Abstract
Enzymes that modify the epigenetic status of cells provide attractive targets for therapy in various diseases. The therapeutic development of epigenetic modulators, however, has been largely limited to direct targeting of catalytic active site conserved across multiple members of an enzyme family, which complicates mechanistic studies and drug development. Class IIa histone deacetylases (HDACs) are a group of epigenetic enzymes that depends on interaction with Myocyte Enhancer Factor-2 (MEF2) for their recruitment to specific genomic loci. Targeting this interaction presents an alternative approach to inhibiting this class of HDACs. We have used structural and functional approaches to identify and characterize a group of small molecules that indirectly target class IIa HDACs by blocking their interaction with MEF2 on DNA.Weused X-ray crystallography and 19F NMRto show that these compounds directly bind to MEF2. We have also shown that the small molecules blocked the recruitment of class IIa HDACs to MEF2-targeted genes to enhance the expression of those targets. These compounds can be used as tools to study MEF2 and class IIa HDACs in vivo and as leads for drug development.
INTRODUCTION
Alterations of epigenetic regulation are a characteristic of many diseases. Small molecules that are being developed as drugs against these diseases often function by modulating the epigenetic control of cellular processes (1). This approach of drug design is best exemplified by the discovery and development of small molecule inhibitors of histone deacetylases (HDACs) that show anti-tumor activity as well as therapeutic effects in neurodegenerative diseases and inflammation (1). HDACs deacetylate histone and non-histone proteins and are a major class of epigenetic regulators of diverse cellular processes. This family of enzymes can be phylogenetically divided into four classes: class I (HDAC1, 2, 3 and 8), class II (HDAC4, 5, 7, 9, 6 and 10) and class IV (HDAC 11), while class III (sirtuins, Sirt1-Sirt7) represents a structurally and functionally distinct family of HDAC enzymes. Most of the currently available HDAC inhibitors target the zinc-containing catalytic domain common to class I, II and IV HDACs, while some of these inhibitors appear to have limited isoform selectivity (2). In general, the broad inhibition of HDACs using active site inhibitors leads to complex cellular responses, which complicate mechanistic analyses and may explain some of the undesired side effects of these drugs in clinical applications (1). Therefore, it is important to develop inhibitors that specifically target a particular member or subset of HDACs to dissect the in vivo functions of HDACs and to explore, and eventually exploit, the full therapeutic potential of HDAC inhibition in a wide range of diseases.
Class II HDACs can be further divided into class IIa (HDAC4, 5, 7 and 9) and IIb (HDAC6 and 10). Class IIa HDACs are highly expressed in muscle cells, neurons and T cells. Extensive genetic studies have demonstrated the important physiological roles of these HDACs in development and adaptive responses of the muscle, nervous and immune systems (3–8) where they participate in calcium-dependent transcriptional responses (9,10). These findings have drawn attention to the functional mechanisms of class IIa HDACs in those systems and their potential as therapeutic targets (11). However, recent studies reveal that class IIa HDACs do not respond to most of the existing HDAC inhibitors (2). In fact, compared with class I HDACs, class IIa HDACs have a catalytic domain that is less active, leading to the hypothesis that class IIa HDACs may function as acetyllysine receptors (2). These observations emphasize the need to develop specific inhibitors of this class for mechanistic studies and therapeutic development. They also raise the question of whether it is more effective to develop subtype-specific inhibitors of class IIa HDACs by focusing on functions other than the catalytic deacetylase activity of the protein.
Class IIa HDACs contain a unique regulatory domain N-terminal to the catalytic domain, which is absent in other HDAC members. This regulatory domain mediates interactions with a variety of other proteins, one of which is the MADS-box family of transcription factor Myocyte Enhancer Factor-2 (MEF2A-D). MEF2 plays a central role in the development and adaptive response of diverse tissues and organs (12); it is also selectively targeted for mutations in several types of cancers (13–16). Class IIa HDACs do not bind to DNA but depend on their interaction with the sequence-specific transcription factor MEF2 for genomic targeting (17,18). This interaction is mediated by a short amphipathic helix conserved in the N-terminal regulatory domain of class IIa HDACs. Crystallography analyses and in vitro biochemical studies reveal that the amphipathic helix binds to a highly conserved hydrophobic groove on the MADS-box/MEF2 domain of MEF2 (19–21). These studies suggest that small molecules binding to the hydrophobic pocket of MEF2 could block the recruitment of class IIa HDACs to DNA, thereby inhibiting the function of class IIa HDACs.
In this study, we used a structure-guided and mechanism-based approach to identify and characterize small molecule compounds that inhibit the MEF2:class IIa HDAC interaction. Our results reveal that these compounds can directly bind the MADS-box domain of MEF2 and prevent the recruitment of class IIa HDACs to the target genes and alter the expression of MEF2 target genes. Therefore, our findings suggest that MEF2 is a potential target for small molecule-based modulation of epigenetic regulation of specific gene expression in specific tissues.
MATERIALS AND METHODS
Drug treatment and luciferase assay
For the two-hybrid assay, cells were treated with 10 µM drug overnight unless indicated otherwise. The amount of DMSO was kept below 0.2% V/V. A luciferase assay was performed according to the manufacturer’s protocol (Promega). The luciferase response was normalized against the Renilla Luciferase as an internal control. The data are presented as a mean ± SD (n = 2) of normalized HDAC4:MEF2 luciferase response against the normalized response values for GAL4-VP16 for each condition to correct for non-specific inhibition of the luciferase signal.
Compound synthesis and preparation
BML-210 was purchased from Enzo Life Sciences. NKL02, NKL08, NKL09, NKL11, NKL13, NKL22, NKL30, NKL54 and NAP1407 were synthesized as shown in Supplementary Figure S2. Stock solutions for BML-210 (Enzo Life Sciences) and analogs were made at 10 mM concentration by dissolving in 100% DMSO and stored at −20°C. TSA (Enzo Life Sciences) was dissolved in DMSO as a 100 µM stock solution and stored at −20°C.
19F NMR
The experimental protocol is similar to that published (22). The NMR samples were prepared in an aqueous (pH 7.66) buffer containing 10 mM HEPES, 250 mM NaCl and 1 mM EDTA. Sample volumes were 700 μL. NKL54 solution was made by serial dilution of 10 mM NKL54 in 100% DMSO. NKL54 at 0.5–5 µM was incubated with MEF2A (1–95) at 0.5–10 µM for 30 min, and the 19F NMR spectra were acquired on a Varian VNMRS 500 Spectrometer at 25°C using a 54° pulse with total acquisition and delay time of 1.6 s for 3000 scans (80 min acquisition time). An internal capillary containing DMSO-d6 was used to obtain a lock signal. A spectral width of 10 ppm was used, and the data were multiplied with an exponential function of 0.2 Hz prior to Fourier transformation. The CF3 resonance of the free NKL54 was at −62.86 ppm and that of the compound bound to MEF2 was at −63.23 ppm. The CF3 resonance of the free compound without any added protein was also observed at −62.86 ppm. Chemical shifts are referenced to trifluoroacetic acid. The final DMSO concentration in the samples was below 0.05%.
Crystallization and data collection
The DNA used in crystallization was the same as previously described (23) and purified as previously reported (21). The MEF2A (1–78)/BML-210/DNA complex was prepared in two steps. First, MEF2A (1–78) at 0.5 mg/ml was mixed with 10% volume of 10 mM BML-210 in DMSO and concentrated using a Biomax 5K cut-off filter (Millipore) to a final concentration of 17 mg/ml. In the second step, the MEF2A(1–78)/BML-210/DNA complex was prepared by mixing the protein/BML-210 complex and DNA at a 2:1 molar ratio. The amount of DMSO was adjusted back to 10% volume to ensure drug solubility before setting crystal trays. Crystals were grown at 18°C by the hanging drop method with a reservoir buffer of 24% PEG 4K, 50 mM Tris HCl, pH 8.18, 3.3% glycerol, 142 mM NaCl, 5 mM MgCl2, 10 mM CaCl2 and 0.004% NaN3. Plate-like crystals grew to 50–100 µm overnight. Crystals were stabilized in a harvest/cryoprotectant buffer (30% PEG 4K, 50 mM Tris HCl, pH 8.18, 1 mM BML-210, 18% glycerol, 142 mM NaCl, 5 mM MgCl2, 10 mM CaCl2, 0.004% NaN3) and flash frozen in liquid nitrogen for storage and data collection. Data were collected at a home X-ray source (copper anode on a Rigaku generator) equipped with confocal mirror, liquid nitrogen cooling system and Raxis IV++ detector (MSC Inc.). The structure of the MEF2A (1-78)/BML-210/DNA complex was solved by molecular replacement using the MEF2A (1–78)/DNA complex as a search model (23) (see Supplementary Data for more details).
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) experiments were performed according to standard procedures with the following exceptions. Two 10 cm dishes of transfected HeLa cells were combined for each ChIP experiment (∼10 × 106 cells). Monoclonal anti-FLAG M2 antibody (5 µg) (Sigma) or normal rabbit IgG, sc-2027 (Santa Cruz), as a control IgG was used for each ChIP. Samples were sonicated in 0.5 ml volume using a 550 sonic dismembrator (Fisher scientific) in 30 s pulses, 90 s rest for 4 min at a setting of 2. A 10 µl sample was kept as input. Purified samples were re-suspended in 62 µl 10 mM Tris HCl, pH 8, and purified input samples were then diluted 1:4 before using 4 µl of each sample as template for real-time PCR. Samples were quantified using an Opticon 2 real-time cycler (Bio Rad). Samples were quantified in triplicate with primers specific for the FXN promoter region (24), using the ΔΔCt method. The data are presented as a mean fold change ± SD of fold change from two independent experiments. Statistical analysis for mean fold change between different conditions was performed using an independent T test (nDMSO = 2, nNKL30 = 2).
Image analysis
Imaging was done on a Bio-Rad MRC-1024 confocal microscope with a ×40 objective at ×1 zoom as a single optical section. Image analysis was performed using ImageJ (NIH). Images used for quantification did not contain saturated pixels. Cells with a clearly defined nucleus, based on MEF2 staining, were chosen for analysis. Background subtracted amounts of MEF2 and HDAC4 within the nucleus was measured by mean fluorescent intensity per pixel within the region marked by nuclear MEF2. Data are presented as mean nuclear HDAC4 intensity per pixel normalized against the mean nuclear MEF2 intensity per pixel ± SD. Statistical analysis was performed using an independent T test(nDMSO = 25 nuclei, nNKL30 = 20 nuclei).
RESULTS
Structure-guided analysis of the HDAC4:MEF2 interaction
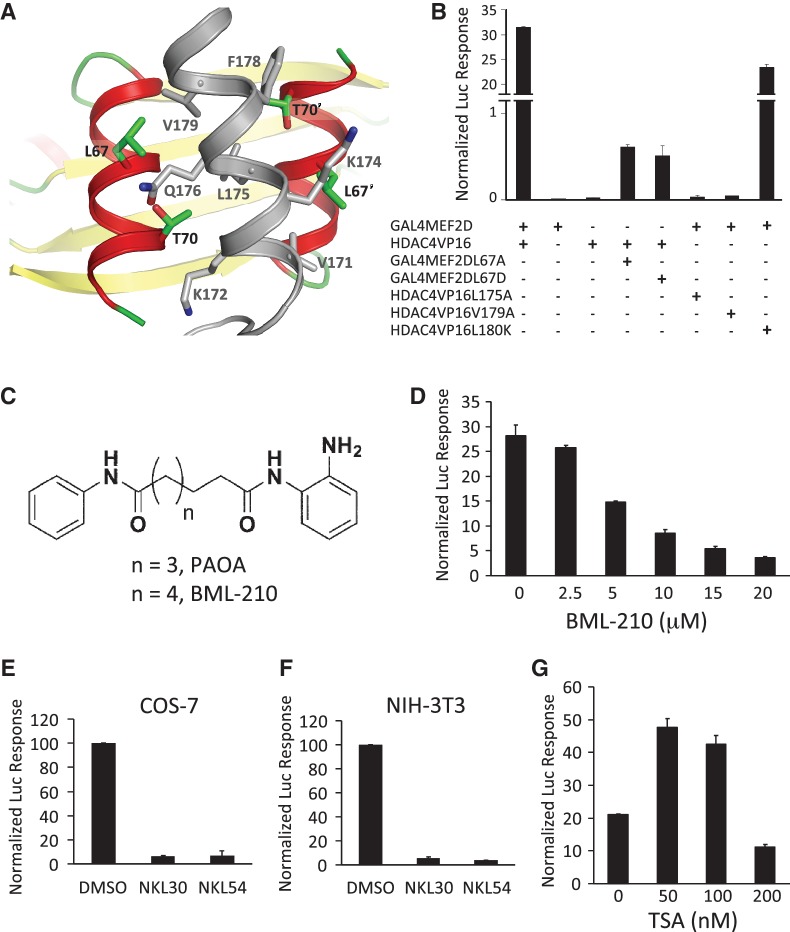
A potential challenge to inhibiting the binding of protein complexes is the large protein–protein interfaces. Some of these interfaces often require multiple mutations to disrupt (25,26). Such interfaces are particularly difficult to inhibit using small molecules. To see if the class IIa HDAC:MEF2 interface is sensitive to localized mutations, we analyzed their binding interaction using structure-guided mutations in a mammalian two-hybrid system. Since the protein–protein interactions involved in the recruitment of class IIa HDACs by MEF2 are highly conserved (19), we used HDAC4 and MEF2D as representative members of their respective families in our studies. The structure of the HDAC4:MEF2 interface (Figure 1A) was modeled after the crystal structure of the closely related HDAC9:MEF2B complex (19). For the mammalian two-hybrid assay (Supplementary Figure S1a), MEF2D was fused with the GAL4 DNA binding domain (GAL4-MEF2D) while the MEF2-binding motif of HDAC4 (AA 155-220) was fused with the viral transactivator VP-16 (HDAC4-VP16). This setup allows us to detect the interaction between HDAC4 and MEF2D with minimal interference from endogenous factors. The HDAC4 fragment also lacks the catalytic domain so that the deacetylase activity was excluded from the assay. Human epithelial carcinoma cells (HeLa) co-transfected with these constructs and the reporter plasmid produced a strong signal comparable to that generated by the positive control of GAL4-VP16 (Supplementary Figure S1b). Structure-guided mutation of a number of residues at the HDAC4:MEF2D interface (Figure 1A), including Leu67Ala and Leu67Asp of MEF2D and Leu175Ala and Val179Ala of HDAC4, had been shown to disrupt HDAC4:MEF2 interaction in vitro (18,19). Each of these mutations also diminished the luciferase signal, demonstrating that the HDAC4:MEF2 interface can be disrupted by site-specific mutations in cells (Figure 1B). Expression levels of the HDAC4 mutants was confirmed by western blot (Supplementary Figure S1c). Mutation of Val180Lys on HDAC4, which weakened the binding of MEF2 by ∼60% in vitro (Kd changed from 0.47 to 0.81 µM) (19), partially reduced the luciferase activity (Figure 1B). These results suggest that the response from the mammalian two-hybrid assay correlates very well with the molecular interaction between HDAC4 and MEF2D. These findings also establish a sensitive and specific method for the detection of HDAC4:MEF2 interaction inside cells.
Figure 1.
Identification of small molecule inhibitors of the HDAC4:MEF2 interaction. (A) A structural model of the binding interface between HDAC4 (gray) and MEF2 (red and yellow). (B) Analyzing the binding interaction between HDAC4 to MEF2 using structure-guided mutations in the mammalian two-hybrid assay in HeLa cells. (C) Chemical structures of PAOA and BML-210. (D) Dosage-dependent inhibition of the reporter signal driven by HDAC4:MEF2 interaction with BML-210 in HeLa cells. (E, F) Effect of BML-210 analogs on the HDAC4:MEF2-mediated luciferase response in COS-7 cells (E) and NIH 3T3 cells (F). (G) Effect of TSA on the luciferase response driven by HDAC4:MEF2 in HeLa cells.
Identification of small molecule inhibitors of the HDAC4:MEF2 interaction
To search for small molecules that bind MEF2 at the HDAC-binding site, we performed virtual screening of the ZINC database: a database of commercially available compounds, using the crystal structure of the HDAC9:MEF2 complex as a guide (27). Based on the docking results, we tested a series of compounds using the mammalian two-hybrid assay (Supplementary Figure S1a). Most of the compounds were either cytotoxic or showed no effect at 10 µM concentration, while some inhibited the HDAC4:MEF2 reporter as well as a control reporter (Supplementary Figure S1d–S1f). The most promising leads came from the pimeloylanilide o-aminoanilide (PAOA) class (28) (Figure 1C). Our subsequent analyses focused on one of the most active members of this family, N-(2-aminophenyl)-N′-phenyloctanediamide, which is commercially available as BML-210. This compound inhibited the HDAC4-VP16-driven reporter signal in a dose-dependent manner with an apparent IC50 of ∼5 µM (Figure 1D). BML-210 did not reduce the expression of HDAC4-VP16 (Supplementary Figure S1g) but did reduce a reporter signal driven by GAL4-VP16 by 5.6 fold at 10 µM (Supplementary Figure S1h), indicating non-specific inhibition by this compound on the expression of luciferase activity under our experimental conditions. However, the same concentration of BML-210 decreased the reporter signal driven by GAL4-MEF2D and HDAC4-VP16 by about 26 fold (Supplementary Figure S1h), suggesting that BML-210 has a specific disruptive effect on the HDAC4:MEF2 interaction beyond its general inhibitory effect. Similarly, analogs of BML-210 inhibited the MEF2:HDAC4-VP16 reporter signal in transfected monkey kidney cells (COS-7) (Figure 1E) and in mouse embryonic fibroblast cells (NIH 3T3) (Figure 1F). None of the cell lines showed inhibitor-induced cell death under our experimental conditions. In contrast, Trichostatin A (TSA), a potent HDAC inhibitor that has been shown to bind the catalytic site of HDAC (29), did not show dose-dependent inhibition of the HDAC4-VP16-mediated reporter signal (Figure 1G) in HeLa cells. TSA reduced the HDAC4-VP16 luciferase response by only 1.4 fold at its IC50 value of 100 nM while reducing the GAL4-VP16 reporter signal by ∼2.7 fold (Supplementary Figure S1i and S1j). These observations suggest that the catalytic inhibition of HDAC is unlikely the cause of the reduction of the luciferase response in the two-hybrid assay. Taken together, our data suggest that BML-210 and its analogs can inhibit HDAC4:MEF2 interaction inside cells.
Biochemical characterization of the BML-210:MEF2 interaction
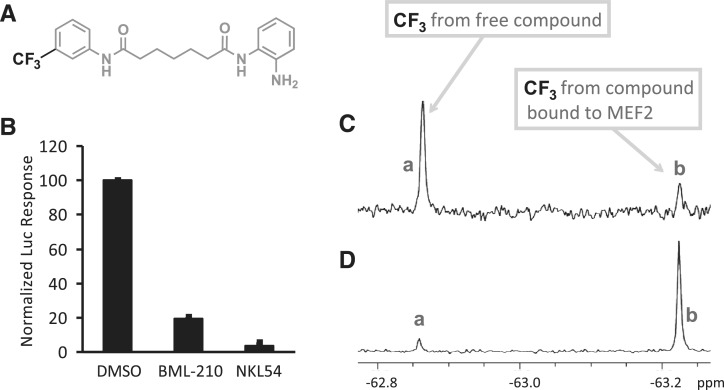
To further analyze the binding interaction between BML-210 and MEF2, we synthesized a number of fluorine-containing analogs of BML-210 for 19F NMR analysis (Supplementary Figure S2a). One of these compounds, termed NKL54 (Figure 2A), inhibited the HDAC4:MEF2-driven luciferase signal (Figure 2B) and showed a characteristic peak in the 19F NMR spectrum. Incubation of NKL54 with MEF2A (1–95) produced a new peak at the upper field (Figure 2C), while increasing the concentration of MEF2 resulted in a larger peak at the upper field and correspondingly a smaller peak at the lower field (Figure 2D). Based on titration analyses using lower protein and ligand concentrations (Supplementary Figure S2b), the Kd for the binding of NKL54 to MEF2A (1–95) was estimated to be 0.5 µM, which is close to the equilibrium-binding constant of HDAC4 to MEF2 (Kd ∼ 0.47 µM) (19). We also analyzed the effect of BML-210 on the binding of HDAC4 to MEF2 in vitro using two different methods. The first was surface plasmon resonance (SPR) on a Biacore instrument. Here, HDAC4 (AA 155–220) was immobilized on a CM5 sensor chip, and purified MEF2A (1–95) was used as the analyte. The binding of MEF2A at various concentrations to the immobilized HDAC4 generated a series of well-defined sensograms (Supplementary Figure S3a). Pre-incubation of MEF2 with increasing amounts of BML-210 decreased the binding signals (Supplementary Figure S3b). In a second approach, an HDAC4 fragment (AA 155–218) was labeled by 5-Iodoacetamidofluorescein (5-IAF) via Cys194; the binding of MEF2A (1–95) to HDAC4 (155–218) was monitored by fluorescence anisotropy (Supplementary Figure S4a). Incrementally increased amounts of BML-210 caused a gradual decay of fluorescence anisotropy, suggesting the displacement of HDAC4 from MEF2 by the small molecule (Supplementary Figure S4b). Low solubility of BML-210 in aqueous solution in both Biacore and fluorescence anisotropy assays made quantitative analysis difficult (Supplementary Data). Nevertheless, taken together, the in vitro binding data strongly suggest that BML-210 and its analogs bind MEF2 directly and competitively with HDAC4.
Figure 2.
Detecting the binding of a fluorinated analog of BML-210 to MEF2 by 19F NMR. (A) Structure of the fluorinated analog NKL54. (B) Similar to BML-210, NKL54 inhibits the HDAC4-VP16-driven reporter signal. (C) 19F NMR spectrum when the free fluorinated compound is in excess. (D) 19F NMR spectrum when the MEF2 protein is in excess.
Structural analysis of the BML-210:MEF2:DNA complex
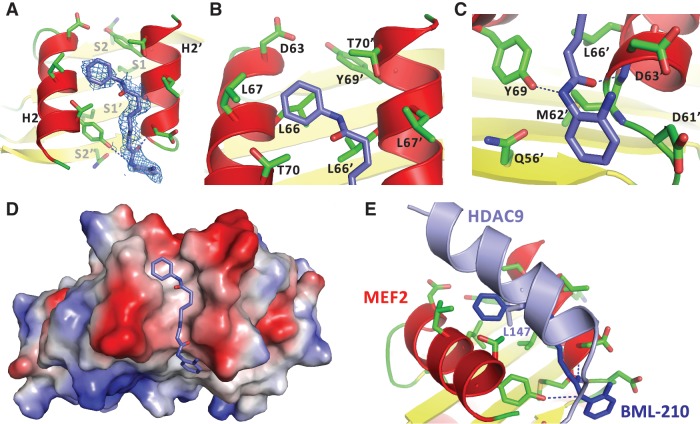
To analyze the structural details of the binding interaction, we co-crystallized BML-210 with MEF2A bound to DNA. The crystals diffracted to 2.4 Å. The structure was solved by molecular replacement using the MEF2A:DNA complex as the search model (23) (Supplementary Table S1). After initial rounds of rigid body refinement, electron density matching the shape of BML-210 was clearly visible in the hydrophobic pocket of MEF2 (Figure 3A). Since BML-210 has a nearly symmetric structure (Figure 1C), its orientation in the complex could not be uniquely assigned at the current resolution. However, since the ring-shaped region at one end of the electron density is surrounded by a number of hydrophobic residues including Leu66, Leu67, Thr70, Leu66′, Leu67′ and Thr70′ (the prime sign denotes residues from the other monomer), we assigned this density to the phenyl group (Figure 3B). At the other end, the ring-like electron density is in a more hydrophilic environment surrounded by Asn73, Gln56′, Asp61′ and Asp63′. We assigned this density to the 2-aminophenyl group, which makes van der Waals contacts and potential hydrogen bonding interactions with residues of MEF2 (Figure 3C). It is also possible that BML-210 adopts two alternative orientations in the crystal and the observed electron density represents a weighted average of the two populations. The methylene groups of the octanediamide fit snugly between helix H2 of the two MEF2 monomers, making numerous contacts to the main chain and side chain of MEF2 residues, mostly of hydrophobic nature. Overall, BML-210 adopts an extended conformation to bind the surface groove of MEF2 (Figure 3D). Interestingly, a comparison with MEF2 complexes bound by Cabin1 and HDAC9 shows that BML-210 shares some binding features with natural ligands (19,20) (Figure 3E). For example, the binding of the phenyl ring of BML-210 to the central hydrophobic pocket formed by Leu66, Leu67 and Thr70 from each monomer is analogous to that of Leu147 of HDAC9 (Figure 3E). Modeling studies also suggest that some MEF2-binding proteins, including members of the myocardin family, use an identical phenyl ring from phenylalanine to bind this pocket (21,30).
Figure 3.
Structural characterization of the binding of BML-210 to MEF2A. (A) Electron density (blue mesh) matching the shape of BML-210 (blue stick) was identified in the hydrophobic pocket of MEF2 (red and yellow ribbon). (B) The phenyl group of BML-210 is surrounded by a number of hydrophobic residues of MEF2. (C) The 2-aminophenyl group of BML-210 interacts with a number of residues on MEF2. (D) A surface representation showing that the methylene groups of the octanediamide fit snugly between helix H2 of the two MEF2 monomers and that BML-210 adopts an extended conformation to bind the surface groove of MEF2. Positive and negative surface potentials are indicated by red and blue, respectively. (E) Structural superposition using MEF2 as the reference showing that BML-210 and HDAC9 share the same binding site on MEF2 and that the synthetic compound mimics some of the binding interactions of the natural ligands.
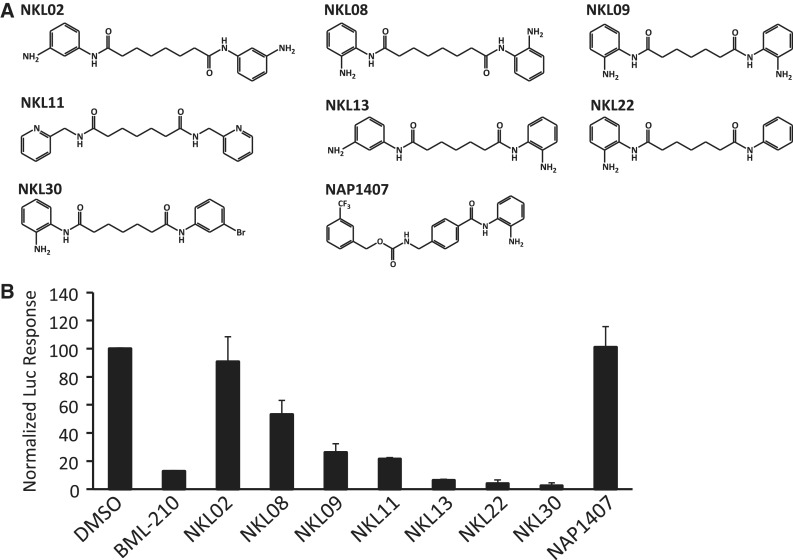
The crystal structure can be used to guide the synthesis of BML-210 derivatives to improve the affinity and pharmacological properties. For example, the binding of BML-210 to the hydrophobic pocket of Leu66, Leu67, Thr70, Leu66′, Leu67′ and Thr70′ is largely dependent on the phenyl group that does not completely fill the pocket, suggesting that substitutions on the phenyl ring could be an attractive strategy for optimization. The 2-aminophenyl group of BML-210, however, engages in numerous hydrogen-bond interactions with MEF2, which imposes more constraints on synthetic modifications. Another important factor is the methylene chain whose length is limited by the groove on MEF2. The narrow width of the groove also suggests that substitutions on the methylene chain may be highly limited. Considering these factors, we synthesized a series of new BML-210 analogs (Figure 4A, Supplementary Figure S2a). These compounds were designed to systematically explore the effect of ring substitutions and the length of the methylene linker on inhibiting the MEF2:HDAC4 interaction in the two-hybrid assay (Figure 4B, Supplementary Figure S2c). As shown in Figure 4B, substitutions on the phenyl rings and variations of the linker length indeed showed strong effect on inhibition of the MEF2:HDAC4 interaction. For example, compounds with a five-carbon linker were generally more active than those with a six-carbon linker. Most notably, NKL11 that lacks the putative zinc chelating 2-amino group also showed significant inhibition in the two-hybrid assay. By contrast, NAP1407, a close analog of the benzamide HDAC inhibitor MS275, is completely inactive. These observations suggest that BML-210 and its derivitives act outside the catalytic site of HDAC to inhibit the MEF2:HDAC4 interaction.
Figure 4.
Development of BML-210 analogs. (A) Chemical structures of BML-210 analogs. (B) Effect of BML-210 analogs on the HDAC4:MEF2-mediated luciferase response. Response is indicated as mean percentage from the DMSO luciferase response ± SD (n = 2).
BML-210 blocks MEF2-dependent recruitment of HDAC4 to DNA
BML-210 has been shown to enhance the expression of frataxin in Friedreich’s ataxia (FRDA), a neurodegenerative disease associated with trinucleotide repeat expansion that apparently leads to epigenetic silencing of the FXN gene (24,31). Although the mechanism seems to involve induced histone acetylation, more potent and non-discriminatory HDAC inhibitors such as SAHA did not activate the expression of frataxin, suggesting that BML-210 may target a specific complex or complexes involved in frataxin expression (24).
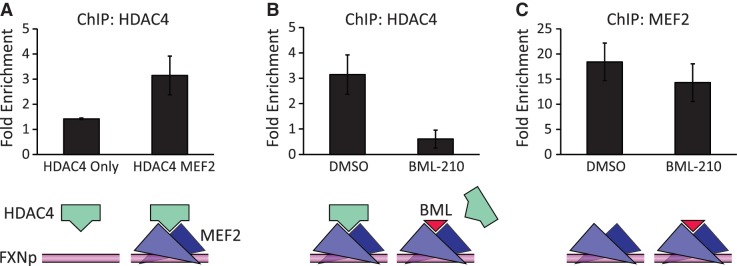
To further explore the role of MEF2 in frataxin expression, we analyzed the promoter region of the FXN gene for putative MEF2 binding sites. We found four highly conserved A/T rich elements that resemble the consensus MEF2-binding sequence (5′-CTA(A/T)4TAG-3′) approximately 200 bp upstream of the transcription start site (Supplementary Figure S5a). These putative MEF2 sites are located in the MIR (mammalian-wide interspersed repeats) region of the frataxin promoter that had been previously reported to contribute significantly to the activity of the frataxin promoter (32). Recent studies also show that SRF, another MADS-box transcription factor that shares a similar DNA binding site with MEF2, regulates the expression of Frataxin in human cells (33). We detected the enrichment of MEF2 at the FXN promoter in the FRDA cells using ChIP (Supplementary Figure S5b). Therefore, to see if BML-210 could mediate its effect on frataxin expression by disrupting MEF2/co-repressor complexes, we analyzed the binding of MEF2 and HDAC4 to the frataxin promoter using ChIP in transfected cells. We showed that MEF2 could bind and recruit HDAC4 to the frataxin promoter (Figure 5A). Treatment of the cells with BML-210 blocked this recruitment, evident by diminished binding of HDAC4 (Figure 5B), but had little effect on the occupancy of the locus by MEF2 (Figure 5C). This effect is not due to reduced expression of HDAC4 (Supplementary Figure S5c), but consistent with BML-210-mediated inhibition of HDAC4 binding to MEF2. The results presented here suggest that BML-210 could block the binding of HDAC4 to MEF2-targeted promoters in vivo.
Figure 5.
BML-210 inhibits MEF2-dependent recruitment of HDAC4 to the frataxin promoter. ChIP assays were performed with chromatin from HeLa cells transfected with FLAG-tagged HDAC4 and GFP and/or FLAG-tagged MEF2C. Drug-treated and control-treated chromatin were immunoprecipitated with anti-FLAG antibody and normal rabbit IgG as negative control, and precipitated genomic DNA was amplified with primers flanking the MEF2 site on the endogenous FXN promoter. (A) Co-transfection of HeLa cells with both MEF2C and HDAC4 resulted in enhanced enrichment of HDAC4 on the FXN promoter. (B) 6 h treatment with 10 µM BML-210 diminished the MEF2C-mediated enrichment of HDAC4 on the Frataxin promoter (P < 0.04). (C) The BML-210 treatment did not significantly affect the MEF2C enrichment on the FXN promoter. Lower panels are schematic interpretations of the ChIP data. FXNp: frataxin promoter (magenta line); MEF2 dimer (blue triangles); HDAC4 (green bar); BML-210 (red triangle).
BML-210 analogs alter the cellular localization of HDAC4 in vivo
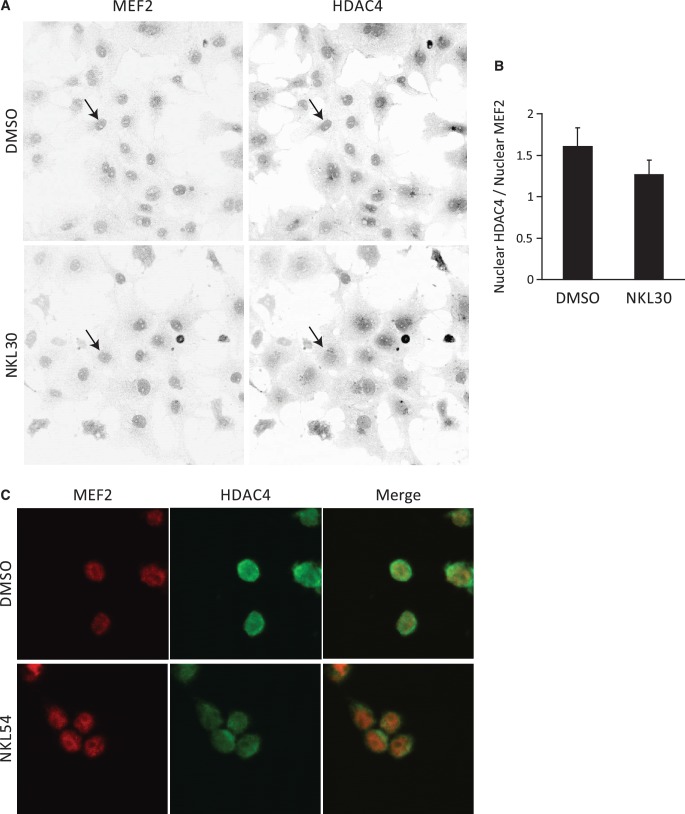
Previous studies have shown that direct binding to MEF2 is required for nuclear targeting of HDAC4 and mutation of the key MEF2-binding residue Leu175 in HDAC4 resulted in a diffusive distribution of HDAC4 in the cytoplasm (18). To see if BML-210 and its analogs can mimic this effect in vivo, we analyzed the cellular localization of HDAC4 in COS-7 cells and FRDA lymphoid cells using immunocytochemistry (Figure 6A, C). Since the compounds NKL30 and NKL54 seem to be more soluble than BML-210 and more potent in inhibiting the MEF2 and HDAC4 interaction in the luciferase assay (Figure 4B), these compounds were used for the immunocytochemistry experiments. Cells were treated with 10 µM NKL30 overnight (ON) and immunostained with antibodies against MEF2 and HDAC4. Drug treatment did not result in significant cytotoxicity under the assay conditions. As shown in Figure 6A, NKL30 treatment clearly induced a much more diffusive localization pattern of HDAC4 (lower right panel) compared to the vehicle (upper right panel). Since the amount of nuclear MEF2 did not change significantly (P > 0.25) between drug-treated and vehicle-treated samples (Supplementary Figure S6a and S6b), MEF2 was used for staining the nucleus and also for normalizing the nuclear HDAC4 level for quantitative analysis. The data indicated that the NKL30-treated cells have 21% less HDAC4 in the nucleus (P < 0.001) (Figure 6B). We also detected the effect of NKL54 on the cellular distribution of HDAC4 in the FRDA lymphoid cell line (GM15850) (Figure 6C). NKL54 decreased the co-localization of MEF2 and HDAC4 compared to the vehicle. There was no apparent effect on the nuclear localization of MEF2. Although we could not quantify the effect of NKL54 on the cellular localization of HDAC4 in the FRDA cells due to technical difficulties, this result, together with the more diffusive distribution of HDAC4 in the COS-7 cells, suggests that BML-210 and its analogs can disrupt MEF2:HDAC4 co-localization in vivo, presumably through the inhibition of MEF2:HDAC4 interaction.
Figure 6.
BML-210 analog NKL30 disrupts MEF2: HDAC4 co-localization in vivo. (A) Analysis of the cellular localization of MEF2 and HDAC4 in COS-7 cells treated with DMSO and NKL30 using immunocytochemistry, showing that NKL30 induced a delocalization of HDAC4 but showed no apparent effect on the cellular distribution of MEF2. The arrows point at the nucleus of a DMSO treated COS-7 cell and an NKL30 treated COS-7 cell. (B) Quantitative image analysis showing that NKL-30 reduced the amount of HDAC4 in the nucleus by 21% (P < 0.001). (C) Immunocytochemistry showing cellular localization of MEF2 (red) and HDAC4 (green) in FRDA lymphoid cell line (GM15850) treated with DMSO and NKL54. NKL54 induced delocalization of HDAC4. There was no apparent effect on the cellular distribution of MEF2 in the FRDA cells.
DISCUSSION
In this study, we have demonstrated that the interaction between class IIa HDACs and MEF2 can be inhibited by small molecules. We have identified a class of compounds belonging to the PAOA family that bind MEF2 at the same site as class IIa HDACs and inhibit their binding interactions. We have also shown that these compounds can block the recruitment of class IIa HDACs to MEF2 targeted promoters without affecting the occupancy of MEF2 on DNA. Our findings suggest that the observed effect of PAOA compounds on the level of histone acetylation and gene expression (24) may include an indirect effect on the inhibition of MEF2 and class IIa HDAC interaction.
BML-210 or PAOA-like compounds were originally discovered as HDAC inhibitors that selectively induced acetylation of histone but not tubulin (28,34), presumably through the inhibition of HDACs other than HDAC6, a tubulin-specific deacetylase (35,36). The molecular mechanisms by which these benzamide derivatives inhibit HDAC function have remained unclear. The evidence of direct binding to MEF2 presented here suggests a previously unknown mechanism for the action of these molecules that does not involve targeting the active site of HDAC, but rather the protein–protein interactions between HDAC and its functional partners. In fact, these results provide the first example of subtype-selective inhibition of HDACs by targeting the protein complex between class IIa HDACs and MEF2. We have shown that these compounds, consistent with this model, can disrupt the co-localization of a class IIa HDAC and MEF2 in vivo.
Additionally, a number of our observations suggest that the FXN gene, a previously unknown target of MEF2, might be regulated by MEF2. Close examination of the FXN promoter revealed a number of conserved MEF2 binding sites in an enhancer region previously shown to be important for frataxin expression (32). ChIP analyses have shown that MEF2 binds this region in vivo. In fact, in transiently transfected cells, the class IIa HDAC member HDAC4 was shown to bind this region and the binding was enhanced in MEF2 co-transfected cells, suggesting a MEF2 mediated recruitment. Although our efforts as well as others to analyze the role of endogenous HDACs on the frataxin promoter in FRDA cells were unsuccessful (37), our data suggest that BML-210-like compounds activate FXN expression in Friedreich’s ataxia through MEF2-dependent mechanisms. It is possible that BML-210 like compounds may activate the expression of the FXN gene by blocking MEF2-mediated recruitment of transcription co-repressors other than HDACs, such as Cabin1 (20). This model provides a plausible explanation for the intriguing observations that a number of compounds, such as 7b, 10b, and 14b described by Hermann et al., are potent inducers of the FXN gene, while they lack the characteristic functional groups of HDAC inhibitors (24). These compounds showed little or no inhibition on HDAC activity yet enhanced the expression of the FXN gene substantially (24). Characterizing the details of the FXN induction mechanisms will require further studies. The compounds developed here can be used for dissecting the function of MEF2 and class IIa HDACs in FXN regulation and other cellular processes.
Recent studies reveal that MEF2 is highly mutated in non-Hodgkin lymphoma (15,16). Most of the mutations are amino acid substitutions at or near the cofactor recruitment sites defined by our crystal structures (15,19,20,38,39), suggesting that deregulated interactions between MEF2 and its co-activators (CBP/p300) and/or co-repressors (class IIa HDACs) may be involved in tumorigenesis. Supporting this hypothesis are observations that CBP/p300 and class IIa HDACs are also frequently mutated or deregulated in lymphoma/leukaemia and other types of cancers (11,16,39). Remarkably, BML-210 has been shown to induce growth inhibition, apoptosis and differentiation in several leukeamia cell lines (40). Whether BML-210 acts through MEF2 to exert its anti-tumor effect will require further studies. The structure and biochemical assays established here helped us develop more specific and potent small molecule inhibitors that can be used to address this question and analyze the function of MEF2 and class IIa HDACs. We have synthesized a series of new compounds that inhibit the interaction between MEF2 and class IIa HDACs. Some of these compounds were several folds more potent than BML-210 in inhibiting the interaction. Therefore, our results and newly introduced compounds provide leads for developing potential epigenetic therapies against multiple diseases in which deregulation of MEF2 and class IIa HDACs is implicated.
ACCESSION NUMBERS
RCSB Protein Data Bank: coordinates and structural factors have been deposited under the accession code 3MU6.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–6, Supplementary Table 1, Supplementary Methods and Supplementary References [19,21,32,41–45].
FUNDING
National Institute of Health challenge grant (RC1 DA028790 to L.C. and N. A. P., R01 HL076334 to L.C.); two grants for NMR spectrometers by National Science Foundation (DBI-0821671); National Institute of Health (IH S10-RR25432). Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Z. Chen and Dr. S. Li for help with protein preparation; Dr. M. Goodman, Dr. N. Chelyapov and USC NanoBiophysics Core Facility for assistance in Biacore and fluorescence anisotropy studies; Dr. T. Williams for assistance in the NMR studies; Dr. D. Arnold and V. Balasanyan for help with immunocytochemistry; Dr. X. Chen, R. Kalhor and Dr. G. Tao for helpful discussions; Dr. X. J. Yang and Dr. X. Liu for providing MEF2 and HDAC4 plasmids; Dr. J. Tower, Dr. N. Arnheim, Dr. N. Chen and J. Brown for help with expression and ChIP analyses; ALS BCSB staff members for help with data collection.
REFERENCES
- 1.Paris M, Porcelloni M, Binaschi M, Fattori D. Histone deacetylase inhibitors: from bench to clinic. J. Med. Chem. 2008;51:1505–1529. doi: 10.1021/jm7011408. [DOI] [PubMed] [Google Scholar]
- 2.Bradner JE, West N, Grachan ML, Greenberg EF, Haggarty SJ, Warnow T, Mazitschek R. Chemical phylogenetics of histone deacetylases. Nat. Chem. Biol. 2010;6:238–243. doi: 10.1038/nchembio.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang CL, McKinsey TA, Chang S, Antos CL, Hill JA, Olson EN. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 2002;110:479–488. doi: 10.1016/s0092-8674(02)00861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tao R, de Zoeten EF, Ozkaynak E, Chen C, Wang L, Porrett PM, Li B, Turka LA, Olson EN, Greene MI, et al. Deacetylase inhibition promotes the generation and function of regulatory T cells. Nat. Med. 2007;13:1299–1307. doi: 10.1038/nm1652. [DOI] [PubMed] [Google Scholar]
- 5.Mottet D, Bellahcene A, Pirotte S, Waltregny D, Deroanne C, Lamour V, Lidereau R, Castronovo V. Histone deacetylase 7 silencing alters endothelial cell migration, a key step in angiogenesis. Circ. Res. 2007;101:1237–1246. doi: 10.1161/CIRCRESAHA.107.149377. [DOI] [PubMed] [Google Scholar]
- 6.Renthal W, Maze I, Krishnan V, Covington HE, III, Xiao G, Kumar A, Russo SJ, Graham A, Tsankova N, Kippin TE, et al. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron. 2007;56:517–529. doi: 10.1016/j.neuron.2007.09.032. [DOI] [PubMed] [Google Scholar]
- 7.Tsankova NM, Berton O, Renthal W, Kumar A, Neve RL, Nestler EJ. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat. Neurosci. 2006;9:519–525. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- 8.Bolger TA, Yao TP. Intracellular trafficking of histone deacetylase 4 regulates neuronal cell death. J. Neurosci. 2005;25:9544–9553. doi: 10.1523/JNEUROSCI.1826-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bertos NR, Wang AH, Yang XJ. Class II histone deacetylases: structure, function, and regulation. Biochem. Cell. Biol. 2001;79:243–252. [PubMed] [Google Scholar]
- 10.Verdin E, Dequiedt F, Kasler HG. Class II histone deacetylases: versatile regulators. Trends Genet. 2003;19:286–293. doi: 10.1016/S0168-9525(03)00073-8. [DOI] [PubMed] [Google Scholar]
- 11.Clocchiatti A, Florean C, Brancolini C. Class IIa HDACs: from important roles in differentiation to possible implications in tumourigenesis. J. Cell. Mol. Med. 2011;15:1833–1846. doi: 10.1111/j.1582-4934.2011.01321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Development. 2007;134:4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- 13.Yuki Y, Imoto I, Imaizumi M, Hibi S, Kaneko Y, Amagasa T, Inazawa J. Identification of a novel fusion gene in a pre-B acute lymphoblastic leukemia with t(1;19)(q23;p13) Cancer Sci. 2004;95:503–507. doi: 10.1111/j.1349-7006.2004.tb03240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prima V, Gore L, Caires A, Boomer T, Yoshinari M, Imaizumi M, Varella-Garcia M, Hunger SP. Cloning and functional characterization of MEF2D/DAZAP1 and DAZAP1/MEF2D fusion proteins created by a variant t(1;19)(q23;p13.3) in acute lymphoblastic leukemia. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, U.K. 2005;19:806–813. doi: 10.1038/sj.leu.2403684. [DOI] [PubMed] [Google Scholar]
- 15.Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, Johnson NA, Severson TM, Chiu R, Field M, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, Trifonov V, Kasper LH, Lerach S, Tang H, Ma J, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471:189–195. doi: 10.1038/nature09730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu J, McKinsey TA, Zhang CL, Olson EN. Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol. Cell. 2000;6:233–244. doi: 10.1016/s1097-2765(00)00025-3. [DOI] [PubMed] [Google Scholar]
- 18.Wang AH, Yang XJ. Histone deacetylase 4 possesses intrinsic nuclear import and export signals. Mol. Cell Biol. 2001;21:5992–6005. doi: 10.1128/MCB.21.17.5992-6005.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han A, He J, Wu Y, Liu JO, Chen L. Mechanism of recruitment of class II histone deacetylases by myocyte enhancer factor-2. J. Mol. Biol. 2005;345:91–102. doi: 10.1016/j.jmb.2004.10.033. [DOI] [PubMed] [Google Scholar]
- 20.Han A, Pan F, Stroud JC, Youn HD, Liu JO, Chen L. Sequence-specific recruitment of transcriptional co-repressor Cabin1 by myocyte enhancer factor-2. Nature. 2003;422:730–734. doi: 10.1038/nature01555. [DOI] [PubMed] [Google Scholar]
- 21.Wu Y, Dey R, Han A, Jayathilaka N, Philips M, Ye J, Chen L. Structure of the MADS-box/MEF2 domain of MEF2A bound to DNA and its implication for myocardin recruitment. J. Mol. Biol. 2010;397:520–533. doi: 10.1016/j.jmb.2010.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papeo G, Giordano P, Brasca MG, Buzzo F, Caronni D, Ciprandi F, Mongelli N, Veronesi M, Vulpetti A, Dalvit C. Polyfluorinated amino acids for sensitive 19F NMR-based screening and kinetic measurements. J. Am. Chem. Soc. 2007;129:5665–5672. doi: 10.1021/ja069128s. [DOI] [PubMed] [Google Scholar]
- 23.Santelli E, Richmond TJ. Crystal structure of MEF2A core bound to DNA at 1.5 A resolution. J. Mol. Biol. 2000;297:437–449. doi: 10.1006/jmbi.2000.3568. [DOI] [PubMed] [Google Scholar]
- 24.Herman D, Jenssen K, Burnett R, Soragni E, Perlman SL, Gottesfeld JM. Histone deacetylase inhibitors reverse gene silencing in Friedreich's ataxia. Nat. Chem. Biol. 2006;2:551–558. doi: 10.1038/nchembio815. [DOI] [PubMed] [Google Scholar]
- 25.Chen L, Glover JN, Hogan PG, Rao A, Harrison SC. Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA. Nature. 1998;392:42–48. doi: 10.1038/32100. [DOI] [PubMed] [Google Scholar]
- 26.Wu Y, Borde M, Heissmeyer V, Feuerer M, Lapan AD, Stroud JC, Bates DL, Guo L, Han A, Ziegler SF, et al. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell. 2006;126:375–387. doi: 10.1016/j.cell.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 27.Irwin JJ, Shoichet BK. ZINC—a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wong JC, Hong R, Schreiber SL. Structural biasing elements for in-cell histone deacetylase paralog selectivity. J. Am. Chem. Soc. 2003;125:5586–5587. doi: 10.1021/ja0341440. [DOI] [PubMed] [Google Scholar]
- 29.Finnin MS, Donigian JR, Cohen A, Richon VM, Rifkind RA, Marks PA, Breslow R, Pavletich NP. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature. 1999;401:188–193. doi: 10.1038/43710. [DOI] [PubMed] [Google Scholar]
- 30.Creemers EE, Sutherland LB, Oh J, Barbosa AC, Olson EN. Coactivation of MEF2 by the SAP domain proteins myocardin and MASTR. Mol. Cell. 2006;23:83–96. doi: 10.1016/j.molcel.2006.05.026. [DOI] [PubMed] [Google Scholar]
- 31.Rai M, Soragni E, Jenssen K, Burnett R, Herman D, Coppola G, Geschwind DH, Gottesfeld JM, Pandolfo M. HDAC inhibitors correct frataxin deficiency in a Friedreich ataxia mouse model. PLoS One. 2008;3:e1958. doi: 10.1371/journal.pone.0001958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greene E, Entezam A, Kumari D, Usdin K. Ancient repeated DNA elements and the regulation of the human frataxin promoter. Genomics. 2005;85:221–230. doi: 10.1016/j.ygeno.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 33.Li K, Singh A, Crooks DR, Dai X, Cong Z, Pan L, Ha D, Rouault TA. Expression of human frataxin is regulated by transcription factors SRF and TFAP2. PLoS One. 2010;5:e12286. doi: 10.1371/journal.pone.0012286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haggarty SJ, Koeller KM, Wong JC, Butcher RA, Schreiber SL. Multidimensional chemical genetic analysis of diversity-oriented synthesis-derived deacetylase inhibitors using cell-based assays. Chem. Biol. 2003;10:383–396. doi: 10.1016/s1074-5521(03)00095-4. [DOI] [PubMed] [Google Scholar]
- 35.Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 36.Haggarty SJ, Koeller KM, Wong JC, Grozinger CM, Schreiber SL. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc. Natl Acad. Sci. USA. 2003;100:4389–4394. doi: 10.1073/pnas.0430973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu C, Soragni E, Chou CJ, Herman D, Plasterer HL, Rusche JR, Gottesfeld JM. Chemical probes identify a role for histone deacetylase 3 in Friedreich's ataxia gene silencing. Chem. Biol. 2009;16:980–989. doi: 10.1016/j.chembiol.2009.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He J, Ye J, Cai Y, Riquelme C, Liu JO, Liu X, Han A, Chen L. Structure of p300 bound to MEF2 on DNA reveals a mechanism of enhanceosome assembly. Nucleic Acids Res. 2011;39:4464–4474. doi: 10.1093/nar/gkr030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mullighan CG, Zhang J, Kasper LH, Lerach S, Payne-Turner D, Phillips LA, Heatley SL, Holmfeldt L, Collins-Underwood JR, Ma J, et al. CREBBP mutations in relapsed acute lymphoblastic leukaemia. Nature. 2011;471:235–239. doi: 10.1038/nature09727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Savickiene J, Borutinskaite VV, Treigyte G, Magnusson KE, Navakauskiene R. The novel histone deacetylase inhibitor BML-210 exerts growth inhibitory, proapoptotic and differentiation stimulating effects on the human leukemia cell lines. Eur. J. Pharmacol. 2006;549:9–18. doi: 10.1016/j.ejphar.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 41.Chan JK, Sun L, Yang XJ, Zhu G, Wu Z. Functional characterization of an amino-terminal region of HDAC4 that possesses MEF2 binding and transcriptional repressive activity. J. Biol. Chem. 2003;278:23515–23521. doi: 10.1074/jbc.M301922200. [DOI] [PubMed] [Google Scholar]
- 42.Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Meth. Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 43.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kleywegt G, Jones TA. In: From First Map to Final Map Model. Bailey S, Hubbard R, Walker D, editors. Warrington: Daresbury Laboratory; 1994. [Google Scholar]
- 45.Lewis TL, Jr, Mao T, Svoboda K, Arnold DB. Myosin-dependent targeting of transmembrane proteins to neuronal dendrites. Nat. Neurosci. 2009;12:568–576. doi: 10.1038/nn.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.