Abstract
DNA mimic proteins occupy the DNA binding sites of DNA-binding proteins, and prevent these sites from being accessed by DNA. We show here that the Neisseria conserved hypothetical protein DMP19 acts as a DNA mimic. The crystal structure of DMP19 shows a dsDNA-like negative charge distribution on the surface, suggesting that this protein should be added to the short list of known DNA mimic proteins. The crystal structure of another related protein, NHTF (Neisseria hypothetical transcription factor), provides evidence that it is a member of the xenobiotic-response element (XRE) family of transcriptional factors. NHTF binds to a palindromic DNA sequence containing a 5′-TGTNAN11TNACA-3′ recognition box that controls the expression of an NHTF-related operon in which the conserved nitrogen-response protein [i.e. (Protein-PII) uridylyltransferase] is encoded. The complementary surface charges between DMP19 and NHTF suggest specific charge–charge interaction. In a DNA-binding assay, we found that DMP19 can prevent NHTF from binding to its DNA-binding sites. Finally, we used an in situ gene regulation assay to provide evidence that NHTF is a repressor of its down-stream genes and that DMP19 can neutralize this effect. We therefore conclude that the interaction of DMP19 and NHTF provides a novel gene regulation mechanism in Neisseria spps.
INTRODUCTION
The Neisseria spp. are Gram-negative β-protobacteria that colonize the mucosal surfaces of many animals. Of the 11 species that colonize humans, only Neisseria meningitidis and Neisseria gonorrhoeae are pathogens. N. meningitidis is an important cause of septicaemia and meningitis in childhood and N. gonorrhoeae is responsible for the sexually transmitted infection gonorrhea (1–3). The determination of the complete genome sequences of these two Neisseria spp. provided good starting points for the study of the pathogenesis of meningitis and gonorrhea (4,5).
To date, there are still around 900 uncharacterized genes in N. meningitidis (6). Among these genes, N. meningitidis NMB0541 is a conserved Neisseria hypothetical protein that has no functional annotation. Microarray analysis has shown that this gene is conserved in the major hypervirulent lineages and serogroups of N. meningitidis that cause human disease (6,7). In the present article, we determined the crystal structure of this 19 kDa protein and used proteomic approaches to investigate its function and the factors with which it interacts. We found that NMB0541 acts as a DNA mimic, and thus refer to this protein as DMP19. Generally, DNA mimic proteins imitate the shape and charge of dsDNA using the similarly arranged negative charges (from acidic Asp and Glu amino acids) on the surface of the protein. DNA mimic proteins usually function through simple competitive inhibition by occupying the DNA-binding sites of DNA-binding proteins, thus preventing these sites from being accessed by DNA (8,9).
The DNA mimic proteins often target DNA regulatory proteins, such as restriction enzymes, DNA gyrase, TATA-box-binding protein, prokaryotic nucleoid-associated protein and histone proteins (8,10). Here, we found that DMP19 interacts with NMB1204, which was subsequently identified to be a Neisseria hypothetical transcription factor that we refer to as NHTF. We also determined the crystal structure of NHTF and characterized its DNA-binding activity. In a competitive DNA-binding assay, we found that DMP19 could prevent NHTF from binding to a 5′-TGTNAN11TNACA-3′ recognition box on its 5′-untranslated region. An operon analysis suggests NHTF controls the expression of the nitrogen-response protein (Protein-PII) uridylyltransferase. Using an in situ gene regulation assay, we further demonstrate that when NHTF bound to its target DNA-binding site, the expression of a down-stream reporter gene was repressed. We then show that the addition of DMP19 neutralized this effect. Therefore, the interaction between NHTF and DMP19 constitutes a novel gene regulation in the nitrogen response in Neisseria spps.
MATERIALS AND METHODS
Preparation and purification of recombinant DMP19 and NHTF
The full length DMP19 gene (NMB0541, amino acid residues 1–165) was amplified by PCR and ligated into pET 16 b expression vector (Novagen). After induction with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), the recombinant DMP19 protein with its N-terminal His10-tag was expressed by Escherichia coli BL21 (DE3) cells at 20°C for 16 h. Soluble DMP19 was purified by immobilized metal–ion chromatography with a Ni-NTA column, followed by gel filtration using Superdex 200 pg (Amersham Biosciences). The His10-tag was then removed by FactorXa cleavage (Novagen).
Recombinant NHTF was expressing an exactly similar way expect that the expression vector was pET21b (Novagen). After purification immobilized metal–ion chromatography, the C-terminal His6-tagged NHTF was immediately dialyzed overnight at 4°C against acid buffer (50 mM sodium citrate pH 5.8, 300 mM NaCl) in which it is stable. Using the same acid buffer, Superdex 75 gel filtration (Amersham Biosciences) was used to further improve the purity of this protein.
Recombinant DMP19 crystallization, data collection and structure determination
Purified DMP19 was dialyzed overnight at 4°C against crystallization buffer (100 mM NaCl, 20 mM Tris pH 8.0), and concentrated to 30 mg/ml. For crystallization, 2 µl of the DMP19 solution was mixed with 2 µl of a reservoir containing 2.0 M ammonium sulfate, 200 mM potassium sodium tartrate tetrahydrate and 100 mM sodium citrate tribasic dihydrate pH 5.4 as a precipitant, and equilibrated with the reservoir by the sitting drop method. Prior to flash-cooling, crystals were rinsed with a cryoprotectant solution of 20% ethylene glycol and 80% reservoir. After soaking the DMP19 crystals with 1 mM AuCl3 (Sigma-Aldrich) for 48 h, three-wavelength multi-wavelength anomalous dispersion (MAD) X-ray diffraction data were collected on beamline 44XU at the Spring-8 Synchrotron Radiation Research Center in Hyogo, Japan. The data were processed using HKL2000 (11). The space group of the AuCl3 soaked-DMP19 crystals was P21, with unit cell dimensions of a = 43 Å, b = 66 Å, c = 59 Å and β = 98° (Table 1).
Table 1.
Data collection, phase determination and refinement statistics of DMP19 crystals
| Data collection | Edge | Peak | High remote | Native |
|---|---|---|---|---|
| Wavelength (Å) | 1.0400 | 1.0397 | 1.0229 | 1.5418 |
| Space group | P21 | P21 | ||
| Unit cell a, b, c (Å) | 43.2, 66.3, 58.9 | 43.9, 65.3, 59.1 | ||
| β (°) | 97.6 | 99.7 | ||
| Resolution (Å) | 50–2.47 (2.56–2.47) | 50–2.47 (2.56–2.47) | 50–2.43 (2.52–2.43) | 25–2.0 (2.07–2.0) |
| Unique reflections | 11 840 (1168) | 11 907 (1198) | 12 453 (1219) | 22 353 (2218) |
| Redundancy | 3.7 (3.3) | 7.4 (6.7) | 3.7 (3.2) | 9.6 (9.3) |
| Completeness (%) | 99.4 (98.4) | 99.6 (99.4) | 99.3 (97.4) | 100.0 (100.0) |
| I/σ(I) | 36.8 (4.4) | 53.8 (7.4) | 35.1 (3.8) | 64.8 (11.8) |
| Rmerge (%) | 5.4 (34.5) | 6.4 (32.1) | 6.5 (38.7) | 5.2 (30.9) |
| Phasing | Refinement | |||
|---|---|---|---|---|
| Resolution (Å) | 30–2.5 | No. of reflections | 11201 (1046)a | 22063 (2126) |
| Initial number of sites | 1 | Rwork (%) | 24.4 (33.9) | 16.3 (19.3) |
| Initial Fig. of merit | 0.51 | Rfree (%) | 30.8 (44.3) | 20.2 (23.6) |
| Initial Z-score | 10.8 | Bond r.m.s.d. (Å) | 0.013 | 0.016 |
| Final number of sites | 6 | Angle r.m.s.d. (°) | 1.65 | 1.7 |
| Final Fig. of merit | 0.54 | Most favored (%) | 89.8b | 97.2 |
| Final Z-score | 27.1 | Allowed (%) | 9.5b | 2.8 |
| Amino acids built | 212 | Protein B/atoms | 51.6/2532 | 30.2/2596 |
| Rigid-body R-value | 0.49 | Solvent B/atoms | 71.6/6c | 46.3/349 |
aThree data sets were merged for 30–2.5 Å resolution using SCALEPACK with an Rmerge of 5.7%.
bThe remaining 0.7% was due to Ser39 in a loop, and probably due to the strong NCS constraints.
cOne gold had occupancy of 0.98, two had 0.76 and the other three had 0.37, all with similar B-values.
The DMP19 structure was determined using the MAD phasing method and the programs SOLVE and RESOLVE (12,13). Three data sets in the range of 50–2.47 Å resolution collected at the wavelengths of 1.0397 Å (peak), 1.0229 Å (high energy remote) and 1.0400 Å (edge) were used. The space group was identified to be P21, and six Au sites were located in an asymmetric unit. After solvent flattening and phase extension to 2.0 Å by the Crystallography and NMR System (CNS) (14) using the native data set, the resulting electron density map was clear. This allowed automatic building of the model with the program BUCCANEER (15), and revealed that each asymmetric unit contained two DMP19 molecules. The program COOT (16) and Refmac (17) were used in the subsequent refinement. Statistics for the data collection, MAD phasing and refinement are shown in Table 1. The CCP4 package (18) and Pymol program (19) were used for the structural analyses and also for figure production.
Recombinant NHTF crystallization, data collection and structure determination
To obtain NHTF crystals, 2 µl purified C-terminal His6-tagged NHTF (20 mg/ml in a buffer containing 300 mM NaCl, 50 mM sodium citrate tribasic dihydrate, pH 5.8) were mixed with a 2 µl reservoir of buffer (0.72 M sodium formate, 9% PEG8000, 9% PEG1000 and 100 mM sodium acetate, pH 5.5) as a precipitant, and equilibrated with the reservoir by the sitting drop method. During data collection, 10% ethylene glycol was used as cryoprotectant. Native X-ray diffraction data from the NHTF crystals were collected on beamline 13B at the National Synchrotron Radiation Research Center in Hsinchu, Taiwan. After data processing by using HKL2000 (11), the space group of the NHTF crystals was C2 and the unit cell dimensions were a = 75 Å, b = 109 Å, c = 44 Å and β = 115.6° (Table 2).
Table 2.
Data collection and refinement statistics of NHTF crystals
| Data collection | Native |
|---|---|
| Wavelength (Å) | 1.5418 |
| Space group | C2 |
| Unit cell a, b, c (Å) | 75.0, 108.9, 44.1 |
| β (°) | 115.6 |
| Resolution (Å) | 25–2.0 (2.07–2.0) |
| Unique reflections | 21 471 (2125) |
| Redundancy | 7.5 (7.5) |
| Completeness (%) | 99.7 (100.0) |
| I/σ(I) | 47.1 (6.8) |
| Rmerge (%) | 5.9 (35.6) |
| Refinement | |
| No. of reflections | 20 865 (2018) |
| Rwork (%) | 17.2 (25.7) |
| Rfree (%) | 21.7 (32.6) |
| Bond r.m.s.d. (Å) | 0.019 |
| Angle r.m.s.d. (°) | 1.7 |
| Most favored (%) | 98.8 |
| Allowed (%) | 1.2 |
| Protein B/atoms | 37.4/2113 |
| Solvent B/atoms | 52.3/282 |
The sequence alignment results showed that Neisseria NHTF transcription factor is similar to the restriction–modification system control factor C.BclI (2B5A.pdb) (20) with 40% sequence identity (Supplementary Figure S1). Because of the sequence similarity, the crystal structure of C.BclI was used as the search model by the program Molrep (21). An initial map was produced after preliminary refinement using Refmac (17), and the procedures for model building, refinement and structural analysis were similar to those used for DMP19. Statistics for the data collection and structural refinement are shown in Table 2.
His-pulldown assay to identify proteins that interact with DMP19
For the His-pulldown assay, the soluble proteins of N. meningitidis MC58 were extracted by bacterial protein extraction reagents (B-PER; Thermo Scientific) following the standard protocol. Briefly, a pellet of N. meningitidis MC58 (1 g) was resuspended in 4 ml of B-PER that contained Benzonase (Novagen, 10 U/ml). After incubation for 15 min at room temperature, the lysate was centrifuged at 15000 ×g for 10 min to separate the soluble and insoluble proteins. The soluble protein fraction (200 µg) was then mixed with purified N-terminal His10-tagged DMP19 (1 mg). After adjusting to a final volume of 1 ml with binding buffer (1 × PBS with 20 mM imidazole), the reaction mixture was incubated for 16 h at 4°C. Incubation was then continued with Ni-NTA beads (50 μl) for another 1 h. After incubation, the beads were washed 100 times with the same binding buffer (5 ml). The proteins that bound to the beads were eluted by the addition of buffer containing 500 mM imidazole and resolved by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE). Protein bands of interest were manually excised from the gels. After in-gel trypsin digestion followed by LC–nanoESI-MS/MS, the search program MASCOT was used to search for matches. Variable modifications considered were methionine oxidation and cysteine carboxyamidomethylation. Significant hits (as defined by Mascot probability analysis) were regarded as positive identification. In another His-pulldown assay that was used to confirm the interaction between DMP19 and NHTF, purified C-terminal His6-tagged NHTF (final concentration: 1 μM) was mixed with 3 μM DMP19 without any tag (final concentration: 3 μM). The same His-pulldown procedure was used in this assay.
Bissulfosuccinimidyl suberate (BS3) cross-linking assay to confirm the interaction between DMP19 and NHTF
For the BS3 assay, 0.4 ml of 3 μM purified C-terminal His6-tagged NHTF was mixed with 0.4 ml of 3 μM DMP19, a Bovine serum albumin (BSA) control or DMP12, which is another conserved hypothetical protein found in Neisseria (NMB2106). After adjusting to a final volume of 1 ml with a binding buffer (1 × PBS), the reaction mixture was incubated for 30 min at 25°C. Following the addition of BS3 to a final concentration of 0.5 mM, incubation was then continued for another hour. The reactions were quenched by 50 mM Tris–HCl. After the BS3 cross-linking, a His-pulldown assay was used to identify the proteins that cross-linked to the C-terminal His6-tagged NHTF. All reactions were resolved by SDS–PAGE, and western blot analysis using DMP19 antibody was used to detect the DMP19.
Gel filtration to determine the native molecular weight of target proteins
To evaluate the approximate molecular weight of DMP19, NHTF and the DMP19/NHTF complex, each protein was analyzed separately using a Superdex 75 HR 10/30 gel filtration column (Amersham Biosciences). For DMP19 and DMP19/NHTF, the buffer was 20 mM Tris–base pH 7.2. For NHTF, the buffer was 50 mM sodium citrate pH 5.8, because in the absence of DMP19 or DNA substrate, NHTF at concentrations higher than 50 µM will precipitate out of buffers with a neutral pH. Gel filtration standard proteins (conalbumin [75 kDa], ovalbumin [43 kDa], carbonic anhydrase [29 kDa] and RNase A [13.7 kDa]) were used to calibrate the column. For each protein, the logarithm of molecular weight (log MW) was plotted against Kav, which was calculated as follows: Kav = (Ve − Vo)/(Vt − Vo), where Ve is the elution volume, Vo is the column void volume using blue dextran 2000, and Vt is the total column bed volume (24 ml for the Superdex 75 HR 10/30 gel filtration column).
DNA-binding assays (EMSA assays)
These assays used either PCR products or short dsDNA fragments as DNA substrates. The PCR products of the NHTF coding and 5′ untranslated regions were prepared by standard PCR protocol using the corresponding primer sets (Supplementary Table S1). The short dsDNA fragments were prepared by mixing equal volumes and concentrations of complementary oligonucletides (Supplementary Table S1), incubating the reaction mixtures at 95°C for 3–5 min, and then lowering the temperature to RT for another 30 min. For the NHTF–DNA-binding assays, different amounts of purified recombinant C-terminal His6-tagged NHTF were mixed with DNA substrates. The reaction mixtures were adjusted to a volume of 15 μl in binding buffer (1 × PBS) and incubated at 37°C for 15 min. For the DMP19/DNA competitive binding assays, purified recombinant C-terminal His6-tagged NHTF was mixed with different amounts of DMP19. The reaction mixtures were adjusted to a volume of 14 μl in binding buffer (1 × PBS) and pre-incubated at 37°C for 1 h. Finally, 1 μl of binding buffer containing the DNA substrate was added and incubation continued for 15 min. The reaction products were run on a 0.8% agarose gel and stained with SYBR Green I (Sigma-Aldrich).
In situ gene regulation assay
A synthesized gene fragment containing the T7 promoter, lac operator, NHTF-binding site and the green fluorescent protein (GFP) gene was ligated into a PUC 57 plasmid (Genscript) to produce the construct pNHTFbs-GFP. Cell-free protein synthesis was performed using an EasyXpress protein synthesis kit (QIAgen). For the gene regulation assay, NHTF was mixed with or without DMP19. The reaction mixtures were adjusted to a volume of 10 μl in binding buffer (1 × PBS) such that the final concentrations of NHTF and DMP19 were both 30 µM. After pre-incubation at 37°C for 1 h, the mixture was added to the protein synthesis reaction buffer (provided by the kit and containing E. coli extract). The reaction was initiated by adding 5 nM pNHTFbs–GFP and 0.25 mM IPTG. After 1 h of incubation at 37°C, the expressed GFP was detected by excitation at 395 nm and emission at 509 nm.
RESULTS
The crystal structure of DMP19
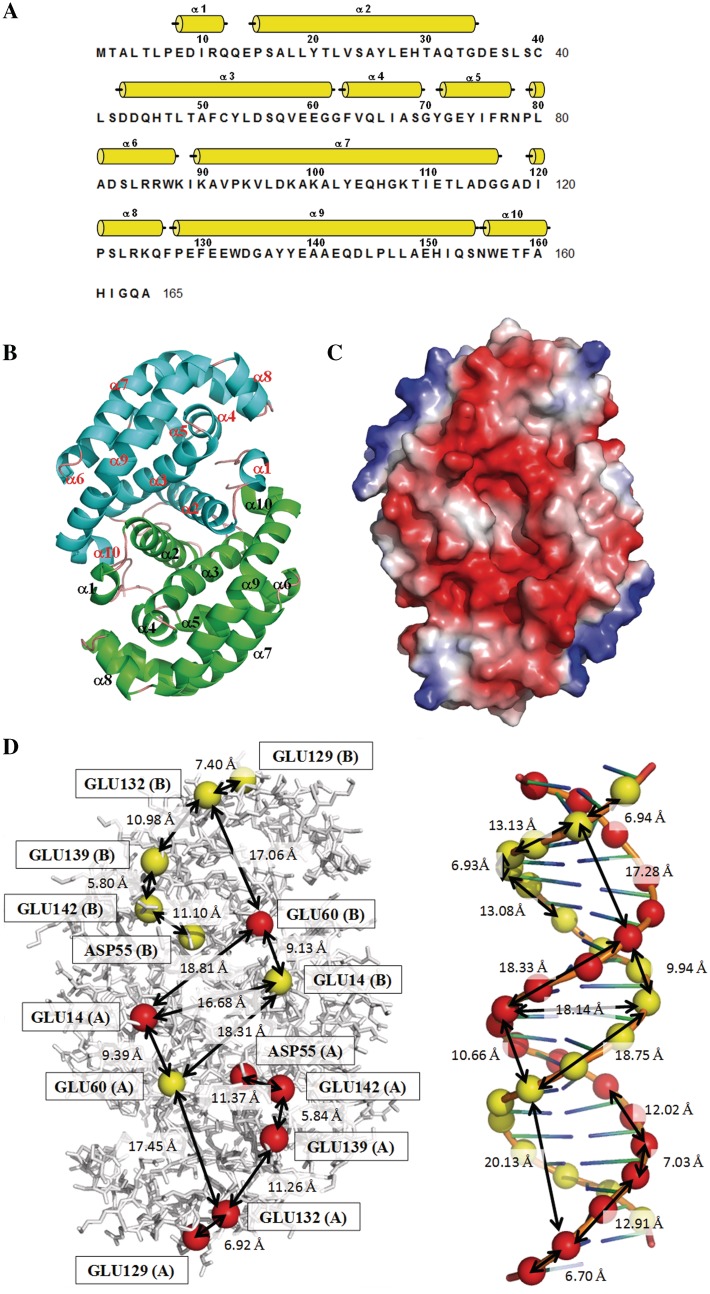
The protein structure of DMP19 from N. meningitides MC58 was determined by applying the MAD method to an AuCl3-soaked crystal. DMP19 contains only α-helices (α1–α10; Figure 1A). The refined structure contains two DMP19 molecules arranged as a dimer in each asymmetric unit (Figure 1B). The refinement statistics are listed in Table 1. A structural homology search using the DALI website showed that the DMP19 structure has no similarity to any other known structure, indicating a novel fold.
Figure 1.
The negative charge distribution of DMP19 resembles the B-form DNA helix. (A) The secondary structural elements are shown above the amino acid sequence, with yellow cylinders representing α-helices. (B) A ribbon diagram of the DMP19 dimer. The two monomers are colored green and cyan, respectively. (C) The molecular surfaces of the DMP19 dimer. Color-coding is by Pymol, with red to blue representing the electrostatic potential from −90 kBT to +90 kBT. A DNA-like negative charge distribution can be seen on one-dimensional surface of DMP19. (D) Comparison of the negative charge distribution of DMP19 and dsDNA. The spheres denote the DMP19 acidic residues that correspond to the negatively charged phosphate groups on the dsDNA. The approximate distances between spots are also shown.
The DMP19 protein is an acidic protein (calculated pI < 4.5), which means that it is negatively charged under physiological conditions. Its surface-charge distribution reveals a dsDNA-like topology (Figure 1C and D), with a dsDNA (from PDB 3CRO) shown at the same scale for comparison. The patches of negatively charged amino acids are arranged into two rows on the DMP19 surface, separated by 17 Å (Glu14 to the same amino acid on the other monomer), which is comparable to the interphosphate distance in the opposite strands of dsDNA (18 Å). Additionally, the distances between the DMP19 carboxyl groups Glu-14, Asp-55, Glu-60, Glu-129, Glu-132, Glu-139 and Glu-142 are similar to that between two adjacent phosphate groups in dsDNA. These results suggest that DMP19 mimics the negative charge distribution of dsDNA.
Identification of the Neisseria proteins that may interact with DMP19
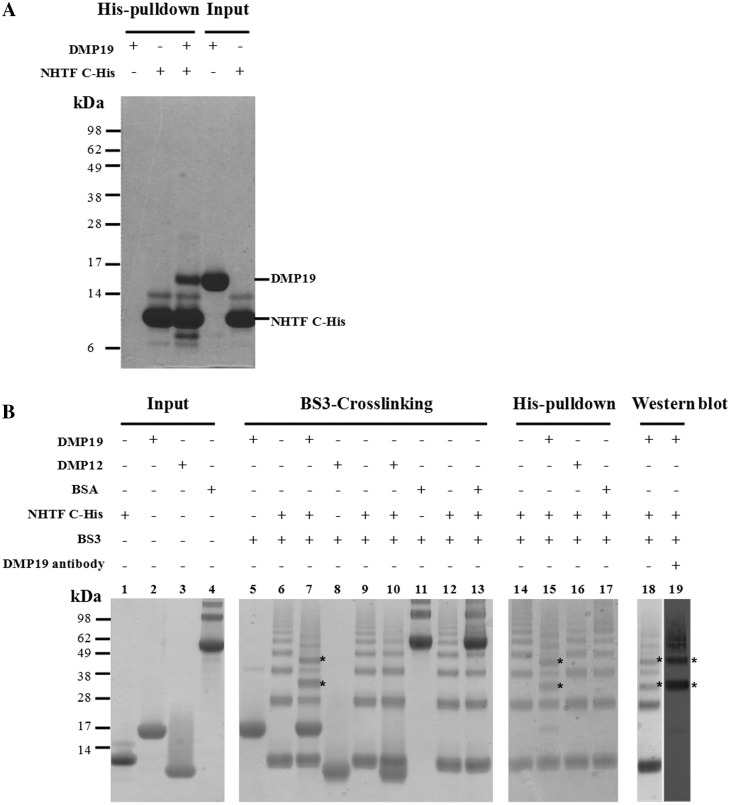
If DMP19 is a DNA mimic, then at least part of its function would likely involve direct interaction with DNA-binding protein(s). Using extracted Neisseria proteins as prey and the N-terminal His10-tagged DMP19 as a bait, the His-pulldown assay results revealed six Neisseria proteins that appear to interact with DMP19. The biological roles of these proteins are: (a) DNA-directed RNA polymerase β subunit, (b) pyruvate dehydrogenase subunit E1, (c) molecular chaperone DnaK, (d) elongation factor Tu, (e) DNA-directed RNA polymerase α subunit and (f) Neisseria hypothetical transcription factor (NHTF; Supplementary Figure S2 and Table S2). In the present study, only NHTF was selected for further investigation. The interaction between DMP19 and NHTF was confirmed by another pulldown assay (Figure 2A) which showed that DMP19 was pulled down by C-terminal His6-tagged NHTF (NHTF C-His). A BS3 cross-linking assay was used to further clarify this interaction. As Figure 2B shows, the addition of DMP19 caused a shift in the NHTF C-His bands. The shifted bands are indicated by asterisks and confirm the interaction of these two proteins.
Figure 2.
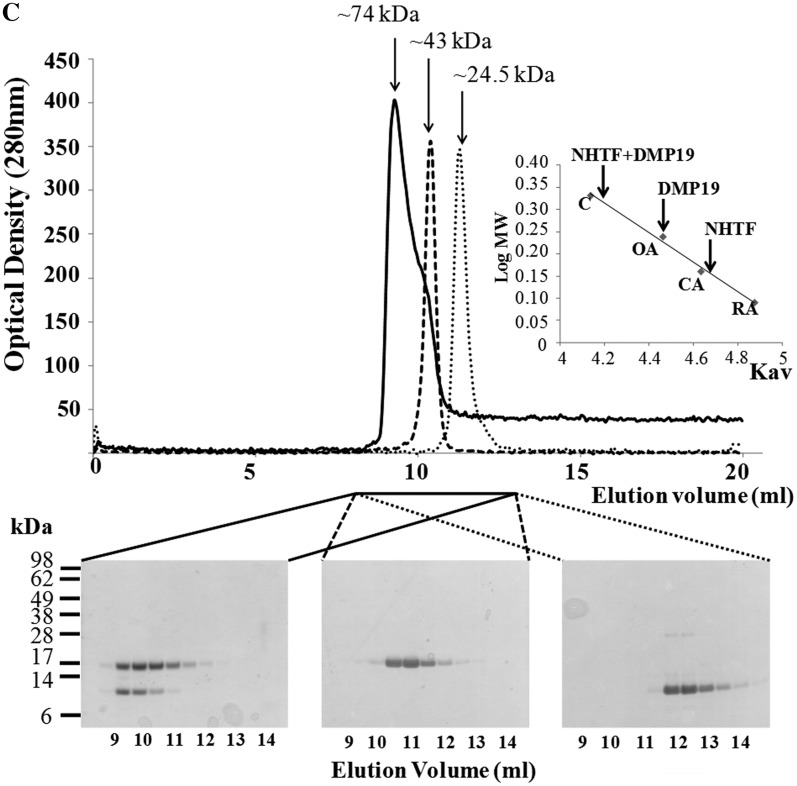
His-pulldown, BS3 cross-linking assays, and gel filtration all confirm that DMP19 and NHTF interact. (A) In this His-pulldown assay, DMP19 without any tag was used as prey and C-terminal His6-tagged NHTF transcription factor (NHTF C-His) was used as bait. DMP19 was pulled-down by NHTF C-His. (B) A BS3 cross-linking assay was carried out to further confirm the interaction between DMP19 and NHTF C-His. NHTF C-His was co-incubated separately with DMP19 and two negative controls (BSA, pI 5.8 and DMP12, pI 4.5). After the addition of BS3, shifted bands that indicated protein–protein cross-linking were only seen in the DMP19/NHTF C-His reactions (lanes 7, 15, 18 and 19); the shifted bands are marked with an asterisk. NHTF C-His was pulled down using Ni-NTA beads, and cross-linked DMP19/NHTF C-His was found in lane 15. The presence of DMP19 in the shifted bands was confirmed by using anti-DMP19 antibody (lane 19). The presence of DMP19 and NHTF C-His in the same shifted bands of this experiment was further confirmed by western blot analysis using anti-DMP19 and anti-NHTF antibodies (Supplementary Figure S3). (C) The molecular weights of DMP19, NHTF and DMP19/NHTF complex were measured by gel filtration on a Superdex 75 HR 10/30 column monitored at 280 nm. The standard proteins conalbumin (C; 75 kDa), ovalbumin (OA; 43 kDa), carbonic anhydrase (CA; 29 kDa) and RNase A (RA; 13.7 kDa) were fractionated on the same column and used to generate a plot of Kav against log MW (inset). The Kav of each target protein (DMP19, NHTF and DMP19/NHTF complex) was then used to find its molecular weight.
Gel filtration chromatography was used to investigate the native molecular weights of DMP19, NHTF and the DMP19/NHTF complex (Figure 2C). Based on the Kav versus log MW values of protein standards that were fractionated in the same column (Figure 2C, inset), the apparent molecular weights of the target proteins were calculated to be 43, 24.5 and 74 kDa, respectively. The predicted molecular weights of DMP19 and NHTF are a good match to their theoretical dimer molecular weight (37 and 26 kDa), and the approximate molecular weight of the DMP19/NHTF complex is also close to the sum of the theoretical molecular weights of the DMP19 and NHTF dimers. An analytical ultracentrifugation analysis of DMP19/NHTF complex formation also provided similar results (Supplementary Figure S4 and Supplementary Table S3). All of these data suggest that DMP19 forms a dimer to dimer complex with NHTF.
The crystal structure of hypothetical transcription factor NHTF from Neisseria meningitides MC58
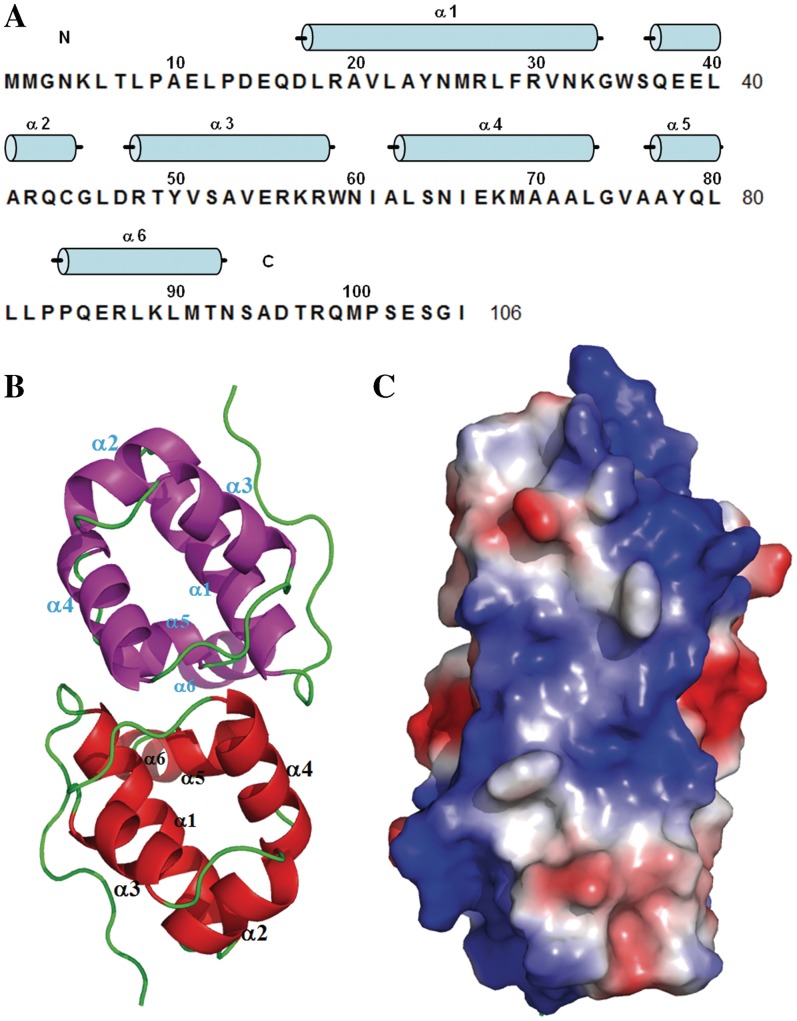
Like DMP19, NHTF consists entirely of α-helices (α1–α6; Figure 3A). A sequence homology search predicted that α2 and α3 formed helix-turn-helix (HTH) motif (Supplementary Figure S1). The crystal structure of NHTF (4–92) was determined by the molecular replacement (MR) method (Figure 3B) using XRE transcription factor C.BclI (PDB 2B5A) as the search model. The refinement statistics are listed in Table 2. The asymmetric unit of the crystal comprised three NHTF molecules made up of one dimer and one monomer from another dimer situated on a dyad axis. A structural homology search using the DALI website showed that the NHTF structure can be assigned to the XRE family of transcription factors. Superposition of the core structure (α1–α4) from NHTF and three other XRE transcription factors [restriction–modification controller protein C.BclI (20), C.AhdI (22) and 434 Cro repressor (23)] confirmed the presence of the predicted classical HTH motif (α2 and α3; Supplementary Figures S1 and S5A), while the location of the DNA-binding region of NHTF was suggested by the structural superposition of NHTF and the phage 434 CRO/DNA complexes (Figure 3C and Supplementary Figure S5B).
Figure 3.
The crystal structure and the proposed DNA-binding region of Neisseria NHTF transcription factor. (A) The secondary structural elements are shown above the amino acid sequence, with blue cylinders representing α-helices. (B) A ribbon diagram of the NHTF dimer. The two monomers are colored red and magenta. Because the electron density maps were not clear in the N and C-terminal regions, only residues 4–92 were included in this model. (C) The molecular surfaces of the NHTF dimer. The color-coding is by Pymol, with red to blue representing the electrostatic potential from −74 kBT to +74 kBT.
DMP19 prevents the DNA binding of Neisseria transcriptional factor NHTF
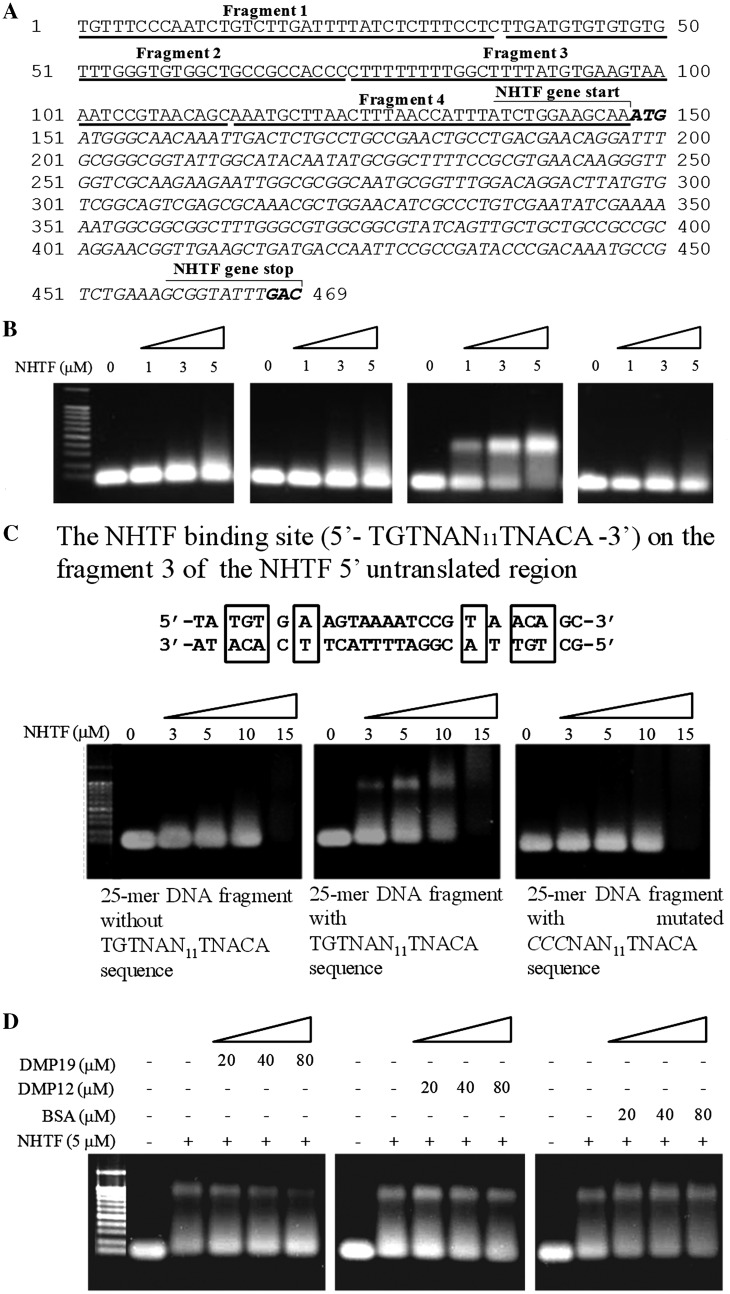
A number of XRE transcription factors, including C.BclI (20) and C.AhdI (22), regulate their own expression by means of a control region located in their 5′-untranslated sequence. Here, we used an electrophoresis mobility shift assay (EMSA) to test whether NHTF also binds to the 5′-untranslated region of the NHTF gene. A preliminary EMSA study showed that although there was no evidence of specific binding to the NHTF coding region, there was a clear DNA shift of the NHTF 5′-untranslated region at low concentration (1–3 µM; Supplementary Figure S6). We hypothesized that the 5′-untranslated region of the NHTF gene may include a site that is specifically recognized by NHTF. This possibility was further investigated by EMSA assays with four DNA fragments from different parts of the 5′-untranslated region (Figure 4A and B). The NHTF protein bound specifically to the fragment 3, but not to the other three fragments. Fragment 3 contains two repeated sequences related by a pseudo-dyad axis, 5′-TGTNAN11TNACA-3′, which is similar to the binding site of Bacillus subtilis transcription factor TnrA (5′-TGTNAN7TNACA-3′). A subsequent EMSA result showed that specific shifting was only observed when the 5′-TGTNAN11TNACA-3′ sequence was present and that mutation of this pseudo-dyad axis sequence prevented NHTF from binding to this fragment (Figure 4C).
Figure 4.
NHTF binding to the NHTF gene cluster and effect of DMP19 on its specificity. (A) The NHTF gene cluster encompasses a 5′-untranslated region along with the coding region. (B) The NHTF 5′-untranslated region was cut into four fragments with similar length (∼40-mer) and incubated with NHTF. Only fragment 3 was shifted after incubation with NHTF. (C) EMSA results showed that within fragment 3, only the 25-mer DNA fragment that included the un-mutated TGTNAN11TNACA pseudo-dyad axis sequence produced the specific DNA shift. (D) Addition of the DNA mimic DMP19 reduces the specific DNA binding of NHTF in a dosage-dependent manner. The addition of the other two negatively charged proteins BSA and DMP12 had no effect. The DNA concentration used in all of the EMSA assays is 1 µM.
If DMP19 is a DNA mimic, then it may prevent DNA from binding to the NHTF. To investigate this, competitive EMSA assays were used to investigate the effect of DMP19 on the DNA binding of NHTF. The dosage-dependent decrease in the shifted bands when DMP19 was pre-incubated with NHTF shows that DMP19 prevented the specific binding of NHTF to its specific 25-mer target (i.e. 5′-TGTNAN11TNACA-3′) (Figure 4D). Pre-incubation with the negatively charged BSA(pI 4.7) and DMP12 (NMB2106, pI 4.5) controls had no such effect.
NHTF down-regulates down-stream gene expression and DMP19 neutralizes this effect
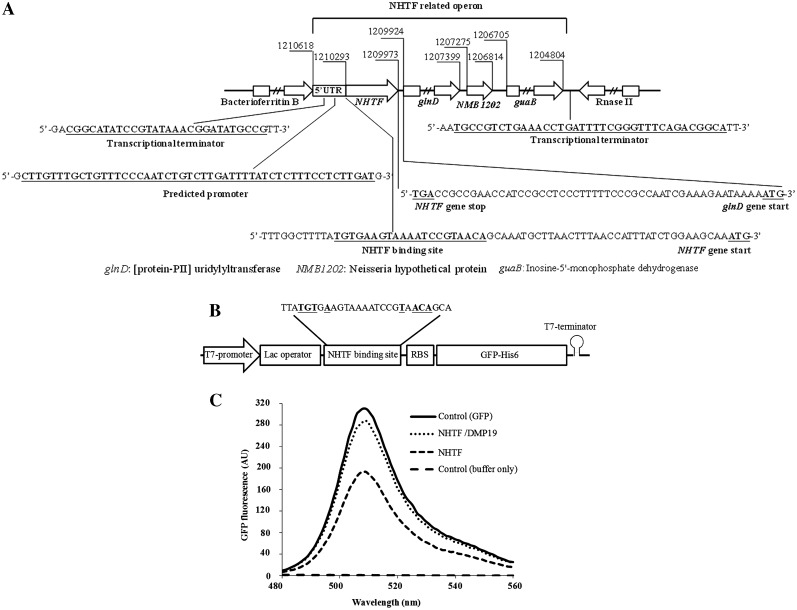
An operon analysis of the NHTF gene region shows that the NHTF and glnD genes are separated by a 49-base pair intergenic region that lacks a terminator structure. Furthermore, no putative promoter sequence has been found in this intergenic region. This suggests that the glnD gene of Neisseria is most likely co-transcribed with NHTF, and we hypothesize that NHTF may therefore control the expression of both the NHTF gene and the downstream (Protein-PII) uridylyltransferase gene (glnD) by binding to its 5′-untranslated region (Figure 5A). To test whether NHTF can regulate the expression of a down-stream gene, we designed an in situ gene regulation assay using GFP as the reporter gene. This assay uses cell-free protein synthesis and a plasmid (pNHTFbs–GFP) that contains the operon shown in Figure 5B. Figure 5C shows that the addition of 30 µM NHTF decreased the GFP fluorescent signal to 0.63 ± 0.09-fold compared to the GFP control (from three independent assays). Further, this gene repression effect was neutralized by the addition of the same concentration of DMP19 (the GFP fluorescent signal was restored to 0.97 ± 0.01 compared to the GFP control). We therefore conclude that NHTF acts as a gene repressor of its down-stream genes and that DMP19 is able to neutralize this effect.
Figure 5.
NHTF represses down-stream gene expression and DMP19 prevents this effect. (A) Operon analysis of the chromosomal NHTF-glnD region of N. meningitides MC58. The BDGP (24) and ARNold programs (25–27) were used to predict the promoters and transcription terminators for the intergenic regions. (B) Schematic of the operon used in the pNHTFbs-GFP construct for the in situ gene regulation assay. The sequence of this operon is included in Supplementary Figure S7. (C) GFP fluorescence emission spectra (480–560 nm) after cell-free protein synthesis. NHTF alone reduces the fluorescence signal to ∼63%, while the further addition of DMP19 restores the signal to ∼97%. The addition of DMP19 alone had no effect on GFP signal strength (data omitted for clarity).
DISCUSSION
In this report, we used structural and proteomic analyses to determine the relationship between two hypothetical proteins, DMP19 and NHTF, from the pathogen N. meningitidis MC58. Structural analysis of DMP19 revealed a dsDNA-like charge distribution on the DMP19 dimer (Figure 1C and D), suggesting that it might act as a DNA mimic protein. DNA mimic proteins are hard to identify using bioinformatic searches because their amino acid sequences lack homology to other known proteins and because their structures are so divergent (8,9). However, these proteins are important to living cells, and to date, DNA mimic proteins have been found in prokaryotes, eukaryotes, bacteriophages and recently in a eukaryotic virus (8,10). These known DNA mimic proteins are involved in many DNA-regulation mechanisms, such as DNA repair (28), restriction (29,30), DNA packaging (10,31,32), topology (33), transcription (34), single-strand DNA binding (35) and recombination (36).
The second protein identified in this report, NHTF, is a hypothetical transcription factor that interacted with DMP19 (Figure 2). The crystal structure of NHTF provides evidence that it contains a HTH motif, and that it is a member of the XRE transcriptional factor family (Figure 3B and Supplementary Figure S5). Members of the XRE family are involved in a wide range of gene regulation, including plasmid copying, restriction and modification systems, bacteriophage transcription control and stress responses (37). Among the reported XRE family proteins, NHTF is most similar to the restriction and modification system (RMS) controller proteins (C proteins) C.BclI (20) and C.AhdI (22). In bacteria, the RMS system acts as a defense mechanism against phage infection and other types of DNA invasion, but it is a system that must be tightly controlled to avoid cell death via autorestriction (38–41), while still providing protection against the invading viral DNA.
Given the similarity in structure and amino acid sequence between NHTF and the C proteins, it is possible that NHTF may also be involved in the same restriction and modification system. However, our analysis of the NHTF gene region on the Neisseria chromosome (Figure 5A) suggests another possible function. The NHTF gene is located upstream of the Neisseria (Protein-PII) uridylyltransferase gene (glnD) in all known Neisseria genomes including those of N. meningitidis, N. gonorrhoeae and N. lactamica. No transcriptional terminator structure or predicted promoter could be found in any of the intergenic regions between the NHTF and glnD genes. That suggests the glnD gene of Neisseria is most likely to be co-transcribed with NHTF and regulated by the binding of NHTF to the target DNA sequence on its 5′-untranslated region (5′-UTR; Figure 4). The gene product of glnD is (Protein-PII) uridylyltransferase, a protein which plays an important role in the regulation of nitrogen assimilation by controlling the uridylylation state of the PII signal transduction protein in response to intracellular signals (42–46). We further note that the NHTF target DNA sequence (5′-TGTNAN11TNACA-3′) is similar to the binding site of TnrA (5′-TGTNAN7TNACA-3′), a transcription factor that is required for global nitrogen regulation in B. subtilis (47). All of these data suggest that NHTF is an auto-regulated transcription factor that is involved in the response to nitrogen levels in Neisseria spp.
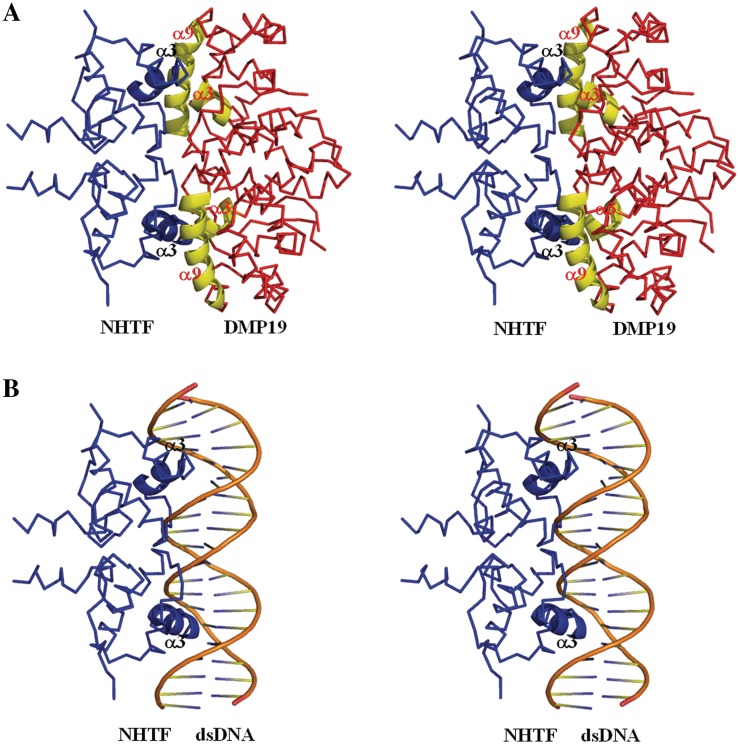
The competitive EMSA results showed that DMP19 prevented the specific binding of NHTF to its specific 25-mer target (Figure 4D). We attribute this preventative effect mainly to the specific charge–charge interaction between the complementary surface charges of DMP19 and the DNA-binding surface of NHTF (Supplementary Figure S8). A proposed model for this DMP19/NHTF interaction is shown in Figure 6A. In this model, the positively charged α-helix 3 of NHTF (i.e. the proposed recognition helix; Figure 6B and Supplementary Figure S5B) fits into the negatively charged cavity formed by the α-helices 3 and 9 of DMP19 with no steric clashes. Although this model has not yet been verified by structural analysis of the real complex, the interaction between DMP19 and NHTF would be biologically meaningful in the context of the nitrogen regulation system of Neisseria spp. The in situ gene regulation assay (Figure 5C) provides further evidence that DMP19 acts to prevent the gene repressor activity of NHTF. Assuming that NHTF is indeed an auto-repressor that down-regulates its own expression as well as the expression of down-stream genes such as glnD, there are likely to be critical situations where more control would be needed. Our data suggest that DMP19 plays a role in controlling this auto-feedback mechanism by temporarily interfering with the binding of NHTF to its specific target and thus restoring the expression of the down-stream genes expression.
Figure 6.
The proposed models of (A) NHTF/DMP19 binding and (B) NHTF/DNA binding. The diagrams are shown in stereo.
In summary, we conclude that together, DMP19 and NHTF constitute a novel system for the transcriptional control of (Protein-PII) uridylyltransferase, a key enzyme in the nitrogen regulation system. Since the NHTF-glnD operon is distinct from the operons in other known bacteria, such as E. coli (44), Rhizobium leguminosarum (45) and Gluconacetobacter diazotrophicus (46), this system would provide a new mechanism of nitrogen control in Neisseria spp.
ACCESSION NUMBERS
PDB ID of DMP19 is 3VJZ and NHTF is 3VK0.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–8 and Supplementary Reference [48].
FUNDING
Funding for open access charge: Arademia Sinica and National Science Council grants (Grants NSC96-3114-P-001-004, NSC97-3114-P-001-001, NSC99-3113-B-001-001 and 23-23-1005335).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge the staff of the beamline BL13B1 at the Radiation Research Center (NSRRC) in Hsinchu, Taiwan; and the staff of the beamline 44XU at the Spring-8 Synchrotron Radiation Research Center in Hyogo, Japan; for their help in X-ray crystal data collection. We also thank the Core Facilities for Proteomics Research at the Institute of Biological Chemistry, Academia Sinica for their help in Proteomic mass spectrometry analyses.
REFERENCES
- 1.Aspholm M, Aas FE, Harrison OB, Quinn D, Vik A, Viburiene R, Tønjum T, Moir J, Maiden MC, Koomey M. Structural alterations in a component of cytochrome c oxidase and molecular evolution of pathogenic Neisseria in humans. PLoS Pathog. 2010;6:e1001055. doi: 10.1371/journal.ppat.1001055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ren J, Sainsbury S, Nettleship JE, Saunders NJ, Owens RJ. The crystal structure of NGO0477 from Neisseria gonorrhoeae reveals a novel protein fold incorporating a helix-turn-helix motif. Proteins. 2010;78:1798–1802. doi: 10.1002/prot.22698. [DOI] [PubMed] [Google Scholar]
- 3.Snyder LA, Davies JK, Ryan CS, Saunders NJ. Comparative overview of the genomic and genetic differences between the pathogenic Neisseria strains and species. Plasmid. 2005;54:191–218. doi: 10.1016/j.plasmid.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 4.Parkhill J, Achtman M, James KD, Bentley SD, Churcher C, Klee SR, Morelli G, Basham D, Brown D, Chillingworth T, et al. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature. 2000;404:502–506. doi: 10.1038/35006655. [DOI] [PubMed] [Google Scholar]
- 5.Tettelin H, Saunders NJ, Heidelberg J, Jeffries AC, Nelson KE, Eisen JA, Ketchum KA, Hood DW, Peden JF, Dodson RJ, et al. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science. 2000;287:1809–1815. doi: 10.1126/science.287.5459.1809. [DOI] [PubMed] [Google Scholar]
- 6.Exley RM, Sim R, Goodwin L, Winterbotham M, Schneider MC, Read RC, Tang CM. Identification of meningococcal genes necessary for colonization of human upper airway tissue. Infect. Immun. 2009;77:45–51. doi: 10.1128/IAI.00968-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stabler RA, Marsden GL, Witney AA, Li Y, Bentley SD, Tang CM, Hinds J. Identification of pathogen-specific genes through microarray analysis of pathogenic and commensal Neisseria species. Microbiology. 2005;151:2907–2922. doi: 10.1099/mic.0.28099-0. [DOI] [PubMed] [Google Scholar]
- 8.Dryden DT. DNA mimicry by proteins and the control of enzymatic activity on DNA. Trends Biotechnol. 2006;24:378–382. doi: 10.1016/j.tibtech.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 9.Putnam CD, Tainer JA. Protein mimicry of DNA and pathway regulation. DNA Repair. 2005;4:1410–1420. doi: 10.1016/j.dnarep.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 10.Wang HC, Wang HC, Ko TP, Lee YM, Leu JH, Ho CH, Huang WP, Lo CF, Wang AH. White spot syndrome virus protein ICP11: A histone-binding DNA mimic protein that disrupts nucleosome assembly. Proc. Natl. Acad. Sci. USA. 2008;105:20758–20763. doi: 10.1073/pnas.0811233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 12.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Cryst. D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Terwilliger TC. Maximum likelihood density modification. Acta Cryst. D. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR System: a new software suite for structure determination. Acta Cryst. D. 1997;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 15.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Cryst. D. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 16.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 17.Vagin AA, Steiner RS, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. REFMAC5 dictionary: organisation of prior chemical knowledge and guidelines for its use. Acta Cryst. D. 2004;60:2284–2295. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 18.Collaborative Computational Project Number 4. Acta Cryst. D. 1994;50:760–763. [Google Scholar]
- 19.DeLano WL. The PyMOL Molecular Graphics System. Palo Alto, CA, USA: DeLano Scientific LLC; 2008. [Google Scholar]
- 20.Sawaya MR, Zhu Z, Mersha F, Chan SH, Dabur R, Xu SY, Balendiran GK. Crystal structure of the restriction-modification system control element C.Bcll and mapping of its binding site. Structure. 2005;13:1837–1847. doi: 10.1016/j.str.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 21.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 1997;30:1022–1025. [Google Scholar]
- 22.McGeehan JE, Streeter SD, Papapanagiotou I, Fox GC, Kneale GG. High-resolution crystal structure of the restriction-modification controller protein C.AhdI from Aeromonas hydrophila. J. Mol. Biol. 2005;346:689–701. doi: 10.1016/j.jmb.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 23.Beamer LJ, Pabo CO. Refined 1.8 A crystal structure of the lambda repressor-operator complex. J. Mol. Biol. 1992;227:177–196. doi: 10.1016/0022-2836(92)90690-l. [DOI] [PubMed] [Google Scholar]
- 24.Gautheret D, Lambert A. Direct RNA motif definition and identification from multiple sequence alignments using secondary structure profiles. J. Mol. Biol. 2001;313:1003–1011. doi: 10.1006/jmbi.2001.5102. [DOI] [PubMed] [Google Scholar]
- 25.Lesnik EA, Sampath R, Levene HB, Henderson TJ, McNeil JA, Ecker DJ. Prediction of rho-independent transcriptional terminators in Escherichia coli. Nucleic Acids Res. 2001;29:3583–3594. doi: 10.1093/nar/29.17.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macke T, Ecker D, Gutell R, Gautheret D, Case DA, Sampath R. RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res. 2001;29:4724–4735. doi: 10.1093/nar/29.22.4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waibel AH, Hanazawa T, Hinton GE, Shikano K, Lang KJ. Phoneme recognition using time-delay neural networks. IEEE Trans. Acoustic Speech Signal Process. 1989;37:328–339. [Google Scholar]
- 28.Putnam CD, Shroyer MJ, Lundquist AJ, Mol CD, Arvai AS, Mosbaugh DW, Tainer JA. Protein mimicry of DNA from crystal structures of the uracil-DNA glycosylase inhibitor protein and its complex with Escherichia coli uracil-DNA glycosylase. J. Mol. Biol. 1999;287:331–346. doi: 10.1006/jmbi.1999.2605. [DOI] [PubMed] [Google Scholar]
- 29.McMahon SA, Roberts GA, Johnson KA, Cooper LP, Liu H, White JH, Carter LG, Sanghvi B, Oke M, Walkinshaw MD, et al. Extensive DNA mimicry by the ArdA anti-restriction protein and its role in the spread of antibiotic resistance. Nucleic Acids Res. 2009;37:4887–4897. doi: 10.1093/nar/gkp478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Walkinshaw MD, Taylor P, Sturrock SS, Atanasiu C, Berge T, Henderson RM, Edwardson JM, Dryden DT. Structure of Ocr from bacteriophage T7, a protein that mimics B-form DNA. Mol. Cell. 2002;9:187–194. doi: 10.1016/s1097-2765(02)00435-5. [DOI] [PubMed] [Google Scholar]
- 31.Parsons LM, Yeh DC, Orban J. Solution structure of the highly acidic protein HI1450 from Haemophilus influenzae, a putative double-stranded DNA mimic. Proteins. 2004;54:375–383. doi: 10.1002/prot.10607. [DOI] [PubMed] [Google Scholar]
- 32.Parsons LM, Liu F, Orban J. HU-alpha binds to the putative double-stranded DNA mimic HI1450 from Haemophilus influenzae. Protein Sci. 2005;14:1684–1687. doi: 10.1110/ps.041275705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hegde SS, Vetting MW, Roderick SL, Mitchenall LA, Maxwell A, Takiff HE, Blanchard JS. A fluoroquinolone resistance protein from Mycobacterium tuberculosis that mimics DNA. Science. 2005;308:1480–1483. doi: 10.1126/science.1110699. [DOI] [PubMed] [Google Scholar]
- 34.Liu D, Ishima R, Tong KI, Bagby S, Kokubo T, Muhandiram DR, Kay LE, Nakatani Y, Ikura M. Solution structure of a TBP-TAF(II)230 complex: protein mimicry of the minor groove surface of the TATA box unwound by TBP. Cell. 1998;94:573–583. doi: 10.1016/s0092-8674(00)81599-8. [DOI] [PubMed] [Google Scholar]
- 35.Bochkareva E, Kaustov L, Ayed A, Yi GS, Lu Y, Pineda-Lucena A, Liao JC, Okorokov AL, Milner J, Arrowsmith CH, et al. Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc. Natl. Acad. Sci. USA. 2005;102:15412–15417. doi: 10.1073/pnas.0504614102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramirez BE, Voloshin ON, Camerini-Otero RD, Bax A. Solution structure of DinI provides insight into its mode of RecA inactivation. Protein Sci. 2000;9:2161–2169. doi: 10.1110/ps.9.11.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santos CL, Tavares F, Thioulouse J, Normand P. A phylogenomic analysis of bacterial helix-turn-helix transcription factors. FEMS Microbiol. Rev. 2009;33:411–429. doi: 10.1111/j.1574-6976.2008.00154.x. [DOI] [PubMed] [Google Scholar]
- 38.De Backer O, Colson C. Transfer of the genes for the StyLTI restriction-modification system of Salmonella typhimurium to strains lacking modification ability results in death of the recipient cells and degradation of their DNA. J. Bacteriol. 1991;173:1328–1330. doi: 10.1128/jb.173.3.1328-1330.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ives CL, Nathan PD, Brooks JE. Regulation of the BamHI restriction-modification system by a small intergenic open reading frame, bamHIC, in both Escherichia coli and Bacillus subtilis. J. Bacteriol. 1992;174:7194–7201. doi: 10.1128/jb.174.22.7194-7201.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ives CL, Sohail A, Brooks JE. The regulatory C proteins from different restriction-modification systems can cross-complement. J. Bacteriol. 1995;177:6313–6315. doi: 10.1128/jb.177.21.6313-6315.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vijesurier RM, Carlock L, Blumenthal RM, Dunbar JC. Role and mechanism of action of C. PvuII, a regulatory protein conserved among restriction-modification systems. J. Bacteriol. 2000;182:477–487. doi: 10.1128/jb.182.2.477-487.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arcondéguy T, Jack R, Merrick M. P(II) signal transduction proteins, pivotal players in microbial nitrogen control. Microbiol. Mol. Biol. Rev. 2001;65:80–105. doi: 10.1128/MMBR.65.1.80-105.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Merrick MJ, Edwards RA. Nitrogen control in bacteria. Microbiol. Rev. 1995;59:604–622. doi: 10.1128/mr.59.4.604-622.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Heeswijk W, Kuppinger O, Merrick M, Kahn D. Localization of the glnD gene on a revised map of the 200-kilobase region of the Escherichia coli chromosome. J. Bacteriol. 1992;174:1702–1703. doi: 10.1128/jb.174.5.1702-1703.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schlüter A, Nöhlen M, Krämer M, Defez R, Priefer UB. The Rhizobium leguminosarum bv. viciae glnD gene, encoding a uridylyltransferase/uridylyl-removing enzyme, is expressed in the root nodule but is not essential for nitrogen fixation. Microbiology. 2000;146:2987–2996. doi: 10.1099/00221287-146-11-2987. [DOI] [PubMed] [Google Scholar]
- 46.Perlova O, Nawroth R, Zellermann EM, Meletzus D. Isolation and characterization of the glnD gene of Gluconacetobacter diazotrophicus, encoding a putative uridylyltransferase/uridylyl-removing enzyme. Gene. 2002;297:159–168. doi: 10.1016/s0378-1119(02)00881-8. [DOI] [PubMed] [Google Scholar]
- 47.Wray LV, Ferson AE, Rohrer K, Fisher SH. TnrA, a transcription factor required for global nitrogen regulation in Bacillus subtilis. Proc. Natl. Acad. Sci. USA. 1996;93:8841–8845. doi: 10.1073/pnas.93.17.8841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ortega A, Amorós D, García de la Torre J. Prediction of hydrodynamic and other solution properties of rigid proteins from atomic- and residue-level models. Biophys. J. 2011;101:892–898. doi: 10.1016/j.bpj.2011.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.