Abstract
In Escherichia coli, the SeqA protein binds specifically to GATC sequences which are methylated on the A of the old strand but not on the new strand. Such hemimethylated DNA is produced by progression of the replication forks and lasts until Dam methyltransferase methylates the new strand. It is therefore believed that a region of hemimethylated DNA covered by SeqA follows the replication fork. We show that this is, indeed, the case by using global ChIP on Chip analysis of SeqA in cells synchronized regarding DNA replication. To assess hemimethylation, we developed the first genome-wide method for methylation analysis in bacteria. Since loss of the SeqA protein affects growth rate only during rapid growth when cells contain multiple replication forks, a comparison of rapid and slow growth was performed. In cells with six replication forks per chromosome, the two old forks were found to bind surprisingly little SeqA protein. Cell cycle analysis showed that loss of SeqA from the old forks did not occur at initiation of the new forks, but instead occurs at a time point coinciding with the end of SeqA-dependent origin sequestration. The finding suggests simultaneous origin de-sequestration and loss of SeqA from old replication forks.
INTRODUCTION
When growing cells divide they need to copy their genetic material and distribute it to ensure that each daughter cell receives one copy. This is a challenging task especially considering the enormous length of the DNA compared to cell size. During DNA replication, organization of the chromosomes is even more demanding, since replication forks continuously produce new DNA. In eukaryotic cells, sister chromatids are held together by cohesin protein complexes during ongoing replication. At anaphase onset, a protein called separase is activated to cleave a subunit of the cohesin complex allowing separation of the two sister chromatids and subsequent transport to the cell poles. In prokaryotic cells, DNA replication and segregation are not temporally separated processes. Some evidence suggests that in E. coli newly synthesized DNA is continuously segregated to opposite cellular positions (1,2). Other work indicates that some parts of segregation may be more abrupt and domain specific (3,4). Coordination of DNA replication and chromosome segregation is complicated by the ability of E. coli to grow with overlapping replication cycles (5,6). Whereas during slow growth, chromosomes are replicated in a simple pattern with only one pair of forks; replication during fast growth occurs with one pair of old and two pairs of new forks on one chromosome. (Forks are considered to be ‘old forks’ as soon as new forks appear at initiation.) Depending on the exact conditions, a cell can have four copies of the multi-fork chromosome and a total of 24 replication forks per cell (7,8). How the cell meets the obvious need for efficient organization during such extensive replication is largely unknown. However, the SeqA protein is one of the strongest candidates to contribute (9,10). Loss of SeqA leads to severe growth impairment during rapid but not slow growth (11). Biochemical studies established that SeqA binding is specific for the sequence GATC with high preference for hemimethylated over fully methylated DNA (12–15). Hemimethylation occurs at newly replicated GATC sites which have not yet been re-methylated by the Dam methylase. A transient hemimethylation after passage of the replication fork was found in an analysis of 10 individual GATC sites (16). Similarly, transient binding of SeqA was detected at seven genomic sites with multiple GATC sequences (17). Multiple DNA-bound SeqA dimers can oligomerize to form a higher order structure (18,19). The above findings suggested that a SeqA complex follows the replication forks, potentially in a treadmilling fashion, growing at the leading end and diminishing at the tailing end. The reduction of the SeqA bound region at the most replisome-distant GATCs would come about through the activity of Dam which turns these sites into non-targets for SeqA by its methylation activity. In this study, we provide strong support for the above model and visualize the proposed SeqA bound region and its hemimethylation for the first time.
MATERIALS AND METHODS
Cell growth and temperature shift experiments
All strains used were E. coli MG1655 derivatives (Table 1). Cells were grown at 37°C to OD600 of about 0.15 in LB-glucose or AB medium supplemented with 10 µg ml−1 thiamin, 25μg ml−1 uridine and either 0.2% glucose and 0.5% casamino acids (CAA) or 0.4% sodium (Na) acetate. Antibiotic selection was used at the following concentrations: ampicillin 100 µg ml−1, chloramphenicol 30 µg ml−1 and tetracycline 10 µg ml−1.
Table 1.
Strains used in this study
| Strain | Relevant characteristic(s) | Source or reference |
|---|---|---|
| E. coli MG1655 | F-λ-rph-1 (wild-type) | (45,46) |
| MG1655dnaC2 | F-λ-rph-1 thr::Tn10 dnaC2 (tet) | (47) |
| TB16 | MG1655 GATC-cluster::tnaA (cm) | (26) |
| TWX30 | MG1655 GATC-cluster::srlA (cm) | This study |
| TWX31 | MG1655 GATC-cluster::hisA (cm) | This study |
| TWX34 | MG1655 GATC-cluster::ter (cm) | This study |
| TWX48 | MG1655dnaC2/pMQ430 (tet, amp) (25) | This study |
For synchronization of cells, according to the initiation of DNA replication, overnight cultures of MG1655 dnaC2 or dnaC2/pMQ430 (TWX48) were diluted 1/1000 in AB glucose-CAA or LB medium, grown exponentially to OD450 ∼0.07 at 30°C (permissive temperature) before shifting to 39°C (non-permissive temperature) for 70 min. Replication was initiated by returning the cultures to 30°C and adding appropriate amounts of 4°C medium (time point 0). At different time points after downshift, 50 ml of the culture was cross-linked for subsequent ChIP analysis or 5 ml of the culture for DNA isolation and subsequent methylation analysis.
Strain construction
The GATC-cluster insertion strains were constructed as described with primers hisa_im_rev/fwd for the hisA insertion, srla_mg_rev/fwd for the srlA insertion and lsr_mbnrn_rev/fwd for the terC insertion (Table 2). Genomic positions of the integration according to the genome annotation (NC_000913.2) were 2093653–2093795 (hisA), 2823883–2825244 (srlA) and 1596580–1596641 (terC). P1vir transduction and introduction of plasmids into competent cells by heat-shock was used for construction of other strains.
Table 2.
Oligonucleotides used in this study
| Name | Sequence |
|---|---|
| Cluster-fw | CTGACTGATGAGATCCAACGA |
| Cluster-rv | CTGGTGCTACGCCTGAATAA |
| Cluster-p | AAATTCGACCCGGCTGTCGC (5′ FAM–3′ TAMRA) |
| 3923874fw | GCCCTGTGGATAACAAGGAT |
| 3923874rv | CCTCATTCTGATCCCAGCTT |
| 3923874p | CGGTCCAGGATCACCGATCATTC (5′ FAM – 3′ TAMRA) |
| Ter-fw | TCCTCGCTGTTTGTCATCTT |
| Ter-rv | GGTCTTGCTCGAATCCCTT |
| Ter-p | CATCAGCACCCACGCAGCAA (5′ FAM – 3′ TAMRA) |
| MseIlong | AGTGGGATTCCGCATGCTAGT |
| MseIshortnewNO | TAACTAGCATGC |
| hisa_im_fwd | CTGGTGGAAACCTATCTGCCCGTCGGCCTGAAA CATGTGCGCGCCGAATAAATACCTGTGACGG |
| hisa_im_rev | AGTAATGCCCGACCAACTATTACGCCGCGCACACC AGTGCCCGTCCTGTGGATCCACTGAATTATG |
| srla_mg_fwd | GGAGAGAACAATGATAGAAACCATTACTCATGGTGC AGAGGCGCCGAATAAATACCTGTGACGG |
| srla_mg_rev | GACGCGAACCGTGTCCTGACGGGCTTCCGCCAGCGA CAAACCGTCCTGTGGATCCACTGAATTATG |
| lsr_mbnrn_fwd | AAGCTGGCTTTTAACAAGCCAGCTCTAAAAGAAGGG AAATCCGTCCTGTGGATCCACTGAATTATG |
| lsr_mbnrn_rev | TGGACTGACGACGTCGTTATGGAAAGCGCCTGGGTT ATAGGCGCCGAATAAATACCTGTGACGG |
Quantitative RT-PCR
Reactions were carried out as described (20) with primers cluster-fw/rv/p for the GATC-cluster and 3923874fw/rv/p for oriC analysis and ter-fw/rv/p for the terminus (Table 2). Standard deviation of triplicate reactions was around 3.5%.
ChIP-Chip and data analysis
Preparation of cell extracts, IP, labelling of DNA, array hybridization and data processing was as described in our optimized protocol (20). All experiments were carried out in duplicate but for synchronized cells only one data set is shown in the figures. Raw as well as processed data are available at the Genome Omnibus Database, accession number GSE28280.
Determination of SeqA bound regions for synchronized cells was based on ChIP-Chip data from time point 17 min. Probes were filtered based on two criteria. First, the probe and both neighbouring probes have an enrichment value >3. Second, the enrichment ratio of the three probes is >50% of the value of SeqA grown unsynchronized in LB (20). A total of 4419 data points passed this constrains. As expected, these points were restricted to the region ∼700 kb to the left and right of oriC as suggested by the whole chromosome plots (Figure 2, one exception was omitted from further analysis). Neighbouring probes were fused to yield one block if less than 500 bp distant. This gave 385 blocks of which 102 were removed because they consisted of one or two probes only resulting in 283 SeqA binding blocks (Supplementary Table S1). For analysis of the Dam effect on SeqA binding (Figure 4B), only the 251 inner blocks were considered (Supplementary Table S1). The GATC number given is the number of GATCs in the respective region plus 500 bp up- and down-stream. The relative GATC number is the number of GATCs divided by the number of bp in this region.
Figure 2.
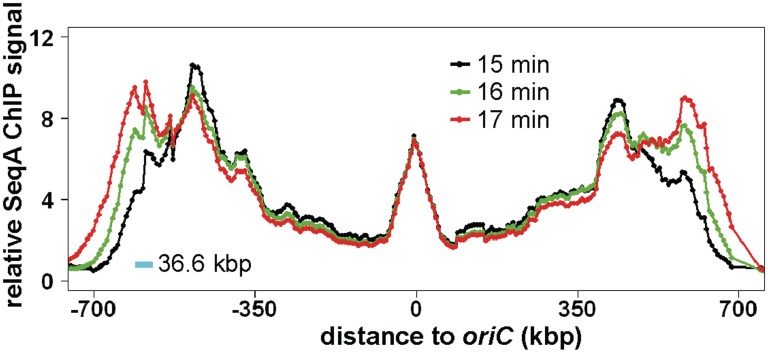
SeqA binding in synchronized E. coli dnaC2 cells at 15, 16 and 17 min after initiation. The experiment and data processing was as described (Figure 1). Relative values for the three time points were aligned at the oriC region based on the finding that SeqA binding there is relatively stable. Only the SeqA bound part of the chromosome is shown with oriC in the middle and the distance to oriC as indicated (for whole genome plot of 15 min time point see Figure 1). Values for the different time points are colored according to legend. The blue scale bar indicates the number of base-pairs replicated per minute in the used system according to (21).
Figure 4.
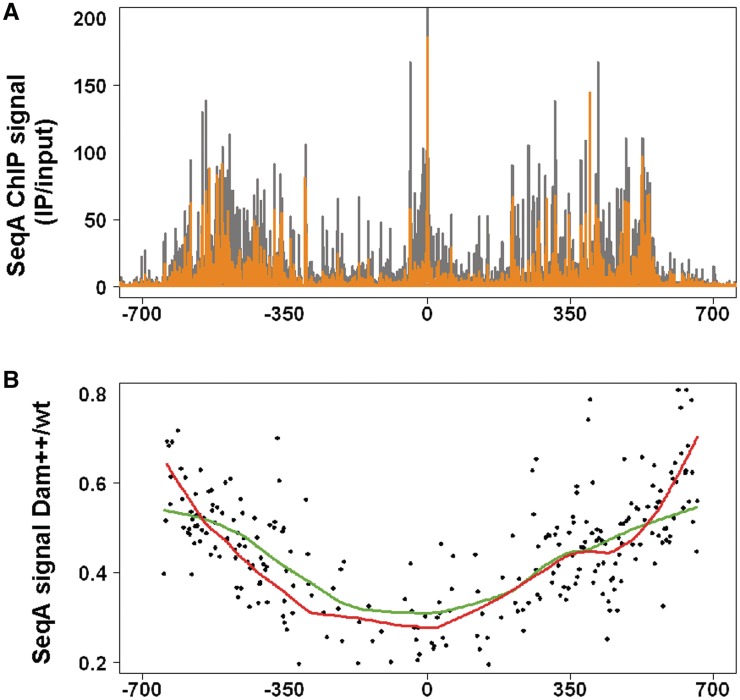
SeqA and Dam compete for GATC sites. (A) ChIP-Chip binding profile of SeqA binding in cells overproducing Dam methylase (orange) compared to cells with wt level of Dam (grey). Temperature shift was as above with cells grown in LB medium (+/− 0.2 % L-arabinose) and cross-linking 15 min after synchronization. Dam induction was through the pBAD18 derivative pMQ430 (25). (B) The average SeqA ChIP signal for 249 SeqA binding blocks was calculated (see ‘Material and Methods’ section for details). Ratios of SeqA signals in Dam overproducing to wt cells were plotted versus chromosomal position. Lowess trendlines (f = 0.3) are given for high (green) and low (red) GATC density regions.
Methylation analysis
Five hundred nanograms of chromosomal DNA were digested with 10 U of MseI (NEB) in 10 μl volume for 3 h at 37°C and heat inactivated for 20 min at 65°C. To prepare adapters, 100 pmol of MseIlong and MseIshortnewNo were annealed in 8 µl ddH2O by heating to 95°C for 3 min and then cooled to 70°C and subsequently to 15°C with 1°C per min. At 15°C, 10 µl MseI digested DNA, 2 µl ligase buffer and 400 U T4-ligase (NEB) were added and ligated over night. Ligase was inactivated at 65°C for 10 min. One half of the ligation mixture was digested with 20 U DpnI for 2 h at 37°C in a volume of 50 µl and the other half treated similarly with water instead of DpnI as control. Five microlitres of the DNA was amplified in a 50 µl PCR reaction with 0.2 mM dNTPs, 0.5 µM primer MseIlong, 10 µl Phusion HF buffer and 1 U Phusion DNA polymerase (Finnzymes) with the program 30 s 98°C, 20× (30 s 98°C, 30 s 62°C, 60 s 72°C), 10 min 72°C. DNA was purified with a Qiagen PCR cleanup kit and ∼200 ng used for labeling with Cy3- or Cy5-dCTP using the BioPrime kit from Invitrogen. Hybridization was for about 36 h at 55°C to custom E. coli microarrays from Oxford Gene Technology. The arrays have a probe length of 60 bases and contain one probe for each possible MseI cut fragment with a size of 60–1000 bp.
Cell cycle analysis
Three cultures of E. coli MG1655 grown in LB-glucose were analyzed by flow cytometry as described (8). Generation times were determined from regression lines of the growth curves with a standard deviation of 0.8 min. The C period was derived from quantitative RT-PCR with primers specific for the origin and the terminus (Table 2) using the formula ori/ter = 2C/τ. The initiation age was calculated from the ratio of 8- to 16-chromosome cells after treatment with rifampicin and cephalexin (standard deviation 6.5%) as described (8).
RESULTS
The SeqA binding tract follows the replication fork
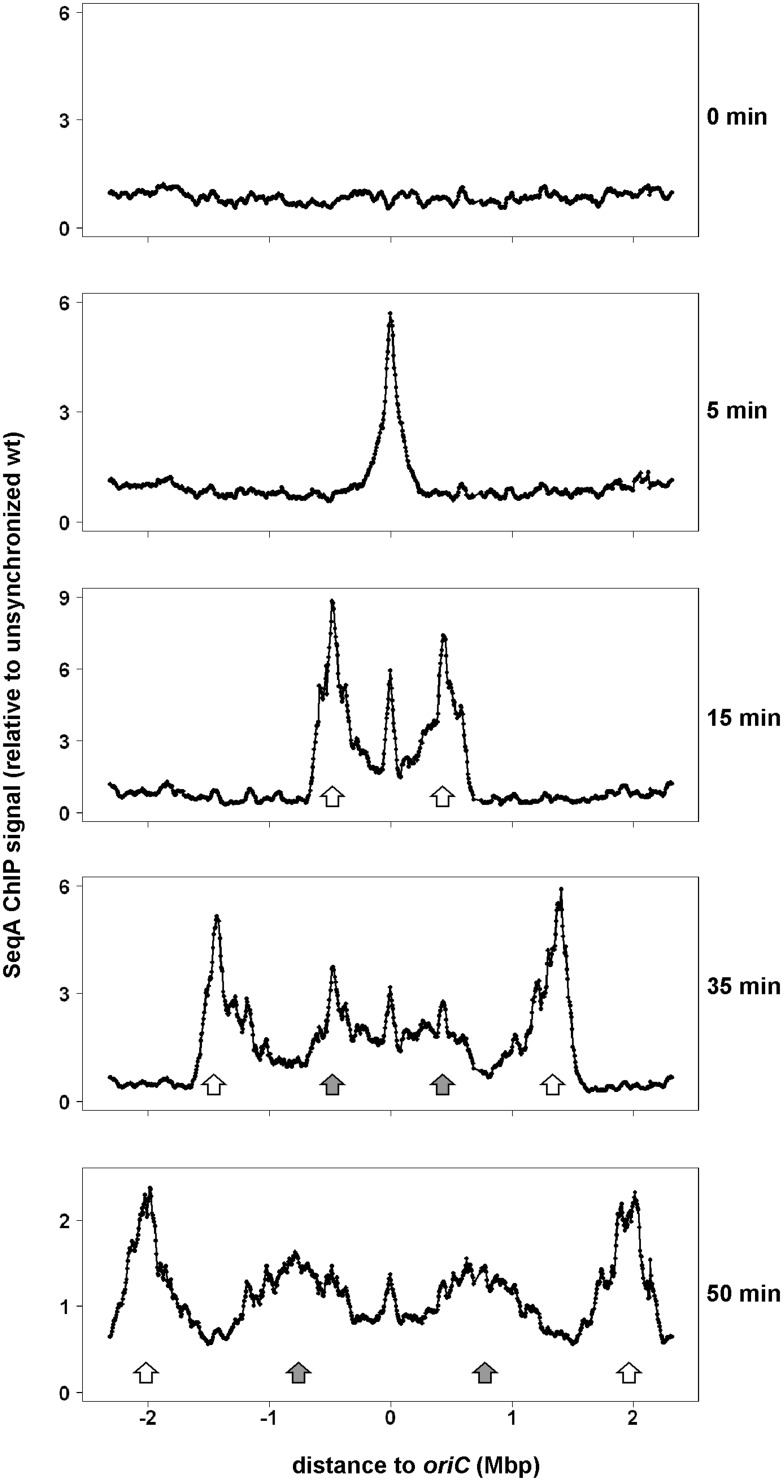
In E. coli, a pair of replication forks moves bidirectionally from oriC towards the replication terminus. We asked how SeqA binding relates to the movement of the replication fork. To this end, we applied ChIP-Chip to E. coli cells synchronized with regard to DNA replication initiation using the temperature sensitive strain MG1655dnaC2. After temperature up-shift, initiation is inhibited but ongoing replication cycles will be completed leading to cells with fully replicated chromosomes. Upon temperature down-shift, the cells initiate DNA-replication in synchrony. Samples of the culture were taken at 0, 5, 15, 35 and 50 min after replication restart and analyzed by ChIP on Chip with a SeqA specific antibody. Binding profiles of data relative to unsynchronized cells are shown in Figure 1. At time 0, no SeqA is bound to the chromosome, which is expected to be fully methylated at this time point. Five minutes after initiation of replication, SeqA binding was detected at an approximately 400 kbp wide region peaking at oriC. At the 15 min time point, two distinct SeqA binding peaks appear ∼0.5 Mbp to the left and right of oriC (Figure 1, white arrows). Notably, strong SeqA binding was also detected in the oriC region. At later time points, the two main SeqA binding peaks moved symmetrically towards the terminus region (Figure 1, white arrows). This is consistent with SeqA binding following a pair of replication forks. Two additional SeqA binding peaks were observed at 35 min after initiation (Figure 1, grey arrows). The peaks are, in a region, covered by SeqA at the 15 min time point. This suggests that a second initiation event occurred approximately 20 min after the first and that the observed SeqA binding follows a second pair of replication forks. These second SeqA-bound regions move bidirectionally towards the terminus region as indicated by their positions after 50 min.
Figure 1.
Whole genome SeqA binding in synchronized E. coli dnaC2 cells. Cells were grown exponentially in AB medium supplemented with glucose and CAA at 30°C and shifted to 39°C for 70 min. Synchronous initiation of replication was induced by a downshift to 30°C. Parts of the culture (60 ml) were removed and subjected to cross-linking at the indicated time points after downshift. ChIP-Chip was performed with a SeqA specific antibody and enrichment values are relative to values of an unsynchronized wt culture (LB) with a moving window of 60 kbp and a step size of 6 kb (20). Data are plotted as a function of their chromosomal position with oriC in the middle and the distance to oriC indicated. The first pair of SeqA peaks/replication forks and the second pair after reinitiation are indicated by white and grey arrows, respectively.
The SeqA binding peaks were asymmetric, i.e. unlike a normal distribution. Instead, the fork-proximal side seems to be steeper than the side nearer oriC. Such a pattern would suggest that SeqA binds fast to newly replicated DNA and leaves the DNA in a more random reaction. To get a higher time resolution, we applied ChIP-Chip to cells also 16 and 17 min after initiation (Figure 2). Earlier reports have shown that in 1 min the replication forks move about 36.6 kbp in the system we used (21) (Figure 2, blue scale bar). This distance corresponds well with the difference in the leading edge of SeqA binding at 16 as compared to 15 min, and at 16 as compared to 17 min. The finding shows that the new GATC sites are quickly bound by SeqA. It is noteworthy that the appearance of SeqA binding as two or more side-by-side peaks for the left forks as well for the right forks is due to the distribution of GATC sites. While SeqA binding at the replication fork side was highest at 17 min and lowest at 15 min, this was opposite on the side of the peak facing oriC. This is consistent with SeqA continuously leaving the ‘older’ GATC sites and leads to a reduction of SeqA bound at the 15 min time point 1 and 2 min later. The reduction per time was not as high as the increase of SeqA binding per time near the forks. It is not clear whether this is due to a surplus of SeqA protein in the relatively large synchronized cells, or whether the phenomenon is found also in asynchronously growing cells.
SeqA binding correlates with hemimethylation
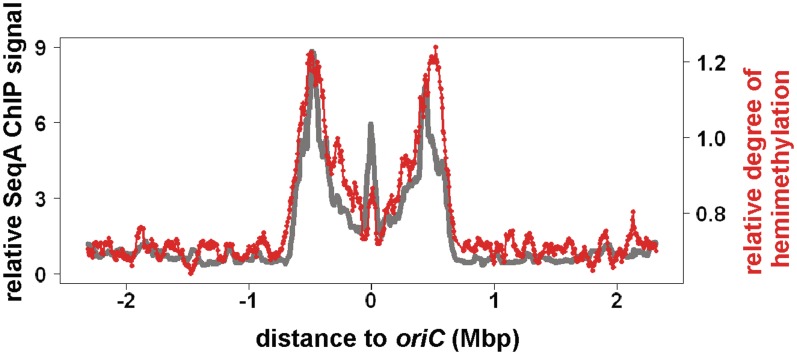
Biochemical experiments have shown that SeqA has a high affinity for hemimethylated as compared to fully methylated DNA. This prompted us to analyze if SeqA binding correlates with the level of hemimethylation also in vivo. To this end, we developed a method to analyze genome-wide methylation in bacteria. The basic principle of this new method is the same as has been applied to analyze cytosine methylation in eukaryotic cells (22). Chromosomal DNA was first digested with MseI to yield fragments of an average size around 250 bp. The fragments were then ligated to universal linkers and cut with the methylation-specific endonuclease DpnI. Since DpnI cuts only fully methylated DNA, fragments with hemimethylated GATC sites will remain intact. In the subsequent PCR amplification with linker-specific primers, only these intact fragments will be amplified. We applied this method to DNA isolated from synchronized cells 15 min after initiation of DNA replication. The processed DNA was labelled and hybridized to a custom DNA microarray with probes specific for MseI-derived fragments. DNA which was not cut with DpnI functioned as hybridization control. A whole-genome plot shows that hemimethylation is restricted to an extended region around oriC with peaks at the origin and at two symmetrical sites on both chromosome arms (Figure 3). The overall pattern resembles that of SeqA binding under similar conditions (Figure 3, grey line). At oriC, the ratio between the hemimethylation signal and SeqA binding appears smaller compared to the replication fork regions. However, because the methylation analysis is considered semi-quantitative only, it remains to be seen if this finding is significant.
Figure 3.
Methylation of synchronized E. coli dnaC2 cells at 15 min after initiation. Cell growth was as described (Figure 1). 15 min after synchronous initiation of replication cells were collected and chromosomal DNA isolated. For details on the methylation analysis, see ‘Results’ and ‘Materials and Methods’ sections. Values shown are average enrichment factors for a moving window of 60 kbp, step size of 6 kb. Red, methylation analysis. Grey, SeqA binding data from Figure 2 at similar time point.
The competition of Dam methylase and SeqA for GATC sites
Newly synthesized hemimethylated DNA is a target also for Dam methylase. In fact, Dam and SeqA have been suggested to be in competition for hemimethylated DNA (14). Experiments with unsynchronized cells indicate that the sequestration period becomes shorter upon Dam overexpression (23,24). SeqA binding is largely limited to hemimethylated DNA, and the action of Dam will, therefore, transform DNA into a non-target for SeqA. To characterize the competition, we overproduced Dam methylase using an established vector system (25) and analyzed the effects on SeqA binding in E. coli cells 15 min after replication initiation as above (Figure 4). SeqA was able to bind DNA despite Dam overproduction. The overall binding pattern was similar to the wt control (Figure 4A, compare orange with grey bars). This result is consistent with in vitro findings showing that SeqA bound to DNA was not actively dissociated by Dam methylase (14). The same study showed that the SeqA protein spontaneously dissociated from bound DNA after some minutes in vitro and that re-binding to the same site was inhibited by methylation (14). We reasoned that in our in vivo system the effect of Dam overproduction on SeqA re-binding should increase with increasing distance from replication forks towards oriC. This is because the longer a SeqA molecule was bound to the DNA the more likely is its dissociation. Such an effect might be too small to be observed by visual comparison of SeqA binding patterns. To check this prediction, we calculated the average SeqA ChIP signal for 249 SeqA binding blocks (see ‘Material and Methods’ section). Ratios of SeqA signals in Dam overproducing to wt cells were plotted versus chromosomal position (Figure 4B). Ratios were high near the forks and decreased towards the origin in accordance with our prediction. This shows that Dam and SeqA are in continuous competition for GATC sites in vivo with SeqA being the considerably stronger competitor.
Since SeqA has been shown to bind better to DNA regions with more densely packed GATC sites, we speculated that such regions would allow SeqA to be better in competing against Dam than do regions with fewer GATC sites. We calculated the average GATC content of the 249 SeqA binding blocks and divided the data accordingly in two halves, namely regions with high- and low-GATC density. They show that low-GATC density regions have relatively high-SeqA binding near the forks in Dam overproducing cells and low-SeqA binding near oriC (Figure 4B). For high-GATC density regions, the curve is similar but appears somewhat more even. The data indicate that differences in GATC density have only minor impact on the competition of SeqA against Dam methyltransferase.
New replication forks bind SeqA more frequently than old forks
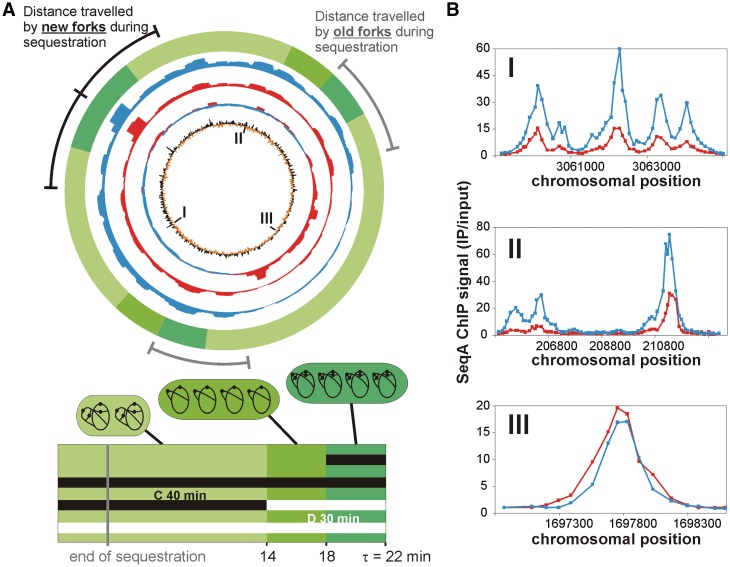
When cells grow slowly the replication fork pattern is quite simple; two forks move in opposite directions from the origin. During rapid growth, four new forks are started on the same chromosome before the two old ones are finished. Old forks are thus called old forks when new forks appear at initiation. We have investigated whether there is any difference in the binding of SeqA to newly replicated DNA in the two situations. ChIP-Chip experiments were performed with extracts from E. coli cells grown in AB-acetate medium at 37°C. Under the slow growth conditions, DNA replication cycles are completed before the next round starts. This data was compared to SeqA ChIP-Chip analysis of E. coli grown in LB-glucose medium (20). The rich medium leads to overlapping DNA replication cycles with multiple forks per chromosome. The overall binding patterns of SeqA appeared similar for both conditions, with good correlation to the density of GATC sites (Figure 5A). Thus, in both poor and rich medium, there is little binding of SeqA to terminus-proximal DNA (Figure 5A, red and blue circles). To compare the SeqA binding in the two situations, the data sets were divided by one another (Figure 5A blue/red circle). This revealed that the SeqA binding was higher in the terminus region in cells with the simple replication pattern relative to cells with overlapping replication cycles. In contrast, most of the rest of the chromosome was more frequently bound by SeqA when replication cycles were overlapping. This pattern could also be seen at individual binding sites (Figure 5B). It is important to note that the ChIP on Chip experiments as performed here give a value of enrichment relative to the chromosomal DNA. The different copy numbers of oriC- and ter-proximal DNA in cells with overlapping replication can thus not explain the above findings.
Figure 5.
Chromosome-wide SeqA binding varies according to the replication pattern. (A) Top panel: Whole-genome plot of the SeqA binding in unsynchronized, slow-growing E. coli MG1655 cells (AB-acetate) with no overlapping replication (red) and unsynchronized, fast-growing E. coli MG1655 cells (LB-glucose) with overlapping replication (blue). The sum of SeqA ChIP signals in windows of 60 kbp are shown (step size 1 kbp; only values >1 were included). Red/blue second circle from inside: mean centred line graph with ratios of AB-acetate (red) and LB-glucose (blue). Inner circle: mean centred line graph of number of GATC sites in windows of 5 kbp (step size 1 kbp). Outer lines indicate the region replicated by the new (black) and old (grey) forks during the origin sequestration time found to be 1/3 of the cell cycle (16). Outer green circle (see below for explanation): regions replicated by new and old forks at different stages of the cell cycle presented with the same colour code as in the bottom panel. Bottom panel: Schematic of the replication pattern of the cells grown in LB-glucose medium. The width of the rectangle represents one generation with the different greens indicating the different events in the life of the average cell: the period from cell birth to termination (0–14 min), from termination to initiation (14–18 min) and from initiation to cell division (18–22 min). Examples of cells (rods in shades of green) are shown above each of the three intervals with chromosomes (black lines) and origins (black dots). Horizontal lines represent the C period (black) and D period (white line; the total duration of which spans more than three generations) and should be read from top to bottom (see ‘Materials and Methods’ section for calculation details). Initiation of chromosome replication occurs in 18 min old cells leading to 16 copies of oriC (dark green). After cell division at an age of 22 min the cells contain eight oriC copies (light green). The chromosomes are then replicated with four old and eight new forks (light green cell). Old forks finish replication at 14 min cell age. (B) Examples of SeqA binding to chromosomal sites in AB-acetate (red) and LB-glucose medium (blue). Values are mean values of two replicates. Chromosomal positions of sites are marked by roman numbers in A.
The main difference between the replication forks of slowly growing and rapidly growing cells is that the former have no competition from new forks when replicating the terminus region of the chromosome, whereas forks of rapidly growing cells do. To get a picture of whether competition from new forks is a reasonable explanation for the observation, we calculated where the old and new replication forks are situated relative to each other during growth in LB-glucose medium based on cell cycle analysis (Figure 5A outer green circle and lower panel; see ‘Materials and Methods’ section). At initiation, cells contained four chromosomes with two replication forks and two origins on each chromosome. The old replication forks had then replicated about 55% of the chromosome. However, the terminus area of low SeqA binding is not encountered until after about 70–75% is replicated. Thus, the old forks do not enter this area at the time of initiation but instead 7–8 min later, which means that there is a substantial interval between the large increase in numbers of replication forks at initiation of DNA replication and the point where SeqA is lost from the old forks. Interestingly, the duration of this interval is similar to that of the origin sequestration period of about 1/3 of the cell cycle according to (16) (Figure 5A, grey and black lines in top panel). The result indicates that although competition from new forks might be involved in the low SeqA binding in the terminus area of rapidly growing cells, this cannot be the only explanation. There seems to be a delay corresponding to the origin sequestration period before SeqA is lost from the old forks.
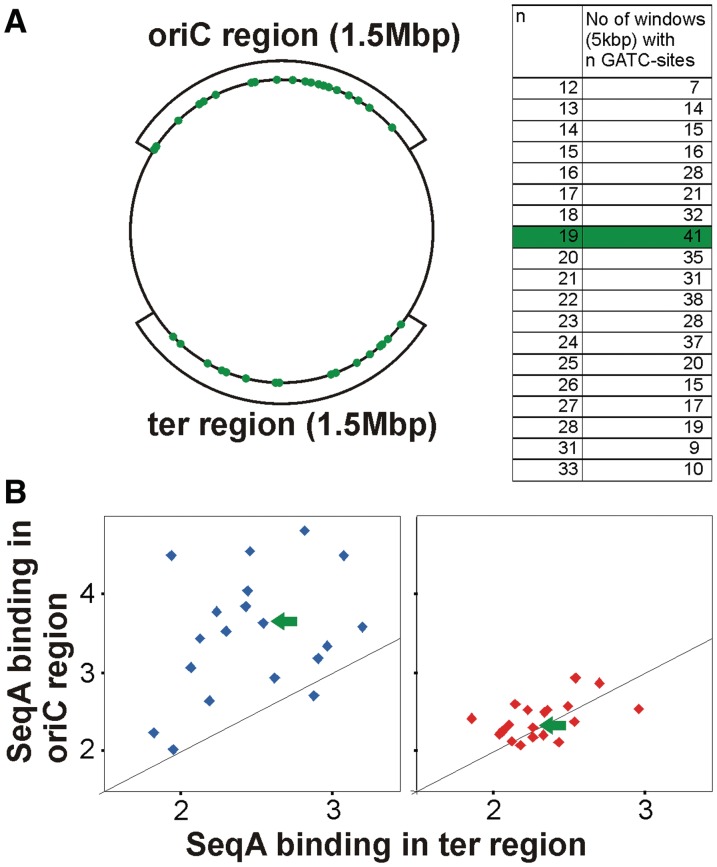
It is also possible that the difference in SeqA binding under fast compared to slow growth does not have to do with organization of new and old forks, but, for instance, with events specific to termination of replication in slow growing cells. To exclude this possibility, the fast growth data set was analyzed separately comparing the binding signal in regions replicated by new forks (oriC proximal) and old forks (ter proximal; Figure 6A). Such a direct comparison is made difficult by the unsystematic distribution of GATC sites. To overcome this problem, we divided the chromosome in 5 kbp windows and grouped them according to their GATC content (Figure 6A table). We then calculated the average SeqA binding in windows with the same numbers of GATC sites for the oriC and the ter regions, respectively. A plot of these values showed that there is significantly higher binding of SeqA in the region around oriC (replicated by new forks) compared to the ter region (replicated by old forks; Figure 6B left panel). This finding is supported by the finding that there was no significant difference in SeqA binding to the two regions when the slow growth data set was analyzed in a similar way (Figure 6B right panel).
Figure 6.
New replication forks are more frequently bound by SeqA. (A) The chromosome was divided in 5 kbp windows and grouped according to their GATC content and the number of respective windows in two 1.5 Mbp regions at the origin and terminus was calculated as indicated. An example of 41 windows with 19 GATCs is marked in green. (B) The average SeqA ChIP signal was calculated for all windows in the ter or oriC region with the same number of GATC sites. Values were plotted versus each other for data derived from cells grown in LB-glucose (blue) and AB-acetate (red). The black line indicates the theoretical pattern if binding is the same in the oriC and the ter region.
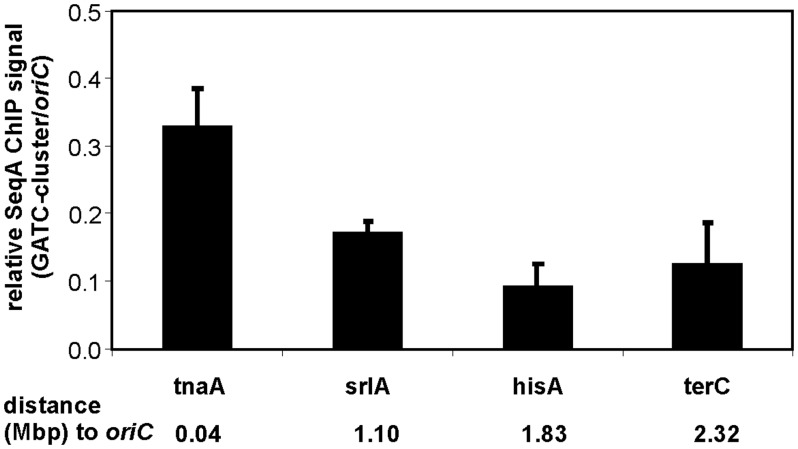
To further verify that the relative SeqA binding around the chromosome during rapid growth is higher at new forks compared to old forks, we made use of a synthetic cluster of GATC sites with the same GATC distribution as oriC with random sequences between the sites (26), and investigated binding of SeqA to the cluster at four different positions (tnaA, srlA, hisA and terC) around the chromosome in fast growing cells. For each cluster-insertion strain ChIP-qPCR experiments were performed with relevant primers (Table 1). SeqA enrichment at the GATC cluster at tnaA was found to be 0.33 relative to oriC whereas when the GATC cluster was situated at the terminus, it bound much less SeqA with 0.12-fold enrichment relative to oriC (Figure 7). Similar low SeqA enrichment (0.09 fold) was also found when the GATC cluster was inserted at hisA. This insertion site is further away from the terminus but still in the chromosomal region replicated by old forks (Figure 7). At the GATC cluster inserted into srlA, at about halfway between oriC and the terminus, SeqA enrichment was intermediate (0.17-fold). The results support the above finding that replication forks outside the terminus zone are more frequently bound by SeqA.
Figure 7.
SeqA binding to the GATCs cluster at different insertion sites. E. coli strains carrying cluster insertions at indicated sites were grown in LB batch culture. SeqA ChIP enrichment was quantified by qPCR with primers specific for the GATC cluster or oriC. Respective ratios of three experiments are shown with the standard deviation. The distance of the cluster insertion to oriC are indicated in Mbps.
Cooperativity of SeqA binding
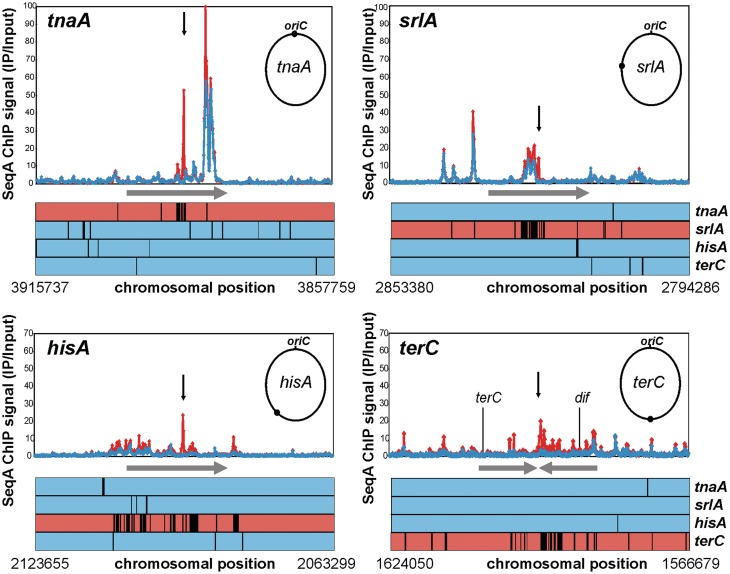
Considering the dynamic binding of SeqA, we asked what effect introduction of a strong binding site such as the GATC cluster has on neighbouring binding sites. To answer this question, we performed SeqA ChIP-Chip with extracts of the four GATC-cluster-insertion strains used above grown in a LB-glucose batch culture. The overall binding pattern resembled that of wild-type E. coli (data not shown). However, the insertion of the GATC cluster at terC increased the SeqA binding at sites up to ∼25 kb away (Figure 8). A similar effect was also seen for the other insertions, although less pronounced (Figure 8). Since the general level of ChIP enrichment can vary from experiment to experiment, normalization is needed for better comparison of data sets. To this end, we performed a ranked based data analysis selecting the 500 most enriched probes in an insertion strain relative to the other three (see ‘Materials and Methods’ section). Results for the insertion regions are shown as line plots in Figure 8. It appears that there is a significant increase in SeqA binding at sites in vicinity of the cluster-insertion.
Figure 8.
Insertion of a GATC cluster affects SeqA binding to neighbouring sites. E. coli strains with a cluster inserted at indicated regions (arrow) of the chromosome were grown in LB-glucose batch culture. SeqA ChIP-Chip was performed in duplicate for each strain. ChIP signals are plotted for a region of 250 probes up and down stream of the insertion (∼60 kbp). Red, values for the strain with the insertion. Blue, average of the respective three other strains. Line plots below the diagrams show a rank based data analysis. Every black line marks one data point out of the 500 most enriched in the respective strain compared to the three others. Colour code as above.
Interestingly, the amount of SeqA binding at the cluster seemed to be negatively correlated with its effect on neighbouring binding sites (compare Figures 7 and 8). While SeqA binding at the cluster inserted at terC and hisA was low the neighbouring binding sites bound more SeqA. On the other hand, SeqA binding to the cluster inserted at srlA and tnaA was relatively high with limited effects on neighbouring binding sites.
DISCUSSION
Why does SeqA bind more frequently to new replication forks?
In contrast to many other bacteria, E. coli is capable of overlapping replication with one pair of old and two pairs of new forks on each chromosome when growing fast. As a consequence, the chromosome is divided into a region near oriC which is replicated by new/double pairs of forks and a region around the terminus which is replicated by old/single pairs of forks. We found here that SeqA binding differs in these two regions with stronger/more frequent binding at new forks and weaker/less frequent binding at the old forks. Fluorescence microscopy experiments have demonstrated that SeqA protein may be visualized in cells as a number of distinct foci that co-localize with BrdU labelled, newly synthesized DNA (7,27,28). In LB medium, 4–8 SeqA structures can be detected per cell (8). In this medium, initiation of replication and generation of new forks occurs when the old forks have travelled about halfway to the termini (Figure 5). At this point, in time, there is no abrupt increase in the numbers of SeqA structures per cell. This has led to the suggestion that the forks are recruited to existing SeqA structures. We find here that there is relatively little binding of SeqA to the terminus-proximal quarter of the chromosome in rapidly growing cells compared to slowly growing cells (Figure 5). This might suggest that the new DNA synthesized at forks replicating the terminus-proximal quarter of the chromosome is no longer bound to SeqA structures in rapidly growing cells. It therefore seems that the SeqA that binds the new DNA behind the replication forks in effect is transferred from old forks to new forks on the same chromosome. Since new forks appear when the old forks have come about half way, but old forks do not show low SeqA binding until about a quarter is left to replicate, there seems to exist a period with both new and old forks in the same SeqA tract or structure.
Whether the above explanation involving a period where the newly replicated DNA at both old and new forks are found in SeqA structures is correct, requires further investigation. It implies that it is not the competition from the new forks that leads to the loss of SeqA at the old forks. If so, something else must drive the loss of old forks from the SeqA structures. One possibility is that re-organization occurs when origins are released from sequestration (or that sequestration ends because re-organization occurs). The sequestration period lasts for about a third of a generation which would in this case represent 7.3 min which is about the same period found from initiation of new forks to the loss of SeqA from old forks (Figure 5). Since no change in the numbers of SeqA fluorescens foci is seen at the time of initiation it has been suggested that the SeqA that remains at an origin throughout the sequestration period is part of a main SeqA structure or that it at least remains close enough to not be resolved by a light microscope. It is not known whether the release of origins from sequestration is an active process. Experiments comparing the binding of SeqA to the GATC cluster situated either in tnaA or in datA with SeqA binding at oriC, indicate that sequestration of these areas may not last for as well-defined a period of time (26,29). Thus, it may be that the end of origin-sequestration involves a reorganization of DNA and/or an active removal of SeqA.
Alternatively, or in addition, it is possible that special segregation requirements of the terminus-proximal parts of the chromosome impose removal of this part of the chromosome from SeqA structures (30,31). Interestingly, the nucleoid occlusion proteins Noc (in B. subtilis) and SlmA (in E. coli) show a similar binding pattern to their respective chromosomes with binding only outside an extended region around the terminus (32–34). Both proteins inhibit cell division by interaction with FtsZ. A similar mechanism was found for MipZ in Caulobacter crescentus (35). While the above-mentioned proteins do not bind to the ter region the MatP protein shows the reverse distribution of binding to the E. coli chromosome with exclusive binding in the ter macrodomain (36). All in all, these examples indicate that there are requirements for segregation and organization of the terminus region of the chromosome that are distinct from the rest of the chromosome and reflected in the distribution of DNA binding proteins (37).
Cooperativity of SeqA binding
In vitro experiments have shown SeqA binding to be cooperative. Cooperativity in this context means that binding of one SeqA dimer to a pair of GATCs increases the likelihood of a second dimer binding to nearby sites. This mode of action was attributed to the ability of SeqA to oligomerize. In the work presented here, we found cooperativity of SeqA binding also in vivo (Figure 8). Importantly, this cooperativity is for far more distant sites than that found in vitro as insertion of a GATC cluster near terC had a positive effect at SeqA binding on sites more than 20 kbp away (Figure 8). This cooperativity is likely to involve long-range interactions mediated by DNA loop formation. This is a common principle used, for example, by the λ repressor or enhancers in higher eukaryotes (38,39). One crucial point is that long-range interactions involve multiple protein-DNA contacts (40), a prerequisite fulfilled by SeqA. One way to explain the enhancement of SeqA binding at sites neighbouring the GATC cluster without multimerization is to propose an effect of local concentration. SeqA molecules dissociating from the GATC-cluster might represent a pool for rebinding to nearby GATC-sites that have lost their SeqA protein. Such a dissociation and secondary binding was also observed in vitro and could be inhibited by methylation of the SeqA binding sites by Dam methyltransferase or a large excess of hemimethylated DNA (14). Interestingly, the positive effect on SeqA binding was found on both sides of the GATC cluster insertion. This indicates that the SeqA structure is highly dynamic and SeqA molecules constantly go on and off the DNA also at inside positions and not exclusively at the ends. This is opposing a strict treadmilling model with the SeqA structure growing at the leading end and shrinking in the trailing end. However, our experiments with synchronized cell cultures clearly illustrate that the treadmilling model in general holds true (Figures 1–3). Synchronized cells have been analyzed with SeqA ChIP-Chip before (41). At the two analyzed time points, unexpected binding of SeqA to the ter region was found before replication restart and binding of SeqA at sites throughout the chromosome 6 min after restart of DNA replication (41). The discrepancy to our results might be due to methodology differences as discussed elsewhere (20).
A method for genome-wide methylation analysis
Here, we present the first method to analyze hemimethylation on a chromosome-wide scale based on protection against DpnI digestion. We used this method to extend the studies of Yamazoe et al. (17) who had shown the correlation of hemimethylation and SeqA binding in synchronized cells for individual sites (Figure 4).
The increasing awareness of the importance of epigenetics for various cellular processes and regulatory circuits has motivated the development of various analytic techniques over the last years (42). For eukaryotic systems, the addition of methyl groups to the N5 position cytosines in CpG dinucleotides is most relevant. The most common analysis method is bisulfite modification treatment of DNA which converts cytosine residues to uracil, but leaves 5-methylcytosine residues unaffected. Detection of modified nucleotides can subsequently be done by sequencing (43). Thus, a base pair resolution analysis of methylation is straight forward. For prokaryotic systems, the N6 methylation of adenines, as introduced by dam methylase, is the most interesting (44). However, the bisulfite modification is not applicable. An additional challenge is that it is desirable to analyze hemimethylated DNA which excludes ChIP based methods with a methylation-dependent antibody. Also, there is no restriction enzyme which is specific for hemimethylated DNA. The traditional way to quantify hemimethylated DNA is by cutting the DNA with methylation sensitive enzymes which overlap GATC sites (16). For example, the restriction enzyme HphI has the recognition site GGTGA. If this site overlaps a GATC site HphI would cut only if the respective half of the site is not methylated. Consequently, such methylation analysis is limited to such ‘diagnostic sites’ and not suitable to global methylation analysis. Although, the whole-genome method we developed is considered a great advance, it has to be pointed out that the resolution is limited. One reason is that the dependence on regions flanked by naturally occurring MseI restriction sites has the consequence that multiple GATC site are analyzed together. We suggest that this limitation could be overcome by adapting the introduced protocol to next-generation sequencing which should in principle make the need for MseI cutting obsolete.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table S1.
FUNDING
Norwegian Research Council FUGE program and German Research Foundation (Grant number WA 2713/1-1). Funding for open access charge: Oslo University Hospital.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Martina Bock for help with construction and analysis of GATC-cluster insertion strains. We are grateful to Ole Skovgaard for devising a way to find DNA fragments with the highest density of GATC sites and for many fruitful discussions. We thank Martin Marinus for providing plasmid pMQ430. We are grateful for the support from the Helse Sør-Øst/University of Oslo Microarray Core Facility. We thank Erik Boye and members of the Skarstad research group for critical reading of the manuscript.
REFERENCES
- 1.Nielsen HJ, Li Y, Youngren B, Hansen FG, Austin S. Progressive segregation of the Escherichia coli chromosome. Mol. Microbiol. 2006;61:383–393. doi: 10.1111/j.1365-2958.2006.05245.x. [DOI] [PubMed] [Google Scholar]
- 2.Reyes-Lamothe R, Possoz C, Danilova O, Sherratt DJ. Independent positioning and action of Escherichia coli replisomes in live cells. Cell. 2008;133:90–102. doi: 10.1016/j.cell.2008.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bates D, Kleckner N. Chromosome and replisome dynamics in E. coli: loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell. 2005;121:899–911. doi: 10.1016/j.cell.2005.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Espeli O, Mercier R, Boccard F. DNA dynamics vary according to macrodomain topography in the E. coli chromosome. Mol. Microbiol. 2008;68:1418–1427. doi: 10.1111/j.1365-2958.2008.06239.x. [DOI] [PubMed] [Google Scholar]
- 5.Cooper S, Helmstetter CE. Chromosome replication and the division cycle of Escherichia coli B/r. J. Mol. Biol. 1968;31:519–540. doi: 10.1016/0022-2836(68)90425-7. [DOI] [PubMed] [Google Scholar]
- 6.Skarstad K, Boye E, Steen HB. Timing of initiation of chromosome replication in individual Escherichia coli cells. EMBO J. 1986;5:1711–1717. doi: 10.1002/j.1460-2075.1986.tb04415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fossum S, Crooke E, Skarstad K. Organization of sister origins and replisomes during multifork DNA replication in Escherichia coli. EMBO J. 2007;26:4514–4522. doi: 10.1038/sj.emboj.7601871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morigen, Odsbu I, Skarstad K. Growth rate dependent numbers of SeqA structures organize the multiple replication forks in rapidly growing Escherichia coli. Genes Cells. 2009;14:643–657. doi: 10.1111/j.1365-2443.2009.01298.x. [DOI] [PubMed] [Google Scholar]
- 9.Waldminghaus T, Skarstad K. The Escherichia coli SeqA protein. Plasmid. 2009;61:141–150. doi: 10.1016/j.plasmid.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 10.Lu M, Campbell JL, Boye E, Kleckner N. SeqA: a negative modulator of replication initiation in E. coli. Cell. 1994;77:413–426. doi: 10.1016/0092-8674(94)90156-2. [DOI] [PubMed] [Google Scholar]
- 11.Boye E, Stokke T, Kleckner N, Skarstad K. Coordinating DNA replication initiation with cell growth: differential roles for DnaA and SeqA proteins. Proc. Natl Acad. Sci. USA. 1996;93:12206–12211. doi: 10.1073/pnas.93.22.12206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brendler T, Abeles A, Austin S. A protein that binds to the P1 origin core and the oriC 13mer region in a methylation-specific fashion is the product of the host seqA gene. EMBO J. 1995;14:4083–4089. doi: 10.1002/j.1460-2075.1995.tb00080.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brendler T, Austin S. Binding of SeqA protein to DNA requires interaction between two or more complexes bound to separate hemimethylated GATC sequences. EMBO J. 1999;18:2304–2310. doi: 10.1093/emboj/18.8.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang S, Lee H, Han JS, Hwang DS. Interaction of SeqA and Dam methylase on the hemimethylated origin of Escherichia coli chromosomal DNA replication. J. Biol. Chem. 1999;274:11463–11468. doi: 10.1074/jbc.274.17.11463. [DOI] [PubMed] [Google Scholar]
- 15.Slater S, Wold S, Lu M, Boye E, Skarstad K, Kleckner N. E. coli SeqA protein binds oriC in two different methyl-modulated reactions appropriate to its roles in DNA replication initiation and origin sequestration. Cell. 1995;82:927–936. doi: 10.1016/0092-8674(95)90272-4. [DOI] [PubMed] [Google Scholar]
- 16.Campbell JL, Kleckner N. E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell. 1990;62:967–979. doi: 10.1016/0092-8674(90)90271-f. [DOI] [PubMed] [Google Scholar]
- 17.Yamazoe M, Adachi S, Kanaya S, Ohsumi K, Hiraga S. Sequential binding of SeqA protein to nascent DNA segments at replication forks in synchronized cultures of Escherichia coli. Mol. Microbiol. 2005;55:289–298. doi: 10.1111/j.1365-2958.2004.04389.x. [DOI] [PubMed] [Google Scholar]
- 18.Guarne A, Brendler T, Zhao Q, Ghirlando R, Austin S, Yang W. Crystal structure of a SeqA-N filament: implications for DNA replication and chromosome organization. EMBO J. 2005;24:1502–1511. doi: 10.1038/sj.emboj.7600634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Odsbu I, Klungsoyr HK, Fossum S, Skarstad K. Specific N-terminal interactions of the Escherichia coli SeqA protein are required to form multimers that restrain negative supercoils and form foci. Genes Cells. 2005;10:1039–1049. doi: 10.1111/j.1365-2443.2005.00898.x. [DOI] [PubMed] [Google Scholar]
- 20.Waldminghaus T, Skarstad K. ChIP on Chip: surprising results are often artifacts. BMC Genomics. 2010;11:414. doi: 10.1186/1471-2164-11-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Breier AM, Weier HU, Cozzarelli NR. Independence of replisomes in Escherichia coli chromosomal replication. Proc. Natl Acad. Sci. USA. 2005;102:3942–3947. doi: 10.1073/pnas.0500812102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfister S, Schlaeger C, Mendrzyk F, Wittmann A, Benner A, Kulozik A, Scheurlen W, Radlwimmer B, Lichter P. Array-based profiling of reference-independent methylation status (aPRIMES) identifies frequent promoter methylation and consecutive downregulation of ZIC2 in pediatric medulloblastoma. Nucleic Acids Res. 2007;35:e51. doi: 10.1093/nar/gkm094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boye E, Lobner-Olesen A. The role of dam methyltransferase in the control of DNA replication in E. coli. Cell. 1990;62:981–989. doi: 10.1016/0092-8674(90)90272-g. [DOI] [PubMed] [Google Scholar]
- 24.Skarstad K, Lobner-Olesen A. Stable co-existence of separate replicons in Escherichia coli is dependent on once-per-cell-cycle initiation. EMBO J. 2003;22:140–150. doi: 10.1093/emboj/cdg003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calmann MA, Marinus MG. Regulated expression of the Escherichia coli dam gene. J. Bacteriol. 2003;185:5012–5014. doi: 10.1128/JB.185.16.5012-5014.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bach T, Skarstad K. An oriC-like distribution of GATC sites mediates full sequestration of non-origin sequences in Escherichia coli. J. Mol. Biol. 2005;350:7–11. doi: 10.1016/j.jmb.2005.04.055. [DOI] [PubMed] [Google Scholar]
- 27.Adachi S, Fukushima T, Hiraga S. Dynamic events of sister chromosomes in the cell cycle of Escherichia coli. Genes Cells. 2008;13:181–197. doi: 10.1111/j.1365-2443.2007.01157.x. [DOI] [PubMed] [Google Scholar]
- 28.Molina F, Skarstad K. Replication fork and SeqA focus distributions in Escherichia coli suggest a replication hyperstructure dependent on nucleotide metabolism. Mol. Microbiol. 2004;52:1597–1612. doi: 10.1111/j.1365-2958.2004.04097.x. [DOI] [PubMed] [Google Scholar]
- 29.Bach T, Morigen, Skarstad K. The initiator protein DnaA contributes to keeping new origins inactivated by promoting the presence of hemimethylated DNA. J. Mol. Biol. 2008;384:1076–1085. doi: 10.1016/j.jmb.2008.09.042. [DOI] [PubMed] [Google Scholar]
- 30.Lau IF, Filipe SR, Soballe B, Okstad OA, Barre FX, Sherratt DJ. Spatial and temporal organization of replicating Escherichia coli chromosomes. Mol. Microbio.l. 2003;49:731–743. doi: 10.1046/j.1365-2958.2003.03640.x. [DOI] [PubMed] [Google Scholar]
- 31.Wang X, Possoz C, Sherratt DJ. Dancing around the divisome: asymmetric chromosome segregation in Escherichia coli. Genes Dev. 2005;19:2367–2377. doi: 10.1101/gad.345305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cho H, McManus HR, Dove SL, Bernhardt TG. Nucleoid occlusion factor SlmA is a DNA-activated FtsZ polymerization antagonist. Proc. Nat. Acad. Sci. USA. 2011;108:3773–3778. doi: 10.1073/pnas.1018674108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tonthat NK, Arold ST, Pickering BF, Van Dyke MW, Liang S, Lu Y, Beuria TK, Margolin W, Schumacher MA. Molecular mechanism by which the nucleoid occlusion factor, SlmA, keeps cytokinesis in check. EMBO J. 2011;30:154–164. doi: 10.1038/emboj.2010.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu LJ, Ishikawa S, Kawai Y, Oshima T, Ogasawara N, Errington J. Noc protein binds to specific DNA sequences to coordinate cell division with chromosome segregation. EMBO J. 2009;28:1940–1952. doi: 10.1038/emboj.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thanbichler M, Shapiro L. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell. 2006;126:147–162. doi: 10.1016/j.cell.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 36.Mercier R, Petit MA, Schbath S, Robin S, El Karoui M, Boccard F, Espeli O. The MatP/matS site-specific system organizes the terminus region of the E. coli chromosome into a macrodomain. Cell. 2008;135:475–485. doi: 10.1016/j.cell.2008.08.031. [DOI] [PubMed] [Google Scholar]
- 37.Dame RT, Kalmykowa OJ, Grainger DC. Chromosomal macrodomains and associated proteins: implications for DNA organization and replication in gram negative bacteria. PLoS Genet. 2011;7:e1002123. doi: 10.1371/journal.pgen.1002123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dodd IB, Shearwin KE, Perkins AJ, Burr T, Hochschild A, Egan JB. Cooperativity in long-range gene regulation by the lambda CI repressor. Genes Dev. 2004;18:344–354. doi: 10.1101/gad.1167904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matthews KS. DNA looping. Microbiol. Rev. 1992;56:123–136. doi: 10.1128/mr.56.1.123-136.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oehler S, Muller-Hill B. High local concentration: a fundamental strategy of life. J. Mol. Biol. 395:242–253. doi: 10.1016/j.jmb.2009.10.056. [DOI] [PubMed] [Google Scholar]
- 41.Sanchez-Romero MA, Busby SJ, Dyer NP, Ott S, Millard AD, Grainger DC. Dynamic distribution of seqa protein across the chromosome of Escherichia coli K-12. MBio. 2010;1:e00012–10. doi: 10.1128/mBio.00012-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weitao T, Nordstrom K, Dasgupta S. Mutual suppression of mukB and seqA phenotypes might arise from their opposing influences on the Escherichia coli nucleoid structure. Mol. Microbiol. 1999;34:157–168. doi: 10.1046/j.1365-2958.1999.01589.x. [DOI] [PubMed] [Google Scholar]
- 43.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl Acad. Sci. USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wion D, Casadesus J. N6-methyl-adenine: an epigenetic signal for DNA-protein interactions. Nat. Rev. Microbiol. 2006;4:183–192. doi: 10.1038/nrmicro1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guyer MS, Reed RR, Steitz JA, Low KB. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harb. Symp. Quant. Biol. 1981;45(Pt 1):135–140. doi: 10.1101/sqb.1981.045.01.022. [DOI] [PubMed] [Google Scholar]
- 46.Jensen KF. The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J. Bacteriol. 1993;175:3401–3407. doi: 10.1128/jb.175.11.3401-3407.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Withers HL, Bernander R. Characterization of dnaC2 and dnaC28 mutants by flow cytometry. J. Bacteriol. 1998;180:1624–1631. doi: 10.1128/jb.180.7.1624-1631.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.