Abstract
Regulation of the positive transcription elongation factor, P-TEFb, plays a major role in controlling mammalian transcription and this is accomplished in part by controlled release of P-TEFb from the 7SK snRNP that sequesters the kinase in an inactive state. We demonstrate here that a similar P-TEFb control system exists in Drosophila. We show that an RNA previously suggested to be a 7SK homolog is, in fact, associated with P-TEFb, through the action of a homolog of the human HEXIM1/2 proteins (dHEXIM). In addition, a Drosophila La related protein (now called dLARP7) is shown to be the functional homolog of human LARP7. The Drosophila 7SK snRNP (d7SK snRNP) responded to treatment of cells with P-TEFb inhibitors and to nuclease treatment of cell lysates by releasing P-TEFb. Supporting a critical role for the d7SK snRNP in Drosophila development, dLARP7 and dHEXIM were found to be ubiquitously expressed throughout embryos and tissues at all stages. Importantly, knockdown of dHEXIM was embryonic lethal, and reduction of dHEXIM in specific tissues led to serious developmental defects. Our results suggest that regulation of P-TEFb by the d7SK snRNP is essential for the growth and differentiation of tissues required during Drosophila development.
INTRODUCTION
The highly orchestrated pattern of gene expression driving cellular differentiation and tissue development is to a large extent controlled at the level of transcription, and regulation of the elongation phase of transcription plays an important role. RNA polymerase II elongation control starts with the default action of negative factors including DRB sensitivity inducing factor (DSIF) and negative elongation factor (NELF) that block the movement of initiated polymerases into the body of genes (1). These promoter proximal paused polymerases are poised for a regulated release into productive elongation by the positive transcription elongation factor, P-TEFb (2). The cyclin-dependent kinase activity of P-TEFb (3) coordinates the modification and exchange of factors associated with the elongation complex. The large subunit of DSIF, Spt5, as well as the NELFe subunit is phosphorylated by P-TEFb triggering the release of NELF from the complex (4–6). DSIF remains in the transcription complex and is joined by factors that dramatically change the rate of elongation from essentially zero to an average rate of ∼3.8 kb/min (6,7). The P-TEFb-mediated transition into productive elongation is a singular event occurring near every gene’s 5′-end that commits the engaged polymerase to complete an mRNA.
A large body of evidence points to RNA polymerase II elongation control as a general process required for the biogenesis of essentially all mRNAs. Treatment of cells with P-TEFb inhibitors blocks mRNA production (8) and most transcription by RNA polymerase II in nuclei isolated from the cells (9) and the process is reproduced utilizing in vitro systems derived from Drosophila (10) and mammalian nuclear extracts (2,11) regardless of the identity of the promoter used. Strong support for the generality of the process was found in the results of ChIP-Seq analyses that pinpointed the position of RNA polymerase II across mammalian and Drosophila genomes (12). Promoter proximal paused polymerases were found on a large number of Drosophila genes (13,14) and on most mammalian genes (6,15,16). These included not only genes expressed at moderate to high levels of expression, but also genes with very low expression. The implication of these studies is that P-TEFb mediated release of the poised polymerases into productive elongation could be the rate limiting step of transcription on a large fraction of genes. Together all evidence points to P-TEFb not only being required for mRNA production, but also suggest that directed P-TEFb action could be a principle regulated step (17). In fact, c-myc which is a major regulator of many genes has been demonstrated to function at the level of elongation (6).
Because of the critical role that P-TEFb plays in regulating gene expression metazoans have developed a complex regulatory system that involves controlled sequestration and release of P-TEFb from an inhibitory complex (18,19). This complex is built on a 7SK snRNA scaffold (20) that constitutively contains a La related protein, LARP7 (21–23). 7SK is one of a few snRNAs that are capped by the addition of a single methyl group on the gamma phosphate on the 5′-end of the RNA (24). The methyl phosphate capping enzyme MEPCE responsible for the modification is also an integral part of the 7SK snRNP (21,25,26). In HeLa cells, about half of the 7SK snRNP contains these two proteins along with a heterogeneous array of hnRNP proteins (21,27,28). In the other half of the 7SK snRNPs, the hnRNPs are replaced by a double-stranded RNA-binding protein, HEXIM1 or HEXIM2 and this protein interacts with and inhibits P-TEFb (29–32). Both of the 7SK snRNPs distinguish themselves from all other snRNPs by being readily extracted from mild detergent treated nuclei at low salt indicating that they are not tightly bound to chromatin (33). The P-TEFb not in the 7SK snRNP, on the other hand, is only extracted by higher salt, indicating that it is associated with chromatin and suggesting that it is actually engaged in functional interactions (33). These biochemical properties suggest that P-TEFb could be extracted from the 7SK snRNP at the precise time and location needed to activate expression of specific genes.
Two proteins have been demonstrated to directly cause release of P-TEFb from the 7SK snRNP. One is HIV-1 Tat, a virally encoded transactivator which was the first protein found to functionally interact with P-TEFb (34). A crystal structure of Tat bound to P-TEFb revealed an extensive, stable interface between the viral and host protein (35). Tat also has an RNA-binding domain with specificity toward the nascent HIV transcript, TAR, which ultimately results in recruitment of P-TEFb and activation of viral transcription (36). Expression of Tat alone in human cells leads to the release of P-TEFb from the 7SK snRNP (37). This release does not require any modifications of Tat or the proteins or RNA in the 7SK snRNP because recombinant Tat can extract P-TEFb from highly purified 7SK snRNPs immunoprecipitated from cell extracts (38). Although Tat interacts directly with the 7SK RNA and the RNA-binding domain of Tat has a modest effect on the release of P-TEFb from the 7SK snRNP, the P-TEFb-binding domain is sufficient for efficient release (37,38). The second protein, Brd4, is a bromodomain-containing protein that binds to acetylated histones normally associated with active regions of transcription (39). Brd4 can be found in complex with P-TEFb and this is mediated by a small domain found in the C-terminus of Brd4 (40). Just like Tat, this domain leads to the release of P-TEFb from the 7SK snRNP when expressed in human cells (38). Furthermore, it can also extract P-TEFb directly from the immunoprecipitated 7SK snRNPs (38). Because Brd4 does not have an RNA-binding domain it is clear that P-TEFb binding is the key property required. A number of other transcription factors can interact with P-TEFb and some of them may also have the ability to extract P-TEFb from the 7SK snRNP (1). The fact that at least some factors have the ability to extract P-TEFb provides additional support for a model in which the P-TEFb used to activate a gene is taken directly from the 7SK snRNP. This would guard against accidental activation of poised polymerases on genes that should be silent while maintaining a relatively high level of P-TEFb in a potentially active state.
Drosophila is an excellent model system to examine the role that RNA polymerase II elongation control takes in regulating development. P-TEFb was first identified in Drosophila (2) and fly HSP70 genes were among the first genes shown to have promoter proximal paused polymerase (41). Elegant studies from several labs have demonstrated that these poised polymerases are prevalent across the fly genome and are especially enriched on developmentally controlled genes (13,14,42,43). The rapid induction provided by poised polymerases is useful in synchronous activation of particular genes across a population of cells (43,44) and they also can act as insulators when placed between enhancers and another promoter (45). Unfortunately, only a little is known about the 7SK snRNP in Drosophila. One study identified a potential candidate for 7SK RNA (46) and another demonstrated that the MEPCE homolog, Bin3, was associated with and stabilized the RNA (26). Importantly, it was not determined if P-TEFb was associated with the RNA. We provide conclusive evidence here that the RNA identified is indeed 7SK and that a Drosophila 7SK snRNP containing homologs of human P-TEFb, HEXIM1/2 and LARP7 exists. We also show that dHEXIM is essential for development and that the snRNP is widely expressed. Taken altogether, this suggests an important role of snRNP in development.
MATERIALS AND METHODS
Cell lines and compounds
Drosophila melanogaster Kc cells were maintained in T-150 tissue culture flasks in Hyclone SFX-Insect (Thermo Scientific) at 25°C between 1 × 106 and 1 × 107 cells/ml. In some cases, they were transferred to spinner culture, maintained between 2 × 106 and 1 × 107 cells/ml, and used for experiments at 4–6 × 106 cells/ml. DRB was purchased from Sigma. Stock solutions were 50 mM in EtOH and cells were treated at a final concentration of 100 µM. Stocks of 10 mM flavopiridol were prepared in DMSO and cells were treated at a final concentration of 500 nM. All stocks were aliquoted at stored at −80°C.
Antibodies
Antibodies to Cyclin T, dHEXIM, and dLARP7 were generated in sheep by Elmira Biologicals, Inc. (Iowa City, IA, USA) using the corresponding recombinantly expressed and purified proteins. Following testing of pre-immune, first and second bleed antisera for each antibody, they were affinity purified using the purified recombinant proteins coupled to actigel-ALD resin according to the manufacturer’s protocol (Sterogene).
Cloning and expression of the C-terminal region of Cyclin T (751–1097)
Following PCR amplification of the C-terminal region of Drosophila Cyclin T (amino acids 751–1097) using primers to incorporate a 5′ NdeI site (CATATGCGAGAAAAGCTTCGCAAGCTGG) and a 3′ XhoI site (CTCGAGCTTCTTGTTGTAGACGGGCATGGG), the insert was subcloned into the pCR2.1-TOPO PCR cloning vector (Invitrogen). Subsequent restriction digest using NdeI and XhoI and ligation into the pET21a vector yielded the pET21a-CycT(751–1097) expression vector, which was verified by sequencing.
His-tagged Cyclin T (751–1097) protein used for generation of antibodies was expressed in Escherichia coli BL21 (DE3). After induction of 4 × 1 l cultures with IPTG and overnight growth at 18°C, cells were sonicated in PBS with 1% Triton X-100, 0.1% saturated solution of PMSF in isopropanol, 5 mM imidazole and protease inhibitors (Roche), and cleared by spinning for 50 min at 200 000 g. The lysate was brought to 1 M NaCl and then added to 2 ml of Ni-NTA resin (Qiagen) that had been pre-equilibrated with lysis buffer containing 1 M NaCl. After a 1 h incubation at 4°C, the flow-through was collected, and the resin was washed with 10 volumes of High Salt Wash buffer [10 mM Tris (pH 7.8), 1 M NaCl, 1% Triton X-100, 0.1% PMSF, 35 mM imidazole] and 10 volumes of Low Salt Wash buffer [10 mM HEPES (pH 7.8), 70 mM KCl, 1% Triton X-100, 0.1% PMSF, 35 mM imidazole]. The column was eluted with 4 volumes of Elution Buffer [10 mM HEPES (pH 7.8), 1% Triton, 70 mM KCl, 250 mM imidazole, 0.1% freshly added PMSF saturated in isopropanol]. The protein was then loaded onto a 1 ml Mono S column, which was eluted with a 20 ml linear gradient from 70 mM to 1 M HKEDP [25 mM HEPES (pH 7.6), 70–1000 mM KCl, 0.1 mM EDTA, 1 mM DTT, 1 mM PMSF] and fractions were stored at −80°C.
Cloning and expression of dHEXIM proteins
A pOT2 plasmid containing the cDNA for dHEXIM (CG3508, isoform A) was obtained from the Drosophila Genomics Resource Center (Bloomington, IN, USA). Following PCR amplification of the dHEXIM gene using primers to incorporate a 5′ NdeI site (CATATGGCTGAAGCTGTAAAAAATGAAAGTG) and a 3′ XhoI site (CTCGAGTTTGACAACTGCATCGGCTGGC), the insert was subcloned into the pSC-A PCR cloning vector (Stratagene). Subsequent restriction digest using NdeI and XhoI and ligation into the pET21a vector yielded the pET21a-dHEXIM expression vector, which was verified by sequencing.
dHEXIM containing a mutation in the RNA-binding domain (KHRR to ILAA) was created using the QuikChange site-directed mutagenesis kit (Stratagene). Forward (CGGGAATGCCGAAGAGGATACTTGCGGCGGGAAAAAAATCCAAGATGCAGCCC) and reverse (GGGCTGCATCTTGGATTTTTTTCCCGCCGCAAGTATCCTCTTCGGCATTCCCG) primers targeting the region were designed to introduce six mutations changing the KHRR residues to ILAA. These primers were added to 50 ng of pET21a-dHEXIM template and 18 cycles of PCR using Pfu Turbo were used to generate plasmid with the desired mutations. Following digestion of methylated parental pET21a-dHEXIM by DpnI, pET21a-dHEXIM(ILAA) was amplified and isolated from E. coli.
His-tagged dHEXIM and dHEXIM(ILAA) proteins used for generation of antibodies and EMSA were expressed in E. coli BL21 (DE3) using the same protocol described above for the purification of Cyclin T (751–1097) except that the low salt wash buffer contained 25 mM instead of 35 mM imidazole.
Cloning and expression of dLARP7
The clone, LD09531, for Drosophila MXC (dLARP7) protein was obtained from the Drosophila Genomics Research Center. The dLARP7 coding region was cloned into a pET21a (Novagene) expression vector with a C-terminal histidine tag and the clone was verified by sequencing. His-tagged dLARP7 used for generation of antibodies was expressed in E. coli BL21 (DE3) using the same protocol described above for the purification of Cyclin T (751–1097) except that the protein flowed through Mono S. The Mono S flowthrough was loaded onto a 1 ml Mono Q column, which was eluted with a linear gradient from 70 mM to 1 M HKEDP [25 mM HEPES (pH 7.6), 70–1000 mM KCl, 0.1 mM EDTA, 1 mM DTT, 1 mM PMSF] and fractions were stored at −80°C.
Cloning and T7 transcription of d7SK
Kc cell genomic DNA was isolated using the Genomic-tip 500 kit (Qiagen). The d7SK gene was PCR-amplified from 100 ng of genomic DNA using primers targeting the predicted 5′ (TAATACGACTCACTATAGGAAGTGTATTCTGTGATTG) and 3′ (AAAAGAATGGGCAAATTGCTCGGCA) ends of the 444 nt d7SK gene and incorporating a 5′ T7 promoter, and subsequently cloned into the PCR2.1-TOPO PCR cloning vector (Invitrogen) and confirmed by sequencing.
For in vitro transcription, the T7-d7SK DNA template was amplified from the PCR2.1-d7SK plasmid using the cloning primers. Transcription was carried out using the T7 transcription kit from Promega following the manufacturer’s protocols. For cold d7SK RNA, 3 µg of DNA template were mixed with 0.5 mM NTPs and 40 U of T7 RNA polymerase, and incubated for 2 h at 37°C. For high specific activity RNA probes used for EMSAs, 1 µg of DNA template was mixed with 0.5 mM rATP, rTTP, and rGTP, 12 µM cold rCTP, 20 µCi of α-32P-rCTP, and 20 U of T7 RNA polymerase, and incubated for 1 h at 37°C. Following transcription, the DNA template was degraded using DNase I and the RNA was purified using the RNeasy kit from Qiagen.
Western blotting
Samples for western blotting were resolved by 9% polyacryalmide SDS–PAGE and the proteins were then electroblotted to a 0.45 µm nitrocellulose membrane using an Owl semi-dry transfer apparatus. After being blocked in 10% non-fat dry milk for 30 min, the membranes were incubated in 2.5% milk with 1:1000 dilutions of affinity-purified α-Cyclin T, α-dHEXIM, or α-dLARP7 as indicated overnight at 4°C. Following incubation in 2.5% milk containing a 1:15 000 dilution of HRP-conjugated α-sheep secondary antibody (Sigma) for 2 h at room temperature, the blots were developed with Super Signal Dura West (Pierce) and imaged with a cooled CCD camera (UVP). Quantitation of western blots was accomplished using LabWorks software (UVP).
Electrophoretic mobility shift assay
For generation of the labeled dsRNA probe, complementary ssRNA oligos (derived from human TTF2 coding region) were purchased from IDT (Iowa City, IA, USA): CAAGCAAGAGGCAGAGAUCCCAGGU and ACCUGGGAUCUCUGCCUCUUGCUUG. These oligos were then annealed in a 1:1 ratio and 50 pmol of dsRNA oligo were end-labeled with 50 pmol of γ-32P-ATP (6000 Ci/mmol, Perkin-Elmer) using 20 U of T4 polynucleotide kinase for 1 h at 37°C. Following labeling, the oligo was purified using Nuc-Away spin columns (Ambion).
Binding reactions were carried out in 10 µl reactions in 25 mM HEPES (pH 7.6), 15% glycerol, 60 mM KCl, 0.1 mM EDTA, 0.1 mg/ml BSA, 10 mM DTT, 0.01% NP-40 and 50 ng of tRNA non-specific competitor. The indicated amounts of each protein were added to the reactions containing constant amounts of labeled RNA and were incubated for 15 min at room temperature. Complexes were resolved on a 4% polyacrylamide/0.5 × Tris–glycine native gel and visualized by autoradiography.
Northern analysis of d7SK
A d7SK anti-sense DNA oligo targeting nucleotides 229–268 (AUACGGCUUCCGGUCGAGACCAGAAUCCGCGGGGUGGUGG) was purchased from IDT. A total of 50 pmol of oligo were end-labeled with 50 pmol of γ-32P-ATP (6000 Ci/mmol, Perkin-Elmer) using 20 U of T4 polynucleotide kinase for 1 h at 37°C. Following labeling, the oligo was purified using Nuc-Away spin columns (Ambion). RNA from immunoprecipitations or from 100 µl of glycerol gradient fractions were extracted using Trizol reagent (Invitrogen), ethanol precipitated and resuspended in RNA loading buffer. The RNA was then resolved on a 6% polyacrylamide/1 × TBE/6 M urea denaturing RNA gel. Following EtBr staining to visualize total RNA, the RNA was electro-transferred to NytranN nylon membrane (Sigma), UV crosslinked to the membrane and pre-hybridized in UltraHyb (Ambion) for 2 h at 37°C. End-labeled anti-sense d7SK was then added to the UltraHyb to a final concentration of 1 × 106 cpm/ml and the membrane was incubated overnight at 37°C. Finally, the membrane was washed twice in 2 × SSC/0.1% SDS for 5 min at room temperature and once for 5 min at 37°C, with subsequent visualization by autoradiography.
Glycerol gradient ultracentrifugation
Kc cell lysates from 50 ml spinner flask cultures at 4 × 106 cells/ml were prepared in 500 µl Buffer A [10 mM KCl, 10 mM MgCl2, 10 mM HEPES (pH 7.6), 1 mM EDTA, 1 mM DTT, 0.1% PMSF, EDTA-free complete protease inhibitor cocktail from Roche, 20 µg/ml E-64, 2 µg/ml Pepstatin A, and 1.5 U/µl RNaseOut] containing 150 mM NaCl and 0.5% NP-40 as previously described (31,47). Following incubation on ice for 10 min, the lysates were clarified by centrifugation at 14 000 rpm for 10 min at 4°C in a microfuge. A total of 200 µl of the supernatant was used as input for 4.8 ml, 5–45% glycerol gradients, in Buffer A with 150 mM NaCl. The gradients were spun at 45 000 rpm for 16 h in a SW-55Ti rotor in a L7-55 Beckman ultracentrifuge and were then fractionated into 300 µl aliquots that were subsequently analyzed by western or northern blot.
Co-immunoprecipitation of components of the d7SK snRNP
Antibodies to dLARP7, dHEXIM, or Cyclin T were used to immunoprecipitate the proteins from Kc cell extracts. The cells were grown to 90% confluence, harvested, washed and lysed as for glycerol gradient sedimentation. Ten micrograms of affinity purified antibody was then bound overnight to 50 µl of paramagnetic protein G Dynabeads (Invitrogen) that were blocked and washed with PBS containing 0.5% BSA. The beads were then concentrated and resuspended in 200 µl of Kc cell lysate (one T150 flask worth of cells) and incubated with rotation at 4°C for 2 h. After the incubation, the beads were collected and washed four times with 500 µl immunoprecipitation wash buffer [10 mM HEPES (pH 7.6, 150 mM NaCl, 2 mM MgCl2, 10 mM KCl, 0.1% NP-40, 0.5 mM EDTA]. For western analysis, the beads were suspended in SDS sample loading buffer and heated to 90°C for 10 min and for northern analysis the beads were subjected to a Trizol extraction.
RNAi-mediated knockdown of dHEXIM in Kc cells
Double-stranded RNAs targeting dHEXIM exons 2 and 3, respectively, were generated by T7 transcription from a PCR template using the Megascript kit (Ambion). T7 PCR primers were as follows: exon 2: F 5′-GAATTAATACGACTCACTATAGGGAAAGTGGCTCCCAACAACGACC, R 5′-GAATTAATACGACTCACTATAGGGATTGATTCTCACGTGATAGTTGGCG; exon 3: F 5′- GAATTAATACGACTCACTATAGGGACACTTCCTCGACCAGCAGCG, R 5′-GAATTAATACGACTCACTATAGGGATACAATTGGGAGGCGTCGTTTGCAG. Ten micrograms of each dsRNA was added per 1 × 106 cells and incubated for 72 h before making whole-cell extract. Control cells were incubated with 10 ug of LacZ dsRNA per 1 × 106 cells for 72 h.
Fly stocks and transgenes for dHEXIM studies
All stocks were maintained and raised under standard conditions. dHEXIM RNAi transgenic flies used in this work are provided by the Vienna Drosophila RNAi Center (VDRC) and by Bloomington Drosophila Stock Center for all the other strains (rotund-GAL4, engrailed-GAL4, decapentaplegic-GAL4, patched-GAL4, lozenge-GAL4, UAS-2EGFP). The specificity of the dHEXIM RNAi was confirmed by rescuing the HEXIM developmental defects with a UAS-HEXIM strain over-expressing dHEXIM (Table 1 and Supplementary Figure S2). dHEXIM cDNA isoform A was subcloned in the pUAST vector (48) and microinjected into embryos (BestGene). After hatching, the progeny were crossed with w1118 flies to create transgenic strains. Immunostaining of embryos and larvae was performed following standard procedures. A rabbit polyclonal anti-dHEXIM antibody was custom made (Genecust) and used at 1:1000 dilution along with a secondary antibody Alexa fluor® 488 anti-rabbit (1:200, Invitrogen). Confocal images were captured on Nikon Eclipse confocal microscopy, and processed by Adobe Photoshop software. The bin3 mutant fly lines P(SUPor-P)bin3KG03313 and P(EPgy2)CG10365EY08633 were obtained from the Bloomington Stock Center. The balancers were changed to appropriate green balancer chromosomes (CyO or TM3 bearing actin-GFP transgenes) for scoring non-green homozygous mutant larvae for the northern blot analysis.
Table 1.
Rescue of RNAi-mediated HEXIM loss phenotypes by co-overexpressing HEXIM cDNA (29°C)
| UAS-HEX RNAi | UAS-HEX; UAS-HEX RNAi | ||
|---|---|---|---|
| Driver | Domain of expression | Developmental defects | Rescued phenotypes |
| Rotunda | Wing, leg, antenna, haltere discs | 100% undeveloped wings (n = 148) | 22% undeveloped wings (n = 810) |
| 100% undeveloped legs | 10% undeveloped legs | ||
| 0% wild type | 78% wings & 90% legs wild type | ||
| Lozengeb | Eye discs | 95.8% lethal pupae (n = 227) | 8.1% lethal pupae (n = 222) |
| 4.2% viable with mutant eyes | 14.9% viable with mutant eyes | ||
| 0% wild type | 77% wild type | ||
| GMR | Eye discs | 44.2% black-spotted eyes (n = 129) | 2% black-spotted eyes (n = 216) |
| 55.8% rough eyes | 48.5% rough eyes | ||
| 0% wild type | 49.5% wild type | ||
| Patched | Early segmental domains of embryos; wing, leg discs | 100% lethal Larvae stage 2 (n = 73) | 25.4% lethal Larvae stage 2 (n = 252) |
| 0% viable | 74.6% viable |
aThe percentages of developmental defects of halteres and antenna were similar to that of wing appendages.
bThe crosses were preferably done at 25°C in order to optimize the mating conditions due to the characteristics of Lozenge strain. All the same mutant phenotypes were also recorded at 29°C.
n = total number of flies counted for each experiment.
Kinase assay
Assays were performed as described earlier(3) except that human DSIF was used as substrate, reaction time was 15 min and the label was 20 µM 32P-ATP. 7SK snRNP immunoprecipitations were carried out as described above except that beads were washed with 150 mM KCl, 25 mM HEPES (pH 7.6), 0.02% Tween20 and 50 U/ml RNAseOut. After the final wash the beads were resuspended in kinase reaction buffer (-ATP), treated or not for 10 min with 10 µg of RNaseA and then the reaction was started with the addition of ATP. The samples were analyzed by SDS PAGE followed by drying of the gel. The results were quantified using a phosphorimager (Fuji FLA7000).
dLARP7 immunofluoresence
Fly tissues were processed for immunofluorescence microscopy following the standard procedure. In brief, dissected wild type third instar larval tissues and adult ovaries were fixed in 4% paraformaldehyde, washed several times with 1 × PBS and permeabilized with 1% Triton X-100 for 10 min. After two washes in 1 × PBS, samples were blocked for 1 h with 5% NGS in 1 × PBS followed by incubation overnight at 4°C with the primary antibody at 1:100 dilution. Signal was detected using secondary antibody conjugated to Alexa-594. Images were captured using Leica TCS SP5 confocal microscope.
RESULTS
Identification and characterization of dHEXIM
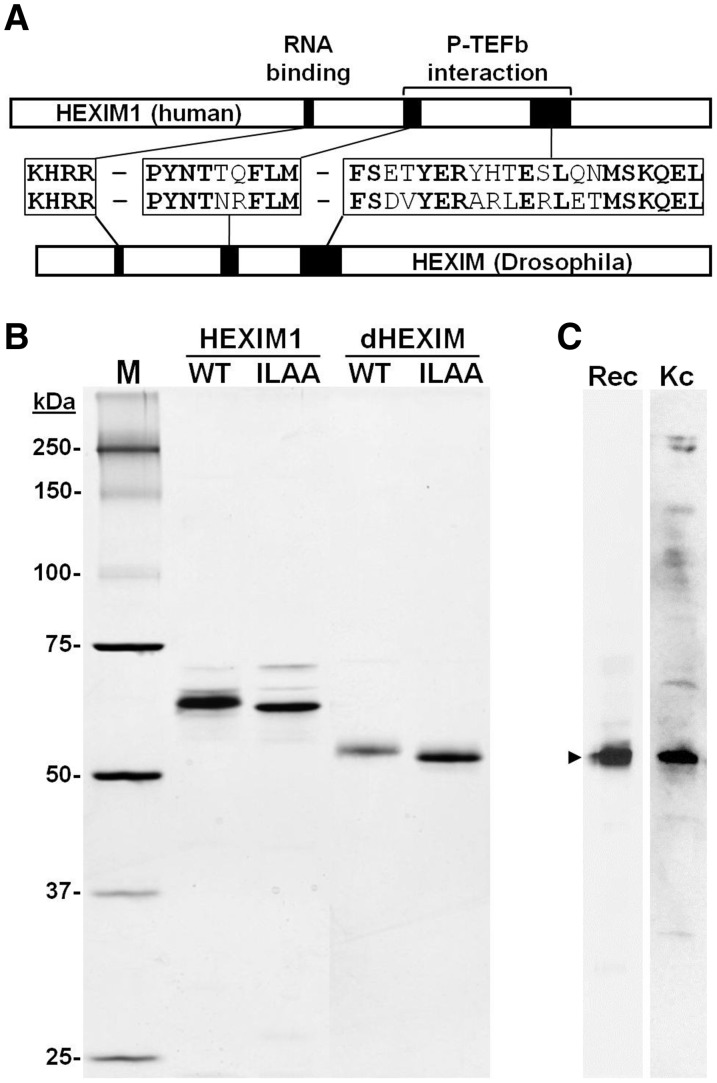
P-TEFb comprised of Cdk9 and Cyclin T was originally identified in Drosophila (2,34,49), but because it was not clear if a large inhibited form of P-TEFb existed in flies, we first searched for potential homologs of the key human inhibitors, HEXIM1 and HEXIM2. A BLAST search identified, two alternative-splicing isoforms of a single, previously uncharacterized gene (CG3508) that contained significant similarity to HEXIM1 (28% identical and 46% conserved) and HEXIM2 (27% identical and 45% conserved). More significantly, this gene encoded a protein containing the important KHRR RNA-binding domain, as well as the PYNT and SDVYER (SETYER in humans) sequences required to bind and inhibit the kinase activity of P-TEFb (29,50) (Figure 1A). To generate tools for biochemical analyses of this protein, the larger isoform of the gene, renamed dHEXIM, and a KHRR to ILAA mutation known in human HEXIM1 to abrogate RNA-binding (29), were subsequently expressed in E. coli and purified (Figure 1B). Sheep polyclonal antibodies were generated to and affinity-purified using the wildtype protein. These antibodies recognized both the recombinant dHEXIM and a similarly sized protein in Kc cell extracts (Figure 1C). This suggests that the putative Drosophila homolog of HEXIM1 is indeed expressed in vivo and may be involved in controlling P-TEFb.
Figure 1.
Purification of recombinant dHEXIM and detection of endogenous dHEXIM. (A) Diagram showing the location of regions highly conserved between human HEXIM1 and Drosophila HEXIM (dHEXIM). (B) Silver-stained SDS PAGE analysis of wildtype (WT) and RNA-binding domain mutant (ILAA) of HEXIM1 and dHEXIM. (C) western blot analysis of recombinant dHEXIM expressed in E. coli (Ec) and in endogenous dHEXIM in Kc cells (Kc) probed with affinity purified anti-dHEXIM antibody.
To determine whether or not dHEXIM might be involved in controlling P-TEFb using the same mechanism as human HEXIM1, we first investigated whether or not it is a general dsRNA-binding protein like HEXIM1 (51). An electrophoretic mobility shift assays (EMSAs) was used to monitor RNA-binding. Purified recombinant wild-type (WT) dHEXIM or the RNA-binding mutant (ILAA) was titrated into reactions containing a constant amount of labeled dsRNA oligo. After 10 min incubation, the complexes were resolved on a Tris–glycine native gel and visualized by autoradiography (Figure 2A). As wild-type dHEXIM was titrated into the reaction, there was a concomitant increase in a single band of decreased mobility in the gel, representing the dHEXIM•RNA complex. In contrast, when equal amounts of mutant dHEXIM were assayed, there was no observable complex formation. These results demonstrate that dHEXIM is also a general dsRNA-binding protein, and that, like the human HEXIM proteins, the KHRR domain is required for this binding.
Figure 2.
Characterization of dHEXIM. (A) EMSA with dsRNA and recombinant dHEXIM proteins. The indicated amounts of wildtype (WT) and KHRR to ILAA mutant dHEXIM were incubated with labeled dsRNA and analyzed on a native gel as described in ‘Materials and Methods’ section. (B) Glycerol gradient analysis of control and RNase A treated Kc cell extracts. A Kc cell lysate was prepared and analyzed by glycerol-gradient sedimentation as described in ‘Materials and Methods’ section. Half of the lysate was treated with RNase A and the other half served as a control. Gradients were fractionated from the top so that increasing fraction number correlates with increasing particle size. Aliquots of each fraction were subjected to western blotting with the indicated antibodies. (C) Quantification of signals in (B).
To determine if dHEXIM and P-TEFb are normally found associated with RNA in vivo, glycerol gradient ultracentrifugation analysis was performed on a Kc cell lysate that was treated or not with RNase A (Figure 2B). Following fractionation of the gradient, western analysis was performed for both Cyclin T and dHEXIM. In the absence of RNase (control), there were two forms of P-TEFb with about half of the P-TEFb in the larger molecular weight fractions (fractions 11–16). A distribution of sizes were also found for dHEXIM; a smaller molecular weight peak (fractions 5–8), and a larger molecular weight peak (fractions 9–13) that overlapped, but did not completely co-fractionate, with the large form of P-TEFb. Upon treatment with RNaseA, P-TEFb shifted up the gradient to smaller size strongly suggesting that the large form of P-TEFb was an RNP. Interestingly, ribonuclease treatment had a less profound effect on the sedimentation of dHEXIM. Quantification of the distribution of Cyclin T and dHEXIM for the four gradients demonstrated that most of the P-TEFb, but only ∼10% of the dHEXIM shifted to the smaller forms after ribonuclease treatment (Figure 2C). It is not clear if the large form of dHEXIM that remains after ribonuclease treatment is due to oligomerization of dHEXIM or association with other proteins. However, after ribonuclease treatment almost all of the high-molecular weight dHEXIM is in fractions without P-TEFb, therefore as found in humans the HEXIM•P-TEFb interaction requires RNA. The glycerol gradient results suggest much of the P-TEFb and a small but significant fraction of dHEXIM is normally bound to RNA.
Identification of a LARP7 Homolog, dLARP7
Our attention turned next to the identification of a Drosophila homolog for LARP7, an RNA-binding protein that plays an important role in the human 7SK snRNP (18). The gene identified by a simple bioinformatic search as the potential homolog was initially called multi sex combs (MXC). Genetic analysis of MXC demonstrated that it was a Polycomb Group protein that was important in development and when mutated caused cancerous transformation of blood cells (52). However, recently, MXC was reassigned to an adjacent gene and the LARP7 homolog was renamed CG42569. The potential LARP7 homolog is 22% identical and 45% similar to the human protein and has an identical domain organization with a La motif and two RNA-binding motifs (RRMs) (Figure 3A). A cDNA encoding the full-length protein was expressed in E. coli; however, only a C-terminal proteolysis product was purified using the C-terminal His tag. An antibody was produced in sheep and affinity purified. This antibody recognized one major band on a western blot of whole cell or nuclear extracts from Kc cells (Figure 3B). It was not unexpected that the apparent molecular weight of the protein recognized (90 kDa) was larger than that predicted (67 kDa) because it is highly charged.
Figure 3.
Characterization of dLARP7. (A) Diagram showing the location of regions highly conserved between human LARP7 and Drosophila HEXIM (dLARP7). (B) Western blot analysis of endogenous dLARP7 in Kc cell whole-cell (W) and nuclear (N) extract probed with affinity purified anti-dLARP7 antibody. Arrow indicates dLARP7. (C) Glycerol-gradient analysis of control and RNase A treated Kc cell extracts. A Kc cell lysate was prepared and analyzed by glycerol gradient sedimentation as described in ‘Materials and Methods’ section. Half of the lysate was treated with RNase A and the other half served as a control. Gradients were fractionated from the top so that increasing fraction number correlates with increasing particle size. Aliquots of each fraction were subjected to western blotting with the indicated antibodies.
Glycerol-gradient sedimentation was performed on Kc cell lysates to examine the size of complexes containing the potential LARP7 homolog. As was done for dHEXIM the cell lysate was either treated with RNase A or left untreated as a control. Antibodies to cyclin T gave the expected patterns demonstrating an RNA dependence on high-molecular weight P-TEFb complexes. The antibodies against the putative LARP7 homolog gave patterns that indicated that almost half of the protein was in the same region of the gradient as the large form of P-TEFb. This material sedimented more slowly after RNase treatment. The sequence similarity to human LARP7 and the co-sedimentation with P-TEFb in an RNase sensitive complex, justifies renaming the gene, LARP7, and the protein, Drosophila LARP7 or dLARP7.
Characterization of d7SK
A key component of the large P-TEFb complex is the structural RNA, 7SK (19). Standard bioinformatics approaches to identify Drosophila 7SK were not successful. However, a putative arthropod 7SK was proposed by Gruber et al. (46), based on the presence of a Pol III promoter elements and structural modeling of the 5′- and 3′-ends of the transcript (46). Based on the sequence similarity of this gene within many Drosophila species, it appears that this gene encodes a 444 nt RNA (Figure 4A). Interestingly, this RNA begins and ends identically to the human 7SK RNA (GG and UUCUUUU, respectively), and, furthermore, contains two AUCUG sequences separated by 8 nt, exactly like human 7SK (37) (Figure 4B, underlined sequences).
Figure 4.
Characterization of the Drosophila 7SK snRNP. (A) Drosophila 7SK gene. The diagram shows the Pol III promoter driving 7SK RNA. The track below the diagram shows conservation of the region across all Drosophila species (from UCSC Genome Browser). (B) Sequence of 7SK RNA. Underlined nucleotides denote a region that is highly conserved between human and Drosophila 7SK. (C) Association of 7SK with LARP7. As described in ‘Materials and Methods’ section, the affinity purified LARP7 antibody was used to pull down LARP7 complexes and the RNA found in the bound ‘B’ and unbound ‘UB’ fractions was detected on a denaturing gel by ethidium bromide staining and by northern blot. (D) EMSA of 7SK RNA with dHEXIM on native gel as described in ‘Materials and Methods’ section. (E) Immunoprecipitation analysis of 7SK snRNP. Antibodies against HEXIM ‘H’, LARP7 ‘L’ and Cyclin T ‘C’ were used in immunoprecipitate the proteins from Kc cell extract. Western blots using the indicated antibodies were used to probe the bound and unbound material from each immunoprecipitation. A northern blot of similar fractions from another set of immunoprecipitations (denoted by an asterisk) was used to examine the presence of 7SK. The immunoprecipitations were not as efficient for this set of reactions. (F) Kinase assay with DSIF substrate. P, P-TEFb; C, Control 7SK snRNP; R, RNaseA treated 7SK snRNP; d, 50 µM DRB. The experiment shown in lanes 3–6 was repeated three times giving an average 1.8-fold stimulation with RNaseA.
The association of the potential Drosophila 7SK (d7SK) with dLARP7 and dHEXIM was examined to gather evidence that it was indeed a part of the large inactive P-TEFb complex. First, immunoprecipitation of LARP7 from Kc cell extracts was carried out and the RNA that was bound and unbound to the antibody beads was analyzed on a denaturing gel. Ethidium bromide staining demonstrated that an RNA that ran between the 500 and 800 nt marker was specifically pulled down (Figure 4C). A smaller RNA (∼300 nt) was also present, but a large amount of similarly sized RNA was present in the unbound fraction with many other unbound RNAs suggesting that the smaller RNA was recovered in the bound fractions due to a weak or non-specific association. The bound and unbound fractions were then subjected to northern blotting using a probe designed from the proposed 7SK. A band of identical size to the large RNA pulled down was detected only in the bound fraction (Figure 4C). We were initially concerned by the apparent large size of the RNA identified, but a 444 nt RNA generated by in vitro transcription of the d7SK gene had a similar aberrant mobility (data not shown). This is likely due to the predicted very high structure content of the RNA which is not completely removed in the TBE/Urea gels used. To determine if dHEXIM would bind to d7SK, labeled in vitro transcribed RNA was used in an RNA mobility shift assay. As increasing amounts of dHEXIM were incubated with the RNA, an RNA protein complex was correspondingly detected in the gel (Figure 4D). Association of 7SK with dLARP7 in vivo and dHEXIM in vitro strongly suggests that d7SK was identified correctly.
To determine if P-TEFb, dHEXIM, dLARP7 and d7SK were all present in the same complex immunoprecipitations from Kc cells extracts with antibodies to each of the proteins were performed (Figure 4E). Significantly, dHEXIM antibodies pulled down Cyclin T, dLARP7 and 7SK. dLARP7 antibodies pulled down Cyclin T and 7SK. dHEXIM was not detected due to the small fraction of dHEXIM bound to 7SK and to interference with detection by heavy chain antibody in that region of the gel. Cyclin T pulled down dLARP7, and 7SK, but again dHEXIM was not detected. These immunoprecipitation results are consistent with the existence of a Drosophila 7SK snRNP containing P-TEFb, dHEXIM, dLARP7 and d7SK.
Finally, to confirm that P-TEFb was held in an inactive state when associated with the 7SK snRNP, a kinase assay was performed. As expected, Drosophila P-TEFb phosphorylated DSIF in a DRB-sensitive manner (Figure 4F, lanes 1, 2). 7SK snRNP was immunoprecipitated using LARP7 antibodies from fractions 11–14 of the control glycerol gradient shown in Figure 2B. Reactions containing equal amounts of the 7SK snRNP were assayed with and without RNaseA treatment in the absence and presence of DRB. A representative of three replicates shown in Figure 4F (lanes 3–6) demonstrates an activation of P-TEFb by RNaseA. Kinase activity was increased an average of 1.8-fold (±0.3). This fold activation is somewhat low due to a small amount of free P-TEFb in that region of the gradient (Figure 2B; RNaseA).
Functional characterization of the Drosophila 7SK snRNP
Since treatment of human cells with P-TEFb inhibitors leads to release of P-TEFb from the 7SK snRNP, glycerol-gradient analysis of Kc cells before and after treatment with either DRB or flavopidirol was performed to further characterize the properties of the d7SK snRNP. The gradients were probed using westerns for Cyclin T, dHEXIM and dLARP7, and using northerns for 7SK (Figure 5). As shown earlier, Cyclin T was found in two peaks across the gradient, a small form (fractions 5–9) and a large form (fractions 10–14). dLARP7 displayed broad sedimentation with a clear peak in fractions 11 and 12. dHEXIM was again also found in two regions of the gradient. In contrast, d7SK was found in a single region of the gradient, the majority of which peaked in fractions 11 and 12 with Cyclin T, and dLARP7.
Figure 5.
Glycerol gradient analysis of lysates from Kc cells treated with P-TEFb inhibitors or HEXIM siRNA. Treatments and generation of lysates is described in ‘Materials and Methods’ section. Antibodies used in westerns are indicated. (A) Control cells. (B) Cells treated with Flavopiridol for 1 h. (C) Cells treated with DRB for 1 h. (D) Effect of knockdown of dHEXIM. Kc cells were treated with RNAi to lacZ ‘L’ as a control or dHEXIM ‘H’ and equal amounts of SDS lysed cells were analyzed by western blot using the dHEXIM antibody. Cell lysates were also prepared and analyzed by glycerol-gradient sedimentation and western analysis using antibodies to Cyclin T.
Identical glycerol-gradient analyses were performed after treatment of Kc cells for 1 h with either DRB or flavopiridol to inhibit P-TEFb (Figure 5B and C). Upon treatment with either drug there was a dramatic shift of P-TEFb as detected by Cyclin T to the small form (fractions 5–9) as it was released from the d7SK snRNP. The most rapidly sedimenting (largest) dHEXIM shifted up the gradient one fraction, as was observed during the RNase treatment, again supporting the hypothesis that at least some of the dHEXIM is in the d7SK snRNP. As expected, the small form of dHEXIM did not shift significantly up the gradient. d7SK and dLARP7 did not display a shift in sedimentation after the treatments as has been found for the human 7SK and LARP7 because human LARP7 is always found associated with 7SK. Because dHEXIM and P-TEFb were observed to leave the complex, this suggests that they are replaced by other proteins that keep the d7SK in the same position within the gradient, consistent with acquisition of hnRNPs as is found in humans (27).
To confirm that dHEXIM is required for the recruitment of P-TEFb into the 7SK snRNP, the effect of knocking down dHEXIM in Kc cells was examined. RNAi was used to knockdown either LacZ as a negative control or dHEXIM before cell lysates were analyzed by glycerol gradient sedimentation. The knockdown was efficient as indicated by a western blot of cell lysates (Figure 5D, left panel). In control cells the normal pattern was observed for Cyclin T (Figure 5D, upper right panel), but after knockdown of dHEXIM all of the P-TEFb shifted to smaller size (Figure 5D, lower right panel). This experiment demonstrates that the large form of P-TEFb is dependent on dHEXIM.
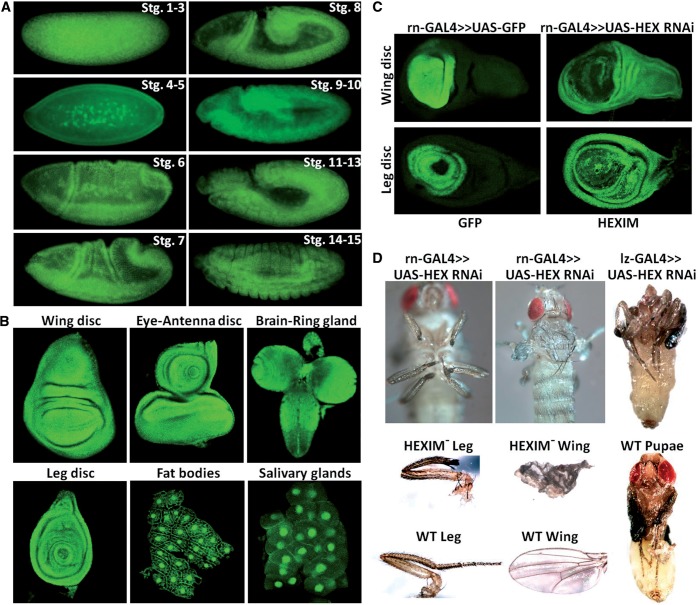
dLARP7 is expressed ubiquitously during fly development
The results described above indicate that Drosophila Kc cells express P-TEFb, dHEXIM dLARP7 and d7SK and these components are assembled into a d7SK snRNP. To extend these results into a physiologically relevant system, we first examined the expression of dLARP7 in whole flies. Consistent with the expected global role for the 7SK snRNP, dLARP7 was expressed at all developmental stages, both in diploid and polyploid tissues (Figure 6). Irrespective of this expression all through development, certain differences in dLARP7 expression are noteworthy. For example, dLARP7 was expressed predominantly in the ommatidial cluster of the eye disc, while expression in the rest of the eye and antennal discs was significantly reduced (Figure 6A). Similar is the case with the wing disc, wherein, including the wing pouch, there was no signal detected above background for dLARP7, except in a cluster of cells in the notum region (Figure 6A). Wherever expression was observed, the protein was always localized to the nucleus, though the intra-nuclear distribution varied depending on the cell type (Figure 6). For example, dLARP7 was pan-nuclear, excluding nucleoli, in larval salivary glands and fat bodies. In contrast, predominant nucleolar localization was found in the cells of gut and gastric caecae (Figure 6B). Because LARP7 is a required partner of 7SK in humans and levels of dLARP7 are variable in different tissues, it is likely that the level of the d7SK snRNP is regulated in a biologically significant way.
Figure 6.
Expression profile of dLARP7 during fly development. All panels show immunofluorescence using confocal microscopy and antibodies to dLARP7 (red) individually or merged with DAPI staining (blue). (A) dLARP7 is differentially expressed in the larval eye-antennal and wing discs. AD, antennal disc; NT, notum; WP, wing pouch. The ommatidial cluster of the eye disc is demarcated by a dashed line and zoomed in the lower panel. (B) dLARP7 is nuclear localized in the larval tissues. Nuclei in the indicated tissues are shown. Arrow heads indicate nucleoli. The inset in the merged image from the Gut shows just the DNA staining of the dashed region to highlight the nucleoli (arrow). (C) Nuclear localization of dLARP7 in the adult ovarian egg chamber.
dHEXIM is necessary for proper fly development
dHEXIM is continuously and ubiquitously expressed during embryogenesis (Figure 7A). A tight correlation between the expression patterns of dHEXIM protein and HEXIM mRNA was detected by in situ hybridization (data not shown). Of note, there was a strong accumulation of dHEXIM before the zygotic expression (stages 1–3) indicative of a maternal origin. The strong and ubiquitous dHEXIM expression persists to latter stages (i.e. third instar larvae) in all the tested tissues and organs (Figure 7B). Importantly, HEXIM is predominately localized in the nucleus, which is consistent with its role in regulation of transcription and with the localization of the d7SK snRNP.
Figure 7.
Expression profile and knockdown of HEXIM during development. (A) Immunostaining of dHEXIM during embryogenesis using confocal microscopy. Developmental stages are indicated on each panel. All embryos are oriented anterior to the left, dorsal uppermost. (B) Immunostaining of dHEXIM during organogenesis (third instar larvae). Top left to bottom right: wing disc, eye-antennal discs, brain and ring gland, leg disc, fat bodies and salivary glands. (C) The expression domain of the rotund-GAL4 driver (rn-GAL4) is revealed by GFP signal in third instar wing and leg discs (left column), and the expression pattern of dHEXIM in wing and leg discs expressing dHEXIM RNAi under the control of the rotund-GAL4 driver (right column). (D) Mutant phenotypes of dHEXIM knockdown with rotund-GAL4 and lozenge-GAL4 (lz-GAL4). In dHEXIM− leg, most of the tarsal segments from 1 to 5 are missing, thus leading to much foreshortened legs (left column). dHEXIM- wing is strongly atrophied and crumpled (middle column). dHEXIM− pharates (right column) are headless and fail to progress to adult stage.
To determine the requirement of dHEXIM in embryogenesis, early development and in the adult, the effects of tissue-specific knockdown of dHEXIM were examined. Knockdown was carried out using the GAL4 enhancer trap system that generates tissue-specific RNAi (48). To this end, UAS-dHEXIM RNAi lines (53) were crossed with lines driving GAL4 expression to a specific tissue and/or developmental stage. UAS-GFP lines, expressing GFP under the control of the GAL4 promoter, were used as a control to visualize the proper expression domain of the GAL4 driver (Figure 7C, left panel). As expected, dHEXIM expression was specifically reduced in the areas of the GAL4 driver expression domain (Figure 7C, right panel). dHEXIM knockdown was further confirmed by western blot analysis on the same flies (data not shown). Moreover, dHEXIM knockdown was systematically rescued by dHEXIM cDNA over-expression (Table 1 and Supplementary Figure S1), thus unambiguously showing the specificity of dHEXIM knockdown. Of note, given that dHEXIM can bind dsRNA and that >90% of dHEXIM is unbound (to the 7SK snRNP), we cannot exclude that the RNAi phenotypes could be partly due to the disruption of additional interactions of dHEXIM with yet-to-be discovered RNAs, other than the 7SK.
dHEXIM knockdowns carried out with the ubiquitously expressed actin- or tubulin-GAL4 drivers were lethal, with development stopping by the third instar larval stage. A collection of GAL4 drivers (engrailed-GAL4, decapentaplegic-GAL4, patched-GAL4), which have a more restricted expression pattern during embryogenesis, also led to developmental arrest, but at earlier times (embryogenesis or first or second instar larvae). When dHEXIM knockdown was targeted to a specific region of any imaginal disc, the corresponding tissue systematically failed to develop properly and was often missing at the adult stage (Figure 7D and data not shown). For example, when dHEXIM RNAi is driven by rotund-GAL4 (expressed in wing and haltere pouch, center of leg, and antenna imaginal discs), adult flies harbored strongly atrophic wings, halteres, legs and antennae. Strikingly, the lozenge-GAL4 driver (expressed in eye-antennal discs) induced headless pharate adults which died during the eclosion process (Figure 7D). More than 10 different organogenesis-specific drivers have been tested and all resulted in developmental arrest or organ defects dependent on the temporal and spatial expression of the corresponding GAL4 drivers. These results demonstrate that dHEXIM is a crucial factor required, in particular, for the proper organ formation, and in general for the entire organism development.
DISCUSSION
We have provided strong evidence that the 7SK snRNP which has been demonstrated to control P-TEFb in mammals is also expressed in Drosophila. Bioinformatic, biochemical and molecular genetic methods led to the identification of a P-TEFb complex containing homologs of HEXIM1/2, LARP7 and 7SK. The d7SK snRNP responds to inhibition of P-TEFb in an immortal cell line by release of P-TEFb and dHEXIM suggesting that it is regulated by similar mechanisms as found in humans. Importantly, having identified most of the components of the d7SK snRNP and developed molecular probes for the proteins and RNA, allows for the complex to be studied during the development of an organism. Toward that end, we demonstrated that components are expressed in a variety of tissues and embryonic developmental stages. Significantly, the key P-TEFb regulator, dHEXIM, is essential for proper Drosophila development.
The fact that the various components of the 7SK snRNP complex are found in both Drosophila and human is unlikely the result of convergent evolution. Rather, this strongly suggests that the complex characterized in this work is a true homolog of the one found in mammals. This is further supported by the fact that the fly 7SK snRNP complex is sensitive to P-TEFb inhibitors in a manner similar to that of human’s. Thus, one would expect the complex to have similar biological function in both Drosophila and human at the molecular, cellular and developmental level.
Although we were able to draw many similarities between the human and Drosophila 7SK snRNP, there were some differences found. Most of the detailed work on the 7SK snRNP in humans has been carried out in HeLa cells, and here, we described the biochemical characterization of complexes obtained from the Kc cell line. Both cell lines are immortal and have similar doubling times. As was found in the human system, P-TEFb is inhibited by the 7SK snRNP. Both human HEXIM1 (51) and dHEXIM (Figures 1 and 2) have the ability to bind to small dsRNAs in addition to 7SK. In HeLa cells, ∼80% of HEXIM1 is found in a low molecular weight form out of the 7SK snRNP, and in Drosophila, 90–95% of dHEXIM is not in the d7SK snRNP. A difference is that most of the ‘free’ dHEXIM is found in a high molecular weight form that is slightly smaller than the 7SK snRNP. We do not know the nature of the rapidly sedimenting form of dHEXIM, but based on a number of our results (gradient analyses and co-immunoprecipitions) it is not part of the 7SK snRNP. In both organisms, all 7SK is bound by LARP7 (21)(Figure 4C), but only in Drosophila is some free LARP7 found (Figure 5). What role LARP7 might play out of the 7SK complex is not clear (Figure 3). Treatment of human (18) or Drosophila (Figure 5) cells with P-TEFb inhibitors leads to release of P-TEFb and HEXIM. While it has been determined that human 7SK undergoes a conformational change upon loss of P-TEFb and HEXIM1 (38), this has not been examined for d7SK. However, the main structural elements of human 7SK are found in d7SK including two highly conserved regions that predict that the 5’-end of the RNA would pair with a region just upstream of the final stem and loop that is required for P-TEFb association with the 7SK snRNP (18,19).
Bin3, the Drosophila homolog of the human methyl phosphate capping enzyme (MEPCE) that methylates the gamma phosphate at the 5′-end of 7SK (21,25,54) was recently demonstrated to be required for the stability of d7SK (26). We performed a similar analysis on flies’ mutant for Bin3 and found that mutation of Bin3 lead to destabilization of d7SK without affecting U6 snRNA that is capped in a similar manner to 7SK snRNA (Supplementary Figure S2). Bin3 was discovered as a protein that interacted with Bicoid, a homeodomain protein that directs early pattern formation (55). Besides playing a role in stabilizing d7SK, Bin3 was found in a complex with bicoid on the 3′-UTR of the caudal mRNA (26). It will be interesting to determine if Bin3’s role in repressing translation of caudal mRNA is related to its role in the d7SK snRNP.
It is of interest that dLARP7 showed differential intra-nuclear distribution in a tissue-specific manner. In tissues like larval salivary glands, fat bodies and the adult ovary, dLARP7 displayed pan-nuclear localization, excluding nucleoli as is found for LARP7 in HeLa cells (22) (Supplementary Figure S3). In contrast, in the larval gut and gastric caecae, it localizes predominantly to the nucleoli, with diffuse staining in the nucleoplasm. Similar intra-nuclear localization was reported for the mammalian 7SK RNA, though the exact reason for the observed discrepancy is not clearly understood. For example, while a number of groups have showed that 7SK is pan-nuclear, colocalizing with splicing factor compartments (56,57,58), the work from the Zhou lab (59) showed that a majority of nuclear 7SK is localized to the nucleolus. While this discrepancy could be due to the age of the culture or the culture conditions applied, it is implicit in these data that the localization of 7SK snRNP is dynamic and might change in response to developmental and physiological needs. Accordingly, dLARP7 also showing these localizations trends reminiscent of 7SK snRNP suggests a greater biological meaning to differential expression and localization.
What have we learned about the involvement of the 7SK snRNP in Drosophila development? Although some tissues seem to have more or less dLARP7 and dHEXIM, both are ubiquitously expressed. Variation in expression may relate to the amount of the 7SK snRNP that is needed for each tissue to supply P-TEFb for its transcriptional program. The most important finding was that dHEXIM is absolutely essential for survival of the embryo and is required for development of all tissues tested. When knocked down early embryos fail to progress past the larval stage. When the early requirement is bypassed by expressing siRNA later in specific tissues, every tissue targeted was dramatically affected yielding severe phenotypes. Our results support the idea that regulation of P-TEFb plays a critical role in Drosophila development which fits with the recent findings indicating that genes controlling development are usually loaded with polymerases poised for the P-TEFb-mediated transition into productive elongation (42,60).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures S1–S3.
FUNDING
National Institutes of Health (grants GM25232 to J.T.L., GM35500 to D.H.P., GM053034 to A.G.M.); Agence Nationale pour la Recherche (ANR-06-BLAN-0072) (to P.U.); Cancéropôle Région Ile-de France Fellowship (to D.N.). Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the VDRC and the Bloomington Drosophila Stock Center for providing fly stocks. We also thank P. Lecorre and O. Fayol for technical assistance.
REFERENCES
- 1.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 2.Marshall NF, Price DH. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem. 1995;270:12335–12338. doi: 10.1074/jbc.270.21.12335. [DOI] [PubMed] [Google Scholar]
- 3.Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J. Biol. Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 4.Yamada T, Yamaguchi Y, Inukai N, Okamoto S, Mura T, Handa H. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol. Cell. 2006;21:227–237. doi: 10.1016/j.molcel.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 5.Fujinaga K, Irwin D, Huang Y, Taube R, Kurosu T, Peterlin BM. Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol. Cell. Biol. 2004;24:787–795. doi: 10.1128/MCB.24.2.787-795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh J, Padgett RA. Rates of in situ transcription and splicing in large human genes. Nat. Struct. Mol. Biol. 2009;16:1128–1133. doi: 10.1038/nsmb.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sehgal PB, Darnell JE, Jr, Tamm I. The inhibition by DRB (5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole) of hnRNA and mRNA production in HeLa cells. Cell. 1976;9:473–480. doi: 10.1016/0092-8674(76)90092-1. [DOI] [PubMed] [Google Scholar]
- 9.Chao SH, Price DH. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J. Biol. Chem. 2001;276:31793–31799. doi: 10.1074/jbc.M102306200. [DOI] [PubMed] [Google Scholar]
- 10.Marshall NF, Price DH. Control of formation of two distinct classes of RNA polymerase II elongation complexes. Mol. Cell. Biol. 1992;12:2078–2090. doi: 10.1128/mcb.12.5.2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marciniak RA, Sharp PA. HIV-1 Tat protein promotes formation of more-processive elongation complexes. EMBO J. 1991;10:4189–4196. doi: 10.1002/j.1460-2075.1991.tb04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nechaev S, Adelman K. Pol II waiting in the starting gates: Regulating the transition from transcription initiation into productive elongation. Biochim. Biophys. Acta. 2011;1809:34–45. doi: 10.1016/j.bbagrm.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat. Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Core LJ, Lis JT. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Price DH. Poised polymerases: on your mark…get set…go! Mol. Cell. 2008;30:7–10. doi: 10.1016/j.molcel.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 18.Diribarne G, Bensaude O. 7SK RNA, a non-coding RNA regulating P-TEFb, a general transcription factor. RNA Biol. 2009;6:122–128. doi: 10.4161/rna.6.2.8115. [DOI] [PubMed] [Google Scholar]
- 19.Peterlin BM, Brogie JE, Price DH. 7SK snRNA: a noncoding RNA that plays a major role in regulating eukaryotic transcription. Wiley interdisciplinary reviews. RNA. 2012;3:92–103. doi: 10.1002/wrna.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marz M, Donath A, Verstraete N, Nguyen VT, Stadler PF, Bensaude O. Evolution of 7SK RNA and its protein partners in metazoa. Mol. Biol. Evol. 2009;26:2821–2830. doi: 10.1093/molbev/msp198. [DOI] [PubMed] [Google Scholar]
- 21.Krueger BJ, Jeronimo C, Roy BB, Bouchard A, Barrandon C, Byers SA, Searcey CE, Cooper JJ, Bensaude O, Cohen EA, et al. LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res. 2008;36:2219–2229. doi: 10.1093/nar/gkn061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Markert A, Grimm M, Martinez J, Wiesner J, Meyerhans A, Meyuhas O, Sickmann A, Fischer U. The La-related protein LARP7 is a component of the 7SK ribonucleoprotein and affects transcription of cellular and viral polymerase II genes. EMBO Rep. 2008;9:569–575. doi: 10.1038/embor.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He N, Jahchan NS, Hong E, Li Q, Bayfield MA, Maraia RJ, Luo K, Zhou Q. A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol. Cell. 2008;29:588–599. doi: 10.1016/j.molcel.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta S, Busch RK, Singh R, Reddy R. Characterization of U6 small nuclear RNA cap-specific antibodies. Identification of gamma-monomethyl-GTP cap structure in 7SK and several other human small RNAs. J. Biol. Chem. 1990;265:19137–19142. [PubMed] [Google Scholar]
- 25.Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C, Therien C, Bergeron D, Bourassa S, Greenblatt J, et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol. Cell. 2007;27:262–274. doi: 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh N, Morlock H, Hanes SD. The Bin3 RNA methyltransferase is required for repression of caudal translation in the Drosophila embryo. Dev. Biol. 2011;352:104–115. doi: 10.1016/j.ydbio.2011.01.017. [DOI] [PubMed] [Google Scholar]
- 27.Barrandon C, Bonnet F, Nguyen VT, Labas V, Bensaude O. The transcription-dependent dissociation of P-TEFb-HEXIM1-7SK RNA relies upon formation of hnRNP-7SK RNA complexes. Mol. Cell. Biol. 2007;27:6996–7006. doi: 10.1128/MCB.00975-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Herreweghe E, Egloff S, Goiffon I, Jady BE, Froment C, Monsarrat B, Kiss T. Dynamic remodelling of human 7SK snRNP controls the nuclear level of active P-TEFb. EMBO J. 2007;26:3570–3580. doi: 10.1038/sj.emboj.7601783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michels AA, Fraldi A, Li Q, Adamson TE, Bonnet F, Nguyen VT, Sedore SC, Price JP, Price DH, Lania L, et al. Binding of the 7SK snRNA turns the HEXIM1 protein into a P-TEFb (CDK9/cyclin T) inhibitor. EMBO J. 2004;23:2608–2619. doi: 10.1038/sj.emboj.7600275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yik JH, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell. 2003;12:971–982. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- 31.Byers SA, Price JP, Cooper JJ, Li Q, Price DH. HEXIM2, a HEXIM1-related protein, regulates positive transcription elongation factor b through association with 7SK. J. Biol. Chem. 2005;280:16360–16367. doi: 10.1074/jbc.M500424200. [DOI] [PubMed] [Google Scholar]
- 32.Yik JH, Chen R, Pezda AC, Zhou Q. Compensatory contributions of HEXIM1 and HEXIM2 in maintaining the balance of active and inactive positive transcription elongation factor b complexes for control of transcription. J. Biol. Chem. 2005;280:16368–16376. doi: 10.1074/jbc.M500912200. [DOI] [PubMed] [Google Scholar]
- 33.Biglione S, Byers SA, Price JP, Nguyen VT, Bensaude O, Price DH, Maury W. Inhibition of HIV-1 replication by P-TEFb inhibitors DRB, seliciclib and flavopiridol correlates with release of free P-TEFb from the large, inactive form of the complex. Retrovirology. 2007;4:47. doi: 10.1186/1742-4690-4-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu Y, Pe'ery T, Peng J, Ramanathan Y, Marshall N, Marshall T, Amendt B, Mathews MB, Price DH. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 1997;11:2622–2632. doi: 10.1101/gad.11.20.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tahirov TH, Babayeva ND, Varzavand K, Cooper JJ, Sedore SC, Price DH. Crystal structure of HIV-1 Tat complexed with human P-TEFb. Nature. 2010;465:747–751. doi: 10.1038/nature09131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kao SY, Calman AF, Luciw PA, Peterlin BM. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature. 1987;330:489–493. doi: 10.1038/330489a0. [DOI] [PubMed] [Google Scholar]
- 37.Sedore SC, Byers SA, Biglione S, Price JP, Maury WJ, Price DH. Manipulation of P-TEFb control machinery by HIV: recruitment of P-TEFb from the large form by Tat and binding of HEXIM1 to TAR. Nucleic Acids Res. 2007;35:4347–4358. doi: 10.1093/nar/gkm443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krueger BJ, Varzavand K, Cooper JJ, Price DH. The mechanism of release of P-TEFb and HEXIM1 from the 7SK snRNP by viral and cellular activators includes a conformational change in 7SK. PLoS ONE. 2010;5:e12335. doi: 10.1371/journal.pone.0012335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou Q, Yik JH. The Yin and Yang of P-TEFb regulation: implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation. Microbiol. Mol. Biol. Rev. 2006;70:646–659. doi: 10.1128/MMBR.00011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bisgrove DA, Mahmoudi T, Henklein P, Verdin E. Conserved P-TEFb-interacting domain of BRD4 inhibits HIV transcription. Proc. Natl Acad. Sci. USA. 2007;104:13690–13695. doi: 10.1073/pnas.0705053104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rougvie AE, Lis JT. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- 42.Levine M. Paused RNA polymerase II as a developmental checkpoint. Cell. 2011;145:502–511. doi: 10.1016/j.cell.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Larschan E, Bishop EP, Kharchenko PV, Core LJ, Lis JT, Park PJ, Kuroda MI. X chromosome dosage compensation via enhanced transcriptional elongation in Drosophila. Nature. 2011;471:115–118. doi: 10.1038/nature09757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boettiger AN, Levine M. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science. 2009;325:471–473. doi: 10.1126/science.1173976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chopra VS, Cande J, Hong JW, Levine M. Stalled Hox promoters as chromosomal boundaries. Genes Dev. 2009;23:1505–1509. doi: 10.1101/gad.1807309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gruber AR, Kilgus C, Mosig A, Hofacker IL, Hennig W, Stadler PF. Arthropod 7SK RNA. Mol. Biol. Evol. 2008;25:1923–1930. doi: 10.1093/molbev/msn140. [DOI] [PubMed] [Google Scholar]
- 47.Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature. 2001;414:322–325. doi: 10.1038/35104581. [DOI] [PubMed] [Google Scholar]
- 48.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 49.Peng J, Marshall NF, Price DH. Identification of a cyclin subunit required for the function of Drosophila P-TEFb. J. Biol. Chem. 1998;273:13855–13860. doi: 10.1074/jbc.273.22.13855. [DOI] [PubMed] [Google Scholar]
- 50.Li Q, Price JP, Byers SA, Cheng D, Peng J, Price DH. Analysis of the large inactive P-TEFb complex indicates that it contains one 7SK molecule, a dimer of HEXIM1 or HEXIM2, and two P-TEFb molecules containing Cdk9 phosphorylated at threonine 186. J. Biol. Chem. 2005;280:28819–28826. doi: 10.1074/jbc.M502712200. [DOI] [PubMed] [Google Scholar]
- 51.Li Q, Cooper JJ, Altwerger GH, Feldkamp MD, Shea MA, Price DH. HEXIM1 is a promiscuous double-stranded RNA-binding protein and interacts with RNAs in addition to 7SK in cultured cells. Nucleic Acids Res. 2007;35:2503–2512. doi: 10.1093/nar/gkm150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Santamaria P, Randsholt NB. Characterization of a region of the X chromosome of Drosophila including multi sex combs (mxc), a Polycomb group gene which also functions as a tumour suppressor. Mol. Gen. Genet. 1995;246:282–290. doi: 10.1007/BF00288600. [DOI] [PubMed] [Google Scholar]
- 53.Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 54.Xue Y, Yang Z, Chen R, Zhou Q. A capping-independent function of MePCE in stabilizing 7SK snRNA and facilitating the assembly of 7SK snRNP. Nucleic Acids Res. 2010;38:360–369. doi: 10.1093/nar/gkp977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu W, Hanes SD. Identification of drosophila bicoid-interacting proteins using a custom two-hybrid selection. Gene. 2000;245:329–339. doi: 10.1016/s0378-1119(00)00048-2. [DOI] [PubMed] [Google Scholar]
- 56.Matera AG, Ward DC. Nucleoplasmic organization of small nuclear ribonucleoproteins in cultures human cells. J. Cell. Biol. 1993;17:715–727. doi: 10.1083/jcb.121.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shav-Tal Y, Blechman J, Darzacq X, Montagna C, Dye BT, Patton JG, Singer RH, Zipori D. Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol. Biol. Cell. 2005;16:2395–2413. doi: 10.1091/mbc.E04-11-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prasanth KV, Camiolo M, Chang G, Tripathi V, Denis L, Nakamura T, Hübner MR, Spector DL. Nuclear organization and dynamics of 7SK RNA in regulating gene expression. Mol. Biol. Cell. 2010;21:4184–4196. doi: 10.1091/mbc.E10-02-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He N, Pezda AC, Zhou Q. Modulation of a P-TEFb functional equilibrium for the global control of cell and differentiation. Mol. Cell. Biol. 2006;26:7068–7076. doi: 10.1128/MCB.00778-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chopra VS, Hong JW, Levine M. Regulation of Hox gene activity by transcriptional elongation in Drosophila. Curr. Biol. 2009;19:688–693. doi: 10.1016/j.cub.2009.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.