicity, which identified miR-19 as the most important oncogenic miRNA in the miR-17–92 cluster (55).
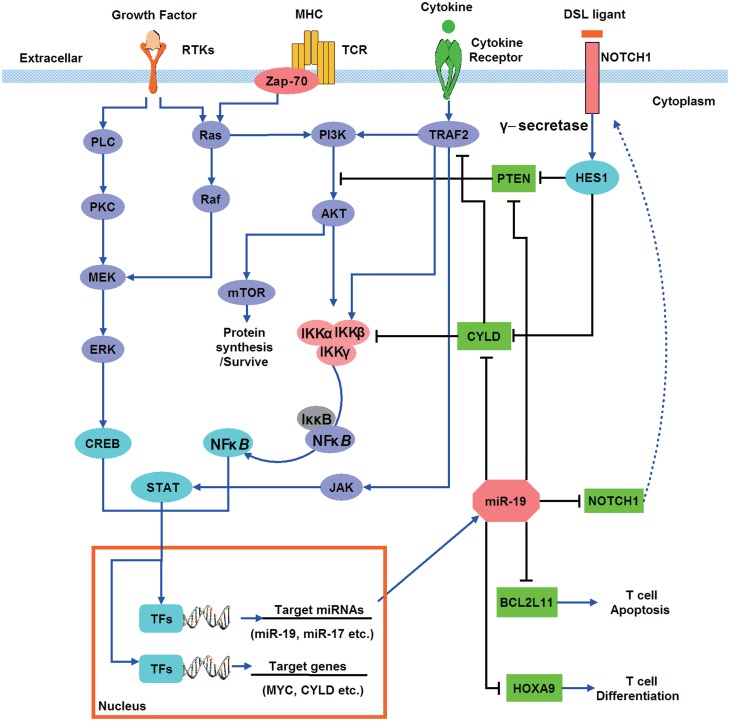
To explore the regulatory mechanism of T-ALL, we proposed a model to demonstrate the regulation of miR-19 in T-ALL-signaling pathways by integrating the results of our miRNA-TF regulatory network with the canonical T-cell development and T-ALL-signaling pathways (Figure 6). In T-ALL patients and hematological malignant cell lines, miR-19 was significantly upregulated, and its target genes such as PTEN, HOXA, BCL2L11, CYLD and NOTCH1 were repressed. PTEN is a tumor suppress gene and its down-regulation will activate the PI3K/AKT signaling pathway in cancers (56,57). In T-ALL patients, post-translational inactivation of PTEN would cause incontrollable proliferation of T cell (58,59). HOXA9 is one of the hub genes in our regulatory network, and it was reported as a leukemogenic homeoprotein in T-ALL (60). BCL2L11 is another hub gene, transgenic studies in mouse suggested that BCL2L11 acted as an essential initiator of apoptosis in thymocyte-negative selection (61). Expression of BCL2L11 can be induced by nerve growth factor, and its dysregulation might lead to leukemia (62,63). NOTCH1 plays a vital role in T-cell development and transformation, and ∼50% T-ALL patients show abnormal expression of NOTCH1 (64). Downstream transcriptional targets of Notch1 include HES1 and MYC, both of which can affect the PI3K/AKT and NF-κB-signaling pathways. CYLD is a promising hub gene in our network, which acts as a negative regulator of NF-κB and JNK signaling (65). In summary, all the evidences suggested that up-regulating of miR-19 will cause the disorder of multiple T-ALL genes and implies strongly carcinogenic potential.
Figure 6.
A model of miR-19 and its targets involving in signaling pathways and regulatory network in T-ALL. Cell surface receptors activate multiple signaling pathways, including Ras/MAPK cascade, PI3K/AKT, JAK and NOTCH signaling. These signal transductions trigger several nucleus TFs such as STAT, NF-κB and CREB to regulate the transcription of miRNAs (e.g. miR-19) and T-ALL genes. Members of miR-17–92 cluster especially miR-19 inhibit tumor suppressor genes PTEN and CYLD. CYLD was a suppressor of NF-κB.
Furthermore, the experimental results confirmed the reliability of our regulatory network. CYLD was found with low expression in T-ALL cell lines and with obvious difference comparing to healthy subjects. Down-regulation of miR-19 can significantly restore the expression of CYLD, and enhanced miR-19 inhibitor concentration would increase CYLD expression (Figure 5A and B). High expression level of CYLD reduced the nuclear translocation of NF-κB significantly and the IL-6 expression, which was dependent on the activation of NF-κB. But the decrease of IL-6 was less than NF-κB (P65), which may ascribe to the balance of a relationship between the classical and non-classical NF-κB pathway (66). Both the TFBS prediction and UCSC ENCODE ChIP-Seq data showed that there were potential NF-κB-binding sites existing on promoters of CYLD and miR-19. Therefore, CYLD, miR-19 and NF-κB participated in a FFL, which provides a new mechanism of the sustained activation of NF-κB in T-ALL and hematologic malignancies.
Inspiringly, our regulatory network provided a powerful tool to investigate mechanisms of T-ALL, through which we may find some potential therapeutic targets. The first potential therapeutic strategy is the inhibition of miR-17–92 cluster, which might result in elevated expression of BCL2L11, PTEN and CYLD, and leukemia cells proliferation would be suppressed and trend to apoptosis. It has been reported that glucocorticoids repress the expression of the microRNA cluster miR-17–92 (67), which may be adopted as a safe mean for inhibition of miR-17–92. The second therapeutic strategy is the inhibition of NOTCH1-signaling pathway. Multiple pathways were regulated by NOTCH1/HES1 in the process of leukemogenesis, γ-secretase inhibitors would repress the NOTCH1 maturation and induce cell-cycle arrest in T-ALL cell lines (64). The third therapeutic approach is the inhibition of oncogenic PI3K/AKT/mTOR pathway. The PI3K/AKT pathway was found high-frequency abnormalities in T-ALL (59), and mTOR inhibitor rapamycin showed promising effects in preclinical models and Jurkat cell line (68,69).
In summary, our miRNA-TF co-regulatory network analysis provided some core regulators and clues for the regulatory mechanisms of T-ALL. Furthermore, we experimentally validated one of the hub miRNAs and hub genes in the network. We confirmed that miR-19 regulated the CYLD expression and the existence of miR-19-CYLD-NF-κB FFL in T-ALL. Consequently, our study may provide potential therapeutic targets in T-ALL and present a new strategy combined bioinformatics analysis with experimental validation to study complex diseases.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–5 and Supplementary Figures 1–4.
FUNDING
National Natural Science Foundation of China (31171271) to A.Y.G., Young Teachers’ Fund for Doctor Stations, Ministry of Education of China (20110142120042), Scientific Research Fund for the Returned Overseas Chinese Scholars, Ministry of Education of China and the State Key Laboratory of Freshwater Ecology and Biotechnology (2012FB02). Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jun Yan, Wei Liu, Zhaowu Ma, Hui Liu and Hong-Mei Zhang for helpful discussion.
REFERENCES
- 1.Pui CH, Relling MV, Downing JR. Acute lymphoblastic leukemia. N. Engl. J. Med. 2004;350:1535–1548. doi: 10.1056/NEJMra023001. [DOI] [PubMed] [Google Scholar]
- 2.Hagemeijer A, Graux C. ABL1 rearrangements in T-cell acute lymphoblastic leukemia. Genes Chromosomes Cancer. 2010;49:299–308. doi: 10.1002/gcc.20743. [DOI] [PubMed] [Google Scholar]
- 3.Goldberg JM, Silverman LB, Levy DE, Dalton VK, Gelber RD, Lehmann L, Cohen HJ, Sallan SE, Asselin BL. Childhood T-cell acute lymphoblastic leukemia: the Dana-Farber Cancer Institute acute lymphoblastic leukemia consortium experience. J. Clin. Oncol. 2003;21:3616–3622. doi: 10.1200/JCO.2003.10.116. [DOI] [PubMed] [Google Scholar]
- 4.Coustan-Smith E, Mullighan CG, Onciu M, Behm FG, Raimondi SC, Pei D, Cheng C, Su X, Rubnitz JE, Basso G, et al. Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol. 2009;10:147–156. doi: 10.1016/S1470-2045(08)70314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Staal FJ, van Dongen JJ, Langerak AW. Novel insights into the development of T-cell acute lymphoblastic leukemia. Curr. Hematol. Malig. Rep. 2007;2:176–182. doi: 10.1007/s11899-007-0024-0. [DOI] [PubMed] [Google Scholar]
- 6.Teitell MA, Pandolfi PP. Molecular genetics of acute lymphoblastic leukemia. Annu. Rev. Pathol. 2009;4:175–198. doi: 10.1146/annurev.pathol.4.110807.092227. [DOI] [PubMed] [Google Scholar]
- 7.Van Vlierberghe P, Pieters R, Beverloo HB, Meijerink JP. Molecular-genetic insights in paediatric T-cell acute lymphoblastic leukaemia. Br. J. Haematol. 2008;143:153–168. doi: 10.1111/j.1365-2141.2008.07314.x. [DOI] [PubMed] [Google Scholar]
- 8.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 9.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 10.Schotte D, Chau JC, Sylvester G, Liu G, Chen C, van der Velden VH, Broekhuis MJ, Peters TC, Pieters R, den Boer ML. Identification of new microRNA genes and aberrant microRNA profiles in childhood acute lymphoblastic leukemia. Leukemia. 2009;23:313–322. doi: 10.1038/leu.2008.286. [DOI] [PubMed] [Google Scholar]
- 11.Lu M, Zhang Q, Deng M, Miao J, Guo Y, Gao W, Cui Q. An analysis of human microRNA and disease associations. PLoS One. 2008;3:e3420. doi: 10.1371/journal.pone.0003420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bousquet M, Harris MH, Zhou B, Lodish HF. MicroRNA miR-125b causes leukemia. Proc. Natl. Acad. Sci. USA. 2010;107:21558–21563. doi: 10.1073/pnas.1016611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schotte D, Lange-Turenhout EA, Stumpel DJ, Stam RW, Buijs-Gladdines JG, Meijerink JP, Pieters R, Den Boer ML. Expression of miR-196b is not exclusively MLL-driven but is especially linked to activation of HOXA genes in pediatric acute lymphoblastic leukemia. Haematologica. 2010;95:1675–1682. doi: 10.3324/haematol.2010.023481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fulci V, Colombo T, Chiaretti S, Messina M, Citarella F, Tavolaro S, Guarini A, Foá R, Macino G. Characterization of B- and T-lineage acute lymphoblastic leukemia by integrated analysis of MicroRNA and mRNA expression profiles. Genes Chromosomes Cancer. 2009;48:1069–1082. doi: 10.1002/gcc.20709. [DOI] [PubMed] [Google Scholar]
- 15.Zanette DL, Rivadavia F, Molfetta GA, Barbuzano FG, Proto-Siqueira R, Silva WA, Jr, Falcão RP, Zago MA. miRNA expression profiles in chronic lymphocytic and acute lymphocytic leukemia. Braz. J. Med. Biol. Res. 2007;40:1435–1440. doi: 10.1590/s0100-879x2007001100003. [DOI] [PubMed] [Google Scholar]
- 16.Mavrakis KJ, Wolfe AL, Oricchio E, Palomero T, de Keersmaecker K, McJunkin K, Zuber J, James T, Khan AA, Leslie CS, et al. Genome-wide RNA-mediated interference screen identifies miR-19 targets in Notch-induced T-cell acute lymphoblastic leukaemia. Nat. Cell Biol. 2010;12:372–379. doi: 10.1038/ncb2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagel S, Venturini L, Przybylski GK, Grabarczyk P, Schmidt CA, Meyer C, Drexler HG, Macleod RA, Scherr M. Activation of miR-17-92 by NK-like homeodomain proteins suppresses apoptosis via reduction of E2F1 in T-cell acute lymphoblastic leukemia. Leuk Lymph. 2009;50:101–108. doi: 10.1080/10428190802626632. [DOI] [PubMed] [Google Scholar]
- 18.Li X, Sanda T, Look AT, Novina CD, von Boehmer H. Repression of tumor suppressor miR-451 is essential for NOTCH1-induced oncogenesis in T-ALL. J. Exp. Med. 2011;208:663–675. doi: 10.1084/jem.20102384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol. Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shalgi R, Lieber D, Oren M, Pilpel Y. Global and local architecture of the mammalian microRNA-transcription factor regulatory network. PLoS Comput. Biol. 2007;3:e131. doi: 10.1371/journal.pcbi.0030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Re A, Corá D, Taverna D, Caselle M. Genome-wide survey of microRNA-transcription factor feed-forward regulatory circuits in human. Mol. Biosyst. 2009;5:854–867. doi: 10.1039/b900177h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim J, Inoue K, Ishii J, Vanti WB, Voronov SV, Murchison E, Hannon G, Abeliovich A. A MicroRNA feedback circuit in midbrain dopamine neurons. Science. 2007;317:1220–1224. doi: 10.1126/science.1140481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu Z, Wang C, Wang M, Li Z, Casimiro MC, Liu M, Wu K, Whittle J, Ju X, Hyslop T, et al. A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation. J. Cell. Biol. 2008;182:509–517. doi: 10.1083/jcb.200801079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brosh R, Shalgi R, Liran A, Landan G, Korotayev K, Nguyen GH, Enerly E, Johnsen H, Buganim Y, Solomon H, et al. p53-Repressed miRNAs are involved with E2F in a feed-forward loop promoting proliferation. Mol. Syst. Biol. 2008;4:229. doi: 10.1038/msb.2008.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friard O, Re A, Taverna D, De Bortoli M, Cora D. CircuitsDB: a database of mixed microRNA/transcription factor feed-forward regulatory circuits in human and mouse. BMC Bioinform. 2010;11:435. doi: 10.1186/1471-2105-11-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Lu M, Qiu C, Cui Q. TransmiR: a transcription factor-microRNA regulation database. Nucleic Acids Res. 2010;38:D119–D122. doi: 10.1093/nar/gkp803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo AY, Sun J, Jia P, Zhao Z. A novel microRNA and transcription factor mediated regulatory network in schizophrenia. BMC Syst. Biol. 2010;4:10. doi: 10.1186/1752-0509-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genom. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sethupathy P, Corda B, Hatzigeorgiou AG. TarBase: a comprehensive database of experimentally supported animal microRNA targets. RNA. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang Q, Wang Y, Hao Y, Juan L, Teng M, Zhang X, Li M, Wang G, Liu Y. miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acids Res. 2009;37:D98–D104. doi: 10.1093/nar/gkn714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 35.Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2010;39:D876–882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 39.Zhao H, Wang D, Du W, Gu D, Yang R. MicroRNA and leukemia: tiny molecule, great function. Crit. Rev. Oncol. Hematol. 2010;74:149–155. doi: 10.1016/j.critrevonc.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 40.Landais S, Landry S, Legault P, Rassart E. Oncogenic potential of the miR-106-363 cluster and its implication in human T-cell leukemia. Cancer Res. 2007;67:5699–5707. doi: 10.1158/0008-5472.CAN-06-4478. [DOI] [PubMed] [Google Scholar]
- 41.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 43.Sun SC. CYLD: a tumor suppressor deubiquitinase regulating NF-kappaB activation and diverse biological processes. Cell Death Differ. 2010;17:25–34. doi: 10.1038/cdd.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lappas M, Permezel M, Georgiou HM, Rice GE. Nuclear factor kappa B regulation of proinflammatory cytokines in human gestational tissues in vitro. Biol. Reprod. 2002;67:668–673. doi: 10.1095/biolreprod67.2.668. [DOI] [PubMed] [Google Scholar]
- 45.Vilimas T, Mascarenhas J, Palomero T, Mandal M, Buonamici S, Meng F, Thompson B, Spaulding C, Macaroun S, Alegre ML, et al. Targeting the NF-kappaB signaling pathway in Notch1-induced T-cell leukemia. Nat. Med. 2007;13:70–77. doi: 10.1038/nm1524. [DOI] [PubMed] [Google Scholar]
- 46.Palomero T, Dominguez M, Ferrando AA. The role of the PTEN/AKT Pathway in NOTCH1-induced leukemia. Cell Cycle. 2008;7:965–970. doi: 10.4161/cc.7.8.5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharma VM, Draheim KM, Kelliher MA. The Notch1/c-Myc pathway in T cell leukemia. Cell Cycle. 2007;6:927–930. doi: 10.4161/cc.6.8.4134. [DOI] [PubMed] [Google Scholar]
- 48.Medyouf H, Ghysdael J. The calcineurin/NFAT signaling pathway: a novel therapeutic target in leukemia and solid tumors. Cell Cycle. 2008;7:297–303. doi: 10.4161/cc.7.3.5357. [DOI] [PubMed] [Google Scholar]
- 49.Leung KT, Li KK, Sun SS, Chan PK, Ooi VE, Chiu LC. Activation of the JNK pathway promotes phosphorylation and degradation of BimEL—a novel mechanism of chemoresistance in T-cell acute lymphoblastic leukemia. Carcinogenesis. 2008;29:544–551. doi: 10.1093/carcin/bgm294. [DOI] [PubMed] [Google Scholar]
- 50.Wong P, Iwasaki M, Somervaille TC, Ficara F, Carico C, Arnold C, Chen CZ, Cleary ML. The miR-17-92 microRNA polycistron regulates MLL leukemia stem cell potential by modulating p21 expression. Cancer Res. 2010;70:3833–3842. doi: 10.1158/0008-5472.CAN-09-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mi S, Li Z, Chen P, He C, Cao D, Elkahloun A, Lu J, Pelloso LA, Wunderlich M, Huang H, et al. Aberrant overexpression and function of the miR-17-92 cluster in MLL-rearranged acute leukemia. Proc. Natl. Acad. Sci. USA. 2009;107:3710–3715. doi: 10.1073/pnas.0914900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sasaki K, Kohanbash G, Hoji A, Ueda R, McDonald HA, Reinhart TA, Martinson J, Lotze MT, Marincola FM, Wang E, et al. miR-17-92 expression in differentiated T cells—implications for cancer immunotherapy. J. Transl. Med. 2010;8:17. doi: 10.1186/1479-5876-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mu P, Han YC, Betel D, Yao E, Squatrito M, Ogrodowski P, de Stanchina E, D’Andrea A, Sander C, Ventura A. Genetic dissection of the miR-17∼92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23:2806–2811. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, Cordon-Cardo C, Li QJ, Lowe SW, Hannon GJ, He L. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009;23:2839–2849. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chalhoub N, Baker SJ. PTEN and the PI3-kinase pathway in cancer. Annu. Rev. Pathol. 2009;4:127–150. doi: 10.1146/annurev.pathol.4.110807.092311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Renner O, Blanco-Aparicio C, Carnero A. Genetic modelling of the PTEN/AKT pathway in cancer research. Clin. Transl. Oncol. 2008;10:618–627. doi: 10.1007/s12094-008-0262-1. [DOI] [PubMed] [Google Scholar]
- 58.Silva A, Yunes JA, Cardoso BA, Martins LR, Jotta PY, Abecasis M, Nowill AE, Leslie NR, Cardoso AA, Barata JT. PTEN posttranslational inactivation and hyperactivation of the PI3K/Akt pathway sustain primary T cell leukemia viability. J. Clin. Invest. 2008;118:3762–3774. doi: 10.1172/JCI34616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gutierrez A, Sanda T, Grebliunaite R, Carracedo A, Salmena L, Ahn Y, Dahlberg S, Neuberg D, Moreau LA, Winter SS, et al. High frequency of PTEN, PI3K, and AKT abnormalities in T-cell acute lymphoblastic leukemia. Blood. 2009;114:647–650. doi: 10.1182/blood-2009-02-206722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dorsam ST, Ferrell CM, Dorsam GP, Derynck MK, Vijapurkar U, Khodabakhsh D, Pau B, Bernstein H, Haqq CM, Largman C, et al. The transcriptome of the leukemogenic homeoprotein HOXA9 in human hematopoietic cells. Blood. 2004;103:1676–1684. doi: 10.1182/blood-2003-07-2202. [DOI] [PubMed] [Google Scholar]
- 61.Hughes P, Bouillet P, Strasser A. Role of Bim and other Bcl-2 family members in autoimmune and degenerative diseases. Curr. Dir. Autoimmun. 2006;9:74–94. doi: 10.1159/000090773. [DOI] [PubMed] [Google Scholar]
- 62.Li WQ, Guszczynski T, Hixon JA, Durum SK. Interleukin-7 regulates Bim proapoptotic activity in peripheral T-cell survival. Mol. Cell. Biol. 2010;30:590–600. doi: 10.1128/MCB.01006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jiang N, Koh GS, Lim JY, Kham SK, Ariffin H, Chew FT, Yeoh AE. BIM is a prognostic biomarker for early prednisolone response in pediatric acute lymphoblastic leukemia. Exp. Hematol. 2011;39:321–329. doi: 10.1016/j.exphem.2010.11.009. 329 e1–e3. [DOI] [PubMed] [Google Scholar]
- 64.Paganin M, Ferrando A. Molecular pathogenesis and targeted therapies for NOTCH1-induced T-cell acute lymphoblastic leukemia. Blood Rev. 2011;25:83–90. doi: 10.1016/j.blre.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Courtois G. Tumor suppressor CYLD: negative regulation of NF-kappaB signaling and more. Cell Mol. Life Sci. 2008;65:1123–1132. doi: 10.1007/s00018-007-7465-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun SC. Non-canonical NF-kappaB signaling pathway. Cell Res. 2011;21:71–85. doi: 10.1038/cr.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Molitoris JK, McColl KS, Distelhorst CW. Glucocorticoid-mediated repression of the oncogenic microRNA cluster miR-17∼92 contributes to the induction of Bim and initiation of apoptosis. Mol. Endocrinol. 2011;25:409–420. doi: 10.1210/me.2010-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao YM, Zhou Q, Xu Y, Lai XY, Huang H. Antiproliferative effect of rapamycin on human T-cell leukemia cell line Jurkat by cell cycle arrest and telomerase inhibition. Acta Pharmacol. Sin. 2008;29:481–488. doi: 10.1111/j.1745-7254.2008.00767.x. [DOI] [PubMed] [Google Scholar]
- 69.Avellino R, Romano S, Parasole R, Bisogni R, Lamberti A, Poggi V, Venuta S, Romano MF. Rapamycin stimulates apoptosis of childhood acute lymphoblastic leukemia cells. Blood. 2005;106:1400–1406. doi: 10.1182/blood-2005-03-0929. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.