Abstract
Oxidative stress-related damage to the DNA macromolecule produces lesions that are implicated in various diseases. To understand damage to DNA, it is important to study the free radical reactions causing the damage. Measurement of DNA damage has been a matter of debate as most of the available methods measure the end product of a sequence of events and provide limited information on the initial free radical formation. We report a measurement of free radical damage in DNA induced by a Cu(II)-H2O2 oxidizing system using immuno-spin trapping supplemented with electron paramagnetic resonance. In this investigation, the short-lived radical generated is trapped by the spin trap 5,5-dimethyl-1-pyrroline N-oxide (DMPO) immediately upon formation. The DMPO adduct formed is initially electron paramagnetic resonance active, but is subsequently oxidized to the stable nitrone adduct, which can be detected and visualized by immuno-spin trapping and has the potential to be further characterized by other analytical techniques. The radical was found to be located on the 2′-deoxyadenosine (dAdo) moiety of DNA. The nitrone adduct was repaired on a time scale consistent with DNA repair. In vivo experiments for the purpose of detecting DMPO–DNA nitrone adducts should be conducted over a range of time in order to avoid missing adducts due to the repair processes.
INTRODUCTION
DNA-centered radicals are implicated as a root cause of carcinogenesis and aging through their initiation of genomic damage. Organisms must maintain the integrity of their DNA in order to remain healthy and propagate. DNA is continuously exposed to exogenous and endogenous mutagens including reactive oxygen and nitrogen species that can alter its integrity (1–3). Indeed, oxidatively generated DNA damage is an inevitable consequence of cellular metabolism. Injury to this macromolecule can have severe biological consequences including mutation, carcinogenic transformation, cell death and reproductive death (4). Considerable attention has been focused on the formation, stability and detection of various DNA damage modifications, which have been widely studied in terms of formation, mutagenesis and repair. Oxidatively generated damage of cellular DNA by free radicals may be a significant factor in human carcinogenesis (5). Extensive mechanistic studies have provided a large amount of information that allows for the identification of the chemical pathways leading to the existence of oxidative stress. The detection and characterization of radical transients are important in understanding the mechanisms involved.
The hydroxyl radical is the most reactive of the several reactive oxygen species and, almost uniquely among free radicals, reacts rapidly with DNA (3). When DNA damage occurs to the extent that it can no longer be repaired, senescence, cell death and carcinogenesis may occur.
Several immunological assays have also been designed for the measurement of oxidatively generated DNA damage (6,7). However, the immunoassays developed to detect DNA damage have been plagued by a lack of specificity. Several attempts have been made to use antibodies raised against 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodGuo, one of the most popular markers of oxidative DNA damage), but it has been difficult to obtain a sufficiently specific antibody. 8-oxodGuo differs from dGuo by a single oxygen atom, which is a challenge for the specificity of the antibody. The applications of this method have been limited by cross reactivity of the antibodies with normal DNA bases and other abundant biological constituents (8). In addition, antibodies to a specific nucleotide oxidation product will not recognize the numerous other oxidation products.
While it is critical to detect and identify the primary DNA radicals in order to understand DNA damage, detection of DNA radicals is difficult due to their short lifetimes. The spin-trapping technique in electron spin resonance (ESR) relies on the addition of the free radical across the double bond of a spin trap to form a more stable adduct than was originally detected by ESR. Although more stable than the original free radical, the radical adduct still has a comparatively short lifetime and can be oxidized to a stable end product, a nitrone, which preserves the chemical bond at the site of high spin density formed by spin trapping. The base, or sugar, where the spin trap is attached can then be determined by MS or by other analytical techniques such as NMR. A method developed in our laboratory, called immuno-spin trapping (IST), detects the stable nitrone after oxidation to combine the specificity of spin trapping with the sensitivity of an antigen–antibody-based assay (9–11).
IST has been very successful in detecting radical adducts in proteins (11–17), DNA (18–21), cells (11,22,23) and tissue (24,25). Its major limitation is that the chemical structure of the free radical is not identified by the IST procedure. Since detection of 5,5-dimethyl-1-pyrroline N-oxide (DMPO)–DNA nitrone adducts is done by dot blots or enzyme-linked immunosorbent assay (ELISA), the identification of a radical as DNA-derived and not a contaminant (protein-derived) depends on the absolute purity of the DNA as achieved by traditional DNA purification methods. The structural identification of DNA-derived radicals detected by ELISA in previous works (18–21,24) is further compromised by the limited investigations of DNA radical adducts by ESR or the corresponding DNA nitrone adducts by mass spectrometry (MS). We have recently been able to combine IST with ESR, MS and MS/MS to detect and identify a DNA radical adduct (26).
In this work we used the sensitive IST technique to detect, visualize and identify primary DNA radical damage free of artifacts because additional nitrone adduct formation is impossible once the DMPO is diluted by DNA extraction, and subsequent nitrone adduct formation is precluded. Dilution of DMPO to 1 mM is sufficient to prevent artifactual nitrone formation because, at DMPO concentrations below this, DNA radicals will decay before they can be trapped in detectable amounts. Here, we have chosen a Cu(II)-H2O2 oxidizing system to induce hydroxyl-free radical damage to DNA. Copper-induced oxidative damage by H2O2 is attributed to the formation of hydroxyl radical by a mechanism analogous to the iron-catalyzed Haber–Weiss cycle, and a Cu(I) intermediate is formed (27). Copper-mediated damage is significant because it can be toxic as in Wilson’s disease (28), which is a copper overload disorder. Furthermore, there is increasing evidence of elevated levels of copper in tumor growth and angiogenesis (29–31). The hydroxyl radical generated reacts at diffusion-controlled rates with virtually any macromolecule, including DNA. Cu(II) bound to DNA undergoes cyclic reduction. The reducing agent in our system is hydrogen peroxide or superoxide radical anion formed by the Haber–Weiss cycle, which reduces the metallic complex yielding Cu(I)-bound DNA. Subsequently, this reduced state can react with H2O2 in the Fenton reaction, yielding the hydroxyl radical which, in turn, causes site-specific damage to nuclear DNA. We examined this damage using ESR and IST and detected and identified a nitrone adduct on the 2′-deoxyadenosine moiety in cellular DNA (26). With confocal microscopy, we were also able to visualize these DNA radical adducts at the nucleus and see that the free radical formation was significantly greater in the nuclear fraction than in the cytosolic fraction. Finally, we examined the effect of DNA damage repair on the DMPO–DNA nitrone adduct and investigated the inhibition of this repair mechanism by using known inhibitors of excision repair pathways.
MATERIALS AND METHODS
Reagents
2′-Deoxycytidine (dCyd), 2′-deoxyguanosine (dGuo), thymidine (Thd) and 2′-deoxyadenosine (dAdo) were purchased from MP Biomedicals (Irvine, CA, USA). Calf thymus DNA was obtained from Sigma Aldrich (St Louis, MO, USA). Methoxyamine (TRC 102) was obtained from Supelco (Bellefonte, PA, USA) and nickel chloride was from Alfa Aesar (Ward Hill, MA, USA). The spin trap DMPO was purchased from Dojindo (Rockville, MD, USA). The DMPO concentration was measured at 228 nm, assuming a molar absorption co-efficient of 7800 M−1 cm−1. Hydrogen peroxide was obtained from Fisher Scientific Company (Fairlawn, NJ, USA). The hydrogen peroxide concentration was verified using UV absorption at 240 nm (ε240 nm = 43.6 M−1 cm−1). All buffers used were treated with Chelex 100 ion exchange resin (Bio-Rad Laboratories, Hercules, CA, USA) to avoid transition metal-catalyzed reactions.
Chemical reactions
Production of the DNA–nitrone adducts in nucleosides and calf thymus DNA
Typically, reaction mixtures contained 2.5–5 mM nucleoside, 300 μM CuCl2, 100 μM H2O2 and 100 mM DMPO in 100 mM Chelex-treated phosphate buffer (pH 7.4) and were incubated for 1 h at 37°C. For 2′-deoxyguanosine experiments, a saturated solution of the nucleoside was used.
Production of the DNA–nitrone adducts in RAW 264.7 cells
The mouse macrophage cell line RAW 264.7 was grown and maintained in Dulbecco’s modified eagle medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum with penicillin and streptomycin (10 000 units penicillin and 10 mg streptomycin per ml in 0.9% NaCl). Upon reaching 80% confluence, the cells were counted and seeded at the needed density. Exponentially growing RAW cells (1–3 million/well) were seeded into 12-well plates and treated with 100 µM CuCl2/100 µM H2O2 in the presence of DMPO for 12–15 h at 37°C in a humidified 5% CO2 environment. After this treatment, cell viability was determined using the tryphan blue exclusion assay. The cells were pelleted and frozen at −80°C until DNA extraction and analysis of the DMPO nitrone adducts.
Cell viability
Exponentially growing RAW cells (1–3 million/well) were seeded into 12-well plates and treated with 100 µM CuCl2/100 µM H2O2 in the presence of DMPO. After treatment, the number of viable cells was determined by tryphan blue dye exclusion and counted manually with a hemocytometer.
Nuclear and cytosolic fractionation
Nuclear/cytosolic fractionation was carried out with treated and untreated cells using a nuclear/cytosolic fractionation kit from BioVision (Mountain View, CA, USA).
Formation and repair of DMPO–DNA nitrone adducts
Exponentially growing RAW cells (1–3 million/well) were seeded into 12-well plates and treated with 100 µM CuCl2 and 100 µM H2O2 in the presence of DMPO. At designated time points, treated cells were harvested and stored at −80°C until DNA extraction and analysis of the DMPO nitrone adducts.
Preparation of cellular lysates for repair studies of DMPO–DNA nitrone adducts
Exponentially growing RAW cells at 80–90% confluence were harvested in ice-cold lysis buffer to release soluble cellular proteins. The cells were scraped from the plates, transferred to centrifuge tubes and placed on ice to ensure efficient lysis. Lysates were centrifuged at 10 000 rpm for 5 min. The supernatant was pooled and added to a prepared DMPO–DNA nitrone adduct reaction mixture.
Preparation of cellular lysates for inhibition of the repair studies of DMPO–DNA nitrone adducts
Two known inhibitors, methoxyamine (TRC 102) of the base excision repair (BER) pathway and NiCl2 of the nucleotide excision repair (NER) pathway, were each added to exponentially growing cells and incubated for 20–24 h. The cells were then harvested and lysed as was done for the repair study above.
DNA extraction procedure
Cell pellets stored at −80°C were re-suspended in digestion buffer with proteinase K, and DNA extraction was carried out as described previously (18,19). The extraction procedure preserves the nitrone adduct covalently bound to the DNA (18,19). DNA concentration and purity were measured from the absorbance at 260 and 280 nm. Pure DNA will exhibit an A260/A280 ratio between 1.8 and 2.
IST analysis by ELISA
A standard ELISA procedure in 96-well plates (Greiner Labortechnik, Frickenhausen, Germany) was used as described in detail (18). Briefly, DNA solutions were diluted to 5 μg ml−1 in 1× PBS. A volume of 25 μl of the DNA solution and 25 μl of Reacti-Bind DNA coating solution were placed in each well of the plate and the plate incubated for 2–4 h at 37°C. The plates were washed once with washing buffer (1× PBS containing 0.05% non-fat dry milk and 0.1% Tween-20) and blocked with blocking buffer (1× PBS containing 3% non-fat dry milk) for 2 h at 37°C or overnight at 4°C. After washing once with the washing buffer, the rabbit anti-DMPO serum (diluted 1:10 000 in the wash buffer) was added and incubated at 37°C for 60 min. After three more washes, 100 μl of the secondary antibody (HRP-conjugated), diluted 1:10 000 in the wash buffer, was added and incubated for 60 min. After three washes, the antigen–antibody complexes were developed using the Immobilon chemiluminescence substrate (Millipore Corp., Billerica, MA, USA) and the light emitted was recorded as arbitrary light units using X-Fluor software (Tecan US, Research Triangle Park, NC, USA).
ESR spin-trapping experiments
Upon mixing, the reaction mixture was immediately transferred to a flat cell and the ESR scan initiated within 1 min after starting the reaction. ESR spectra were obtained with an ELEXSYS E500 ESR spectrometer (Bruker Biospin, Billerica, MA, USA) equipped with an ER4122SHQ cavity operating at 9.76 GHz and at room temperature. The ESR spectrometer settings were as follows: scan range, 100 G; modulation frequency, 100 kHz; modulation amplitude, 1.0 G; microwave power, 20 mW; receiver gain, 2 × 104; time constant, 327 ms; and conversion time, 327 ms.
Confocal microscopy
For confocal microscopy studies, treated RAW cells were allowed to adhere to polylysine-coated plates (MatTek) for 30 min on ice. The cells were fixed with 0.4% paraformaldehyde and permeabilized with 0.01% Surfact-Amps X-100 (Thermo Scientific). After blocking with 0.1% BSA in PBS, DMPO nitrone adducts were stained using anti-DMPO (raised in rabbit) antisera. After 1 h incubation, the cells were washed and the secondary Alexaflour anti-rabbit antibody conjugated with fluorescein (Molecular Probes Invitrogen) was added.
RESULTS
ESR spin trapping of a radical with a single nucleoside and a mixture of nucleosides
ESR studies were carried out with each of the 2′-deoxyribonucleosides (dGuo, dAdo, Thd and dCyt). When 5 mM (or saturated, for dGuo) solutions of the nucleosides reacted with CuCl2 (300 μM), H2O2 (100 μM) and DMPO (100 mM), a radical adduct was detected only with 2′-deoxyadenosine (26).
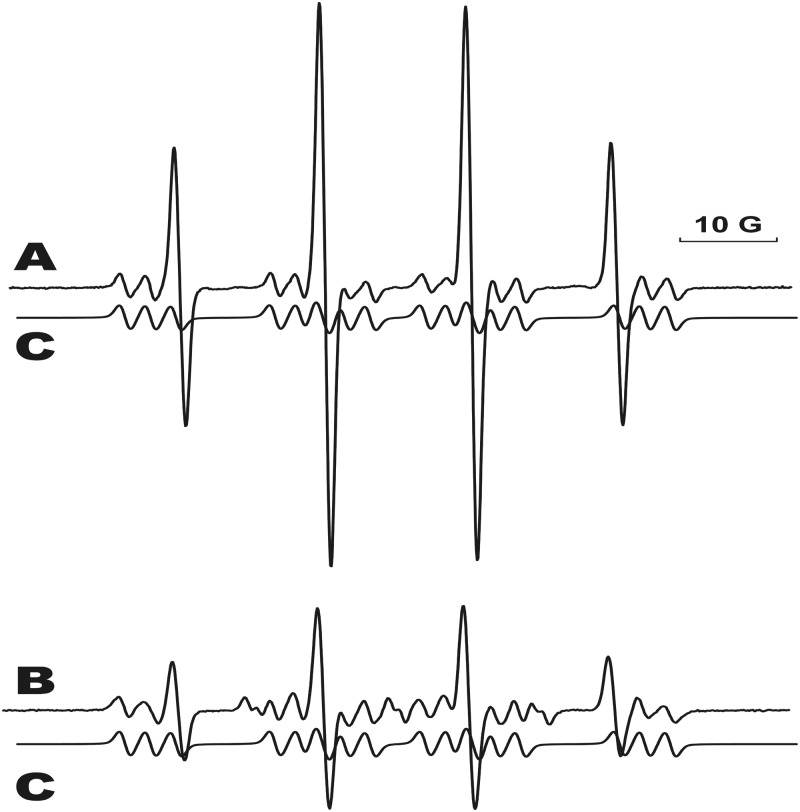
When calf-thymus DNA reacted with CuCl2 (300 μM), H2O2 (100 μM) and 100 mM DMPO, no assignable electron paramagnetic resonance (EPR) spectra were detected (26). Our attempts to digest the DNA to obtain motionally narrowed, sharper isotropic spectra were not successful. Therefore, in order to try to mimic DNA, we treated an equimolar mixture of adenosine, thymidine, cytidine and guanosine with CuCl2 (300 μM), H2O2 (100 μM) and 100 mM DMPO. A spectrum was obtained of a dAdo-derived, nitrogen-centered DMPO radical adduct (Figure 1A) nearly identical to that obtained from the reaction of only dAdo under similar circumstances (26). The presence of dAdo and all other reaction components (CuCl2, H2O2 and DMPO) was necessary for radical adduct formation. Figure 1 shows the comparison of the adduct obtained from the four-nucleoside mixture with the simulated spectra obtained from the dAdo-only reaction mixture (Figure 1B).
Figure 1.
Formation of the radical-derived DMPO adduct from the reaction of an equimolar mixture of all four DNA nucleosides with CuCl2/H2O2 in the presence of DMPO. All components of the reaction must be present for the adduct formation. (A) The mixture contained 1 mM of all four DNA nucleosides, CuCl2 (300 μM), H2O2 (100 μM) and DMPO (100 mM) and was incubated at 37°C for 1 h. Instrumental conditions: modulation amplitude, 1.0 G; time constant, 327 ms; receiver gain, 2 × 104; and microwave power, 20 mW. (B) The EPR spectrum of the dAdo-DMPO radical derived from the reaction of adenosine with CuCl2/H2O2 in the presence of DMPO. (C) The computer simulation of the dAdo-DMPO adduct.
IST analysis of DMPO nitrone adducts
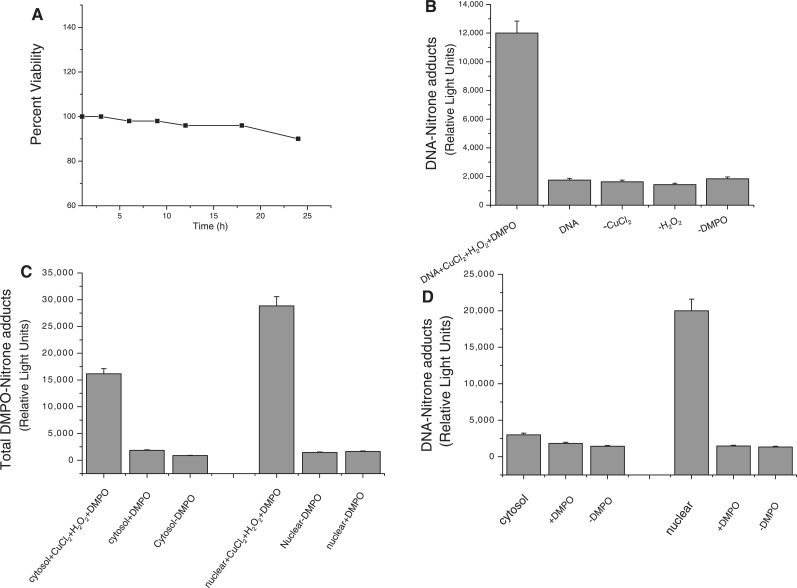
RAW 264.7 macrophage cells were treated with CuCl2 and H2O2 and DNA extracted as outlined earlier (18,19). Viability assays showed that the cells were ∼90% viable even at 22 h post-treatment (Figure 2A). It is known that DNA radicals can be detected in DNA-extracted cells using an anti-DMPO antibody in ELISA experiments (18,22). Here, production of the DNA nitrone adducts required the presence of all components of the reaction mixture (Figure 2B). In order to determine the localization of the DMPO nitrone adducts, we separated the cytosolic and the nuclear fractions using a nuclear/cytosolic fractionation kit from BioVision and analyzed them by ELISA (Figure 2C). Clearly, the nuclear fraction had a higher level of DMPO nitrone adducts. In order to determine whether the observed chemiluminescence was due to DNA adducts and not protein (specifically histone in the nuclear fraction), DNA extraction was carried out from both fractions and concentration determined. The resulting ELISA showed a substantial increase in nitrone adduct in the nuclear fraction as compared to the cytosolic fraction (Figure 2D). No protein–DMPO adducts were detected in Western blots of either the cytosolic or the nuclear fractions (data not shown).
Figure 2.
Immunological detection of the DNA-derived radical adduct from the reaction of cellular DNA obtained from RAW cells treated with CuCl2 and H2O2 in the presence of DMPO at 37°C for 1 h. (A) Shows the time course of cell viability. (B) All components of the reaction must be present for adduct formation. (C) The cytosolic and the nuclear fractions of the RAW cells were separated and the total DMPO adducts (both protein and DNA) determined in both fractions. (D) DNA was extracted from both the cytosolic and the nuclear fractions, concentration determined and DNA–DMPO adduct determined.
Confocal microscopy
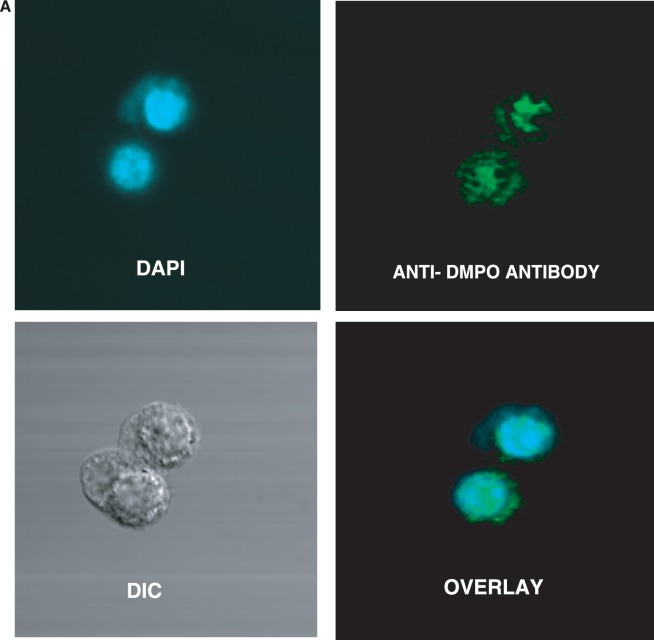
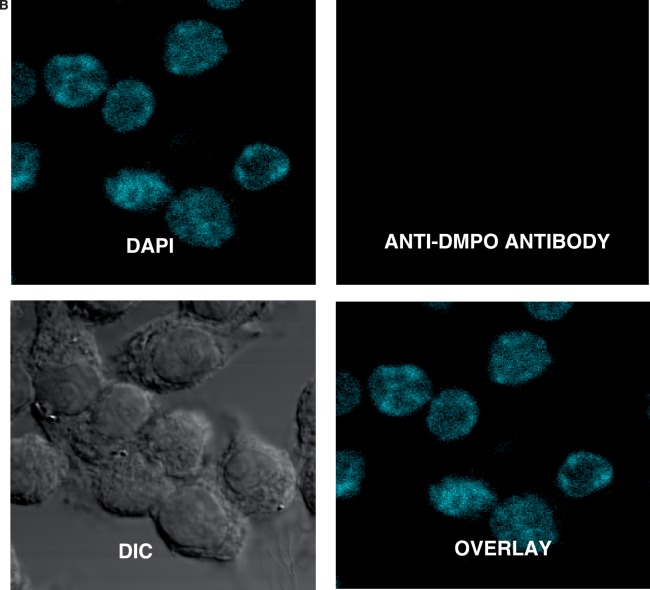
Parallel confocal fluorescence microscopy experiments were also performed to visually determine the localization of the nitrone adducts. After fixation and permeabilization, cells were stained with rabbit anti-DMPO serum followed by Alexa 567 conjugated anti-rabbit antisera to stain the DMPO nitrone adducts. The data showed that the cells exposed to CuCl2/H2O2 had a fluorescent signal localized primarily in the nucleus (Figure 3A). In the absence of CuCl2, H2O2 and/or DMPO (Figure 3B), no such fluorescence was detected. These data indicate that the radical formation is largely localized to the nucleus and is generated by the reaction of copper with H2O2.
Figure 3.
Confocal laser scanning microscopy of DNA-derived radical adducts in RAW 264.7 cells. Adherent RAW 264.7 cells in glass-bottomed 32 mm2 dishes were treated with CuCl2/H2O2 in the presence of 25 mM DMPO for 1 h. Cells were fixed with paraformaldehyde and stained with the anti-DMPO antibody, which was detected with an Alexa 488 conjugated secondary antibody. (A) Cells were imaged and anti-DMPO immunoreactivity (green stain) could be seen primarily in the nucleus (DAPI, blue stain), as evidenced by the co-localization (overlay image). (B) There was no DMPO immunoreactivity in cells that were not treated with the spin trap.
Repair of DMPO–DNA nitrone adducts
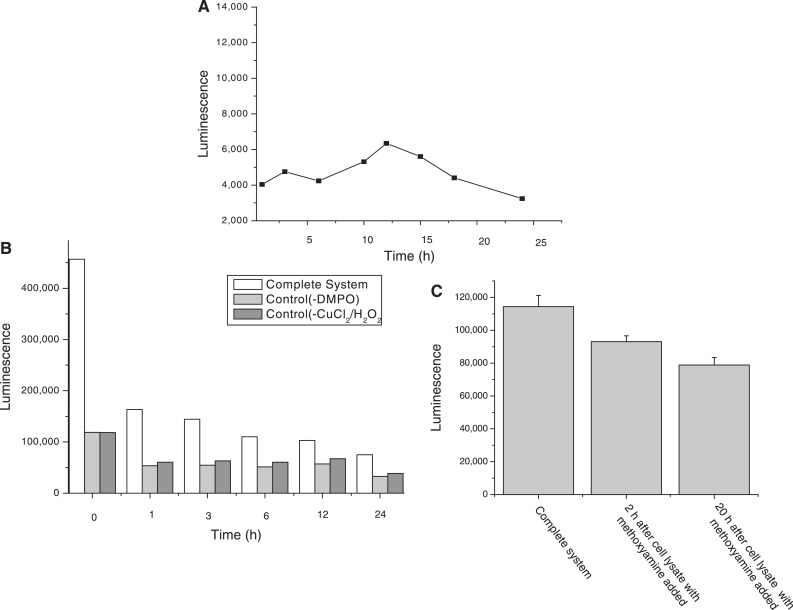
The formation and repair of DMPO–DNA nitrone adducts were assessed over a 24-h time course. Figure 4A shows a time profile of the formation of the adduct. The DMPO–DNA nitrone adduct formed within 30 min, then increased slowly over the next several hours before reaching an adduct maximum at ∼12 h. Thereafter, apparent repair of the DMPO–DNA nitrone adduct occurred, and the adduct formation then dropped to a value of ∼50% of the maximum value. As there exists the possibility of cell replication, causing dilution of the DNA–DMPO due to DNA synthesis during this time frame, we further tested the direct repair of the DMPO–DNA nitrone adduct formation. Preformed DNA–DMPO adduct from calf thymus treated with 300 µM CuCl2 and100 µM H2O2 in the presence of 100 mM DMPO was prepared, and cell lysate from the RAW 267.4 cells added. The DMPO–DNA nitrone adduct was analyzed over a 24-h time frame. The DNA–DMPO adduct formed was clearly repaired over a relatively short period of time (Figure 4B).
Figure 4.
Repair of DNA–DMPO adduct. (A) The DNA–DMPO adduct resulting from treatment of the RAW 264.7 cells with CuCl2 and H2O2 in the presence of DMPO forms within 30 min of treatment, then slowly increases before reaching a maximum at 12 h with a subsequent drop in the adduct level. (B) The DMPO–DNA nitrone adduct was repaired in a few hours upon the addition of the cell lysate to the reaction mixture. (C) The repair of the preformed DMPO–DNA nitrone adduct was significantly inhibited upon the addition of the cell lysate containing methoxyamine even after 20 h.
Inhibition of the repair of DMPO–DNA nitrone adducts
Cell lysates containing inhibitors of the BER and NER pathways were added to preformed DNA adducts (from calf thymus treated with 300 µM CuCl2 and100 µM H2O2 in the presence of DMPO), and repair observed in the above experiment was inhibited when the methoxyamine was added to cell lysate (Figure 4C). NiCl2 added to cell lysate did not have this effect, indicating that the repair of this damage is predominantly by the BER pathway although the repair pathways are overlapping.
DISCUSSION
The measurement of oxidized bases and nucleosides in DNA may be used to gain insights into the nature and importance of chemical reactions generated in DNA by oxidizing agents. Significant improvement for the measurement of oxidatively generated damage to DNA has been obtained by the use of HPLC coupled to MS and MS/MS (3). This method has been gaining prominence and has overcome many of the limitations that previously hindered the measurement of oxidatively generated DNA damage (3). It now represents the usual method of choice for identifying modified DNA bases (1,2,32).
IST, which can be combined with MS and MS/MS, is a relatively new method for studying DNA radical damage (18–21). Our recent investigations with EPR and with IST with MS and MS/MS have allowed the characterization of nitrone adducts formed in Fenton systems by the hydroxyl radical (26). The objective of the present work was to determine the time course of formation and repair of free radical-derived DNA nitrone adduct, and to use confocal microscopy to visualize the damage to the DNA in the nucleus. Separation of cytosolic and nuclear fractions and subsequent DNA purification demonstrated the presence of DNA–DMPO adduct, presumably of the known DMPO/N6-centered adenosine nitrone adduct in the nucleus (26). MS and MS/MS confirmed the DMPO/N6-centered adenosine in the nuclear fraction (data not shown).
Using ESR, MS and MS/MS, we determined that the radical trapped by DMPO was located at N6 of the adenosine (26). Hydroxyl radical can add to the double bond at C4, C5 and C8 of the adenosine (33–37), of which C4 constitutes the most abundant addition site (81%) (11). This adduct undergoes proton or hydrogen transfer, followed by dehydration, to form an N6-dehydrogenated radical. N6 has also been shown elsewhere to be the most likely site of hydrogen abstraction by hydroxyl radical (38,39). However, the absence of detection of other DNA radicals is not proof of their absence. It should be noted that this type of site-specific hydroxyl radical generation should not form as many DNA-derived radicals as ionizing radiation where freely diffusing hydroxyl radical is formed.
When the RAW 267.4 cells were treated with CuCl2 and H2O2, the radical generated was trapped by DMPO immediately upon formation to form a DMPO nitrone adduct. The DMPO radical adduct formed is initially EPR active but subsequently decays to the more stable nitrone adduct, which can be very conveniently detected by the IST method (20,26,30) and confirmed and identified by MS and MS/MS. The radical is trapped spontaneously by DMPO, and artifactual generation of DMPO adducts during the extraction and digestion steps is minimized, if not eliminated. Furthermore, as it is a DMPO adduct that is formed, its identity can be confirmed by MS/MS from its fragmentation to DMPO and the parent radical ion, thereby eliminating the requirement of a pure product for identification.
It should be recognized that the oxidation event does not have to take place inside the cell nucleus, but may occur either within the cytosol or even in the extracellular compartment. However, the damage inflicted by CuCl2/H2O2 Fenton chemistry to the RAW 267.4 macrophages was indeed found to be largely localized to the nuclear DNA. This localization was evident from the confocal data as well as from the immunochemical detection of the cytosolic and nuclear fractions.
We also examined the persistence of the DMPO–DNA adducts formed. When the adducts were monitored over a period of time, they formed detectable amounts within 30 min, then increased slowly over the next several hours before reaching a maximum at ∼12 h. Thereafter, repair of the DMPO–DNA nitrone adduct apparently dominates, accompanied by a drop in the adduct level to a value of ∼50% of the maximum. However, because this effect could also be caused by turnover due to DNA synthesis, we also studied the repair of the preformed DNA–DMPO adduct added to cell lysate. Preformed DMPO–DNA nitrone repair by treatment with cell lysate was largely complete within an hour. Clearly, the DMPO–DNA nitrone adduct concentrations detected were the result of continuous hydroxyl radical attack on DNA balanced against DNA repair processes. DNA repair enzymes modify DNA damage including removal of DNA adducts, and there are multiple and overlapping DNA repair pathways (40). Inhibition of the repair pathways will block this mechanism. We used two known inhibitors, namely methoxyamine (TRC 102) of the BER pathway (41) and NiCl2 of the NER pathway (42). Methoxyamine is a small molecule repair inhibitor (41) and inhibited the repair of the single-base lesions in preformed DMPO–DNA nitrones.
In summary, with our method of combining IST with ESR, we were able to confirm that DMPO traps a radical adduct from DNA in accordance with published ELISA studies (18–21). This methodology provides an unequivocal assignment of the radical formed in DNA damage induced by the hydroxyl radical in a copper/hydrogen peroxide Fenton system (26). The radical produced when cellular DNA was treated with a Fenton-like system was found to be at the adenosine moiety, which may be an intermediate in a sequence of events leading to the formation of 8-oxo-dGuo (43) and other products such as 8-oxo-7,8-dihydro-2′-deoxyadenosine (8-oxodAdo). Although speculative for us, it has been predicted by hole-trapping researchers that the adenine radical cation contributes to the hole-transfer process through A/T sequences and exists as a real chemical intermediate (44–48). One-electron oxidation of DNA results in migration and localization of the electron loss center to guanine (43).
The damage was found to be localized in the nuclear DNA, but was repaired over a period of time, probably through the BER pathway. The repair study is preliminary in nature and makes clear the interfering effect of DNA repair in a biological system. It is necessary that in vivo experiments studying DMPO–DNA nitrone adducts be conducted over a range of times as repair of the DMPO–DNA nitrone adduct does occur.
FUNDING
Funding for open access charge: Intramural Research Program of the National Institutes of Health and the National Institute of Environmental Health Sciences.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We gratefully acknowledge Jean Corbett for her unconditional help during all stages of the project. We are very thankful to Dr. Marilyn Ehrenshaft and Dr. Fiona Summers for reviewing the manuscript. We thank Dr. Ann Motten and Mary Mason for their invaluable help in preparing this manuscript.
REFERENCES
- 1.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Ames BN, Shigenaga MK, Gold LS. DNA lesions, inducible DNA repair, and cell division: three key factors in mutagenesis and carcinogenesis. Environ. Health Perspect. 1993;101(Suppl 5):35–44. doi: 10.1289/ehp.93101s535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cadet J, Douki T, Gasparutto D, Ravanat JL. Oxidative damage to DNA: formation, measurement and biochemical features. Mutat. Res. 2003;531:5–23. doi: 10.1016/j.mrfmmm.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Halliwell B, Gutteridge JMC. Free radicals in biology and medicine. New York: Oxford University Press, Inc.; 1999. [Google Scholar]
- 5.Beckman KB, Ames BN. Oxidative decay of DNA. J. Biol. Chem. 1997;272:19633–19636. doi: 10.1074/jbc.272.32.19633. [DOI] [PubMed] [Google Scholar]
- 6.Musarrat J, Wani AA. Quantitative immunoanalysis of promutagenic 8-hydroxy-2'-deoxyguanosine in oxidized DNA. Carcinogenesis. 1994;15:2037–2043. doi: 10.1093/carcin/15.9.2037. [DOI] [PubMed] [Google Scholar]
- 7.Toyokuni S, Tanaka T, Hattori Y, Nishiyama Y, Yoshida A, Uchida K, Hiai H, Ochi H, Osawa T. Quantitative immunohistochemical determination of 8-hydroxy-2'-deoxyguanosine by a monoclonal antibody N45.1: its application to ferric nitrilotriacetate-induced renal carcinogenesis model. Lab. Invest. 1997;76:365–374. [PubMed] [Google Scholar]
- 8.Cooke MS. A commentary on "Urea, the most abundant component in urine, cross-reacts with a commercial 8-OH-dG ELISA kit and contributes to overestimation of urinary 8-OH-dG". What is ELISA detecting? Free Radic. Biol. Med. 2009;47:30–31. doi: 10.1016/j.freeradbiomed.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 9.Detweiler CD, Deterding LJ, Tomer KB, Chignell CF, Germolec D, Mason RP. Immunological identification of the heart myoglobin radical formed by hydrogen peroxide. Free Radic. Biol. Med. 2002;33:364–369. doi: 10.1016/s0891-5849(02)00895-x. [DOI] [PubMed] [Google Scholar]
- 10.Mason RP. Using anti-5,5-dimethyl-1-pyrroline N-oxide (anti-DMPO) to detect protein radicals in time and space with immuno-spin trapping. Free Radic. Biol. Med. 2004;36:1214–1223. doi: 10.1016/j.freeradbiomed.2004.02.077. [DOI] [PubMed] [Google Scholar]
- 11.Ramirez DC, Chen YR, Mason RP. Immunochemical detection of hemoglobin-derived radicals formed by reaction with hydrogen peroxide: involvement of a protein-tyrosyl radical. Free Radic. Biol. Med. 2003;34:830–839. doi: 10.1016/s0891-5849(02)01437-5. [DOI] [PubMed] [Google Scholar]
- 12.Ehrenshaft M, Mason RP. Protein radical formation on thyroid peroxidase during turnover as detected by immuno-spin trapping. Free Radic. Biol. Med. 2006;41:422–430. doi: 10.1016/j.freeradbiomed.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 13.Deterding LJ, Ramirez DC, Dubin JR, Mason RP, Tomer KB. Identification of free radicals on hemoglobin from its self-peroxidation using mass spectrometry and immuno-spin trapping: observation of a histidinyl radical. J. Biol. Chem. 2004;279:11600–11607. doi: 10.1074/jbc.M310704200. [DOI] [PubMed] [Google Scholar]
- 14.Bhattacharjee S, Deterding LJ, Jiang J, Bonini MG, Tomer KB, Ramirez DC, Mason RP. Electron transfer between a tyrosyl radical and a cysteine residue in hemoproteins: spin trapping analysis. J. Am. Chem. Soc. 2007;129:13493–13501. doi: 10.1021/ja073349w. [DOI] [PubMed] [Google Scholar]
- 15.Bonini MG, Siraki AG, Bhattacharjee S, Mason RP. Glutathione-induced radical formation on lactoperoxidase does not correlate with the enzyme's peroxidase activity. Free Radic. Biol. Med. 2007;42:985–992. doi: 10.1016/j.freeradbiomed.2006.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ranguelova K, Suarez J, Magliozzo RS, Mason RP. Spin trapping investigation of peroxide- and isoniazid-induced radicals in Mycobacterium tuberculosis catalase-peroxidase. Biochemistry. 2008;47:11377–11385. doi: 10.1021/bi800952b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chatterjee S, Ehrenshaft M, Bhattacharjee S, Deterding LJ, Bonini MG, Corbett J, Kadiiska MB, Tomer KB, Mason RP. Immuno-spin trapping of a post-translational carboxypeptidase B1 radical formed by a dual role of xanthine oxidase and endothelial nitric oxide synthase in acute septic mice. Free Radic. Biol. Med. 2009;46:454–461. doi: 10.1016/j.freeradbiomed.2008.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramirez DC, Mejiba SE, Mason RP. Immuno-spin trapping of DNA radicals. Nat. Methods. 2006;3:123–127. doi: 10.1038/nmeth852. [DOI] [PubMed] [Google Scholar]
- 19.Ramirez DC, Gomez-Mejiba SE, Mason RP. Immuno-spin trapping analyses of DNA radicals. Nat. Protoc. 2007;2:512–522. doi: 10.1038/nprot.2007.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kojima C, Ramirez DC, Tokar EJ, Himeno S, Drobná Z, Stýblo M, Mason RP, Waalkes MP. Requirement of arsenic biomethylation for oxidative DNA damage. J. Natl. Cancer Inst. 2009;101:1670–1681. doi: 10.1093/jnci/djp414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogusucu R, Rettori D, Netto LE, Augusto O. Superoxide dismutase 1-mediated production of ethanol- and DNA-derived radicals in yeasts challenged with hydrogen peroxide: molecular insights into the genome instability of peroxiredoxin-null strains. J. Biol. Chem. 2009;284:5546–5556. doi: 10.1074/jbc.M805526200. [DOI] [PubMed] [Google Scholar]
- 22.Bonini MG, Siraki A, Atanassov BS, Mason RP. Immunolocalization of hypochlorite-induced, catalase-bound free radical formation in mouse hepatocytes. Free Radic. Biol. Med. 2007;42:530–540. doi: 10.1016/j.freeradbiomed.2006.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gomez-Mejiba SE, Zhai Z, Gimenez MS, Ashby MT, Chilakapati J, Kitchin K, Mason RP, Ramirez DC. Myeloperoxidase-induced genomic DNA-centered radicals. J. Biol. Chem. 2010;285:20062–20071. doi: 10.1074/jbc.M109.086579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cassina P, Cassina A, Pehar M, Castellanos R, Gandelman M, deLeón A, Robinson KM, Mason RP, Beckman JS, Barbeito L, et al. Mitochondrial dysfunction in SOD1(G93A)-bearing astrocytes promotes motor neuron degeneration: prevention by mitochondrial-targeted antioxidants. J. Neurosci. 2008;28:4115–4122. doi: 10.1523/JNEUROSCI.5308-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chatterjee S, Lardinois O, Bhattacharjee S, Tucker J, Corbett J, Deterding L, Ehrenshaft M, Bonini M, Mason RP. Oxidative stress induces protein and DNA radical formation in follicular dendritic cells of the germinal center and modulates its cell death patterns in late sepsis. Free Radic. Biol. Med. 2011;50:988–999. doi: 10.1016/j.freeradbiomed.2010.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhattacharjee S, Deterding LJ, Chatterjee S, Jiang J, Ehrenshaft M, Lardinois O, Ramirez DC, Tomer KB, Mason RP. Site-specific radical formation in DNA induced by the Cu(II)-H2O2 oxidizing system, using ESR, immuno-spin trapping, LC-MS, and MS/MS. Free Radic. Biol. Med. 2011;50:1536–1545. doi: 10.1016/j.freeradbiomed.2011.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanna PM, Mason RP. Direct evidence for inhibition of free radical Formation from Cu(I) and hydrogen peroxide by glutathione and other potential ligands using the EPR spin-trapping technique. Arch. Biochem. Biophys. 1992;295:205–213. doi: 10.1016/0003-9861(92)90507-s. [DOI] [PubMed] [Google Scholar]
- 28.Roberts EA, Schilsky ML. Diagnosis and treatment of Wilson disease: an update. Hepatology. 2008;47:2089–2111. doi: 10.1002/hep.22261. [DOI] [PubMed] [Google Scholar]
- 29.Gupte A, Mumper RJ. Elevated copper and oxidative stress in cancer cells as a target for cancer treatment. Cancer Treat. Rev. 2009;35:32–46. doi: 10.1016/j.ctrv.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 30.Tisato F, Marzano C, Porchia M, Pellei M, Santini C. Copper in diseases and treatments, and copper-based anticancer strategies. Med. Res. Rev. 2010;30:708–749. doi: 10.1002/med.20174. [DOI] [PubMed] [Google Scholar]
- 31.Wang F, Jiao P, Qi M, Frezza M, Dou QP, Yan B. Turning tumor-promoting copper into an anti-cancer weapon via high-throughput chemistry. Curr. Med. Chem. 2010;17:2685–2698. doi: 10.2174/092986710791859315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cadet J, Douki T, Frelon S, Sauvaigo S, Pouget JP, Ravanat JL. Assessment of oxidative base damage to isolated and cellular DNA by HPLC-MS/MS measurement. Free Radic. Biol. Med. 2002;33:441–449. doi: 10.1016/s0891-5849(02)00820-1. [DOI] [PubMed] [Google Scholar]
- 33.Cheng Q, Gu J, Compaan KR, Schaefer HF. Hydroxyl radical reactions with adenine: reactant complexes, transition states and product complexes. Chem. Eur. J. 2010;16:11848–11858. doi: 10.1002/chem.201001236. [DOI] [PubMed] [Google Scholar]
- 34.Steenken S. Purine-bases, nucleosides and nucleotides—aqueous-solution redox chemistry and transformation reactions of their radical cations and E- and OH adducts. Chem. Rev. 1989;89:503–520. [Google Scholar]
- 35.Vieira AJSC, Steenken S. Pattern of OH radical reaction with adenine and its nucleosides and nucleotides—characterization of 2 types of isomeric OH adduct and their unimolecular transformation reaction. J. Amer. Chem. Soc. 1990;112:6986–6994. [Google Scholar]
- 36.Vieira AJSC, Steenken S. Pattern of OH radical reaction with 6- and 9- substituted purines. Effect of substituents on the rates and activation parameters of unimolecular transformation reactions of two isomeric OH adducts. J. Phys.Chem. 1987;91:4138–4144. [Google Scholar]
- 37.Llano J, Eriksson LA. Oxidation pathways of adenine and guanine in aqueous solution from first principles electrochemistry. Phys. Chem. Chem. Phys. 2004;6:4707–4713. [Google Scholar]
- 38.Naumov S, von Sonntag C. The energetics of rearrangement and water elimination reactions in the radiolysis of the DNA bases in aqueous solution (e−aq and * OH attack): DFT calculations. Radiat. Res. 2008;169:355–363. doi: 10.1667/RR1081.1. [DOI] [PubMed] [Google Scholar]
- 39.Ito T, Kuno S, Uchida T, Fujita S, Nishimoto S. Properties and reactivity of the adenosine radical generated by radiation-induced oxidation in aqueous solution. J. Phys. Chem. B. 2009;113:389–394. doi: 10.1021/jp801976t. [DOI] [PubMed] [Google Scholar]
- 40.Horton JK, Wilson SH. Hypersensitivity phenotypes associated with genetic and synthetic inhibitor-induced base excision repair deficiency. DNA Repair. 2007;6:530–543. doi: 10.1016/j.dnarep.2006.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taverna P, Hwang H, Schupp JE, Radivoyevitch T, Session NN, Reddy G, Zarling DA, Kinsella TJ. Inhibition of base excision repair potentiates iododeoxyuridine-induced cytotoxicity and radiosensitization. Cancer Res. 2003;63:838–846. [PubMed] [Google Scholar]
- 42.Hu W, Feng Z, Tang M. Nickel (II) enhances benzo[a]pyrene diol epoxide-induced mutagenesis through inhibition of nucleotide excision repair in human cells: a possible mechanism for nickel (II)-induced carcinogenesis. Carcinogenesis. 2004;25:455–462. doi: 10.1093/carcin/bgh012. [DOI] [PubMed] [Google Scholar]
- 43.Bamatraf MMM, O'Neill P, Rao BSM. OH radical-induced charge migration in oligodeoxynucleotides. J. Phys. Chem. B. 2000;104:636–642. [Google Scholar]
- 44.Williams TT, Odom DT, Barton JK. Variations in DNA charge transport with nucleotide composition and sequence. J. Am. Chem. Soc. 2000;122:9048–9049. [Google Scholar]
- 45.Henderson PT, Jones D, Hampikian G, Kan Y, Schuster GB. Long-distance charge transport in duplex DNA: the phonon-assisted polaron-like hopping mechanism. Proc. Natl Acad. Sci. USA. 1999;96:8353–8358. doi: 10.1073/pnas.96.15.8353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Giese B. Long-distance electron transfer through DNA. Annu. Rev. Biochem. 2002;71:51–70. doi: 10.1146/annurev.biochem.71.083101.134037. [DOI] [PubMed] [Google Scholar]
- 47.Bixon M, Jortner J. Charge transport in DNA via thermally induced hopping. J. Am. Chem. Soc. 2001;123:12556–12567. doi: 10.1021/ja010018p. [DOI] [PubMed] [Google Scholar]
- 48.Dohno C, Ogawa A, Nakatani K, Saito I. Hole trapping at N6- cyclopropyldeoxyadenosine suggests a direct contribution of adenine bases to hole transport through DNA. J. Am. Chem. Soc. 2003;125:10154–10155. doi: 10.1021/ja035887o. [DOI] [PubMed] [Google Scholar]