Abstract
Nuclear receptors (NRs) regulate gene expression by binding specific DNA sequences consisting of AG[G/T]TCA or AGAACA half site motifs in a variety of configurations. However, those motifs/configurations alone do not adequately explain the diversity of NR function in vivo. Here, a systematic examination of DNA binding specificity by protein-binding microarrays (PBMs) of three closely related human NRs—HNF4α, retinoid X receptor alpha (RXRα) and COUPTF2—reveals an HNF4-specific binding motif (H4-SBM), xxxxCAAAGTCCA, as well as a previously unrecognized polarity in the classical DR1 motif (AGGTCAxAGGTCA) for HNF4α, RXRα and COUPTF2 homodimers. ChIP-seq data indicate that the H4-SBM is uniquely bound by HNF4α but not 10 other NRs in vivo, while NRs PXR, FXRα, Rev-Erbα appear to bind adjacent to H4-SBMs. HNF4-specific DNA recognition and transactivation are mediated by residues Asp69 and Arg76 in the DNA-binding domain; this combination of amino acids is unique to HNF4 among all human NRs. Expression profiling and ChIP data predict ∼100 new human HNF4α target genes with an H4-SBM site, including several Co-enzyme A-related genes and genes with links to disease. These results provide important new insights into NR DNA binding.
INTRODUCTION
Nuclear receptors (NRs) are ligand-dependent transcription factors (TFs) that play important roles in nearly every aspect of human physiology (1). They are linked to human disease, are popular drug targets and play a major role in regulating the expression of genes responsible for drug metabolism (2–4). Therefore, proper elucidation of their target genes has important clinical ramifications. NRs regulate gene expression primarily by first binding specific DNA response elements in the regulatory regions of target genes. Basic rules for NR DNA binding that consist of distinct half sites motifs for steroid (AGAACA) and non-steroid (AGGTCA) receptors and half site configurations—direct (DR, →→), inverted (IR, →←), everted repeats (ER, →←) and non-repeats (nRs) as well as spacing between the repeats—were established early on (5–8). This dogma, the ‘DR rule’, successfully drove the identification of individual NR target genes in the pre-genomic era. However, there remains considerable overlap in the apparent binding specificity of many NRs, which could be due to the high degree of conservation among the NR DNA-binding domains (DBDs) and/or to the fact that NR DNA binding specificity has not been analyzed in a systematic, global fashion.
The magnitude of the complexity and intricacy of DNA binding specificity is illustrated by the fact that for a 13-nt-long motif, such as a direct repeat of AGGTCA with a spacing of 1 nt (DR1, AGGTCAxAGGTCA), there are 413/2 (∼34 million) different potential DNA sequences. If a given NR binds just 0.01% of those sites for a specificity of 1 in 10 000, that still yields 3400 unique sequences, all of which could have just a minor variation on the DR1 consensus. The question then arises as to how many of those different sequences will bind a given NR and whether there are motifs specific to different NRs.
There are three well-characterized subfamilies of receptors that bind DR1s as homodimers—HNF4α (NR2A1), COUPTF2 (NR2F2) and RXRα (NR2B1). They are all very highly conserved, closely related and regulate a variety of metabolic genes in common tissues such as liver, kidney and intestine (9,10). HNF4α is a constitutive activator that binds an endogenous ligand (linoleic acid) (11). It is considered to be a master regulator of liver-specific gene expression, including genes involved in intermediary metabolism as well as xenobiotic and drug metabolism (12–15). HNF4α is linked to several human diseases including diabetes, hemophilia, hepatitis, atherosclerosis and inflammatory bowel disease (16,17).
RXRα is also expressed primarily in the liver, as well as kidney, gut, muscle and skin (10). While it is best known as a heterodimeric partner of other NRs such as PPAR (NR1C), RAR (NR1B), FXR (NR1H), PXR (NR1I2), TR (NR1A) and VDR (NR1I1) (7,18), it binds directly to the synthetic ligand 9-cis retinoid acid and has been shown to activate transcription in the absence of an ectopically expressed partner (19–23). COUPTF2 is also present in the liver and, like RXRα, is fairly ubiquitously expressed. However, it acts primarily as a repressor of transcription (24,25); it remains an orphan receptor in that its endogenous ligand has not yet been identified, although it has been shown to bind and respond to retinoids (26). Like HNF4α, COUPTF2 binds DNA well as a homodimer but unlike HNF4α it can also heterodimerize with RXRα (21,25,27). It was recognized early on that HNF4α, RXRα and COUPTF2 all share common binding sites (that roughly resemble DR1s) in the promoters of certain genes and consequently compete for regulation of those genes (28–30). However, it was also noted that there are a couple of binding sites in other genes that were not bound by all three NRs (28,31,32). While these results suggested the existence of NR-specific binding motifs, common versus unique binding features were never identified and the extent of the overlap remained obscure.
New genome-scale technologies, both in vivo and in vitro, now allow us to address the issue of NR binding specificity in an appropriately global fashion (33). For in vivo binding, chromatin immunoprecipitation followed by deep sequencing (ChIP-seq) enables robust and accurate identification of TF binding regions, but the resolution is not sufficient to identify the exact site to which TFs bind; this must be done by statistical inference (34). For in vitro binding, protein-binding microarrays (PBMs), can assay the binding of TFs to 10 000 s of DNA probes in a high throughput fashion (35). PBM data are extremely useful for mining ChIP-seq data to identify the precise location of TF binding and to predict new target genes by cross-referencing. We recently applied a modified PBM approach to HNF4α and identified 100 s of new direct targets of HNF4α by combining the PBM results with genome-wide location and expression profiling data (13).
Here, we apply the PBM approach to the problem of distinguishing the binding specificity of HNF4α, RXRα and COUPTF2. We show that RXRα homodimers bind DNA very well in the PBM and that they have a specificity nearly identical to COUPTF2 homodimers, with a clear preference for the 3′ half site of a DR1 motif. The PBMs also identified an HNF4α-specific binding motif (H4-SBM) (CAAAGTCCA) that was verified in vivo in ChIP-seq data. Finally, we determine the amino acid residues in the HNF4α DBD (Asp69 and Arg76) responsible for the unique binding specificity of HNF4α and identify ∼100 new human target genes that contain the HNF4–specific motif. These results have important implications not just for HNF4α, RXRα and COUPTF2, but for the entire NR superfamily as well.
MATERIALS AND METHODS
PBM design, assay and data processing
Two different custom arrays in 8 × 15 k format ordered from Agilent Technologies (Santa Clara, CA, USA) were used to test binding specificity (PBM2) (13) and half site preferences (PBM6.1) (see Supplementary Figure S2 for design and Supplementary Table S4 for complete sequences of PBM6.1). Each of the ∼3000 sequences was spotted four (PBM6.1) to five (PBM2) times in each of the eight grids. The PBM assay was performed as previously described (13) with minor modifications: 1.6 µM Cy5 dUTPs (Enzo Life Sciences) was used to label the double-stranded DNA on the slide; ∼0.8–1.2 µg of each full length NR in crude nuclear extracts from transfected Cos-7 cells was applied to the PBM; and bound NRs were detected with primary antibodies for human HNF4α, RXRα, COUPTF2 and RARα from R&D Systems (Catalogue # PP-H1415, PP-K8508, PP-H7147 and PP-H1920-00, respectively) at a 1:100 dilution overnight at room temperature, and then secondary antibody (Dαm-Dylight-Cy3, Jackson ImmunoResearch) at a 1:50 dilution for 1 h. After three washes in PBS-0.1% Tween-20, the slide was dried and scanned at 633 (Cy5) and 543 nm (Cy3). Data extraction and normalization was performed as previously described (13). The binding threshold of each NR was set to two SD from the mean or the quantile 0.95 of the random controls, whichever was higher. DNA motifs (represented using position weight matrices, PWM) were generated using SeqLogo (36) and Weblogo v2.8.2 (37). Hypergeometric tests are performed using phyper in R. While the HNF4α PBM2 was repeated, our previous raw data for the HNF4α PBM2 (13) was used for analysis since the signal intensities were closer to that of RXRα and COUPTF2.
Motif mining of ChIP-seq peaks
DNA sequences in ChIP-seq peak regions were extracted by Cisgenome and analyzed for relative enrichment level (fold enrichment) compared to matched control regions as described in (38). The length of each control region was determined by the average length of all peaks in a data set and the total number of control regions was five times as many as ChIP-seq peaks. If multiple data sets were available for an NR, the average was used as the final score and SD was given (Supplementary Table S1). The empirical P value of the enrichment score for each motif was determined using the enrichment distribution of 1000 6- or 9-nt random genomic sequences. Coordinates of peak centers were either given in the preprocessed BED or WIG files or obtained by reprocessing the data (in the case of ERRβ and Rev-Erbα). Density plots were generated by R; variances between plots were calculated using a two-sided F-test. Motif mining was performed by Gibbs Motif Sampler (39) implemented in Cisgenome, using 3000 iterations and the third-order Markov Chain model. The best AGGTCA-like motif was selected from 10 candidates with the highest scores.
Identification of HNF4-specific target genes
DNA sequences from −10 to + 10 kb of each gene in the human genome (hg19) were downloaded from the Ensembl genome browser (http://uswest.ensembl.org/index.html) using BioPerl API. The occurrences of HNF4-specific binding sequences, obtained from either the PBM data or support vector machine (SVM) prediction (see details in Supplementary Data), were identified by Seqmap (40) using exact match. A total of 3000 random 13-mers were selected randomly from the human genome. The difference between two frequency distributions was calculated using the Student’s t-test in R. The online search engine for HNF4-specific binding sequence was developed using Perl and cgi, and is hosted by the Bioinformatics Core of Institute for Integrative Genome Biology at University of California at Riverside at http://nrmotif.ucr.edu/NRBSScan/H4SBM.htm. HNF4α ChIP-seq peaks from HepG2 cells (41) were examined and only those peaks containing at least one HNF4-specific binding site were selected as HNF4-specific peaks. If a gene had at least one HNF4-specific peak within 10 kb of its transcription start site (TSS), then this gene was considered as an HNF4-specific candidate target. Only down-regulated candidate genes in HNF4 RNAi assay (in HepG2 cells) (13) were chosen as final targets. Gene Ontology (GO) analysis was performed using DAVID (42).
RESULTS
PBMs reveal a polarity in the DR1 motif for RXRα, COUPTF2 and HNF4α
Custom PBMs were used to examine the DNA binding specificity of human HNF4α2 (referred to as HNF4α), RXRα and COUPTF2. They bound 1371, 1285 and 1530 unique DNA sequences, respectively, although there was considerable overlap, especially between RXRα and COUPTF2 (Figure 1A). Notably, while RXRα in the absence of a heterodimeric partner typically does not bind DNA in shift gels (22,23,27), it bound very well in the PBM in both the presence and the absence of its ligand 9-cis retinoid acid; control experiments verified that the amount of endogenous NRs such as RARα was too low to be detected in the PBMs (see Supplementary Figure S1 for details and Supplementary Table S4 for a list of all sequences). There are reports of RXRα activating transcription or binding DNA in the absence of added NR partners (19–23,43–45), and recent ChIP-seq experiments reveal considerable RXR binding in the absence of a PPAR partner (46), suggesting that RXR homodimers bind DNA in vivo as well as in vitro.
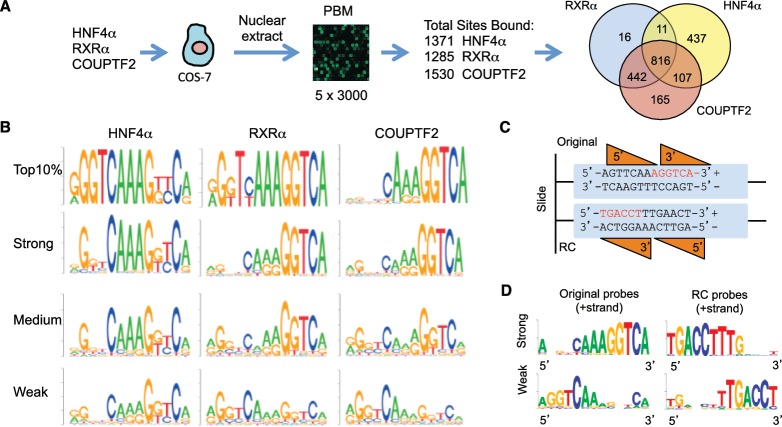
Figure 1.
DNA binding specificity of human HNF4α, RXRα and COUPTF2 determined by PBMs. (A) Work flow and overall results of PBM experiment. Total number of distinct DNA sequences (sites) bound by each NR is given as well as the number of overlapping and unique sequences (Venn diagram). (B) DNA motifs for the top 10% of binders as well as top 33% (strong), middle 33% (medium) and bottom 33% (weak) of binders. (C) Schematic representation of NR binding orientation on original and reverse complement (RC) DNA probes. Orange triangles, DBDs of 5′ and 3′ NR monomers. The canonical DR1 half site AGGTCA (red) is close to the free end of the original probe, while the corresponding sequence in the RC probe is close to the slide in the PBM. (D) Motifs of the 30 strongest and 30 weakest binders of the original or RC probes for COUPTF2 as indicated. (See Supplementary Figures S1B for the results of HNF4α and RXRα, and Figure S2A for details in probe design.).
All bound sequences were ranked according to their normalized signal intensities (binding scores) and DNA motifs were generated for strong, medium and weak binders [binding affinity is linearly correlated with the binding signal from PBMs (47)]. Motifs of the top 10% strongest binders showed that both the 5′ and 3′ half sites of the DR1 motif contain essential positions for binding, reinforcing the notion that these three NRs bind as homodimers on the PBM (Figure 1B). We also noted that the strong binders for RXRα and COUPTF2 had more conserved positions in the 3′ than in the 5′ half site, suggesting that the 3′ half site may be more critical for DNA binding. Since, the 3′ half site is farther from the glass slide in the PBM, it was possible that the observed polarity was an artifact of the PBM design. Therefore, we designed a second PBM in which 30 strong binders of COUPTF2 with AGGTCA half sites on the 3′ side were compared with 30 weak binders with AGGTCA on the 5′ side; a third group of probes had weak AGGTCA-like motifs in both half sites. The reverse compliment (RC) of each probe was also examined (Supplementary Figure S2A). If COUPTF2 DNA binding is affected by the distance from the slide, then the RC probes of corresponding strong binders will become weak binders, and vice versa (Figure 1C). We found that the strongest binders for both the original and the RC probes were the same sequences with a clear preference for AGGTCA in the 3′ half site, even when it was close to the slide; conversely, the weakest binders had the AGGTCA motif in the 5′ half site regardless of the distance to the slide (Figure 1D). Similar results were observed for RXRα and HNF4α (Supplementary Figure S2B). We also examined the effect of the distance of the entire 13-mer from the slide as well as the free end of the probe and found that the strongest binding by all three NRs resulted when the 13-mer is 27 nt from the slide and 5 nt from the free end of the probe (Supplementary Figure S2C and S2D). All together, these results suggest that in vitro RXRα, COUPTF2 and HNF4α prefer the 3′ half site of the DR1 motif.
PBMs identify an HNF4α-specific binding motif
The DNA binding specificity of HNF4α, RXRα and COUPTF2 was further analyzed using scatter plots (Figure 2A). RXRα and COUPTF2 shared very similar binding specificities with an R2 of 0.84 for all bound sequences. In contrast, HNF4α showed a different profile with >200 DNA sequences bound exclusively by HNF4α. The consensus motif of HNF4-specific binders had a subtle yet consistent change at position 10 (p10) and p11, where GT was replaced by TC. Together with seven other conserved positions, it defined the HNF4-specific binding motif (H4-SBM) xxxxCAAAGTCCA (x refers to any nucleotide; the nucleotides that differ from the canonical AGGTCA are underlined) (Figure 2B). Hypergeometric plots using all the binders indicate that TC at p10–p11 are disfavored by both RXRα and COUPTF2 (Figure 2C). Interestingly, the probes that were bound by RXRα and COUPTF2 but not HNF4α strongly resembled a canonical DR1 motif (Figure 2B).
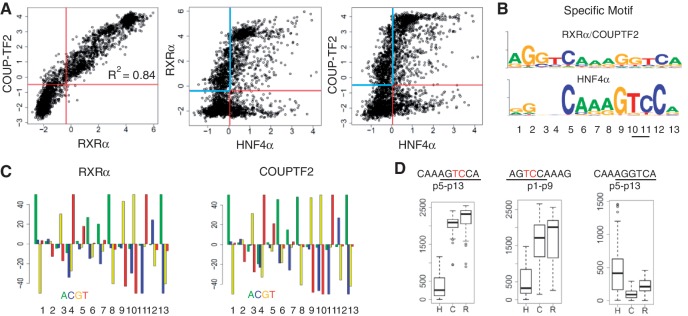
Figure 2.
Identification of an HNF4-specific DNA binding motif. (A) Scatter plots of PBM results of the indicated NRs. Each spot is the average of ∼5 replicates for each unique DNA sequence (∼3000 total) on the PBM; only probes with a coefficient of variation less than 0.3 were used. Each plot is divided into four quadrants by the binding thresholds of the NRs. Common binding sequences of RXRα and COUPTF2 show a linear correlation with a R2 of 0.84. Specific targets of RXRα/COUPTF2 and HNF4α are in the blue and red quadrants, respectively. (B) Shown are motifs derived from the RXRα/COUPTF2 and HNF4α-specific sequences in the blue and red quadrants in the scatter plots in (A), respectively. (C) Hypergeometric plots showing the over- and under-representation for each nucleotide at p1 to p13 (in four colors) of RXRα and COUPTF2 in bound sequences on PBM. Y-axis indicates log(P-value) calculated using phyper test in R. Positive and negative values indicate over-representation and under-representation, respectively. The maximum logP is set as 50. As shown, ‘TC’ at p10–p11 is strongly disfavored by RXRα and COUPTF2. (D) Box plots showing rankings of all PBM probes containing the 9-nt sequence indicated at the top of each plot. The position of the 9-nt sequence in the DR1 motif is indicated; p1–p9 is on the 5′ side and p5–p13 is on the 3′ side. The ‘TC’ in the HNF4-specific half site sequence is in red. Bound probes of HNF4α (H), COUPTF2 (C), and RXRα (R) have scores below 1371, 1530 and 1285, respectively.
We next determined that all 81 of the PBM probes bearing the H4-SBM were in fact HNF4-specific binders; they all bound HNF4α but none bound RXRα or COUTF2 (Figure 2D, left panel). There were two exceptions (AGGTCAAAGTCCA and GGGTCAAAGTCCA) that bound RXRα and COUPTF2 weakly; they are hybrids of the canonical DR1 and the HNF4-specific half site. In contrast, the probes containing AGTCCA only in the 5′ half site were not completely specific for HNF4α (Figure 2D, middle panel) while those with the canonical AGGTCA in the 3′ half site preferred COUPTF2 and RXRα (Figure 2D, right panel). Finally, a similar analysis of 16 H4-SBM-like motifs with all possible permutations at p10 and p11 confirmed that the H4-SBM xxxCAAAGTCCA is the only motif recognized by HNF4α specifically (Supplementary Figure S3).
H4-SBM is bound by HNF4α but not RXRα in vivo
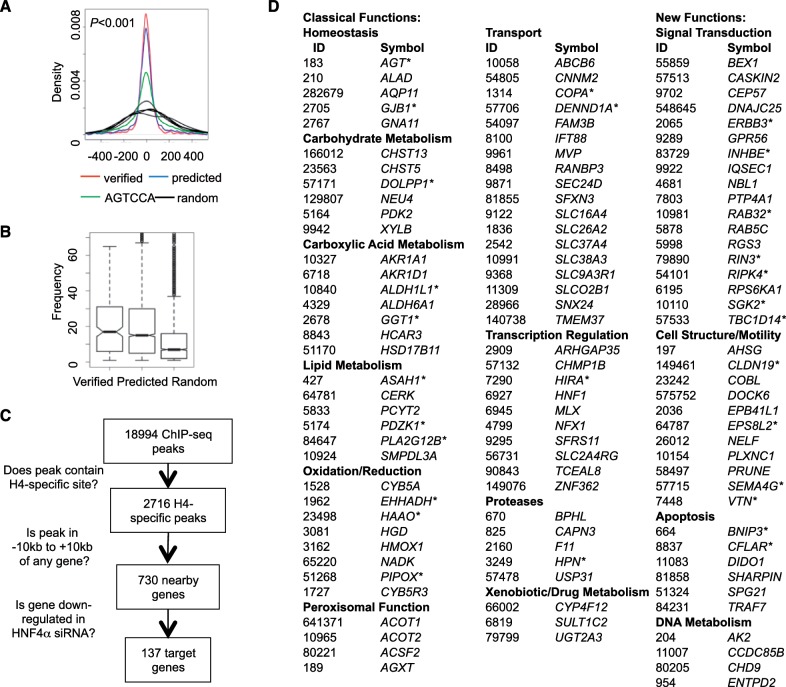
Since the DNA in the PBM does not necessarily have exactly the same conformation as DNA wrapped in nucleosomes, we determined whether the H4-SBM bound by HNF4α in vitro is also bound in vivo by analyzing 26 publicly available genome-wide occupancy profiles for 14 NRs (Supplementary Table S1). Since data from the same tissue/cell type were not available for all the NRs and since we are concerned with binding motifs and not specific target genes, we combined and compared ChIP-seq data for a given NR across different conditions/cell types for greater statistical power. First, we compared HNF4α ChIP’ed from mouse liver (48) and RXRα ChIP’ed from mouse 3T3-L1 (pre-adipocyte) cells (49). In 12 097 peaks unique to HNF4α, the motif mined by Gibbs Motif Sampler (39) was found to be very similar to the H4-SBM, whereas the motif in 4285 peaks unique to RXRα was more similar to the canonical DR1 (Figure 3A). Interestingly, there were 736 ChIP peaks for HNF4α in liver and RXRα in pre-adipocytes that overlapped; the motif mined from those peaks had minor variations from both the H4-SBM and the canonical DR1 but was nearly identical for RXRα and HNF4α.
Figure 3.
In vivo DNA binding specificity of HNF4α and RXRα. (A) Analysis of HNF4α and RXRα ChIP-seq data from mouse liver and pre-adipocyte cells, respectively. Shown are consensus binding motifs in overlapping and non-overlapping ChIP-seq peaks. The dimeric status of RXRα (hetero- versus homodimer) in the ChIP peaks is unknown although only a small portion of peaks overlap with PPARγ peaks (49). Numbers refer to number of ChIP-seq peaks analyzed for each category. (See Supplementary Table S1 for references for the data sets.) (B) Fold enrichment of 16 half site sequences in ChIP-seq peaks of HNF4α and RXRα described in (A). Variations in the half site sequences are underlined. Average enrichment scores of all data sets for the same protein are used and SDs are indicated by error bars. Empirical P values for enrichment levels (*P < 0.05, **P < 0.001, ***P < 0.0001) are estimated by comparison with the enrichment levels of 1000 6-nt random genomic sequences. (C) Enrichment of particular sequences near the center of the HNF4α and RXRα ChIP-seq peaks described in (A). Distribution of the indicated DNA sequences containing AGGTCA (left) or AGTCCA (right) are shown in density plots. The x-axis indicates the distance (in base pairs) from the peak center. Density plots in black lines are derived from five 6-nt random genomic sequences as controls. (D) Binding polarities of HNF4α and RXRα in vivo. Fold enrichment of 9-nt motifs in the HNF4α and RXRα ChIP peaks described in (A) are shown in bar plots. Each 9-nt motif contains a 6-nt canonical DR1 half site or HNF4-specific half site sequence (underlined) and a 3-nt extension (p7-p9) that defines the polarity of the half site [half site sequence on the 3′ side (blue) or the 5′ side (red)]. Error bars indicate SD among multiple data sets.
We next compared the enrichment of the 16 different half site sequences with all possible permutations at p10 and p11 in ChIP-seq peaks of RXRα and HNF4α. The H4-specific half site sequence AGTCCA had a 2-fold higher enrichment level compared to control regions (P = 0.001) in the HNF4α peaks but not the RXRα peaks (Figure 3B). In contrast, the canonical AGGTCA half site showed high and comparable enrichment in both the HNF4α and RXRα peaks. These results suggest that the H4-specific half site is preferred by HNF4α but not RXRα in vivo.
Since ChIP-seq peaks typically cover a region much larger than the size of a TF binding site, it is frequently assumed that the binding site of a protein is close to the peak center (peak summit). Therefore, we compared the distance from the peak center to the canonical and HNF4-specific half site in the ChIP peaks. In both the HNF4α and RXRα peaks, the canonical half site AGGTCA was significantly enriched around the peak center compared to random sequences (P < 0.001); adding 1 nt at a time to the 5′ side of the motif significantly increased the enrichment (Figure 3C, left panels). In contrast, when the HNF4-specific half site was analyzed, the enrichment was observed in HNF4α peaks but not the RXRα peaks (right panels), confirming that the H4-SBM (CAAAGTCCA) is bound in vivo by HNF4α but not RXRα.
Finally, in order to determine whether there was any evidence of half site polarity in vivo, the enrichment levels of 9-mer sequences with the canonical (AGGTCA) and the HNF4-specific (AGTCCA) half site on both the 5′ and 3′ side were compared in the HNF4α and RXRα ChIP-seq peaks. HNF4α had a clear preference for the 3′ side for both half site sequences (Figure 3D, left panel, blue versus red bars). While a similar result was observed for the canonical half site in the RXRα peaks, the HNF4-specific motif was not enriched in either position for RXRα (right panel). These results further confirm that AGTCCA is not bound by RXRα in vivo and that the polarity for the half site preference observed in the PBM for RXRα and HNF4α is also observed in vivo.
H4-SBM is exclusive to HNF4α in vivo
To determine whether the H4-SBM is recognized by any other NR aside from HNF4α, we analyzed ChIP-seq data from 12 additional NRs that are known to be expressed in one or more of the same tissues as HNF4α—PPARγ (NR1C3) (46,49,50), PPARδ (NR1C2) (51), VDR (NR1I1) (52), FXRα (NR1H4) (53,54), PXR (NR1I2) (55), ERRβ (NR3B2) (56), RARα (NR1B1) (57), ERα (NR3A1) (58), LXRβ (NR1H2) (59), LRH-1 (NR5A2) (60), Rev-Erbα (NR1D1) (61). The glucocorticoid receptor (GR, NR3C1) (46,62,63), which prefers a different half site AGAACA (64,65), served as a negative control (Supplementary Figure S4A). Motifs of consensus sequences derived from the 12 NR ChIP-seq peaks did not reveal any binding to the H4-SBM (Supplementary Figure S4B). When 6-mer half site sequences were examined, the H4-specific half site (AGTCCA) was found to be enriched in PXR peaks but not in peaks for the remaining 11 NRs (Figure 4A and Supplementary Figure S5). In contrast, there was an enrichment of the canonical half site AGGTCA in 8 of the 12 NRs (Supplementary Figure S5), which served as a positive control. We also examined the half site sequences as a function of distance from the peak center. There was an enrichment in AGGTCA at the peak center for all tested NRs except GR. The HNF4-specific half site, however, was only enriched in peak centers for HNF4α and three other NRs: Rev-Erbα, FXRα and PXR (Supplementary Figure S6).
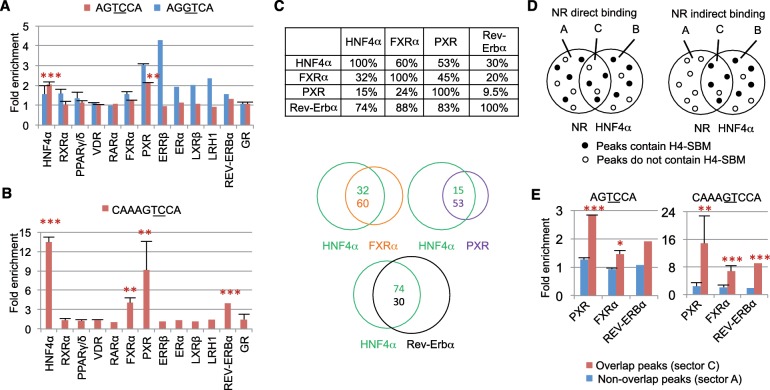
Figure 4.
Comparison of in vivo DNA binding specificity of multiple NRs. (A) Fold enrichment of the canonical DR1 half site sequence (blue) and the HNF4-specific half site sequence (red) in ChIP peaks of 14 NRs. ChIP-seq data for different isoforms of a NR or multiple data sets for the same protein are combined (see Supplementary Table S1 and text for references); SD is indicated by error bars. Empirical P values for enrichment levels (*P < 0.05, ** P < 0.001, *** P < 0.0001) are estimated using enrichment levels compared to 1000 6-nt random genomic sequences. Only P values for AGTCCA are indicated. (See Supplementary Figure S5 for enrichment values for 14 additional half site sequences.). (B) Fold enrichment in ChIP-seq peaks of 14 NRs as in (A) but for the H4-SBM. P values are estimated using 1000 9-nt random genomic sequences. (C) Overlap between ChIP-seq peaks among select NRs in mouse liver. Given is the percent of overlapping ChIP-seq peaks between the NRs indicated on the top and the side. For example, 59.6% of FXRα peaks overlap with HNF4α peaks, while only 32.2% of HNF4α peaks overlap with FXRα peaks. (D) Schematic representation of direct and indirect binding of a NR to the H4-SBM. Left: direct binding to the H4-SBM is characterized by the even distribution of H4-SBM sites in overlapping and non-overlapping peaks (sectors C and A, respectively). Right: indirect binding of a NR to the H4-SBM via HNF4α would yield a greater number of H4-SBM sites in the overlapping peaks. (E) Enrichment of the HNF4-specific half site (left) and the H4-SBM (right) in ChIP-seq peaks for the indicated NRs as described in (C). Blue bars indicate fold enrichment in non-overlapping peaks with HNF4α ChIP-seq peaks (sector A). Red bars indicate fold enrichment in overlapping peaks with HNF4α ChIP-seq peaks (sector C). P values [as in (A)] are estimated using ratios of 1000 6- and 9-nt random genomic sequences, respectively, for two analyzed motifs.
Since searching with the 6-mer (AGTCCA) does not provide any information about its position within a 13-nt motif (i.e. 5′ or 3′ side), the search was repeated with the 9-mer H4-SBM (CAAAGTCCA). We found a 13.5-fold enrichment in HNF4α peaks and a considerable enrichment (>3-fold) in FXRα, PXR and Rev-Erbα peaks; the remaining NRs continued to show no enrichment (Figure 4B). Furthermore, the ChIP-seq peaks for FXRα, PXR and Rev-Erbα ChIP-seq showed considerable overlap with HNF4α peaks (Figure 4C and Supplementary Table S5). Since ChIP cannot distinguish between direct and indirect binding, it was possible that FXRα, PXR and Rev-Erbα were binding to the H4-SBM through HNF4α. To address this, we compared the enrichment level of the HNF4-specific half site (AGTCCA) and the full H4-SBM (CAAAGTCCA) in the peaks of each of the three NRs that overlapped with the HNF4α peaks to those that did not overlap. If a particular sequence is a direct binding target of a NR, then similar enrichment levels are expected in both overlapping (sector C) and non-overlapping peaks (sector A) (Figure 4D, left). On the other hand, if the NR requires HNF4α to bind the sequence, then the overlapping peaks should have a higher enrichment level than the non-overlapping peak (Figure 4D, right). The results show that both AGTCCA and CAAAGTCCA are enriched in the overlapping peaks but not the non-overlapping peaks for all three NRs (Figure 4E). The fold enrichment for H4-SBM was in fact 3- to 6-fold higher in the overlapping peaks compared to the non-overlapping peaks. Furthermore, the HNF4-specific half site was the only 1 of 16 candidate 6-mers that showed such an enrichment for these NRs (Supplementary Figure S7C). A similar analysis with the peaks unique to HNF4α (sector B) verified the methodology (Supplementary Figures S7B and S7D). These results suggest that while PXR, FXRα and Rev-Erbα are localized to regions in the genome that contain the HNF4-specific motif, they may do so only when HNF4α is also present in the same region. While we cannot definitively rule out direct interactions between these NRs and HNF4α, AGGTCA-like motifs were found in about half of the overlapping peaks that contain the H4-SBM (Supplementary Figure S7E–S7H). This suggests that PXR, FXRα and Rev-Erbα may bind canonical AGGTCA-containing motifs cooperatively with HNF4α in regions that contain the H4-SBM. For example, on the Cyp7a1 promoter there are overlapping ChIP-seq peaks for HNF4α, PXR and Rev-Erbα but the only H4-SBM is at the center of the HNF4α peak (Supplementary Figure S7I).
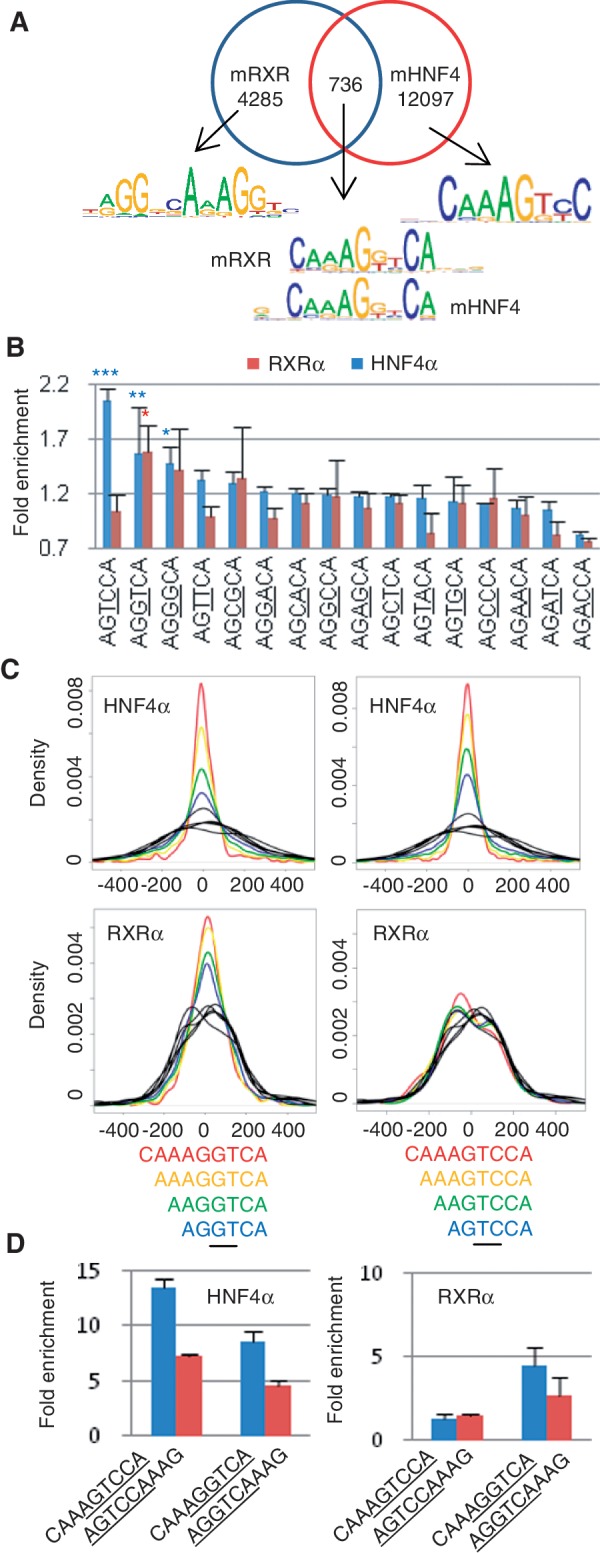
HNF4-specific DNA recognition is mediated by two residues in the DBD, Asp69 and Arg76
To determine the molecular basis of HNF4-specific DNA recognition, protein–DNA interactions were analyzed in the DBD structures of HNF4α and RXRα (66,67). Among all the residues making contacts with the DNA, only four residues differ between RXRα and HNF4α. In HNF4α, two of these residues, Asp69 and Arg76, lie in the first DNA recognition helix of the DBD and interact with side chains of nucleotides at p3 and p4 in both half sites of the DR1 motif (Figure 5A). These two residues are completely conserved in HNF4 genes across all species from human down to Trichoplax, except for Caenorhabditis elegans which contains ∼260 HNF4-like genes (Supplementary Figure S8A). Residues at equivalent positions in RXRα and COUPTF2 are Glu and Lys, respectively (Figure 5B).
Figure 5.
Molecular basis of HNF4-specific DNA recognition. (A) Superimposed crystal structures of HNF4α DBD (3CBB, cyan) and RXRα DBD (1BY4, yellow). In the HNF4α structure, only D69 and R76 interact with the third and fourth base pairs of the AGTCCA half site. The equivalent residues of RXRα are E and K, respectively. (B) Sequence alignment of the indicated human NRs in the DNA recognition helix of the first zinc finger. Numbers 69 and 76 refer to the residues in human HNF4α2. (C) Scatter plots of PBM data as in Figure 2A of the WT and mutant HNF4α compared to WT RXRα. Binding sequences specific to HNF4α WT are indicated as red spots. All the red spots shift to the non-binding quadrant in the HNF4α double mutant D69E/R76K. (D) As in (C) but comparison of HNF4αWT and D69E/R76K mutant to COUPTF2.
To examine the effect of Asp69 and Arg76 on HNF4α binding specificity, single and double point mutations D69E, R76K and D69E/R76K were introduced into the HNF4α DBD to convert it into an RXR/COUPTF2-like DBD. In the PBM, the D69E mutant selectively abolished binding to the H4-SBM while R76K seemed to decrease binding in a non-selective fashion; interestingly, the double mutant D69E/R76K yielded a profile nearly identical to that of D69E (Supplementary Figure S8B), suggesting that both Asp69 and Arg76 are necessary for optimal discrimination between H4-SBM and other sites. Importantly, the HNF4α D69E/R76K double mutant also altered the binding profile to more closely resemble that of RXRα/COUPTF2; none of the HNF4-specific binders were bound by the double mutant (Figure 5C and D). While some common binders of HNF4α and RXRα/COUPTF2 were also affected by the mutations (Supplementary Figure S8C and S8D), most of them bear the AGTTCA motif in the 3′ half site that is also preferred by HNF4α but not RXRα in vivo (Figure 3B). Finally, HNF4α and HNF4γ are the only human NRs with an aspartic acid at residue 69 and an arginine at residue 76 [Figure 5B and (7)], suggesting that the H4-SBM may be truly specific to HNF4 in the entire NR superfamily.
HNF4α activates gene expression using HNF4-specific binding sites
To determine whether the HNF4α binding site preference results in a functional outcome (i.e. gene expression), luciferase reporter assays were performed using a known HNF4α/RXRα response element from the human APOA1 promoter. As predicted, HNF4α activated gene expression using both the wild-type response element (WT-RE) AGGGCAgGGGTCA and an HNF4-specific mutant (MUT-RE) AGTCCAgGGTCCA; in contrast RXRα activated expression only from the WT-RE (Figure 6A). In addition, COUPTF2, a repressor, competed with HNF4α on the WT-RE but not the MUT-RE (Figure 6B). These results verify that HNF4α is capable of activating gene expression using the H4-SBM, while RXRα and COUPTF2 cannot functionally compete with HNF4α for the H4-SBM response element.
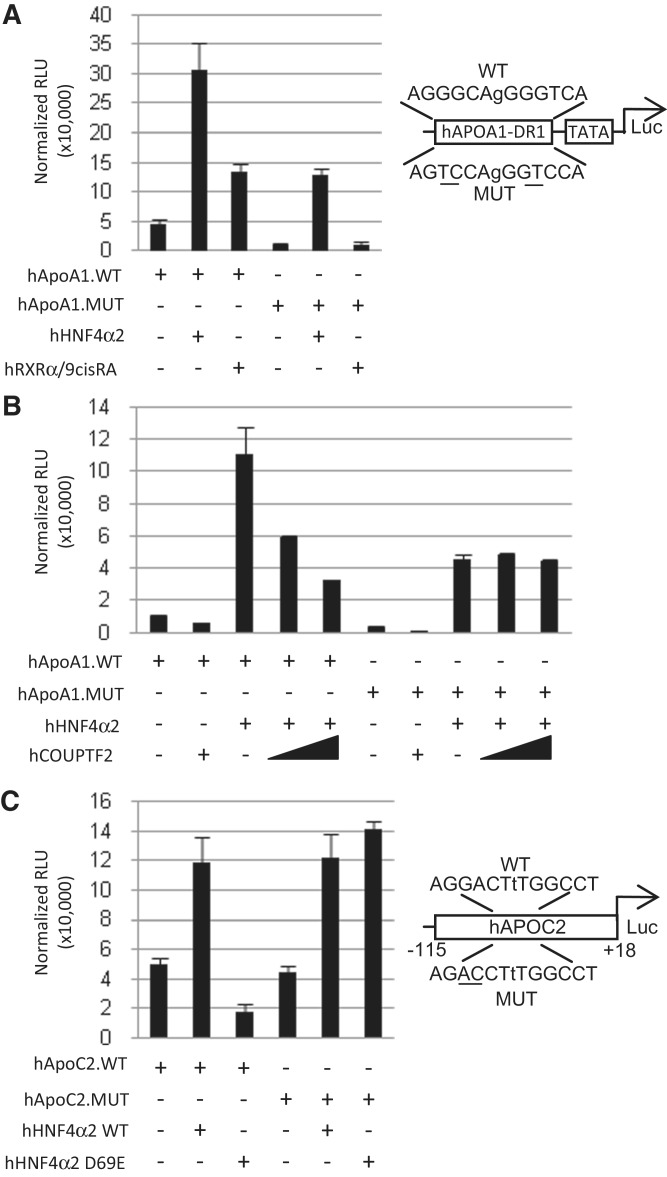
Figure 6.
Specificity of the HNF4-specific binding motif in cell-based reporter gene assays. (A) Luciferase reporter assay in HEK293T cells co-transfected with 500 ng of indicated reporter construct (pGL4.23.hApoA1.DR1.WT or pGL4.23.hApoA1.DR1.MUT) plus 50 ng of expression vector (+, pcDNA3.1.hHNF4α2 or pMT2.hRXRα) or empty vector (−, pcDNA3.1). 9-cis-retinoic acid (1 µM) (or ethanol as vehicle control) was added as indicated. Right: diagram of the wild-type (WT) and mutated (MUT) response elements in the reporter construct for (A) and (B). Shown are relative light units (RLU) normalized to β-galactosidase activity. Bars are average of three replicates. Error bars indicate SEM. (B) As in (A) but with the expression vector (+, pcDNA3.1.hHNF4α) and two different amounts of pMT2.hCOUPTF2 (400 and 800 ng). Empty vector was added to adjust for amount of total DNA transfected. (C) As in (A) but with the indicated reporter constructs (pGL4.10.hAPOC2.WT or pGL4.10.hAPOC2.MUT) plus the expression vector (+, pcDNA3.1hHNF4α or pcDNA3.1hHNF4α.D69E carrying the D69E mutation in its DBD). Right: diagram of the WT and MUT hHNF4α response element in the human APOC2 promoter.
The effect of the D69E mutation was also examined in the luciferase assay using an endogenous promoter from a known HNF4α target gene, APOC2 (31). WT HNF4α successfully activated the expression of the WT promoter containing an AGGCCAaAGTCCT motif whereas the D69E mutant failed to do so (Figure 6C). In contrast, both the WT and D69E mutant activated gene expression when the H4-specific response element was mutated to a more canonical DR1 element AGGCCAaAGGTCT (Figure 6C, reverse complement sequences are shown in the figure to maintain the orientation in the genome). This result confirms that Asp69 is responsible for the HNF4-specific DNA binding.
Functionality of HNF4-specific binding sequences in vivo
In order to determine whether the H4-SBM is functional under physiological conditions, we trained an SVM model on ∼200 verified H4-SBM sites from the PBM to predict all 13-nt sequences that can be bound specifically by HNF4α (variations in the less conserved positions of the consensus H4-SBM, CAAAGTCCA, such as p6-p8 and p13 can be tolerated, Figure 1B). This method generated ∼3000 high confidence, predicted HNF4-specific binding sequences. (See Supplementary Table S3 for a list of all H4-SBM sequences and http://nrmotif.ucr.edu/NRBSScan/H4SBM.htm for a web-based, HNF4-specific binding sequence search tool.)
The predicted H4-SBM sites were first validated by comparing their position in ChIP-seq peaks to the verified H4-SBM sites and the AGTCCA half site (Figure 7A). The predicted and verified H4-SBM sites were then examined across all protein-coding genes in the human genome relative to the TSS (+1), −10 to +10 kb. (HNF4 binding sites are symmetrically distributed on both side of TSS, Supplementary Figure S9). The results show comparable occurrence frequencies for the predicted and verified sites (P > 0.05) that were significantly higher than those of random sequences (P < 0.001) (Figure 7B). Finally, we used the predicted H4-SBM sites to search HNF4α ChIP-seq and expression profiling data to identify potential HNF4α target genes. A total of 730 genes in the human liver cell line HepG2 were found to have at least one predicted H4-SBM site in a HNF4α ChIP-seq peak within −10 to + 10 kb of +1 (Figure 7C). Among these genes, 137 were down-regulated by HNF4α RNA interference (RNAi) in HepG2 cells (13), suggesting that these genes are direct targets of HNF4α. GO analysis showed that the H4-specific genes were enriched in a variety of metabolic processes (e.g. lipid, carbohydrate, xenobiotic/drug metabolic processes; homeostasis; transport), typical of classical HNF4α targets (Supplementary Table S3). There were also genes in some of the new categories of HNF4α targets that we identified in our previous study (13), such as immune response, signal transduction, apoptosis and cell structure (Figure 7D). That study, however, did not identify HNF4-specific motifs or targets. All told, this analysis identified ∼100 new predicted, direct HNF4α target genes that have an H4-SBM in a ChIP peak and are down regulated by an HNF4α RNAi.
Figure 7.
Prediction of HNF4-specific binding sequences and target genes. (A) Distribution of HNF4-specific binding sequences in HNF4α ChIP-seq peaks in HepG2 cells (41) are shown in density plots. Red line, ∼200 HNF4-specific PBM sites; blue line, ∼3000 predicted HNF4-specific sites; green line, the HNF4-specific half site sequence; black line, five 6-nt random genomic sequences used as controls. P value is calculated by the F-test. (B) Box plots of predicted and verified HNF4-specific sites compared to ∼3000 random sequences in the regulatory regions of human genes (−10 to +10 kb from TSS). Both the predicted and verified HNF4-specific sequences occur at a statistically significant high frequency compared to random sequences (P < 0.001 in Student’s t-test). (C) Strategy used to search for potential HNF4-specific target genes. See text for details. (D) Classification of ∼100 predicted human HNF4α target genes based on the analysis in (C). All of these genes have at least one HNF4-specific site in an HNF4α ChIP-seq peak in HepG2 cells (41) and were down regulated by HNF4α RNAi in HepG2 cells (13). See Supplementary Table S4 for a complete list of the 137 genes identified in (C) as well as Gene Ontology results. *, genes previously identified in (13) as predicted HNF4α target genes but with no indication of containing an HNF4-specific binding motif. Nearly all the other genes have not been previously identified as HNF4α targets.
Interestingly, several genes associated with acyl Co-enzyme A metabolism (acyl-CoA synthases, thioesterases, ligases and a co-factor for a desaturase) were identified as putative HNF4α targets with an H4-SBM site (ACSM2B, EHHADH, ACOT2, ACSF2, SLC27A2, CYB5A, AGXT). Others have shown that HNF4α binds not only acyl Co-A binding protein but also fatty acyl thioesters of Co-enzyme A that appear to act as modulators of HNF4α function (68–70). Acyl Co-A is well known for donating an acetate group in acetylation reactions of lipids and proteins. HNF4α itself has been shown to be acetylated in its hinge region (71), as have many of the enzymes encoded by HNF4α target genes—e.g. PEPCK (PCK1), a classical HNF4α target (72) that catalyzes the rate limiting step in gluconeogenesis, and the new H4-SBM target reported here, EHHADH, which encodes enoyl–coenzyme A hydratase/3-hydroxyacyl–coenzyme A that catalyzes two steps in fatty acid oxidation (73). While it is not only possible but very likely that these H4-SBM genes are also regulated by other TFs, including other NRs via other sites in the promoters, these results nonetheless reinforce the notion that there is an important relationship between HNF4α and processes involving Co-enzyme A. Finally, we note that several of the newly identified H4-SBM containing HNF4α target genes are linked to human diseases (LIPA, EHHADH, AGXT, PIPOX, HGD, CYB5A, PDZK1, F11), expanding the clinical relevance of HNF4α. (See Supplementary Table S3 for a complete list of H4-SBM target genes from both a −10 to + 10 kb and a −2 to +1 kb analysis.)
DISCUSSION
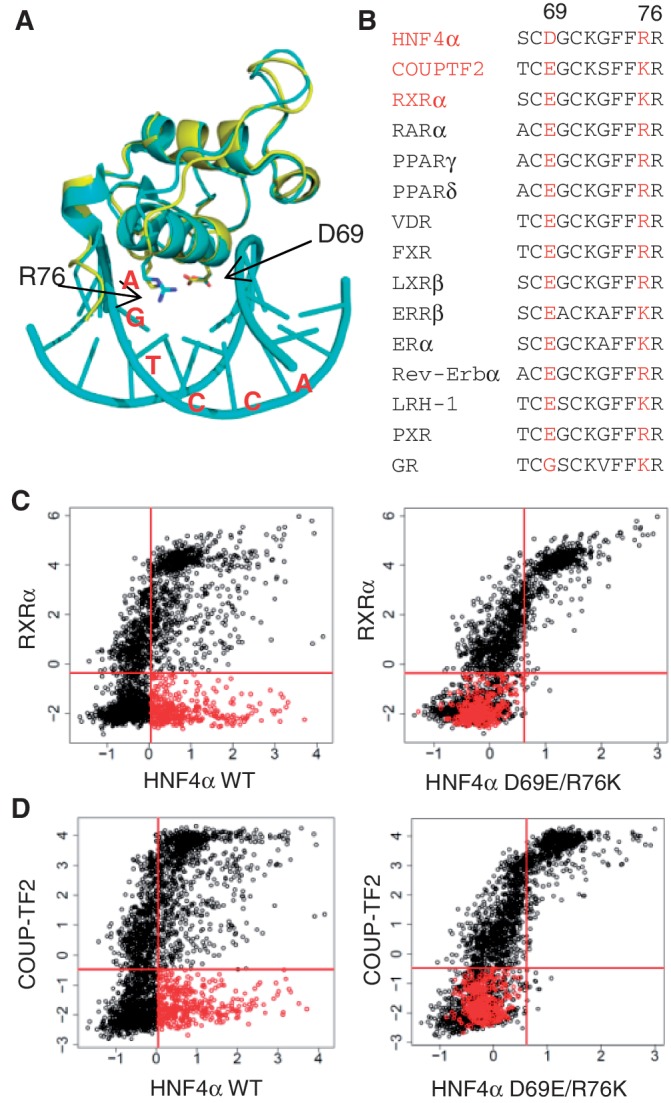
The dogma in the NR field is that NRs recognize DNA targets based on one of two motif modules, AGGTCA (non-steroid receptors plus ER) and AGAACA (all other steroid receptors) (Figure 8A). While there are additional rules for spacing and orientation [as well as some variations on the half site motif, such as AGTTCA (7)] that distinguish the different receptors, those rules do not adequately explain the diversity of NR function in vivo. We addressed this issue by comparing the binding specificity of three highly related NRs—HNF4α, RXRα and COUPTF2—on ∼3000 different variations of a canonical DR1 (AGGTCAxAGGTCA) using PBMs and full length receptors in crude nuclear extracts. We identify a new motif module for HNF4α, AGTCCA and provide additional insight into NR DNA binding (Figure 8A–D).
Figure 8.
New insights into NR DNA binding from PBM and ChIP-seq analysis. (A) Half site motif for the indicated NRs along with the amino acid sequence of the P box region in the DNA recognition helix of the first zinc finger in the DBD showing the residues relevant to this work. GR, MR, AR and PR: glucocorticoid, mineralocorticoid, androgen and progesterone receptors, respectively). The only NRs that have P boxes that are not depicted are TLX (NR2E1) with an Asp (D) and Lys (K) and PNR (NR2E3) with an Asn (N) and Lys (K) at positions 69 and 76, respectively (HNF4α numbering). (B) RXRα, COUPTF2 and HNF4α homodimers all bind classical DR1-like motifs (AGGTCAxAGGTCA) while HNF4α homodimers also bind an HNF4-specific binding motif (H4-SBM, xxxxCAAAGTCCA) that is not bound by RXRα or COUPTF2 in the PBMs, nor 11 other NRs in ChIP-seq. (C) RXRα has a preference for the 3′ half site in a DR1-like motif in RXR:RXR homodimers as well as in RXR:RAR and RXR:PPAR homodimers, suggesting that an exchange between the 5′ RXRα monomer and RAR or PPAR monomers could occur without disrupting the binding of the 3′ RXRα monomer. COUPTF2 also prefers the 3′ half site of the DR1-like motifs, suggesting it could compete with RXRα homo- or heterodimers on these response elements. (D) PXR, FXRα and RevErbα ChIP peaks from mouse liver were all found to be enriched for the H4-SBM but only in the presence of HNF4α, suggesting that they may occupy their own binding sites near HNF4α-bound H4-SBM.
NR DNA binding polarity
In contrast to most gel shift results, including our own (27), we observed excellent binding of RXRα in the PBM in the absence of an ectopically expressed partner. The RXRα binding was nearly identical to that of COUPTF2, and both receptors exhibited a polarity in binding with a preference for the 3′ half site of the DR1 (Figure 8B and C). To our knowledge, this is the first report to examine polarity in DNA binding using PBMs, and the first report of polarity for NR homodimers. Since RXRα homodimers can activate transcription in the presence of the 9-cis retinoic acid and since COUPTF2 generally represses transcription, this suggests that the competition already observed between these two NRs on a few genes may in fact be a much broader phenomenon.
While binding polarities of NR homodimers have not been reported previously, the concept in heterodimers is not new. For example, RXR binds the 5′ half site in DR3, DR4 and DR5 motifs as heterodimers with VDR, TR and RAR, respectively (74–76). This polarity was also noted in crystal and solution structures of RXR heterodimers with RAR and VDR (77,78). However, on DR1 motifs the polarity of the RXR heterodimers is reversed with RXR in heterodimers with RAR and PPAR occupying the 3′ half site in DR1 motifs (78–82). Since our results now show that the RXRα homodimer also prefers the 3′ half site in DR1-like motifs, this suggests that there could be an exchange between the 5′ RXRα monomer in the homodimer and PPAR or RAR monomers (Figure 8C). The net result would be a replacement of the RXRα homodimer with a heterodimeric complex, perhaps while the 3′ RXRα monomer remains bound to the DNA. This would suggest a new paradigm for RXRα dimer exchange and a potential new role for RXRα homodimers as placeholders for RXRα heterodimers on DR1-like motifs, but must be experimentally proven. A thorough analysis of RXR heterodimer DNA binding specificity also needs to be done.
An NR-specific DNA binding motif
Comparison of the HNF4α PBM results to that of RXRα and COUPTF2 allowed us to identify in vitro an HNF4-specific binding motif (xxxxCAAAGTCCA) that we had not identified previously when we analyzed HNF4α alone (13). In vivo analysis showed that ∼8 and 42% of HNF4α ChIP-seq peaks contain CAAAGTCCA and AGTCCA, respectively (motif variations not considered). Comparable results were observed for CAAAGGTCA and AGGTCA (Supplementary Figure S10), suggesting a similar importance of AGGTCA- and AGTCCA-based motifs for HNF4α binding. The H4-SBM was not bound by RXRα or COUPTF2 in the PBM, nor eight other NRs in ChIP-seq data (Figure 8B). However, three NRs (FXRα, PXR and Rev-Erbα) were associated with the H4-SBM in vivo, most likely via their own binding sites, not the H4-SBM (Figure 8D). While there are a few reports of these NRs regulating the same target genes as HNF4α via their own binding sites (83–87), our results suggest that this may be a much broader phenomenon. The considerable overlap between PXR, FXRα, Rev-Erbα and HNF4α peaks in ChIP-seq data also demonstrates the complexity of NR-mediated regulation and the difficulty of precisely identifying binding sites for a given TF in ChIP-seq peaks. This complexity is increased by protein–protein interactions between HNF4α and other NRs such as PXR and FXR and competition for co-regulators (12). Our analysis shows how this problem can be overcome at least in part with a better understanding of binding specificities generated by the PBMs.
While this is the first identification and genome-wide analysis of an HNF4-specific binding motif, there were previous reports using classical methods suggesting that such a motif might exist. A survey of the literature identified 28 response elements that had been examined for responsiveness to HNF4α, RXRα and/or COUPTF2. Six of those elements carry the H4-SBM TC instead of the DR1-like GT at p10 and p11, and all six were responsive to HNF4α but not RXRα or COUPTF2. In contrast, the remaining 22 response elements did not have a TC at p10-11 and were responsive to RXRα or COUPTF2 (Supplementary Table S2). However, since these findings were generated by different groups over a period of years, an HNF4-specific motif was never identified.
In addition to the H4-SBM, we also identified a pair of residues in the HNF4 DBD that is responsible for the H4-SBM binding (Asp69 and Arg76) (Figure 8A). Asp69, which has the greatest effect on HNF4-specific binding, is in the P box which was shown previously to be responsible for the different half site of GR and related receptors (AGAACA) (5,88). Arg 76 is in the helix that contacts the DNA but to our knowledge has not been previously associated with DNA binding specificity. HNF4 is the only human NR with the combination of Asp69 and Arg76 and all HNF4 DBDs, except some of the HNF4-like genes in C. elegans, have these residues, suggesting that the H4-SBM may be truly unique to HNF4. There is only one other NR that has an Asp at position 69, TLX (NR2E1), but it also has a Lys at position 76 instead of an Arg (7). Since the R76K mutant of HNF4 lost most of its DNA binding activity, we assume that TLX would not be able to recapitulate the HNF4-specific binding, although that remains to be determined.
Finally, it is of interest to note that HNF4, RXR and COUPTF are among the most ancient of all the NRs (9). Since the biologically least complex metazoans currently in existence all have at least two NR genes (one HNF4-like and one RXR/COUPTF-like), it has been proposed that the NR family evolved from a now extinct early metazoan that contained a single NR gene, which was most similar to HNF4 (89). Intriguingly, the HNF4 DBDs of primitive metazoans all have the Asp69-Arg76 pair of mammalian HNF4.
In conclusion, our results highlight a complexity of NR DNA binding specificity that was previously under appreciated. They also demonstrate the usefulness of the PBM approach to more accurately define that complexity and thereby more precisely identify NR target genes in vivo. It will be of interest to compare the binding specificity of other NRs in a similar fashion.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–5, Supplementary Figures 1–10, Supplementary Materials and Methods, and Supplementary References [16,27,30,32,39,41,48,53,54,66,67,90–91].
FUNDING
National Institutes of Health (MH087397 to F.M.S. and T.J.) and PhRMA Foundation (to E.B.). Funding for open access charge: National Institutes of Health (MH087397 to F.M.S. and T.J.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr T. Girke (UC Riverside) for bioinformatics input, M. Tsai (Baylor College of Medicine) for Flag.COUPTF2 and J.M. Kurie (M.D. Anderson Cancer Center) for pCDNA6-His-hRARalpha.
REFERENCES
- 1. Laudet,V., Thompson,B., Pratt,W., Claessens,F., Kraus,L., Baniahmad,A., Edwards,D., Trifiro,M., Chrousos,G., Gustafsson,J. et al. (2004) The nuclear receptor superfamily. In: McEwan,I. (ed.), Essays in Biochemistry, Vol. 40. Portland Press Ltd, London.
- 2.Chen T. Nuclear receptor drug discovery. Curr. Opin. Chem. Biol. 2008;12:418–426. doi: 10.1016/j.cbpa.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 3.Overington JP, Al-Lazikani B, Hopkins AL. How many drug targets are there? Nat. Rev. Drug Discov. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 4.Xie W. Nuclear Receptors in Drug Metabolism. Hoboken: Wiley; 2008. [Google Scholar]
- 5.Umesono K, Evans RM. Determinants of target gene specificity for steroid/thyroid hormone receptors. Cell. 1989;57:1139–1146. doi: 10.1016/0092-8674(89)90051-2. [DOI] [PubMed] [Google Scholar]
- 6.Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotnoir-White D, Laperriere D, Mader S. Evolution of the repertoire of nuclear receptor binding sites in genomes. Mol. Cell Endocrinol. 2011;334:76–82. doi: 10.1016/j.mce.2010.10.021. [DOI] [PubMed] [Google Scholar]
- 8.Umesono K, Murakami KK, Thompson CC, Evans RM. Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell. 1991;65:1255–1266. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sladek FM. What are nuclear receptor ligands? Mol. Cell Endocrinol. 2011;334:3–13. doi: 10.1016/j.mce.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006;126:789–799. doi: 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan X, Ta TC, Lin M, Evans JR, Dong Y, Bolotin E, Sherman MA, Forman BM, Sladek FM. Identification of an endogenous ligand bound to a native orphan nuclear receptor. PLoS One. 2009;4:e5609. doi: 10.1371/journal.pone.0005609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hwang-Verslues WW, Sladek FM. HNF4alpha–role in drug metabolism and potential drug target? Curr. Opin. Pharmacol. 2010;10:698–705. doi: 10.1016/j.coph.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolotin E, Liao H, Ta TC, Yang C, Hwang-Verslues W, Evans JR, Jiang T, Sladek FM. Integrated approach for the identification of human hepatocyte nuclear factor 4alpha target genes using protein binding microarrays. Hepatology. 2010;51:642–653. doi: 10.1002/hep.23357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaestner KH. Making the liver what it is: the many targets of the transcriptional regulator HNF4alpha. Hepatology. 2010;51:376–377. doi: 10.1002/hep.23487. [DOI] [PubMed] [Google Scholar]
- 15.Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK, et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bolotin E, Schnabl J, Sladek F. HNF4A (Homo sapiens) Transcription Factor Encyclopedia. 2010 http://www.cisreg.ca/tfe (17 February 2012, date last accessed) [Google Scholar]
- 17. Sladek,F.M. and Seidel,S. (2001) Hepatocyte nuclear factor 4a. In: Burris,T.P. and McCabe,E. 75 (eds), Nuclear Receptors and Genetic Disease. Academic Press, London.
- 18.Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 19.Mangelsdorf DJ, Umesono K, Kliewer SA, Borgmeyer U, Ong ES, Evans RM. A direct repeat in the cellular retinol-binding protein type II gene confers differential regulation by RXR and RAR. Cell. 1991;66:555–561. doi: 10.1016/0092-8674(81)90018-0. [DOI] [PubMed] [Google Scholar]
- 20.IJpenberg A, Tan NS, Gelman L, Kersten S, Seydoux J, Xu J, Metzger D, Canaple L, Chambon P, Wahli W, et al. In vivo activation of PPAR target genes by RXR homodimers. EMBO J. 2004;23:2083–2091. doi: 10.1038/sj.emboj.7600209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kliewer SA, Umesono K, Heyman RA, Mangelsdorf DJ, Dyck JA, Evans RM. Retinoid X receptor-COUP-TF interactions modulate retinoic acid signaling. Proc. Natl Acad. Sci. USA. 1992;89:1448–1452. doi: 10.1073/pnas.89.4.1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kliewer SA, Umesono K, Noonan DJ, Heyman RA, Evans RM. Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature. 1992;358:771–774. doi: 10.1038/358771a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kliewer SA, Umesono K, Mangelsdorf DJ, Evans RM. Retinoid X receptor interacts with nuclear receptors in retinoic acid, thyroid hormone and vitamin D3 signalling. Nature. 1992;355:446–449. doi: 10.1038/355446a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cooney AJ, Tsai SY, O’Malley BW, Tsai MJ. Chicken ovalbumin upstream promoter transcription factor (COUP-TF) dimers bind to different GGTCA response elements, allowing COUP-TF to repress hormonal induction of the vitamin D3, thyroid hormone, and retinoic acid receptors. Mol. Cell. Biol. 1992;12:4153–4163. doi: 10.1128/mcb.12.9.4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai SY, Tsai MJ. Chick ovalbumin upstream promoter-transcription factors (COUP-TFs): coming of age. Endocr. Rev. 1997;18:229–240. doi: 10.1210/edrv.18.2.0294. [DOI] [PubMed] [Google Scholar]
- 26.Kruse SW, Suino-Powell K, Zhou XE, Kretschman JE, Reynolds R, Vonrhein C, Xu Y, Wang L, Tsai SY, Tsai MJ, et al. Identification of COUP-TFII orphan nuclear receptor as a retinoic acid-activated receptor. PLoS Biol. 2008;6:e227. doi: 10.1371/journal.pbio.0060227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang G, Nepomuceno L, Hopkins K, Sladek FM. Exclusive homodimerization of the orphan receptor hepatocyte nuclear factor 4 defines a new subclass of nuclear receptors. Mol. Cell. Biol. 1995;15:5131–5143. doi: 10.1128/mcb.15.9.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakshatri H, Bhat-Nakshatri P. Multiple parameters determine the specificity of transcriptional response by nuclear receptors HNF-4, ARP-1, PPAR, RAR and RXR through common response elements. Nucleic Acids Res. 1998;26:2491–2499. doi: 10.1093/nar/26.10.2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakshatri H, Chambon P. The directly repeated RG(G/T)TCA motifs of the rat and mouse cellular retinol-binding protein II genes are promiscuous binding sites for RAR, RXR, HNF-4, and ARP-1 homo- and heterodimers. J. Biol. Chem. 1994;269:890–902. [PubMed] [Google Scholar]
- 30.Mietus-Snyder M, Sladek FM, Ginsburg GS, Kuo CF, Ladias JA, Darnell JE, Jr, Karathanasis SK. Antagonism between apolipoprotein AI regulatory protein 1, Ear3/COUP-TF, and hepatocyte nuclear factor 4 modulates apolipoprotein CIII gene expression in liver and intestinal cells. Mol. Cell. Biol. 1992;12:1708–1718. doi: 10.1128/mcb.12.4.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kardassis D, Sacharidou E, Zannis VI. Transactivation of the human apolipoprotein CII promoter by orphan and ligand-dependent nuclear receptors. The regulatory element CIIC is a thyroid hormone response element. J. Biol. Chem. 1998;273:17810–17816. doi: 10.1074/jbc.273.28.17810. [DOI] [PubMed] [Google Scholar]
- 32.Crestani M, Sadeghpour A, Stroup D, Galli G, Chiang JY. Transcriptional activation of the cholesterol 7alpha-hydroxylase gene (CYP7A) by nuclear hormone receptors. J. Lipid Res. 1998;39:2192–2200. [PubMed] [Google Scholar]
- 33.Biddie SC, John S, Hager GL. Genome-wide mechanisms of nuclear receptor action. Trends Endocrinol. Metab. 2010;21:3–9. doi: 10.1016/j.tem.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park PJ. ChIP-seq: advantages and challenges of a maturing technology. Nat. Rev. Genet. 2009;10:669–680. doi: 10.1038/nrg2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berger MF, Bulyk ML. Universal protein-binding microarrays for the comprehensive characterization of the DNA-binding specificities of transcription factors. Nat. Protoc. 2009;4:393–411. doi: 10.1038/nprot.2008.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ji H, Vokes SA, Wong WH. A comparative analysis of genome-wide chromatin immunoprecipitation data for mammalian transcription factors. Nucleic Acids Res. 2006;34:e146. doi: 10.1093/nar/gkl803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ji H, Wong WH. TileMap: create chromosomal map of tiling array hybridizations. Bioinformatics. 2005;21:3629–3636. doi: 10.1093/bioinformatics/bti593. [DOI] [PubMed] [Google Scholar]
- 40.Jiang H, Wong WH. SeqMap: mapping massive amount of oligonucleotides to the genome. Bioinformatics. 2008;24:2395–2396. doi: 10.1093/bioinformatics/btn429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wallerman O, Motallebipour M, Enroth S, Patra K, Bysani MSR, Komorowski J, Wadelius C. Molecular interactions between HNF4a, FOXA2 and GABP identified at regulatory DNA elements through ChIP-sequencing. Nucleic Acids Res. 2009;37:7498–7508. doi: 10.1093/nar/gkp823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 43.Chen H, Privalsky ML. Cooperative formation of high-order oligomers by retinoid X receptors: an unexpected mode of DNA recognition. Proc. Natl Acad. Sci. USA. 1995;92:422–426. doi: 10.1073/pnas.92.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanaka T, Suh KS, Lo AM, De Luca LM. p21WAF1/CIP1 is a common transcriptional target of retinoid receptors: pleiotropic regulatory mechanism through retinoic acid receptor (RAR)/retinoid X receptor (RXR) heterodimer and RXR/RXR homodimer. J. Biol. Chem. 2007;282:29987–29997. doi: 10.1074/jbc.M701700200. [DOI] [PubMed] [Google Scholar]
- 45.Tanaka T, De Luca LM. Therapeutic potential of “rexinoids” in cancer prevention and treatment. Cancer Res. 2009;69:4945–4947. doi: 10.1158/0008-5472.CAN-08-4407. [DOI] [PubMed] [Google Scholar]
- 46.Siersbaek R, Nielsen R, John S, Sung MH, Baek S, Loft A, Hager GL, Mandrup S. Extensive chromatin remodelling and establishment of transcription factor ‘hotspots’ during early adipogenesis. EMBO J. 2011;30:1459–1472. doi: 10.1038/emboj.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Egener T, Roulet E, Zehnder M, Bucher P, Mermod N. Proof of concept for microarray-based detection of DNA-binding oncogenes in cell extracts. Nucleic Acids Res. 2005;33:e79. doi: 10.1093/nar/gni079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hoffman BG, Robertson G, Zavaglia B, Beach M, Cullum R, Lee S, Soukhatcheva G, Li L, Wederell ED, Thiessen N, et al. Locus co-occupancy, nucleosome positioning, and H3K4me1 regulate the functionality of FOXA2-, HNF4A-, and PDX1-bound loci in islets and liver. Genome Res. 2010;20:1037–1051. doi: 10.1101/gr.104356.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nielsen R, Pedersen TA, Hagenbeek D, Moulos P, Siersbaek R, Megens E, Denissov S, Borgesen M, Francoijs KJ, Mandrup S, et al. Genome-wide profiling of PPAR:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 2008;22:2953–2967. doi: 10.1101/gad.501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mikkelsen TS, Xu Z, Zhang X, Wang L, Gimble JM, Lander ES, Rosen ED. Comparative epigenomic analysis of murine and human adipogenesis. Cell. 2010;143:156–169. doi: 10.1016/j.cell.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adhikary T, Kaddatz K, Finkernagel F, Schonbauer A, Meissner W, Scharfe M, Jarek M, Blocker H, Muller-Brusselbach S, Muller R. Genomewide analyses define different modes of transcriptional regulation by peroxisome proliferator-activated receptor-beta/delta (PPARbeta/delta) PLoS One. 2011;6:e16344. doi: 10.1371/journal.pone.0016344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramagopalan SV, Heger A, Berlanga AJ, Maugeri NJ, Lincoln MR, Burrell A, Handunnetthi L, Handel AE, Disanto G, Orton SM, et al. A ChIP-seq defined genome-wide map of vitamin D receptor binding: associations with disease and evolution. Genome Res. 2010;20:1352–1360. doi: 10.1101/gr.107920.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thomas AM, Hart SN, Kong B, Fang J, Zhong XB, Guo GL. Genome-wide tissue-specific farnesoid X receptor binding in mouse liver and intestine. Hepatology. 2010;51:1410–1419. doi: 10.1002/hep.23450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chong HK, Infante AM, Seo YK, Jeon TI, Zhang Y, Edwards PA, Xie X, Osborne TF. Genome-wide interrogation of hepatic FXR reveals an asymmetric IR-1 motif and synergy with LRH-1. Nucleic Acids Res. 2010;38:6007–6017. doi: 10.1093/nar/gkq397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cui JY, Gunewardena SS, Rockwell CE, Klaassen CD. ChIPing the cistrome of PXR in mouse liver. Nucleic Acids Res. 2010;38:7943–7963. doi: 10.1093/nar/gkq654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 57.Ross-Innes CS, Stark R, Holmes KA, Schmidt D, Spyrou C, Russell R, Massie CE, Vowler SL, Eldridge M, Carroll JS. Cooperative interaction between retinoic acid receptor-alpha and estrogen receptor in breast cancer. Genes Dev. 2010;24:171–182. doi: 10.1101/gad.552910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, Kutter C, Watt S, Martinez-Jimenez CP, Mackay S, et al. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328:1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heng J-CD, Feng B, Han J, Jiang J, Kraus P, Ng J-H, Orlov YL, Huss M, Yang L, Lufkin T, et al. The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell Stem Cell. 2010;6:167–174. doi: 10.1016/j.stem.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 61.Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, Liu XS, Lazar MA. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331:1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Steger DJ, Grant GR, Schupp M, Tomaru T, Lefterova MI, Schug J, Manduchi E, Stoeckert CJ, Lazar MA. Propagation of adipogenic signals through an epigenomic transition state. Genes Dev. 2010;24:1035–1044. doi: 10.1101/gad.1907110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reddy TE, Pauli F, Sprouse RO, Neff NF, Newberry KM, Garabedian MJ, Myers RM. Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 2009;19:2163–2171. doi: 10.1101/gr.097022.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Strahle U, Klock G, Schutz G. A DNA sequence of 15 base pairs is sufficient to mediate both glucocorticoid and progesterone induction of gene expression. Proc. Natl Acad. Sci. USA. 1987;84:7871–7875. doi: 10.1073/pnas.84.22.7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.La Baer J, Yamamoto KR. Analysis of the DNA-binding affinity, sequence specificity and context dependence of the glucocorticoid receptor zinc finger region. J. Mol. Biol. 1994;239:664–688. doi: 10.1006/jmbi.1994.1405. [DOI] [PubMed] [Google Scholar]
- 66.Rastinejad F, Perlmann T, Evans RM, Sigler PB. Structural determinants of nuclear receptor assembly on DNA direct repeats. Nature. 1995;375:203–211. doi: 10.1038/375203a0. [DOI] [PubMed] [Google Scholar]
- 67.Lu P, Rha GB, Melikishvili M, Wu G, Adkins BC, Fried MG, Chi YI. Structural basis of natural promoter recognition by a unique nuclear receptor, HNF4alpha. Diabetes gene product. J. Biol. Chem. 2008;283:33685–33697. doi: 10.1074/jbc.M806213200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Petrescu AD, Hertz R, Bar-Tana J, Schroeder F, Kier AB. Role of regulatory F-domain in hepatocyte nuclear factor-4alpha ligand specificity. J. Biol. Chem. 2005;280:16714–16727. doi: 10.1074/jbc.M405906200. [DOI] [PubMed] [Google Scholar]
- 69.Petrescu AD, Payne HR, Boedecker A, Chao H, Hertz R, Bar-Tana J, Schroeder F, Kier AB. Physical and functional interaction of Acyl-CoA-binding protein with hepatocyte nuclear factor-4 alpha. J. Biol. Chem. 2003;278:51813–51824. doi: 10.1074/jbc.M303858200. [DOI] [PubMed] [Google Scholar]
- 70.Hertz R, Magenheim J, Berman I, Bar-Tana J. Fatty acyl-CoA thioesters are ligands of hepatic nuclear factor-4alpha. Nature. 1998;392:512–516. doi: 10.1038/33185. [DOI] [PubMed] [Google Scholar]
- 71.Soutoglou E, Katrakili N, Talianidis I. Acetylation regulates transcription factor activity at multiple levels. Mol. Cell. 2000;5:745–751. doi: 10.1016/s1097-2765(00)80253-1. [DOI] [PubMed] [Google Scholar]
- 72.Hall RK, Sladek FM, Granner DK. The orphan receptors COUP-TF and HNF-4 serve as accessory factors required for induction of phosphoenolpyruvate carboxykinase gene transcription by glucocorticoids. Proc. Natl Acad. Sci. USA. 1995;92:412–416. doi: 10.1073/pnas.92.2.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kurokawa R, Yu VC, Naar A, Kyakumoto S, Han Z, Silverman S, Rosenfeld MG, Glass CK. Differential orientations of the DNA-binding domain and carboxy-terminal dimerization interface regulate binding site selection by nuclear receptor heterodimers. Genes Dev. 1993;7:1423–1435. doi: 10.1101/gad.7.7b.1423. [DOI] [PubMed] [Google Scholar]
- 75.Perlmann T, Rangarajan PN, Umesono K, Evans RM. Determinants for selective RAR and TR recognition of direct repeat HREs. Genes Dev. 1993;7:1411–1422. doi: 10.1101/gad.7.7b.1411. [DOI] [PubMed] [Google Scholar]
- 76.Zechel C, Shen XQ, Chambon P, Gronemeyer H. Dimerization interfaces formed between the DNA binding domains determine the cooperative binding of RXR/RAR and RXR/TR heterodimers to DR5 and DR4 elements. EMBO J. 1994;13:1414–1424. doi: 10.1002/j.1460-2075.1994.tb06395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shaffer PL, Gewirth DT. Structural analysis of RXR-VDR interactions on DR3 DNA. J. Steroid Biochem. Mol. Biol. 2004;89–90:215–219. doi: 10.1016/j.jsbmb.2004.03.084. [DOI] [PubMed] [Google Scholar]
- 78.Rochel N, Ciesielski F, Godet J, Moman E, Roessle M, Peluso-Iltis C, Moulin M, Haertlein M, Callow P, Mely Y, et al. Common architecture of nuclear receptor heterodimers on DNA direct repeat elements with different spacings. Nat. Struct. Mol. Biol. 2011;18:564–570. doi: 10.1038/nsmb.2054. [DOI] [PubMed] [Google Scholar]
- 79.Kurokawa R, DiRenzo J, Boehm M, Sugarman J, Gloss B, Rosenfeld MG, Heyman RA, Glass CK. Regulation of retinoid signalling by receptor polarity and allosteric control of ligand binding. Nature. 1994;371:528–531. doi: 10.1038/371528a0. [DOI] [PubMed] [Google Scholar]
- 80.Kurokawa R, Soderstrom M, Horlein A, Halachmi S, Brown M, Rosenfeld MG, Glass CK. Polarity-specific activities of retinoic acid receptors determined by a co-repressor. Nature. 1995;377:451–454. doi: 10.1038/377451a0. [DOI] [PubMed] [Google Scholar]
- 81.Forman BM, Umesono K, Chen J, Evans RM. Unique response pathways are established by allosteric interactions among nuclear hormone receptors. Cell. 1995;81:541–550. doi: 10.1016/0092-8674(95)90075-6. [DOI] [PubMed] [Google Scholar]
- 82.Chandra V, Huang P, Hamuro Y, Raghuram S, Wang Y, Burris TP, Rastinejad F. Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA. Nature. 2008;456:350–356. doi: 10.1038/nature07413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hirokane H, Nakahara M, Tachibana S, Shimizu M, Sato R. Bile acid reduces the secretion of very low density lipoprotein by repressing microsomal triglyceride transfer protein gene expression mediated by hepatocyte nuclear factor-4. J. Biol. Chem. 2004;279:45685–45692. doi: 10.1074/jbc.M404255200. [DOI] [PubMed] [Google Scholar]
- 84.Raspe E, Duez H, Mansen A, Fontaine C, Fievet C, Fruchart JC, Vennstrom B, Staels B. Identification of Rev-erbalpha as a physiological repressor of apoC-III gene transcription. J. Lipid Res. 2002;43:2172–2179. doi: 10.1194/jlr.m200386-jlr200. [DOI] [PubMed] [Google Scholar]
- 85.Noshiro M, Usui E, Kawamoto T, Kubo H, Fujimoto K, Furukawa M, Honma S, Makishima M, Honma K, Kato Y. Multiple mechanisms regulate circadian expression of the gene for cholesterol 7alpha-hydroxylase (Cyp7a), a key enzyme in hepatic bile acid biosynthesis. J. Biol. Rhythms. 2007;22:299–311. doi: 10.1177/0748730407302461. [DOI] [PubMed] [Google Scholar]
- 86.Tirona RG, Lee W, Leake BF, Lan LB, Cline CB, Lamba V, Parviz F, Duncan SA, Inoue Y, Gonzalez FJ, et al. The orphan nuclear receptor HNF4alpha determines PXR- and CAR-mediated xenobiotic induction of CYP3A4. Nat. Med. 2003;9:220–224. doi: 10.1038/nm815. [DOI] [PubMed] [Google Scholar]
- 87.Echchgadda I, Song CS, Oh T, Ahmed M, De La Cruz IJ, Chatterjee B. The xenobiotic-sensing nuclear receptors pregnane X receptor, constitutive androstane receptor, and orphan nuclear receptor hepatocyte nuclear factor 4alpha in the regulation of human steroid-/bile acid-sulfotransferase. Mol. Endocrinol. 2007;21:2099–2111. doi: 10.1210/me.2007-0002. [DOI] [PubMed] [Google Scholar]
- 88.Danielsen M, Hinck L, Ringold GM. Two amino acids within the knuckle of the first zinc finger specify DNA response element activation by the glucocorticoid receptor. Cell. 1989;57:1131–1138. doi: 10.1016/0092-8674(89)90050-0. [DOI] [PubMed] [Google Scholar]
- 89.Bridgham JT, Eick GN, Larroux C, Deshpande K, Harms MJ, Gauthier ME, Ortlund EA, Degnan BM, Thornton JW. Protein evolution by molecular tinkering: diversification of the nuclear receptor superfamily from a ligand-dependent ancestor. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000497. e1000497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chartier FL, Bossu JP, Laudet V, Fruchart JC, Laine B. Cloning and sequencing of cDNAs encoding the human hepatocyte nuclear factor 4 indicate the presence of two isoforms in human liver. Gene. 1994;147:269–272. doi: 10.1016/0378-1119(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 91.Eeckhoute J, Formstecher P, Laine B. Maturity-onset diabetes of the young Type 1 (MODY1)-associated mutations R154X and E276Q in hepatocyte nuclear factor 4alpha (HNF4alpha) gene impair recruitment of p300, a key transcriptional co-activator. Mol. Endocrinol. 2001;15:1200–1210. doi: 10.1210/mend.15.7.0670. [DOI] [PubMed] [Google Scholar]
- 92.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.