Abstract
Adaptation to the host cell environment to efficiently take-over the host cell's machinery is crucial in particular for small RNA viruses like picornaviruses that come with only small RNA genomes and replicate exclusively in the cytosol. Their Internal Ribosome Entry Site (IRES) elements are specific RNA structures that facilitate the 5′ end-independent internal initiation of translation both under normal conditions and when the cap-dependent host protein synthesis is shut-down in infected cells. A longstanding issue is which host factors play a major role in this internal initiation. Here, we show that the functionally most important domain V of the poliovirus IRES uses tRNAGly anticodon stem–loop mimicry to recruit glycyl-tRNA synthetase (GARS) to the apical part of domain V, adjacent to the binding site of the key initiation factor eIF4G. The binding of GARS promotes the accommodation of the initiation region of the IRES in the mRNA binding site of the ribosome, thereby greatly enhancing the activity of the IRES at the step of the 48S initiation complex formation. Moonlighting functions of GARS that may be additionally needed for other events of the virus–host cell interaction are discussed.
INTRODUCTION
Internal Ribosome Entry Site (IRES) elements are specific RNA structures in the 5′-untranslated region (5′-UTR) of mRNAs that recruit components of the cellular translation machinery to govern the cap-independent translation initiation on the RNA. They were initially characterized in the positive-strand genomic RNAs of picornaviruses (1,2) and later also in the RNAs of viruses from other families. The complex picornavirus IRES RNA structures also recruit a variety of other cellular RNA-binding proteins that are thought to be involved in IRES activation and are, at least in part, believed to determine the tropism of picornaviruses to particular mammalian tissues. The translation and replication of poliovirus (PV), a prototype member of the picornaviruses, take place in the gut but can also occur in neuronal cells where it causes degeneration and lysis of cells leading to paralytic poliomyelitis.
The picornavirus IRES-elements are now classified into four types according to their secondary structure: type I [e.g. PV and rhinovirus (HRV)], type II [e.g. encephalomyocarditis virus (EMCV), and foot-and-mouth disease virus (FMDV), type III hepatitis A virus (HAV)] and type IV, the prototype of which is the recently discovered Porcine Teschovirus (PTV) IRES (3–5). The type IV IRESs have a close structural similarity to the IRES of hepatitis C virus (HCV) but differ from the classical picornavirus IRESs (5,6).
The function of type II and IV picornavirus IRES elements has been studied much better than that of types I and III at a molecular level. One of the reasons is that a successful reconstitution of 48S translation initiation complexes from purified components has been attained only for type II and IV IRESs (6–9). In reconstitution assays, researchers have not only identified the minimal set of required canonical initiation factors but also some auxiliary mRNA-binding proteins involved in type II and IV IRES activity (3,4,10).
In contrast, attempts to assemble 48S initiation complexes on IRESs of type I have been unsuccessful. A current explanation is that the PV RNA and RNAs from other type I viruses require a large number of specific mRNA-binding proteins (IRES Trans Acting Factors, ITAFs) to initiate translation. Unlike Type II picornavirus RNAs, the genomic RNAs of type I picornaviruses are poorly and incorrectly translated in rabbit reticulocyte lysate (RRL), a commonly used in vitro-translation extract: addition of HeLa extract to RRL greatly stimulates type I picornavirus translation and abolishes aberrant translation initiation events (11). At present, at least seven ITAFs are known to bind the type I IRESs. These are Poly(C) Binding Protein 2 (PCBP2), SRp20, Upstream of N-Ras (UNR), Polypyrimidine Tract Binding Protein (PTB), La-autoantigen, nucleolin and DRBP76 (12–23). However, the mechanisms of action of these mRNA binding proteins in type I picornavirus RNA translation are not known except for the general assumption that they may modulate the IRES' tertiary structure. Their binding sites are poorly characterized. Somewhat more data is available on PTB and its neuronal version nPTB (24): they are believed to bind at and near the IRES domain V (domV) and affect binding of the key mRNA recruiting factor eIF4G (25).
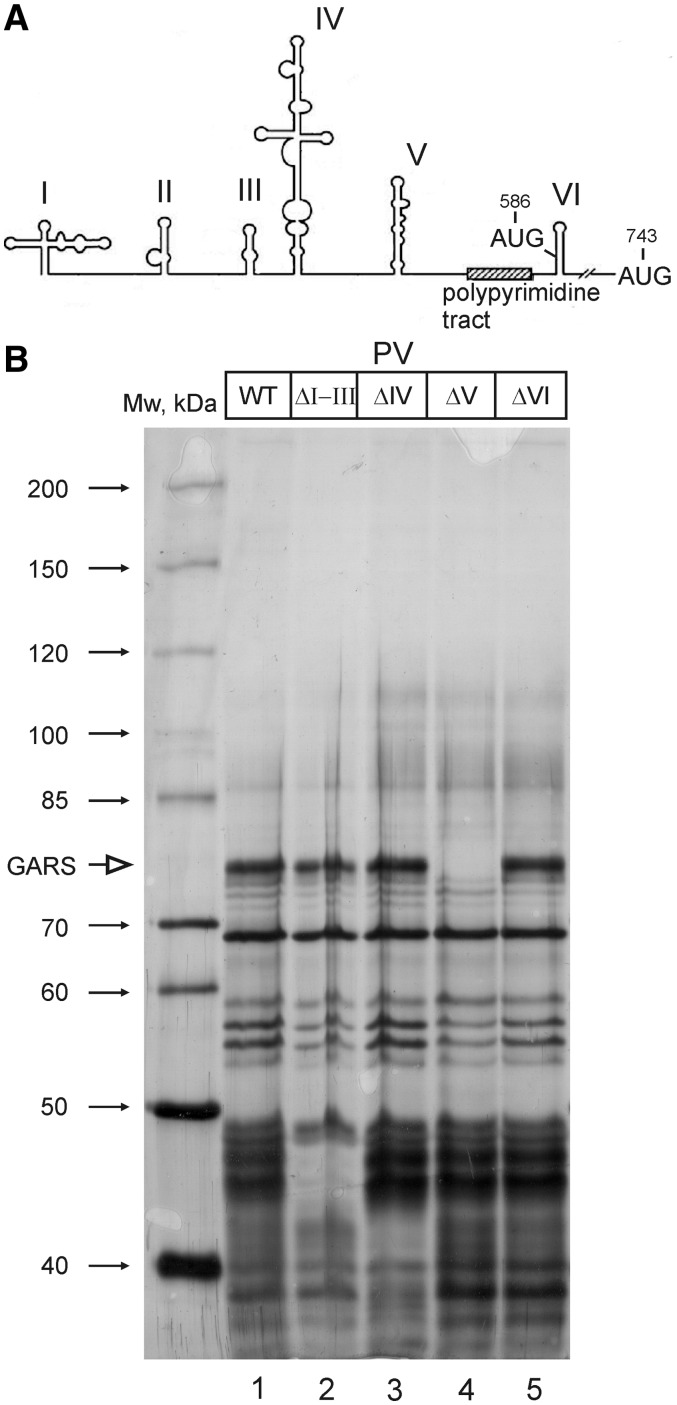
The 5′-UTR of PV RNA contains six stem-looped domains (see Figure 2), of which doms II through VI constitute the IRES. Dom IV and V are thought to play the most important roles in the activity of the PV IRES (3,4).The ‘heart’ of the PV IRES is domV. It binds the complex of eIF4G and eIF4A (26,27), the initiation factors that mediate the entry of the 40S ribosome on the IRES, and eIF4B (28). Mutations within this domain (similar to those in the Sabin live vaccine strains of the oral PV vaccine) attenuate the ability of PV to propagate in neuronal cells (27–33). The current model says that the 40S ribosome loaded with canonical initiation factors and the initiator Met-tRNA enters the PV or rhinovirus IRES at domains V and VI and transiently recognizes the AUG triplet at position 586 of the PV IRES or an equivalent position of rhinovirus IRES. However, the 40S subunit does not initiate translation at this AUG 586 triplet. Instead, it moves further downstream and initiates at the next AUG (743) at a distance of ∼160 nt in enteroviruses and 35 nt in rhinoviruses (34).
Figure 2.
GARS binds to domain V of poliovirus IRES RNA. (A) The secondary structure of the entire 5′-UTR of poliovirus RNA. Stem–loop domains are numbered by Roman numbers. The PV IRES covers the region from domII through domVI. The positions of the non-initiator (AUG586) and authentic (AUG743) start codons (see text) as well as of a polypyrimidine tract characteristic of picornavirus IRES elements are also shown. (B) Proteins interacting with the wt 5′-UTR of poliovirus RNA and the 5′-UTRs with deleted individual domains (ΔI–III, ΔIV, ΔV or ΔVI). The position of GARS is indicated by an arrow.
In spite of the ample literature on the type I IRESs (3,4), we have not yet a clear idea about the events that occur in the course of ribosome entry onto these IRES elements. Moreover, we may not even be sure that we have already identified all participants of the internal entry since no systematic screening for factors involved in this process has ever been performed. That is why we decided to come back to this problem and perform such a screening. As a result, we identified a new essential component that is required for PV RNA translation, glycyl-tRNA synthetase (GARS). Our data show that this house-keeping enzyme binds to the key domain of PV IRES, domV. The binding is highly specific owing to the fact that the apical part of domV, which is adjacent to the binding site of the initiation factor eIF4G, mimics the anticodon stem–loop of tRNAGly. A similar structure can be found in all IRESs of type I. The interaction of GARS with domV is very important for the activity of the PV IRES both in vitro and in cultured cells.
MATERIALS AND METHODS
Plasmids
Plasmid pMPolio wt bearing the complete 5′-UTR of PV type 1 strain Mahoney, and its deletion derivatives lacking domains I to III, IV, V and VI were described in ref. (35). The fragment covering the PV IRES, Bgl II blunted—NarI, was recloned from the pMPolio into pGL3R vector (36) between PvuII and NarI restriction sites. In the resulting construct PV IRES-Fluc, the initiation codon AUG 743 of PV RNA directs translation of Fluc. To prepare the construct PV IRES-Fluc mut [ACC(495–497) → TAG], the reverse P1 and forward P2 primers were used for PCR (For sequences of primers see Supplementary Table S1). All other constructs used for in vitro preparation of mRNAs were described in details in ref. (37). To prepare expression vectors for the human GARS (Gene ID: 2617) and its derivatives, RT–PCR fragments were obtained with the total RNA from HEK293T cells. To this end, a dT16 primed RT reaction was used, followed by PCR with the pair of gene-specific primers P3 and P4 containing BamHI and XhoI sites, respectively. [As the N-terminal 62 amino acids of GARS represent the mitochondrial localization signal (MLS), the PCR product corresponding to the GARS cytoplasmic version without MLS was obtained]. To prepare expression vectors for GARS Δ WHEP (Δ1–119 amino acids) and GARS ΔABD (Δ586–739 amino acids), the PCR-products were obtained using the P5–P6 and P7–P8 pairs of primers, respectively. The resulting PCR products were cloned into pGEX-6p1 vector (GE Healthcare) between BamH1 and XhoI to result in GST-GARS fusions.
GARS expression and purification
pGEX-6p1 GARS, pGEX-6p1 GARS ΔWHEP and pGEX-6p1 GARS ΔABD were expressed in Escherichia coli Rosetta (DE3), and the corresponding proteins were purified using Glutathione–Sepharose 4B and PreScission Protease (Amersham) as recommended by manufacturer, except that PBS was replaced by buffer A100 (20 mM Tris–HCl pH 7.5, 100 mM KCl, 0.1 mM EDTA, 1 mM DTT, 10% glycerol).
Affinity purification of RNA binding proteins from cytoplasmic extracts
Nuclease untreated RRL prepared according to a conventional procedure (39) was a kind gift from L. Ovchinnikov. HeLa cytoplasmic extract was purchased from CilBiotech. RRL (256 µl), 64 µl of HeLa extract, 40 µl of 10 × binding buffer [800 mM KCl, 200 mM Tris–HCl, pH 7.5, 10 mM Mg(CH3COO)2], 40 U of Ribolock RNase inhibitor (Fermentas), 40 µg of biotinylated RNA and H2O up to 400 µl were mixed and incubated at 30°C for 15 min. After that, each sample was diluted with 2 ml of wash buffer A [100 mM KCl, 20 mM Tris–HCl, pH 7.5, 10 mM Mg(CH3COO)2, 1 mM DTT] and complete EDTA-free protease inhibitor cocktail tablets (Roche). Finally, 50 µl of packed pre-washed Streptavidin sepharose high performance (GE Healthcare) were added. The samples were extensively shaken on ice for 1 h, then the beads were washed two times with 2 ml of the wash buffer B [wash buffer A containing 1 mM Mg(CH3COO)2]. Finally, the beads were resuspended in 80 µl of wash buffer B supplemented with 2 mM CaCl2 and 300 U of micrococcal nuclease (Fermentas) and incubated at 37°C for 30 min. Supernatants containing RNA binding proteins were loaded on a SDS–PAGE and the gels stained with Coumassie or silver after electrophoresis.
In vitro transcription
To synthesize monocistronic uncapped RNAs bearing the 5′-UTR (wt or mutated) of PV RNA, the Fluc coding sequence, the vector 3′-UTR and polyA, the PCR products were prepared using the forward primer P9 bearing the T7 promoter sequence and the reverse primer P10. The m7G-capped Rluc mRNA was prepared as described (38). For affinity purification experiments, RNase V1 and T2 protection assays and RelE-printing assays, shorter PCR products were obtained with the same forward primer P9 and the reverse primer P11 annealed to nucleotides 99–119 of Fluc coding sequence. For preparation of isolated domV wt and mut (nucleotides 421–597 of the 5′-UTR of PV RNA), the forward primer P12 bearing the T7 promoter sequence and the reverse primer P13 were used. To prepare a template for the transcription of tRNAGly, the forward primer P14 bearing the T7 promoter and the reverse primer P15 were annealed and filled-in with Taq-polymerase. tRNAPhe from E. coli was a kind gift from I. Boni. The PCR products were then used as templates for in vitro RNA transcription by the T7 RiboMAX Large Scale RNA Production kit (Promega). To prepare biotinylated RNAs, biotin-16 UTP (Roche) was added to the transcription mixture in the ratio 1:10 to UTP. The resulting RNAs were purified by LiCl precipitation (except for tRNAGly which was purified with phenol–chloroform extraction and gel filtration on Sephadex G50 to remove rNTPs) and checked for integrity by PAGE.
In vitro translation
Uncapped polyadenylated RNA (0.1 µg) of interest with Fluc reporter coding sequence was mixed with 1 ng of m7G-capped polyadenylated Rluc RNA, 6 µl RRL (or 4.8 µl RRL and 1.2 µl HeLa extract), 0.4 U of Ribolock RNase inhibitor (Fermentas) and a buffer containing 80 mM KCl, 20 mM Tris–HCl pH 7.5, 1 mM Mg(CH3COO)2 and 8 mM creatine phosphate in a final volume of 10 µl. When required, 0.4 µg of GARS was added (with adjustment of KCl concentration to 80 mM). Samples were incubated at 30°C for 1 h, Fluc and Rluc activities were measured using the Dual Luciferase Assay (Promega).
Cell culture and transfection procedures
Cell cultivation and transfection of reporter mRNAs are described in detail in (37,38). For experiments with siRNAs, GlyRS siRNA (sc-75153, Santa Cruz) and control siRNA (sc-37007, Santa Cruz) were used. HEK293T cells were cultivated in DMEM supplemented with 10% FBS and were replated to 24-well plates. After the cell density reached 50–70%, the transfection of 1μg of either GARS-specific or control siRNAs were performed using RNotion transfection reagent (5 Prime) according to manufacturer's instruction. After 24 h of siRNA transfection, reporter mRNAs were transfected as described previously (37). For western blotting, antibodies anti-GARS (sc-98614, Santa Cruz) and anti-GAPDH (Proteintech Group INC, PTG10494-1-AP) were used.
Toe-printing, enzymatic probing and protection assay, and RelE-printing
To analyze the binary complex GARS•PV IRES by primer extension inhibition, 0.2 µg of RNA bearing wt or mut PV 5′-UTR plus 119 nt of Fluc coding sequence was incubated with 1 µg of GARS in a buffer containing 100 mM KCl, 20 mM Tris–HCl pH 7.5, 1 mM Mg(CH3COO)2, 0.4 U of Ribolock RNase inhibitor (Fermentas) in a final volume of 20 µl. When required, ATP or AMPPNP were added to the final concentration 2 mM and equilibrated with Mg(CH3COO)2. The samples were preincubated for 5 min at 30°C. Then they were either used for reverse transcription (for toeprinting assay) or supplemented with 0.002 U of ribonuclease V1 (Ambion) or 0.5 U of ribonuclease T2 (Invitrogen) and additionally incubated for 10 min at 30°C. Then RNA from samples was purified with phenol/chloroform, and the reverse transcription was performed with the primer P13 annealed to nucleotides 575–597 of the PV 5′-UTR. For other details of the protocol, see ref. (39). The RelE-printing assay was performed as described (40). Briefly, 0.2 µg of RNA was incubated with 12 µl of RRL, 3 µl of HeLa extract, 0.4 U of Ribolock RNase inhibitor (Fermentas) and a buffer containing 80 mM KCl, 20 mM Tris–HCl pH 7.5, 1 mM Mg(CH3COO)2, 8 mM creatine phosphate, 2 mM GMPPNP and 0.2 mM m7GTP [equilibrated with Mg(CH3COO)2] were incubated for 5 min at 30°C. Then, RelE was added to a final concentration of 2 µM, and samples were incubated at 37°C for 10 min. The RNA was purified by phenol/chloroform extraction and the RT reaction was carried out. To detect RelE prints from the 48S complexes at AUG586 and AUG743, the primers P16 (complementary to positions 648–668 of the PV 5′-UTR) and P17 annealed to nucleotides 63–81 of the Fluc coding sequence were used.
RESULTS
The PV IRES-element specifically binds to GARS
To identify essential factors that may have been missed in previous studies, we performed a comprehensive analysis of proteins bound to the PV IRES element using a proteomic approach based on the isolation of mRNPs with biotinylated RNAs. A combined system employing RRL supplemented with 20% (v/v) HeLa extract was used throughout these experiments since the synthesis of PV proteins occurs correctly and efficiently in this system (11). To perform the analysis under more natural competitive conditions, i.e. in the presence of intact cellular mRNAs, neither the RRL nor the HeLa extract were treated with nucleases to remove endogenous RNAs. The biotin-labeled RNA comprising the entire PV 5′-UTR and a part of the firefly luciferase (Fluc) reporter sequence was incubated in the RRL or RRL + HeLa systems and then adsorbed to streptavidin-agarose. After washing off unbound proteins, proteins specifically bound to the PV RNA were brought to solution by RNA digestion using micrococcal nuclease. This protocol resulted in a very low background of irrelevant proteins (proteins adsorbed by the streptavidin–agarose alone). The eluted proteins were separated by SDS–PAGE electrophoresis, the bands of interest were cut out and the corresponding proteins identified by mass spectrometry. As a control RNA, we used a similar construct with the EMCV IRES (a type II picornavirus IRES). It should be noted that the selected extent of biotin-UMP incorporation into RNA did not affect the formation of the 48S preinitiation complex on the PV IRES as determined by sucrose gradient sedimentation analysis (data not shown).
One example of the pattern of proteins bound to the PV 5′-UTR in comparison with the EMCV IRES is shown in Figure 1A. The complete list of proteins interacting with the PV 5′-UTR will be published elsewhere. Their identification is still in progress, but it should be noted here that we found among them the ITAFs that had been previously reported to be implicated in the activity of the PV IRES: UNR, PTB, PCBP, La, nucleolin etc. However, our attention was drawn to the 74-kDa band that was completely absent from the proteins bound to the EMCV IRES (Figure 1A, lane 3), relatively weak among the proteins bound to the PV IRES from RRL (lane 1) and considerably enhanced in the additional presence of HeLa extract (lane 2). This protein was also absent from the proteins bound to the 5′-UTRs of the cellular Apaf-1, Hsp70 and LINE-1 mRNAs (data not shown). Mass spectrometry analysis unambiguously indicated that this prominent band is a house keeping enzyme, GARS.
Figure 1.
Glycyl-tRNA synthetase (GARS) binds the IRES element of poliovirus RNA and stimulates its activity. (A) The proteins bound to PV IRES in comparison with those bound to the EMCV IRES. The position of the 74-kDa band specific for the PV IRES is indicated by an arrow. (B) GARS domain organization. ABD, anticodon binding domain. (C) Effect of addition of the recombinant GARS and its two deletion versions (ΔWHEP and ΔABD) on translation of the PV IRES-Fluc RNA and the control capped Rluc RNA in RRL. The PV IRES-Fluc RNA and the control mRNA were co-translated in the same samples. The amounts of synthesized Fluc and Rluc were normalized to those produced in the control sample, i.e. without addition of GARS or its derivatives. (D) The same for the EMCV-Fluc RNA, the luciferase level of the control without GARS was set as 100% (Rluc values are not showed for space limitations since there is no significant change.).
According to western blot, the amount of GARS in HeLa extracts was ∼4- to 6-fold higher than in RRL (data not shown), so addition of 20% of HeLa extract approximately doubles the level of GARS, this correlates with enhancement of p74 band on the gel.
The anticodon-binding domain of GARS confers a strong stimulation of PV IRES-directed translation
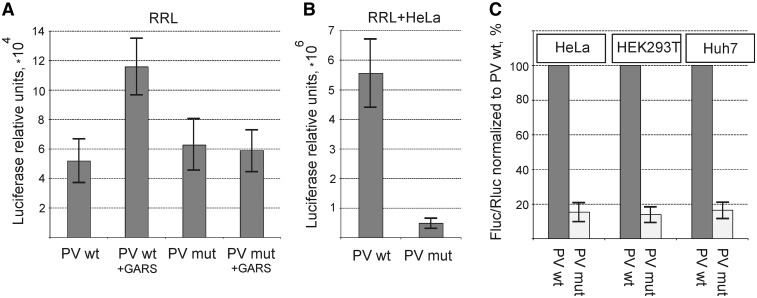
The human GARS belongs to the class II aminoacyl-tRNA synthetases and possesses three structural domains: the so-called WHEP domain, the catalytic core domain and the anticodon binding domain (ABD) (see Figure 1B). To find out whether GARS has any relation to the activity of the PV IRES, we cloned the cDNA coding for the human GARS and produced the recombinant human GARS in E. coli. The recombinant protein corresponds to the cytoplasmic version of GARS, i.e. it does not contain the N-terminal sequence responsible for its import to mitochondria (in this study, this protein lacking the mitochondrial localization sequence is referred to as the wild-type form, wt). Two shortened forms of GARS lacking either its ABD or WHEP domain were prepared along with the wt enzyme (see Figure 1B). The recombinant proteins were purified and tested for their effects on translation of the PV IRES-Fluc RNA in RRL in the absence of HeLa extract. As shown in Figure 1C, addition of the wt GARS or its variant lacking the WHEP domain significantly stimulated the translation of PV IRES-directed mRNA, whereas addition of the GARS protein lacking the ABD did not. The addition of GARS did not affect at all the level of protein synthesis directed by co-translated the capped mRNA encoding the Renilla luciferase (Rluc) and the EMCV-Fluc mRNA (Figure 1D) as well as that of other control reporter mRNAs with the HCV 5′-UTR or m7G and A-capped 5′-UTR from the β-globin mRNA (data not shown).
GARS binds to the apical part of domV of the PV IRES which mimics the anticodon stem–loop of tRNAGly
To locate the binding site of GARS on the 5′-UTR of PV RNA, we compared the patterns of proteins bound to biotinylated IRES variants from which its individual RNA domains were deleted. Four constructs similar to those used by Ochs et al. (28) were used: PV IRES (ΔI + II + III), PV IRES (ΔIV), PV IRES (ΔV) and PV IRES (ΔVI) (Figure 2). The binding of some proteins to the PV IRES was not affected by any of these deletions. Presumably, the corresponding proteins have several binding sites scattered over the IRES or bind to the sequences connecting the IRES domains. In contrast, other bands completely disappeared on deletion of particular domains. In particular, the GARS binding site resides entirely within the IRES domV since its band is completely absent from the pattern of proteins associated with the ΔV PV IRES.
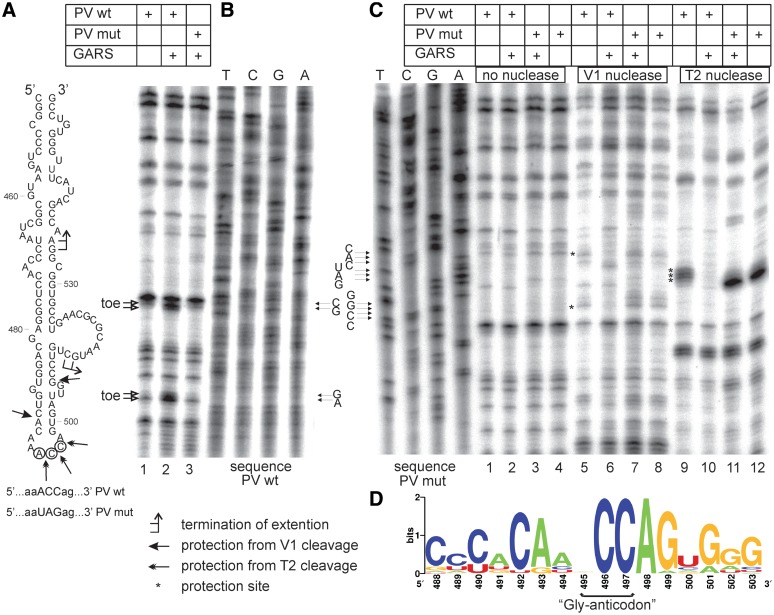
When we looked carefully at the structure of this domain we noticed that its apical part resembled the anticodon stem–loop of tRNAGly with the anticodon ACC with one obvious deviation: domV contained 6 nt residues in the loop instead of seven residues typical of anticodon loops of tRNAs (Figure 3A). Interestingly, this organization of the apical part of domV may be conserved in all type I IRES elements (see below) but is distinct from the functionally analogous domains in the IRES elements of the type II picornavirus IRESs. We hypothesized that this area of domV that is immediately adjacent to the binding site of initiation factors eIF4G + eIF4A (26,27) is the binding site of GARS.
Figure 3.
Mapping of the GARS binding site in the PV IRES by primer extension inhibition and enzymatic foot-printing. (A) The secondary structure of domV of PV IRES. Positions of toeprint stops and the sites of protection by GARS from nuclease attacks are indicated. (B) Primer extension inhibition (toeprinting) performed for the complexes of GARS with the PV IRES wt and PV IRES mut (the IRES in which ACC ‘anticodon’ of domV was mutated to UAG). The stops of reverse transcription (‘toe’) are denoted by arrows. The sequencing lanes obtained using the same oligodeoxynucleotide primer are shown on the right. (C) Foot-printing of the complexes of recombinant GARS with the wt and mutant PV IRES. The nucleotide positions protected by GARS from the attack by nucleases V1 and T2 are marked by asterisks. The sequencing lanes obtained for the mutant IRES using the same primer are shown on the left. (D) Sequence logo generated from aligned sequences of anticodon loop plus flanked residues corresponding to nucleotides 488–503 of PV IRES. The alignment was obtained from Rfam database (ID of alignment—RF00229).
To check this hypothesis, we carried out both a primer extension inhibition assay and a nuclease foot printing using two nucleases—V1 (specific for base-paired regions) and T2 (specific for single stranded loops of stem–loop structures). The reverse transcriptase reaction primed downstream of domV showed two stops in the presence of GARS, at positions 534–535 and 512–513 (Figure 3A and B). Mutation of the putative ‘Gly-anticodon’ ACC to UAG resulted in disappearance of these stops (Figure 3B, compare lane 3 with lane 2). The addition of GARS inhibited cleavages by nuclease V1 in the near-apical stem of domain V (between positions 490–491 and 505–506) and protected the putative anticodon ACC from the T2 nuclease attack (compare lines 9 and 10 in Figure 3C). No inhibition of these cleavages by nuclease V1 or protections within the apical loop was observed for the domV with the mutated ‘anticodon’ (Figure 3A and C, lanes 7, 8 and 11, 12). We concluded that the interaction of GARS with the PV IRES domV requires the ‘Gly-anticodon’-containing loop and presumably adjacent base-paired stems. This structure extends at least to the large side bulge (positions 511–524 in Figure 3A) where the interaction of eIF4G + eIF4A has been proposed (26). These data are in a good agreement with the importance of the ABD domain of the GARS protein for the interaction with the PV IRES described earlier. It may be hypothesized that the mutation of the ‘Gly-anticodon’ of domV does not change significantly its overall conformation as followed from the foot-printing analysis. This correlates with equal activities of the mutant and wt IRESs in RRL (see below) and the data that except for GARS, the pattern of mRNA-binding proteins bound to domV was not affected by the ‘anticodon’ mutation (Supplementary Figure S1).
In order to investigate the conservation of the ‘Gly-anticodon’ in type I IRESes, we analyzed the automatically generated multiple sequence alignment of 2893 sequences of picornaviruses obtained from Rfam database (41). Aligned sequences of the anticodon loop plus flanked residues (corresponding to nucleotides 488–503 of PV IRES) were further processed using Weblogo tool (42) to generate sequence logo (Figure 3D). It is evident that while CC residues are absolutely conserved, any nucleotide can be at the position occupied by A495 in the PV IRES which correlates with the degeneracy of glycine codon.
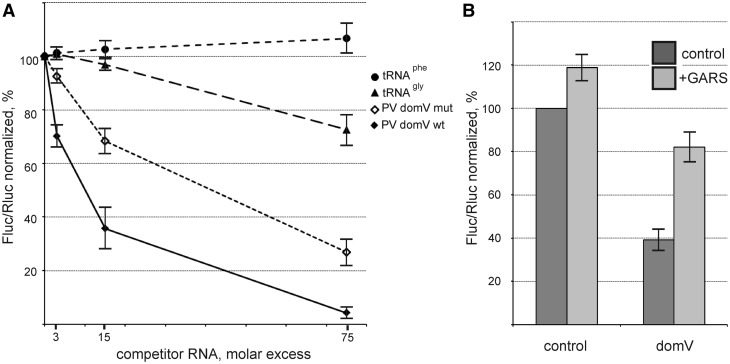
The finding that the upper part of domV mimics the anticodon stem–loop of tRNAGly was additionally confirmed by competition experiments: addition of an excess of in vitro transcribed tRNAGly to the RRL + HeLa cell-free system programmed with the PV IRES-Fluc inhibited the synthesis of Fluc whereas tRNAPhe had no effect (Figure 4A). However, the extent of this inhibition was modest as compared with the inhibitory effect of domV itself. We conclude that GARS interacts with domV significantly stronger than in vitro transcribed tRNAGly. It should be mentioned that the complete dissociation of GARS from the complex with the isolated domV requires a KCl concentration above 0.7 M (data not shown). In addition, domV certainly interacts with other functionally important components. Indeed, although the isolated domV wt suppressed the translation of PV IRES-Fluc more effectively than domV mut, the effect of the latter was also significant (Figure 4A). This is in agreement with the data that apart from GARS the isolated domV additionally binds some canonical and auxiliary factors like eIF4G + eIF4A (26), PTB (25) and presumably La-autoantigen (our preliminary observations). Anyway, a large difference between the inhibitory effects of domV wt and domV mut, especially at lower excesses of competitor RNAs, may be attributed to GARS depletion (Figure 4A). Addition of recombinant GARS to the cell-free system suppressed by the 15-fold molar excess of the competitor domV wt restores the translational efficiency of the PV IRES-Fluc RNA to a significant extent (Figure 4B).
Figure 4.
The GARS-mediated stimulation of translation of PV IRES-Fluc mRNA is accounted for by the interaction of the enzyme with the ‘anticodon’ of domV. (A) Effect of addition of tRNAGly, tRNAPhe, individual domV wt and domV mut (ACC → UAG) on the translation of PV IRES-Fluc in the RRL + HeLa cell-free system. All values for Fluc were normalized to Rluc synthesized from a co-translated capped Rluc mRNA. The Fluc/Rluc value for the PV IRES-Fluc without addition of competitor RNAs was set to 100%. (B) Inhibition of the PV IRES-Fluc translation by the isolated domV wt (15-fold molar excess) in the RRL + HeLa system and its relief by addition of GARS. The Fluc/Rluc value for the PV IRES-Fluc without addition of the competitor RNA and GARS was set to 100% (control).
The contacts of GARS with the ‘anticodon’ loop of domV seem to make a major contribution to the affinity and specificity of the interaction of the enzyme with the PV IRES: the ACC → UAG mutant does not bind GARS at all (Figure 3 and Supplementary Figure S1), though, as discussed earlier, the overall structure of domV does not seem to change noticeably. It should be noted that none of the two ATP binding sites of GARS (see Discussion) appears to be involved in its interaction with the IRES (Supplementary Figure S2).
Mutation of the ‘Gly-anticodon’ of domV dramatically affects the translation potential of the PV IRES both in vitro and in living cells
As expected, the mutation of the ‘anticodon’ in the loop of domV makes the PV IRES-Fluc translation in RRL almost insensitive to the addition of exogenous recombinant GARS. More importantly, without the GARS addition the PV IRES-Fluc mRNAs with wt and mutant domV are translated with rather low but similar efficiency (Figure 5A). This allows us to believe that the mutation of the domV ‘anticodon’ unlikely affects the binding of eIF4G to a significant extent and hence the overall structure of domV does not change significantly as indicated earlier. The low level of translation is accounted for by the insufficiency to express the PV RNA that is inherent to RRL (see ‘Introduction’ section) and by the fact that we use a nuclease untreated RRL system. In such a system, the PV IRES-Fluc RNA experiences a big competition pressure from very efficiently translated capped mRNAs, first of all from α- and β-globin mRNAs that are abundant in RRL.
Figure 5.
Effect of the ‘Gly-anticodon’ mutation of domV (ACC → UAG) on the translation of PV IRES-Fluc in cell-free systems and in cultured cells. (A) Translation in a nuclease untreated RRL. The Fluc values were normalized to those for Rluc. The Rluc values did not vary significantly in parallel assays. (B) Translation in RRL supplemented with 20% (v/v) of cytoplasmic HeLa extract. (C) Expression of Fluc in three human cell lines transfected with the PV IRES-Fluc RNAs harboring either the domV wt or domV mut. The Fluc/Rluc value for the wt RNA construct was set to 100% for each cell line.
This situation changes dramatically when we add 20% (v/v) of the nuclease untreated HeLa extract to the RRL. This supplementation results in a strikingly strong stimulation of PV translation (compare the values of Fluc activity in Figure 5A and B), although no stimulation was observed for the control capped Rluc mRNA (data not shown). Unlike in the RRL alone, in the combined RRL + HeLa system the difference in translation efficiencies between wt and mutant PV IRES elements reached 10-fold (Figure 5B). Thus, even if GARS may not be the only component that makes PV translation that efficient in the RRL + HeLa system, it is supposed to be a very important component for this process. Importantly, the level of GARS in the HeLa extract is 4- to 5-fold higher than in RRL (data not shown), which correlates with a different amount of GARS eluted from the PV IRES after incubation in RRL or RRL plus HeLa in our initial experiment (Figure 1A). Similar results were also obtained in living HEK293T, Huh7 and HeLa cells—a 6- to 7-fold inhibition of the PV translation activity by the ‘anticodon’ mutation in domV (Figure 5C). These results may suggest an essential role of the interaction of GARS with domV for the stimulation of translation of PV IRES-containing mRNA.
To obtain more direct evidence that GARS is required for the efficient translation driven by the PV IRES, we depleted GARS by siRNA interference. This operation turned out to be cytotoxic: we found that for HEK293T cells even a 2-fold GARS depletion resulted in an adverse effect on the cell morphology and viability even at 24 h after siRNA tranfection, and a longer exposure to siRNA resulted in cell death. When we transfected reporter mRNAs in these GARS-depleted cells, we observed a mild inhibition of translation of both PV-Fluc and EMCV-Fluc mRNAs and a 3-fold reduction in the activity of the control capped Rluc mRNA (Supplementary Figure S3). Thus, unfortunately, siRNA interference against GARS is inapplicable way to investigate the function of GARS in the PV IRES mediated translation.
Enhancement of PV IRES activity by GARS occurs at the level of 48S complex formation
The time courses of luciferase synthesis for PV IRES-Fluc mRNAs with the wt and mutated domV diverge from the very first moment of incubation of the RRL + HeLa translation system (data not shown). This suggests that the mutation of the ‘domV anticodon’ impairs translation initiation. To check this hypothesis, the efficiency of recognition of the initiation codons by 40S ribosomes on the PV IRES had to be compared for the wt and mutated PV IRES-Fluc mRNA.
The translation initiation complexes can be directly assembled in vitro in RRL and subsequently analyzed by toeprinting (39,43). Unfortunately, toeprinting in the combined RRL + HeLa system is not possible since cytoplasmic extracts of cultured cells contain an RNase H-like activity which completely destroys duplexes of oligodeoxynucleotide primers and mRNAs. As suggested by Andreev et al. (40), the problem may be solved using RelE printing instead of the toeprint assay. RelE is a bacterial toxin that cleaves mRNA at the nucleotide triplet positioned in the free A-site of the 40S or 80S ribosome if the P-site is already occupied by Met-tRNA or peptidyl-tRNA. Like in the case of toeprinting, also after RelE treatment the positions of reverse transcriptase stops (the sites of cleavage) are determined by primer extension inhibition but the RT reaction is performed with a deproteinized mRNA when the RNase H activity is already removed (40).
Analysis of entry of the ribosomal 40S subunit onto the PV IRES is additionally complicated by the fact that the PV and other type I IRESs contain two AUG codons downstream of the suggested region of the 40S ribosome entry: the AUG at position 586 adjoining the polypyrimidine tract (see Figure 2A) and the authentic AUG 743 that is in a good nucleotide context. The former is now regarded as a preliminary entry codon from which the 40S ribosome is then transferred to AUG 743 by a poorly understood mechanism [for references see (44)]. In fact, this mechanism has never been analyzed in detail. It was based on solid but indirect experiments. For this reason, we inspected by RelE printing both regions, i.e. those surrounding AUG 586 and AUG 743, using two different primers for primer extension. The data are presented in Figure 6A and B.
Figure 6.
RelE-printing of 48S initiation complexes formed in the RRL + HeLa with the wt and mutated PV IRES. The 48S complexes were formed in the presence of GMPPNP. (A) Positions of RelE prints are shown on the left (for AUG586) and right (for AUG743) of the gel. Two different primers were used to detect RelE prints for the AUG586 and AUG743 and hence two separate dideoxynucleotide sequences generated with the corresponding primers were run in parallel. (B) Inhibition of RelE cleavages in the PV IRES-Fluc by addition of the isolated domV wt and its partial relief by the addition of GARS. A 15-fold molar excess of domV wt transcript over the mRNA was added to the RRL + HeLa translation system (see Figure 4B). Intensities of RelE-prints were quantified using Aida Image Analyzer. The intensity of each RelE print was normalized to the overall intensity in the corresponding lane, the RelE print for the control lane without addition of GARS and domain V was set to 100%.
We showed for the first time that the 40S ribosomal subunit recognized both AUG codons, at least in the presence of the GTP analog GMPPNP, since two RelE prints we obtained at positions around 590 and 747 (Figure 6A). The RelE cleavage bands for the mRNA with the mutated domV were very weak and could be seen only after overexposure of the gel (Figure 6A, lanes 1, 3 and 5, 7). These data provide compelling evidence that the mutation of the binding site of GARS in the apical part of domV dramatically impairs the formation of translation initiation complexes at both initiation codons. The essential role of GARS for the correct 40S subunit entry at AUG 586 and AUG 743 could be also demonstrated without mutating the ‘Gly-anticodon’ of domV. A partial sequestration of GARS in the RRL + HeLa system by addition of the isolated domV RNA wt resulted both in inhibition of Fluc synthesis (see Figure 4B) and in weakening of the corresponding RelE cleavages (Figure 6B). In both cases the inhibitory effects could be relieved by addition of recombinant GARS to the system (Figures 4B and 6B). It should be emphasized that for these experiments we intentionally did not use larger excesses of the competitor RNA since this results in a progressive and less specific sequestration of other mRNA binding proteins, including initiation factors.
DISCUSSION
In this study, we identified an essential component of the translation initiation machinery acting on the PV IRES that was missed in previous studies. In contrast to all other ITAFs identified so far which are all mRNA-binding proteins and play important roles in nuclear events, this component is a cytoplasmic house-keeping enzyme. GARS belongs to the class II of aminoacyl-tRNA synthetases (AARSs), functions as a dimer and has a modular organization (45). We show that GARS binds highly specifically to the apical part of the functionally most important domain of the PV IRES, domV: the binding sites of GARS and of the key initiation factors eIF4G + eIF4A are adjacent. Our working hypothesis is that the interaction with GARS is needed for a correct positioning of the 40S ribosome at the PV IRES.
The specificity of the binding of GARS to the IRES domain V is determined by the apical part of this stem–loop structure which mimics the anticodon stem–loop of tRNAGly. We have shown that the ‘anticodon’ loop is fully protected from nucleases in the complex of recombinant GARS with domV. A mutation of the ‘anticodon’ of domV abolishes binding of GARS and results in a dramatic drop of the IRES activity both in the RRL supplemented with a HeLa extract and in transfected cells. The conservation of two C residues in the loop of domain V in known representatives of type I IRES elements is a long standing observation (34,46). In this report, this conserved feature of domV finally gets a solid explanation: these two C residues may be invariant second and third nucleotides of the Gly anticodon, while the first nucleotide (A in the case of PV IRES) is redundant (Figure 3D). These two C residues are known to be essential elements in the cognate tRNAGly for recognition by GARS (45). Thus we suggest that GARS may bind to all type I IRES elements, and our preliminary data show that it also stimulates the translation of mRNA directed by the rhinovirus IRES. It should be noted that GARS has been mentioned by Lin et al. (47) as a result of screening of proteins associated with the 5′-UTR of Enterovirus 71. However, no attention has been given to this fact.
Also the way how the ‘anticodon loop’ is presented is similar in known type I picornavirus IRES elements: the anticodon loop is fixed by a stem of 5 bp (Figure 3A), which in turn is connected to the second 5-bp stem with one unpaired nucleotide at one side and with two bulged nucleotides at the other side.
When the isolated mutated domV is added to the RRL + HeLa cell-free system, it inhibits the translation of a PV IRES-directed mRNA to a significantly lesser extent than the domV wt. The residual inhibitory effect of domVmut may be accounted for by sequestering eIF4G (26) and probably some ITAFs which bind at or close to this domain. In line with this conclusion, the sole addition of the recombinant GARS to RRL (see Figure 5A) is not able to stimulate PV translation to the same level as observed in the RRL + HeLa system (Figure 5B). Therefore, we believe that a concerted action of GARS and other ITAF(s) is needed to fully activate the PV IRES. This activation can indirectly contribute to the ‘correcting function’, as well, since the translation initiation apparatus will be preferentially recruited to the correct initiation site in the viral RNA at the expense of a spurious initiation at aberrant regions. In addition to their function of modulating RNA–RNA and RNA–protein interactions, RNA-binding proteins like PTB and La may also mask spurious initiation sites (48). The versatility of La in its association with RNA (49) suggests that it may bind to multiple sites in the PV IRES. Even though La was demonstrated to bind to the stem–loop VI of the PV IRES (19), no systematic mapping of possible other La binding sites has been reported, and the molecular action of La in the PV translation is not yet understood (50).
The experimental data described in this article allow us to propose a step in the translation initiation on the PV IRES that requires the GARS participation. Before getting to our considerations, it should be reminded that all well documented viral IRESs, including IRESs of picornaviruses, possess two common features (51). They all have a highly specific site(s) to bind one or more key translation initiation components, e.g. eIF4G in case of picornavirus IRESs and the 40S ribosomal subunit and eIF3 for HCV-like IRESs. Such specific sites are needed for the 40S ribosomal subunit entry onto internal regions of mRNAs. However, this feature is mandatory but not sufficient to confer IRES properties to a particular region of RNA. Another feature is the presence of a special structural element(s) in an IRES that ensures the accommodation of its mRNA initiation region in the mRNA binding cleft of the 40S subunit. In the HCV-like IRES elements this accommodation function is performed by domain II and pseudoknot (52–54). The functionally analogous elements in picornavirus IRESs are not yet characterized. They are presumably located in the domains neighboring domV and dom J–K in the picornavirus IRESs of type I and II, respectively.
Our preliminary data indicate that there is no big difference in ribosome loading of the PV IRES wt and that with the ‘mutated Gly codon’. This means that the ribosome recruitment onto the mutant IRES is not significantly impaired. Therefore, our current working model is that GARS via its interaction with other ITAFs or RNA sequences of neighboring IRES domains directs the accommodation of the AUG 586-containing region in the mRNA binding cleft.
It was unexpected to find a ubiquitous house-keeping enzyme performing the same classical function (tRNA aminoacylation) in all cells and in all living organisms as a factor activating translation initiation in mammalian cells by a rather specific mechanism. However, the current literature presents us several curious examples of multi-functionality of proteins, and also for AARSs several moonlighting functions have been described (55). Most of them are not related to translation. As to translation, the classical example is a feedback regulation of the synthesis of AARS in bacteria. The best studied case is that of ThrRS in E. coli. This enzyme binds to its own mRNA near the initiation codon (also employing anticodon stem–loop mimicry) and thereby prevents its excessive production (56). In the yeast Saccharomyces cerevisiae, expression of AspRS is regulated by binding of AspRS to the 5′-UTR of AspRS messenger RNA (57). Exciting results for mammalian systems have also been reported by Paul Fox and his colleagues. They have shown that human glutamylprolyl-tRNA synthetase is able to interact with ribosomal protein L13a, glyceraldehyde-3-phosphate dehydrogenase and NSAP1 to form the GAIT complex that exerts specific translational silencing to regulate inflammation (58,59). In this mechanism, the WHEP domain of Glu-Pro RS rather than its active center needed for tRNA aminoacylation is employed. However, to the best of our knowledge, in all reported cases related to translation, AARS act as highly specific translational repressors. Our work shows for the first time that an aminoacyl-tRNA synthetase is directly involved in stimulating the formation of translation initiation complexes on an mRNA.
The finding that the domV of PV IRES is crowned with GARS looks even more intriguing when we take into account the fact that nothing like that is observed for picornaviruses IRESs of type II. Although type I and type II IRESs differ in secondary structure details, the fundamental aspects of the mechanism of initiation on these classes of IRESs are similar. The type II IRESs have a functionally similar domain (J–K) in the position equivalent to PV domV. Nevertheless, Type II IRESs operate without GARS. To recruit the key initiation factors and the 40S ribosomal subunit to the J–K domain, they employ various hnRNPs, the principal function of which is thought to modulate RNA–RNA and RNA–protein interactions. Therefore, it can be speculated that GARS binds to the top of domV of PV IRES not only to fix or modify its conformation or facilitate binding of other factors. We anticipate that GARS may be additionally needed for other events of the virus–host interaction. In this context it may be interesting to briefly mention the non-canonical functions of GARS.
First of all, GARS is famous for its special relations with motor neurons. Mutations in GARS cause Charcot–Marie–Tooth (CMT2D) diseases which are the most common heritable peripheral neuropathies (60). Although the molecular mechanisms underlying CMT disease are not known, the CMT mutations do not correlate with GARS aminoacylation activity and hence may affect non-canonical functions of GARS. It has been noticed that motor neurons from patients with the CMT syndrome reveal impaired mRNA localization and distribution (61).
The expansion of AARSs non-canonical functionality also can be achieved via their reaction products. For instance, the aminoacylation reaction can be diverted to produce diadenosine tetraphosphate (Ap4A), a universal pleiotropic signaling molecule required for the regulation of cellular pathways (62). GARS is unique among all AARSs: it is able to synthesize Ap4A from two ATP molecules independent of amino acid (glycine) availability (63). Thus, it would be of interest to see whether GARS bound to the PV IRES is still capable of performing moonlighting functions that have some relation to the replication of PV in host cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–3 and Supplementary Table 1.
FUNDING
Russian Foundation of Basic Research (RFBR) (11-04-91335-NNIO_a to I.S.); Deutsche Forschungsgemeinschaft (DFG IRTG 1384, to M.N.) and President of Russian Federation (MK-5309.2011.4 to S.D.). Funding for open access charge: Partially waived by Oxford University Press.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank V.I. Agol and M. Safro for critical reading of the manuscript and valuable advices.
REFERENCES
- 1.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 2.Jang SK, Kräusslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Niepmann M. Internal translation initiation of picornaviruses and hepatitis C virus. Biochim. Biophys. Acta. 2009;1789:529–541. doi: 10.1016/j.bbagrm.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Balvay L, Soto Rifo R, Ricci EP, Decimo D, Ohlmann T. Structural and functional diversity of viral IRESes. Biochim. Biophys. Acta. 2009;1789:542–557. doi: 10.1016/j.bbagrm.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 5.Hellen CU, de Breyne S. A distinct group of hepacivirus/pestivirus-like internal ribosomal entry sites in members of diverse picornavirus genera: evidence for modular exchange of functional noncoding RNA elements by recombination. J. Virol. 2007;81:5850–5863. doi: 10.1128/JVI.02403-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pisarev AV, Chard LS, Kaku Y, Johns HL, Shatsky IN, Belsham GJ. Functional and structural similarities between the internal ribosome entry sites of hepatitis C virus and porcine teschovirus, a picornavirus. J. Virol. 2004;78:4487–4497. doi: 10.1128/JVI.78.9.4487-4497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pilipenko EV, Pestova TV, Kolupaeva VG, Khitrina EV, Poperechnaya AN, Agol VI, Hellen CU. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 2000;14:2028–2045. [PMC free article] [PubMed] [Google Scholar]
- 8.Pestova TV, Hellen CU, Shatsky IN. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pestova TV, Kolupaeva VG, Lomakin IB, Pilipenko EV, Shatsky IN, Agol VI, Hellen CU. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl Acad. Sci. USA. 2001;98:7029–7036. doi: 10.1073/pnas.111145798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorner AJ, Semler BL, Jackson RJ, Hanecak R, Duprey E, Wimmer E. In vitro translation of poliovirus RNA: utilization of internal initiation sites in reticulocyte lysate. J. Virol. 1984;50:507–514. doi: 10.1128/jvi.50.2.507-514.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown EC, Jackson RJ. All five cold-shock domains of unr (upstream of N-ras) are required for stimulation of human rhinovirus RNA translation. J. Gen. Virol. 2004;85:2279–2287. doi: 10.1099/vir.0.80045-0. [DOI] [PubMed] [Google Scholar]
- 13.Boussadia O, Niepmann M, Créancier L, Prats AC, Dautry F, Jacquemin-Sablon H. Unr is required in vivo for efficient initiation of translation from the internal ribosome entry sites of both rhinovirus and poliovirus. J. Virol. 2003;77:3353–3359. doi: 10.1128/JVI.77.6.3353-3359.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blyn LB, Towner JS, Semler BL, Ehrenfeld E. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 1997;71:6243–6246. doi: 10.1128/jvi.71.8.6243-6246.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunt SL, Hsuan JJ, Totty N, Jackson RJ. unr, a cellular cytoplasmic RNA-binding protein with five cold-shock domains, is required for internal initiation of translation of human rhinovirus RNA. Genes Dev. 1999;13:437–448. doi: 10.1101/gad.13.4.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunt SL, Jackson RJ. Polypyrimidine-tract binding protein (PTB) is necessary, but not sufficient, for efficient internal initiation of translation of human rhinovirus-2 RNA. RNA. 1999;5:344–359. doi: 10.1017/s1355838299981414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gosert R, Chang KH, Rijnbrand R, Yi M, Sangar DV, Lemon SM. Transient expression of cellular polypyrimidine-tract binding protein stimulates cap-independent translation directed by both picornaviral and flaviviral internal ribosome entry sites In vivo. Mol. Cell. Biol. 2000;20:1583–1595. doi: 10.1128/mcb.20.5.1583-1595.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merrill MK, Gromeier M. The double-stranded RNA binding protein 76:NF45 heterodimer inhibits translation initiation at the rhinovirus type 2 internal ribosome entry site. J. Virol. 2006;80:6936–6942. doi: 10.1128/JVI.00243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meerovitch K, Pelletier J, Sonenberg N. A cellular protein that binds to the 5′-noncoding region of poliovirus RNA: implications for internal translation initiation. Genes Dev. 1989;3:1026–1034. doi: 10.1101/gad.3.7.1026. [DOI] [PubMed] [Google Scholar]
- 20.Hellen CU, Witherell GW, Schmid M, Shin SH, Pestova TV, Gil A, Wimmer E. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc. Natl Acad. Sci. USA. 1993;90:7642–7646. doi: 10.1073/pnas.90.16.7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Izumi RE, Valdez B, Banerjee R, Srivastava M, Dasgupta A. Nucleolin stimulates viral internal ribosome entry site-mediated translation. Virus Res. 2001;76:17–29. doi: 10.1016/s0168-1702(01)00240-4. [DOI] [PubMed] [Google Scholar]
- 22.Anderson EC, Hunt SL, Jackson RJ. Internal initiation of translation from the human rhinovirus-2 internal ribosome entry site requires the binding of Unr to two distinct sites on the 5′ untranslated region. J. Gen. Virol. 2007;88:3043–3052. doi: 10.1099/vir.0.82463-0. [DOI] [PubMed] [Google Scholar]
- 23.Hellen CU, Pestova TV, Litterst M, Wimmer E. The cellular polypeptide p57 (pyrimidine tract-binding protein) binds to multiple sites in the poliovirus 5′ nontranslated region. J. Virol. 1994;68:941–950. doi: 10.1128/jvi.68.2.941-950.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guest S, Pilipenko E, Sharma K, Chumakov K, Roos RP. Molecular mechanisms of attenuation of the Sabin strain of poliovirus type 3. J. Virol. 2004;78:11097–11107. doi: 10.1128/JVI.78.20.11097-11107.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kafasla P, Morgner N, Robinson CV, Jackson RJ. Polypyrimidine tract-binding protein stimulates the poliovirus IRES by modulating eIF4G binding. EMBO J. 2010;29:3710–3722. doi: 10.1038/emboj.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Breyne S, Yu Y, Unbehaun A, Pestova TV, Hellen CU. Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc. Natl Acad. Sci. USA. 2009;106:9197–9202. doi: 10.1073/pnas.0900153106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ochs K, Zeller A, Saleh L, Bassili G, Song Y, Sonntag A, Niepmann M. Impaired binding of standard initiation factors mediates poliovirus translation attenuation. J. Virol. 2003;77:115–122. doi: 10.1128/JVI.77.1.115-122.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ochs K, Saleh L, Bassili G, Sonntag VH, Zeller A, Niepmann M. Interaction of translation initiation factor eIF4B with the poliovirus internal ribosome entry site. J. Virol. 2002;76:2113–2122. doi: 10.1128/jvi.76.5.2113-2122.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Campbell SA, Lin J, Dobrikova EY, Gromeier M. Genetic determinants of cell type-specific poliovirus propagation in HEK 293 cells. J. Virol. 2005;79:6281–6290. doi: 10.1128/JVI.79.10.6281-6290.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Svitkin YV, Cammack N, Minor PD, Almond JW. Translation deficiency of the Sabin type 3 poliovirus genome: association with an attenuating mutation C472—U. Virology. 1990;175:103–109. doi: 10.1016/0042-6822(90)90190-3. [DOI] [PubMed] [Google Scholar]
- 31.Ren RB, Moss EG, Racaniello VR. Identification of two determinants that attenuate vaccine-related type 2 poliovirus. J. Virol. 1991;65:1377–1382. doi: 10.1128/jvi.65.3.1377-1382.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.La Monica N, Racaniello VR. Differences in replication of attenuated and neurovirulent polioviruses in human neuroblastoma cell line SH-SY5Y. J. Virol. 1989;63:2357–2360. doi: 10.1128/jvi.63.5.2357-2360.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Agol VI, Drozdov SG, Ivannikova TA, Kolesnikova MS, Korolev MB, Tolskaya EA. Restricted growth of attenuated poliovirus strains in cultured cells of a human neuroblastoma. J. Virol. 1989;63:4034–4038. doi: 10.1128/jvi.63.9.4034-4038.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Belsham GJ, Jackson RJ. Translation initiation on picornavirus RNA. In: Sonenberg N, Hershey JWB, Mathews MB, editors. Translational Control of Gene Expression. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press; 2000. pp. 869–900. [Google Scholar]
- 35.Ochs K, Saleh L, Bassili G, Sonntag VH, Zeller A, Niepmann M. Interaction of translation initiation factor eIF4B with the poliovirus internal ribosome entry site. J. Virol. 2002;76:2113–2122. doi: 10.1128/jvi.76.5.2113-2122.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoneley M, Paulin FE, Le Quesne JP, Chappell SA, Willis AE. C-Myc 5′ untranslated region contains an internal ribosome entry segment. Oncogene. 1998;16:423–428. doi: 10.1038/sj.onc.1201763. [DOI] [PubMed] [Google Scholar]
- 37.Andreev DE, Dmitriev SE, Terenin IM, Prassolov VS, Merrick WC, Shatsky IN. Differential contribution of the m7G-cap to the 5′ end-dependent translation initiation of mammalian mRNAs. Nucleic Acids Res. 2009;37:6135–6147. doi: 10.1093/nar/gkp665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dmitriev SE, Andreev DE, Terenin IM, Olovnikov IA, Prassolov VS, Merrick WC, Shatsky IN. Efficient translation initiation directed by the 900-nucleotide-long and GC-rich 5′ untranslated region of the human retrotransposon LINE-1 mRNA is strictly cap dependent rather than internal ribosome entry site mediated. Mol. Cell. Biol. 2007;27:4685–4697. doi: 10.1128/MCB.02138-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dmitriev SE, Pisarev AV, Rubtsova MP, Dunaevsky YE, Shatsky IN. Conversion of 48S translation preinitiation complexes into 80S initiation complexes as revealed by toeprinting. FEBS Lett. 2003;533:99–104. doi: 10.1016/s0014-5793(02)03776-6. [DOI] [PubMed] [Google Scholar]
- 40.Andreev D, Hauryliuk V, Terenin I, Dmitriev S, Ehrenberg M, Shatsky I. The bacterial toxin RelE induces specific mRNA cleavage in the A site of the eukaryote ribosome. RNA. 2008;14:233–239. doi: 10.1261/rna.693208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gardner PP, Daub J, Tate J, Moore BL, Osuch IH, Griffiths-Jones S, Finn RD, Nawrocki EP, Kolbe DL, Eddy SR, et al. Rfam: Wikipedia, clans and the “decimal” release. Nucleic Acids Res. 2011;39:D141–D145. doi: 10.1093/nar/gkq1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dmitriev SE, Terenin IM, Dunaevsky YE, Merrick WC, Shatsky IN. Assembly of 48S translation initiation complexes from purified components with mRNAs that have some base pairing within their 5′ untranslated regions. Mol. Cell. Biol. 2003;23:8925–8933. doi: 10.1128/MCB.23.24.8925-8933.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaminski A, Pöyry TA, Skene PJ, Jackson RJ. Mechanism of initiation site selection promoted by the human rhinovirus 2 internal ribosome entry site. J. Virol. 2010;84:6578–6589. doi: 10.1128/JVI.00123-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Safro MG, Moor NA. [Codases: fifty years after] Mol. Biol. 2009;43:230–242. [PubMed] [Google Scholar]
- 46.Jackson RJ, Hunt SL, Gibbs CL, Kaminski A. Internal initiation of translation of picornavirus RNAs. Mol. Biol. Rep. 1994;19:147–159. doi: 10.1007/BF00986957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin JY, Li ML, Huang PN, Chien KY, Horng JT, Shih SR. Heterogeneous nuclear ribonuclear protein K interacts with the enterovirus 71 5′ untranslated region and participates in virus replication. J. Gen. Virol. 2008;89:2540–2549. doi: 10.1099/vir.0.2008/003673-0. [DOI] [PubMed] [Google Scholar]
- 48.Svitkin YV, Ovchinnikov LP, Dreyfuss G, Sonenberg N. General RNA binding proteins render translation cap dependent. EMBO J. 1996;15:7147–7155. [PMC free article] [PubMed] [Google Scholar]
- 49.Maraia RJ, Bayfield MA. The La protein-RNA complex surfaces. Mol. Cell. 2006;21:149–152. doi: 10.1016/j.molcel.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 50.Costa-Mattioli M, Svitkin Y, Sonenberg N. La autoantigen is necessary for optimal function of the poliovirus and hepatitis C virus internal ribosome entry site in vivo and in vitro. Mol. Cell. Biol. 2004;24:6861–6870. doi: 10.1128/MCB.24.15.6861-6870.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shatsky IN, Dmitriev SE, Terenin IM, Andreev DE. Cap- and IRES-independent scanning mechanism of translation initiation as an alternative to the concept of cellular IRESs. Mol. Cells. 2010;30:285–293. doi: 10.1007/s10059-010-0149-1. [DOI] [PubMed] [Google Scholar]
- 52.Filbin ME, Kieft JS. HCV IRES domain IIb affects the configuration of coding RNA in the 40S subunit's decoding groove. RNA. 2011;17:1258–1273. doi: 10.1261/rna.2594011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Locker N, Easton LE, Lukavsky PJ. HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J. 2007;26:795–805. doi: 10.1038/sj.emboj.7601549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Easton LE, Locker N, Lukavsky PJ. Conserved functional domains and a novel tertiary interaction near the pseudoknot drive translational activity of hepatitis C virus and hepatitis C virus-like internal ribosome entry sites. Nucleic Acids Res. 2009;37:5537–5549. doi: 10.1093/nar/gkp588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guo M, Yang XL, Schimmel P. New functions of aminoacyl-tRNA synthetases beyond translation. Nat Rev Mol Cell Biol. 2010;11:668–674. doi: 10.1038/nrm2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Romby P, Springer M. Bacterial translational control at atomic resolution. Trends Genet. 2003;19:155–161. doi: 10.1016/S0168-9525(03)00020-9. [DOI] [PubMed] [Google Scholar]
- 57.Frugier M, Giege R. Yeast aspartyl-tRNA synthetase binds specifically its own mRNA. J. Mol. Biol. 2003;331:375–383. doi: 10.1016/s0022-2836(03)00767-8. [DOI] [PubMed] [Google Scholar]
- 58.Jia J, Arif A, Ray PS, Fox PL. WHEP domains direct noncanonical function of glutamyl-Prolyl tRNA synthetase in translational control of gene expression. Mol. Cell. 2008;29:679–690. doi: 10.1016/j.molcel.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mukhopadhyay R, Jia J, Arif A, Ray PS, Fox PL. The GAIT system: a gatekeeper of inflammatory gene expression. Trends Biochem. Sci. 2009;34:324–331. doi: 10.1016/j.tibs.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Motley WW, Talbot K, Fischbeck KH. GARS axonopathy: not every neuron's cup of tRNA. Trends Neurosci. 2010;33:59–66. doi: 10.1016/j.tins.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nangle LA, Zhang W, Xie W, Yang XL, Schimmel P. Charcot-Marie-Tooth disease-associated mutant tRNA synthetases linked to altered dimer interface and neurite distribution defect. Proc. Natl Acad. Sci. USA. 2007;104:11239–11244. doi: 10.1073/pnas.0705055104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kisselev LL, Justesen J, Wolfson AD, Frolova LY. Diadenosine oligophosphates (Ap(n)A), a novel class of signalling molecules? FEBS Lett. 1998;427:157–163. doi: 10.1016/s0014-5793(98)00420-7. [DOI] [PubMed] [Google Scholar]
- 63.Guo RT, Chong YE, Guo M, Yang XL. Crystal structures and biochemical analyses suggest a unique mechanism and role for human glycyl-tRNA synthetase in Ap4A homeostasis. J. Biol. Chem. 2009;284:28968–28976. doi: 10.1074/jbc.M109.030692. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.