Abstract
We have determined the X-ray structure of the complex between the DNA quadruplex d(5′-GGGG-3′)4 and daunomycin, as a potential model for studying drug–telomere interactions. The structure was solved at 1.08 Å by direct methods in space group I4. The asymmetric unit comprises a linear arrangement of one d(GGGG) strand, four daunomycin molecules, a second d(GGGG) strand facing in the opposite direction to the first, and Na and Mg cations. The crystallographic 4-fold axis generates the biological unit, which is a 12-layered structure comprising two sets of four guanine layers, with four layers each of four daunomycins stacked between the 5′ faces of the two quadruplexes. The daunomycin layers fall into two groups which are novel in their mode of self assembly. The only contacts between daunomycin molecules within any one of these layers are van der Waals interactions, however there is substantial π–π stacking between successive daunomycin layers and also with adjacent guanine layers. The structure differs significantly from all other parallel d(TGGGGT)4 quadruplexes in that the 5′ guanine adopts the unusual syn glycosyl linkage, refuting the widespread belief that such conformations should all be anti. In contrast to the related d(TGGGGT)/daunomycin complex, there are no ligand–quadruplex groove insertion interactions.
INTRODUCTION
Telomeres are DNA sequences containing multiple repeats of guanine-rich oligonucleotide segments at the 3′ end of chromosomes. For example, the human telomere contains repetitive tracts of [d(TTAGGG)]n, whereas the sequence is [d(TGGGGT)]n in ciliates and [d(GGGGTTTTGGGG)]n in Oxytricha nova (1). The biological role of telomeres has been extensively reviewed (2,3), and is of high-current interest (e.g. Blackburn, Greider and Szostak were awarded the 2009 Nobel Prize in Physiology or Medicine for their discovery of how chromosomes are protected by telomeres and the enzyme telomerase). Regardless of the species, much of the telomeric region is double-helical, but a variable-length single strand protrudes at the extreme 3′ end. Guanines in this single strand can associate together (either intra- or inter-molecularly) to create what is known as a G-4 quadruplex which consists of stacked layers of four guanines, each layer being internally held together by eight hydrogen bonds. The stacks are stabilized by monovalent metal cations, each of which coordinates to eight guanine oxygens, four in the plane above, four in the plane below (4).
Every time normal cells divide some of the telomeric G-rich repeats are lost because DNA polymerase is unable to replicate that end of the telomere. This limits the number of times normal cells can divide before cellular senescence occurs. Telomerase is a reverse transcriptase enzyme which is able to rebuild those telomeric repeats lost during replication (2,3,5,6). Since it was discovered that telomerase is not active in most normal cells, but it is actively expressed in 80–85% of tumour cells (7), a massive effort has been undertaken to exploit this difference as a targeted cancer treatment protocol (8,9). Telomerase activity requires that the 3′ end of the telomere be single-stranded, and it is therefore anticipated that formation of a G-4 quadruplex will disrupt processes that allow immortalization of cancer cells. Attention has focussed on designing suitable ligands which will selectively target and stabilize the telomere quadruplex (10,11). X-ray and NMR studies have revealed many diverse structures for G-4 quadruplexes derived from short telomeric guanine-rich sequences. These may be intermolecular (tetramolecular or bimolecular) or intramolecular (monomolecular), parallel or anti-parallel, with variable intervening oligonucleotide sequences folded into diagonal, lateral or chain-reversal loops (12,13). Established crystal and solution structures of quadruplexes are available in the Nucleic Acid and Protein Data Bases (X-ray and NMR structures are deposited with the Nucleic Acids Database at http://ndbserver.rutgers.edu, and the Protein Data Bank at http://www.rcsb.org). Readers are referred to the comprehensive reviews of references (4,12,14) for descriptions of both general features and detailed properties of quadruplex topology and structure. Quadruplexes derived from short RNA sequences are also now becoming available (11,15,16), and over recent years, oligonucleotide sequences in promoter regions of cancer-related genes have been identified as forming non-telomeric quadruplex structures by folding of guanine-rich tracts within the DNA itself (4). These quadruplexes too offer exciting opportunities for the design of sequence-specific ligands capable of down-regulating gene expression (17). However, successful modelling of potential quadruplex-stabilising ligands continues to be hindered by the paucity of detailed structural information presently available.
The Tetrahymena DNA telomere quadruplex as d(TGGGGT) has proven to be the most amenable to crystallization. The crystal structure of native d(TGGGGT) was first solved in 1994 and to higher resolution in 1997 (18). It is a parallel-stranded right-handed helix stabilized by sodium ions. Other researchers have since investigated the role of alternative metal ions in quadruplex structures, notably Tl (19,20), Li (21), K (22,23) and Ca (24). Crystal structures of quadruplex/drug complexes were unknown prior to the publications in 2003 of d(TGGGGT) with daunomycin (25) and d(GGGGTTTTGGGG) with a disubstituted aminoalkylamido acridine ligand (26). Since then, an increasing number of ligand/quadruplex complexes have been examined, displaying a wide diversity of drug binding modes—face, edge, loop and simultaneous end stacking and loop groove interaction (14). To these modes must now be added in-groove binding as exemplified by the very recent NMR structure of distamycin A with d(TGGGGT) (27).
In quadruplex structures, the terminal G-quartet offers a large surface area onto which ligands containing planar aromatic chromophores can bind by π–π interactions. In d(TGGGGT)/daunomycin, the surface area is large enough to accommodate three coplanar-independent daunomycin molecules (25). It was observed that the protonated daunosamine side-chain of one daunomycin molecule inserted partially into one of the quadruplex grooves, forming favourable hydrogen-bonding interactions with both sides of the groove. The low-symmetry environment precluded any similar groove interactions by the other crystallographically independent daunosamines in that structure, and it was not immediately obvious whether groove interactions played any significant role in stabilising the ligand–quadruplex complex. In all parallel-stranded d(TGGGGT) crystal structures to date the thymines are not securely held in position. They are readily able to bend away from the primary core of the quadruplex, frequently to the point where the thymine atoms cannot be definitively located in electron density maps. In contrast, in quadruplexes constructed from longer telomeric repeat sequences, such as d(G4T4G4), d(TAG3T2AG3T) and d(TAG3 (T2AG3)3) (containing either parallel or antiparallel strands), the thymines are held in close proximity to the ends of the central stack by looping between guanines across the quadruplex face. Researchers are vigorously investigating the interactions between ligand and thymine loop structures as an indicator of quadruplex stability (28–30).
We have now co-crystallized daunomycin with the simpler truncated oligonucleotide d(GGGG), in order to investigate how the ligand binds to the quadruplex face in the absence of thymine interference, and to further examine the potential of groove binding by the basic substituent of the daunomycin side-chain for stabilizing the quadruplex. Sodium was retained in the crystallising medium, in order to provide the most direct comparison with the native d(TGGGGT) quadruplex crystal structure (18) and that of the d(TGGGGT)/daunomycin complex (25).
MATERIALS AND METHODS
Crystallization
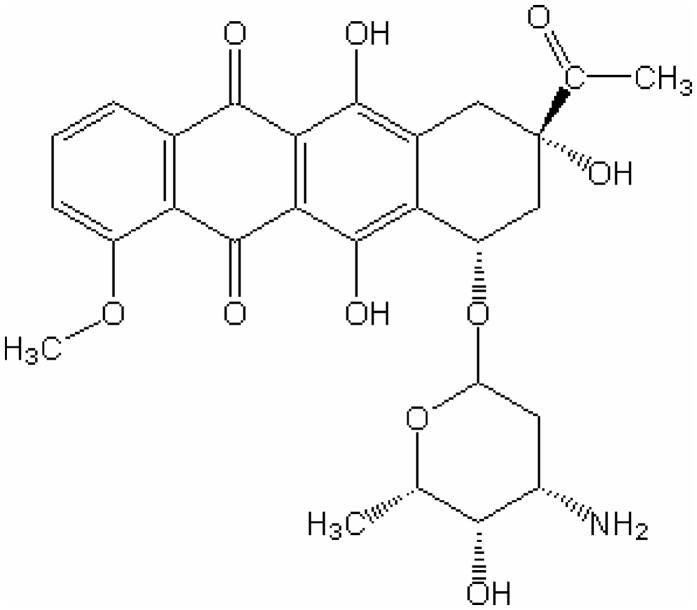
The DNA employed for the crystallizations was sourced from Oswell DNA Services, University of Southampton (now Eurogentec (Belgium), eurogentec.com), as a white HPLC-purified powder. It was dissolved to 3 mM in 30 mM sodium cacodylate buffer at pH 7.0 and annealed by slow cooling from 358 K to room temperature. Daunomycin hydrochloride (chemical structure depicted in Figure 1) from Merck was dissolved to 6 mM in 30 mM sodium cacodylate buffer at pH 7.0.
Figure 1.
Chemical structure of daunomycin.
Crystal growth was achieved using Natrix (nucleic acid matrix) solutions from Hampton Research (hamptonresearch.com), with the best crystals coming from NR46 containing 0.005 M magnesium sulphate, 0.05 M Tris–HCl buffer at pH 8.5 and 35% w/v 1,6-hexanediol solution as precipitant. Actual quantities used in the crystallizations were 2 μl each of the DNA and daunomycin stock solutions and 5 μl of NR46 solution. The reservoir contained 1 ml of the same NR46 solution. Crystals were produced from sitting drops at 4°C, with deep orange-red crystals forming over 3 months.
Data collection and structure determination
No cryoprotectant was necessary for freezing the crystals prior to data collection. The crystals diffracted strongly, and intensity data were initially collected to 1.5 Å at the School of Biological Sciences, The University of Auckland, on a Rigaku RU-H3R diffractometer. The data were processed using the HKL program suite (31), and efforts were made to solve the structure by traditional molecular replacement methods using AMoRe (32). Many models were tried, using partial geometries for the oligonucleotide sequence from the previously solved native quadruplex d(TGGGGT)4 and d(TGGGGT)4/daunomycin structures, but all were unsuccessful.
Crystals were then sent to the Stanford Synchrotron Radiation Laboratory in California where two high-resolution data sets were collected, one covering the range 3.5–1.0 Å and the other 40.0–1.1 Å. The two data sets were processed and merged using Scalepack from the HKL suite. After examining the data processing statistics, it was decided to limit the data resolution to 1.08 Å. Details of the data collection and processing are given in Table 1. Note that the high-symmetry space group I4 yielded a greater average redundancy than other previously published quadruplex crystal structures.
Table 1.
Data collection and refinement statistics
| Data collection | |
|---|---|
| Cell dimensions [a, b, c (Å)] | 40.209, 40.209, 31.865 |
| Crystal system | Tetragonal |
| Space group | I4 |
| All data | |
| Maximum resolution (Å) | 1.08 |
| Total measurements | 1 432 793 |
| Unique reflections | 21 428 |
| Rsym | 0.072 |
| Average I/σ(I) | 81.79 |
| Completeness (%) | 99.7 |
| Redundancy | >19 |
| Outer shell | |
| Resolution range (Å) | 1.13–1.08 |
| Rsym | 0.273 |
| Average I/σ(I) | 15.14 |
| Completeness (%) | 99.2 |
| Refinement | |
| No. of reflections | 14 501 [Fobs > 4σ(Fobs)] |
| Rwork/Rfree (%) | 0.161, 0.199 |
| Cations (occupancy) | 8 Na (0.25), 1 Mg (0.25), 1 Mg (0.0625) |
| Water molecules | 112 (10 with partial occupancy) |
| Mean B-factor (Å2) | |
| DNA atoms | 9.67 |
| Daunomycin | 10.97 |
| Metal ions | Na 18.86, Mg 32.72 |
| Waters with full occupancy | 30.29 |
| RMS deviations | |
| Bond lengths (Å) | 0.019 |
| Bond angles (°) | 1.310 |
Further attempts to solve the structure using the new high-resolution data were made using molecular replacement methods, as before, and again these were unsuccessful. As the data were near to atomic resolution, direct methods were then attempted. The structure finally was solved using SHELXD (33)—but only on run 974 of 1000! This run returned a final correlation coefficient (CC) of 78.75%, high enough to virtually guarantee that the structure was correct. In the ensuing map, no less than 164 out of 170 DNA atoms and 138 out of 152 ligand atoms in the asymmetric unit could be readily located. A subsequent map revealed positions for all remaining DNA and ligand atoms. Metal ions were clearly present in the central column of the stacked quadruplexes.
With hindsight we can readily see why all our attempted molecular replacement strategies were unsuccessful: the DNA configuration and geometries are very significantly different from any of the previous structures which we employed as trial models.
Refinement
Refinement of atomic parameters was carried out using SHELXL97 (33). All DNA and ligand atoms were given full occupancy, and were ultimately assigned anisotropic thermal parameters. Along the quadruplex axis lie one magnesium and eight sodium cations, plus another peak, which we have assigned as a partially occupied magnesium ion. We also refined 102 water molecules with full occupancy, and a further 10 with partial occupancy. The final R factor is 16.1%. Details of the least-squares refinement are presented in Table 1. Included in the supplementary data are the final 2Fo − Fc electron density map of guanine residue G1 (Supplementary Figure S1), and similar maps for the daunomycin ligand D9 (Supplementary Figures S2a and S2b). They clearly show the exceptionally high resolution of the crystal structure.
RESULTS
Description of the crystal structure
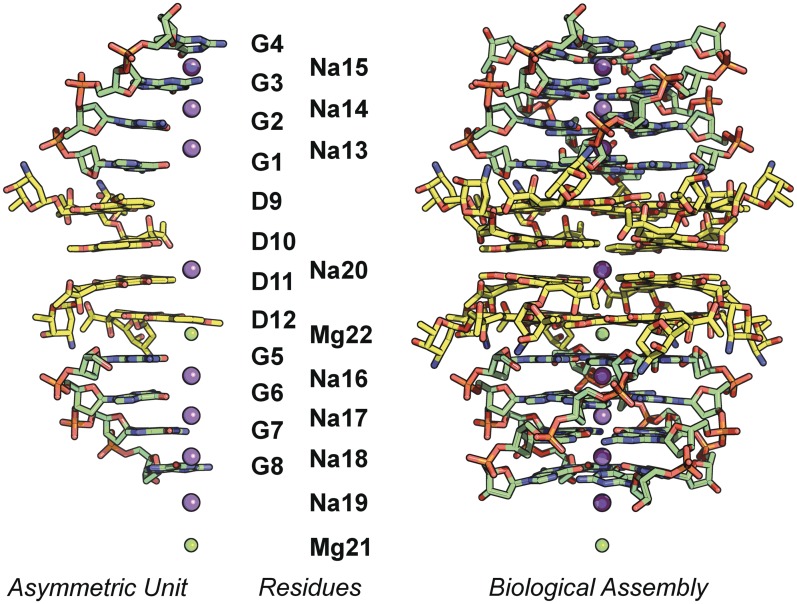
The crystal structure exhibits high symmetry, with the crystallographic 4-fold axis running through the centre of the quadruplex stack. The asymmetric unit contains two d(GGGG) strands separated by four daunomycin molecules. The stacking arrangement is d[(3′-GGGG-5′) DDDD d(5′-GGGG-3′)], where D's represent individual daunomycins. The guanine residues are labelled G1–G8, the daunomycins D9–D12, the metal ions Na13–Na20 and Mg21–Mg22. The asymmetric unit, residue numbering, and full biological assembly generated by the crystallographic tetrad axis, are displayed in Figure 2.
Figure 2.
Asymmetric unit, residue numbering, and full biological assembly generated by the crystallographic tetrad axis.
The two G-4 quadruplexes in the present structure are almost identical (the RMS deviation between the two crystallographically independent G4 strands is only 0.066). However, the glycosyl orientations are syn, anti, anti, anti, with the syn at the 5′ end, and in this respect, the DNA geometry is significantly different from those in all previous d(TGGGGT) structures and d(TGGGGT)/daunomycin, where the glycosyl orientations are consistently anti, anti, anti, anti (18–25). This difference is reflected in the RMS deviations. For example, comparing with TGGGGT/daunomycin, overlay of just guanines 2, 3 and 4 gives an RMS value of 0.059 between the two structures, while overlay of all four guanosines gives 1.436. Guanine 1 at the 5′ end accounts for almost all of this difference.
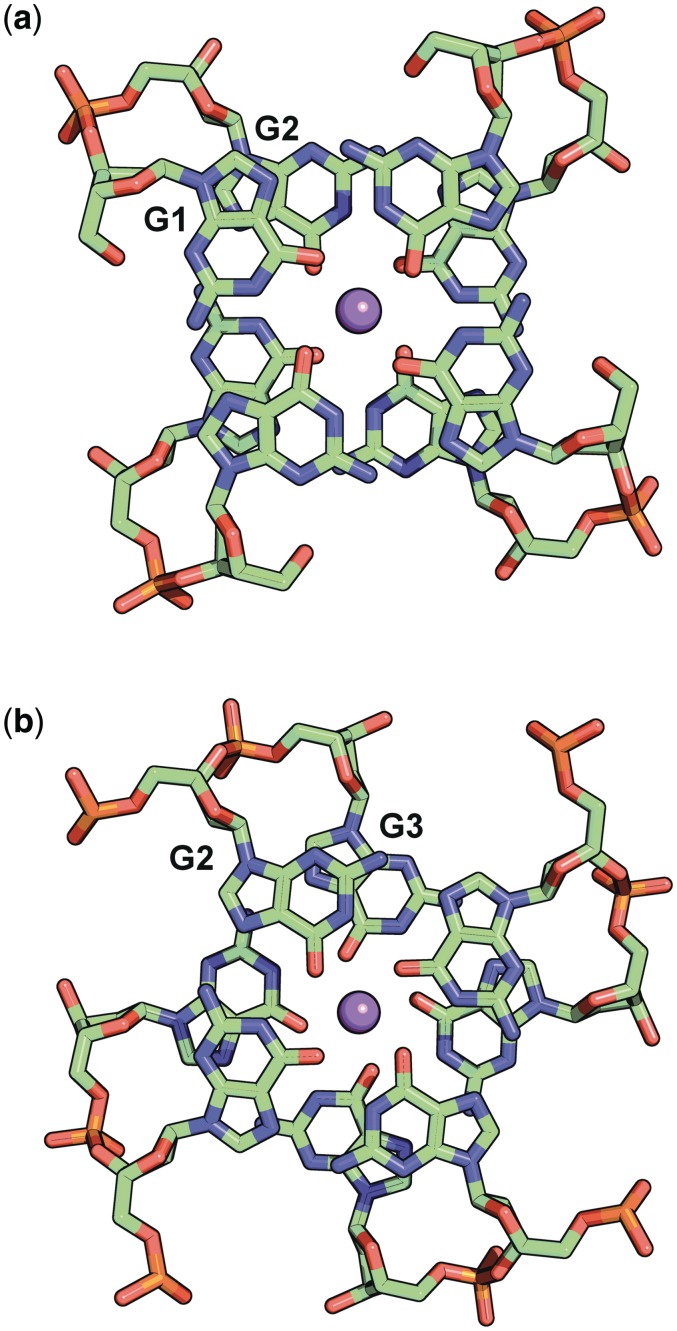
The rotations between successive guanine layers are also atypical (Table 2). Normal values are in the range 30–40°. Views of the differing overlaps between guanine layers G1/G2 and G2/G3 are depicted in Figure 3.
Table 2.
Quadruplex geometries
| Guanine residue | Sugar pucker | Glycosyl torsion |
|---|---|---|
| G1 | C2′ endo | Syn |
| G2 | C1′ exo | Anti |
| G3 | C2′ endo | Anti |
| G4 | C2′ endo | Anti |
| G5 | C2′ endo | Syn |
| G6 | C1′ exo | Anti |
| G7 | C2′ endo | Anti |
| G8 | C1′ exo | Anti |
| Layers | Interlayer rotation |
|---|---|
| G1/G2 | −93 |
| G2/G3 | +35 |
| G3/G4 | +22 |
| G5/G6 | −89 |
| G6/G7 | +34 |
| G7/G8 | +23 |
G1 and G5 are the closest quartets to the daunomycin layers.
Figure 3.
Double guanine layer overlap (a) G1 G2 and (b) G2 G3.
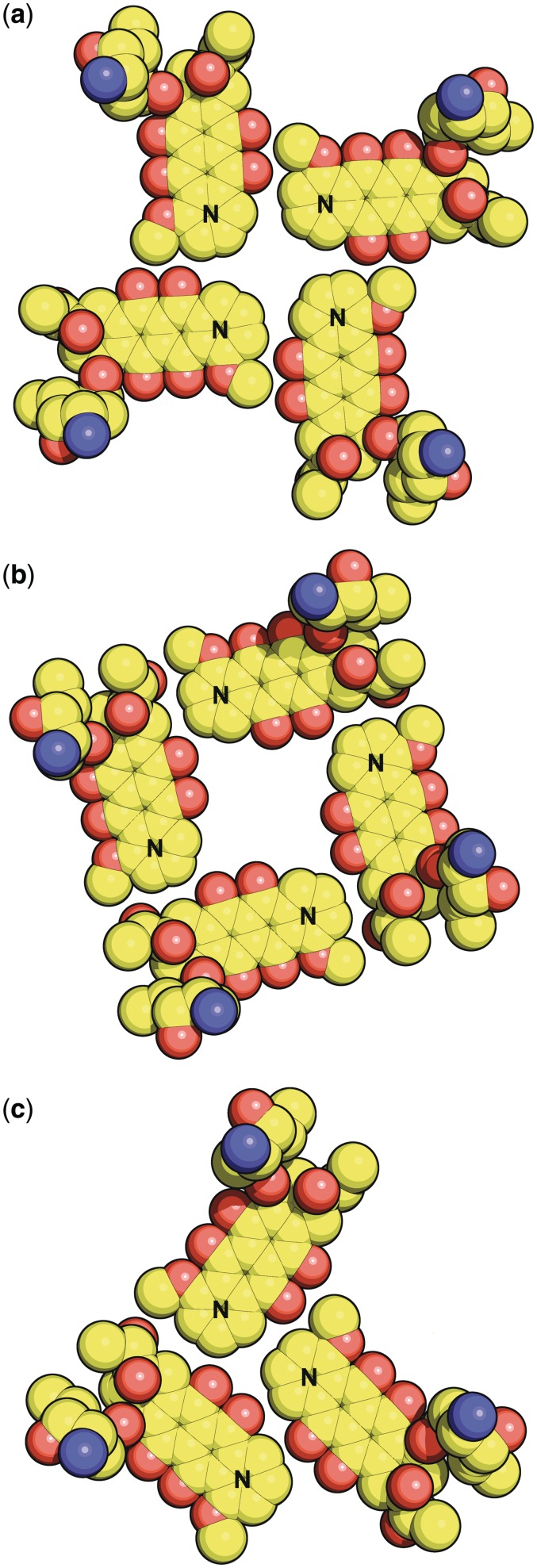
We must also consider the arrangements of the daunomycins. There are four daumonycin layers, each consisting of four symmetry-related molecules disposed about the crystallographic tetrad axis. However, there are two very different types of daunomycin layer formation. If the methoxy end of the daunomycin molecule is described as the nose, and the acetyl end is described as the tail, then, in the outer two daunomycin layers, the four symmetry-related molecules in each layer are aligned ‘noses to noses’ (adopted by layers of D9 and D12), whereas in the inner two layers the four symmetry-related molecules in each layer are aligned ‘noses to tails’ (adopted by layers of D10 and D11). These arrangements are displayed in Figure 4a and b. Note that the letter ‘N’ denotes the ‘nose’. Only van der Waals contacts exist between the planar aromatic chromophores of the molecules within any of the layers, and crystal stability is gained by extensive π–π stacking between successive layers. Supplementary Figure S3a and S3b shows the stacking overlap between daunomycin layers D9/D10 and D10/D11 respectively. There is also extensive π–π stacking at the overlap between outer daunomycin layers with guanine layers (D9 layer/G1 layer, and D12 layer/G5 layer). The daunosamine side-chains project approximately at right angles to the planes of the respective layers. The cluster of four daunomycin layers, each of four daunomycins, stacks between the 5′ ends of the two structurally similar but crystallographically independent guanine quadruplexes. This pattern is fundamentally different from that in the d(TGGGGT)/daunomycin structure (25) where the crystallographic symmetry is lower, and there are only three daunomycin molecules in each daunomycin layer. That arrangement is displayed in Figure 4c for direct comparison with the present structure.
Figure 4.
(a) Daunomycin layer D9 described as ‘nose to nose’. The letter ‘N’ denotes the ‘nose’ (see text). Please refer back to Figure 1 for the chemical structure of daunomycin. Layer D12 is similar to D9. (b) Daunomycin layer D10 described as ‘nose to tail’. Layer D11 is similar to D10. (c) Daunomycin layer arrangement in the TGGGGT/daunomycin crystal structure (20).
Grooves
The crystallographically imposed symmetry demands that the four grooves are equivalent within each quadruplex. This is in marked contrast to the TGGGGT/daunomycin structure (25) where there was no such symmetry requirement, and the four groove widths and depths were all different, and the three independent daunomycins necessarily had different interactions with the grooves. Groove asymmetry is a recurring feature in all other native TGGGGT structures (18–21,25).
Ligand–quadruplex groove interactions
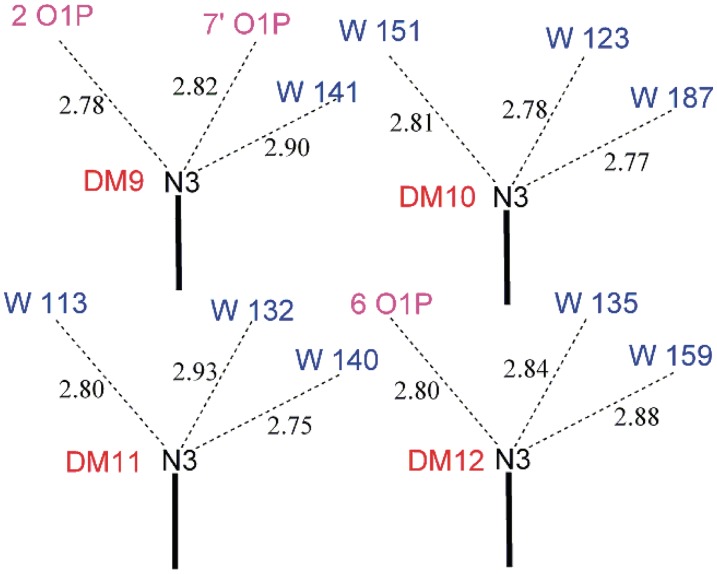
There are no daunomycin/groove insertion interactions in the present structure. If insertion had occurred, the protonated amines N3 of the daunosamine moieties would lie partially or fully inside the hollow of the groove, and would make favourable hydrogen-bonding contacts with phosphate oxygen atoms on either side of the groove. Instead, the two daunomycin molecules which are adjacent to the quadruplex stack (D9 and D12) are positioned, so that their protonated amines point towards the outside edge of the groove, while daunomycin molecules D10 and D11 are remote from any quadruplex (see Supplementary Figure S4). Figure 5 is a schematic diagram showing the contact distances between the protonated nitrogen (N3) of each daunomycin and its hydrogen-bonding acceptor atoms (phosphate oxygens or water molecules). D9 and D12 each make a hydrogen bond to phosphate oxygens O1P on the outside rim of the groove wall (2.78 Å and 2.80 Å respectively). D9, but not D12, makes an additional inter-molecular hydrogen bond of 2.82 Å to phosphate oxygen O1P of a neighbouring quadruplex. Water molecules complete their full hydrogen-bonding complements. The daunosamines of D10 and D11 are involved only in the extended hydrogen-bonding water network.
Figure 5.
Daunosamine–DNA groove contacts (Schematic). Distances are in Angstrom units.
The difference in hydrogen-bonding pattern between D9 and D12 explains why there is no crystallographic symmetry operation perpendicular to the tetrad axis.
Metal ion coordination
A recurring feature of G-4 quadruplexes is the sequence of three monovalent metal ions residing along each quadruplex axis. These ‘classical’ metal ions lie sandwiched between two adjacent G-4 layers and coordinate directly to eight carbonyl oxygen atoms, four from each layer. The typical structure has the middle Na of an individual G-4 quadruplex centrally positioned between its guanine layers with all eight Na–O distances approximately 2.7 Å. As the guanines in a G-4 quadruplex are typically rotated by about 30–40° in moving up the oligonucleotide, that middle Na coordination geometry can be described as a twisted square anti-prism. The other two Na atoms are not centrally located between the layers, but lie closer to the outside of the stack, with four Na–O distances approximately 3.2 Å and four approximately 2.4 Å. The coordination geometry could be described as either square planar or severely distorted square anti-prismatic. There has been considerable discussion regarding the role of metal ions in stabilizing the quadruplex stack (18–25).
The present structure differs in a number of ways from the other analogues. First, all 10 metal ions lie on the crystallographic 4-fold axis, and their coordinating atoms must therefore be disposed symmetrically about that axis. Secondly, the ‘classical’ trios of sodium ions are no longer typical. For the first trio of Na13, Na14 and Na15, the middle Na14 is not centrally positioned between the layers, and for Na15 the relative guanine rotation is reduced to +22°. A much more significant difference is that Na13 lies nearly midway between guanine layers G1 and G2, and the relative rotation between the G1 and G2 layers is −93° instead of the expected +35°. The metal coordination geometry of Na13 is therefore best described as that of a square prism. This change is brought about by the syn glycosyl orientation of guanine residue G1. The pattern of the second trio of ‘classical’ metal ions Na16, Na17 and Na18 replicates that described above for the first trio. Thirdly, we find additional metal ions (hydrated) residing along the central axis of the extended biological unit—a square-planar Na20 occupying the cavity between daunomycin layers D10 and D11, a well-resolved Mg22 in a square pyramidal coordination between the D12 and G5 layers, a square planar Na19 beyond the G8 layer, and a partially occupied Mg21 in a square planar coordination. All these latter, ‘non-classical’ metal ions bind to water molecules which are part of the extended hydrogen-bonding network, and thus they appear to be predominantly crystal-stabilizing rather than quadruplex-stabilizing. The unexpectedly wide ranges of coordination geometries are set out in Table 3.
Table 3.
Metal ion coordination geometry
| Metal ion | Guanine layers | Coordinating atoms | Bond lengths (Å) | Twist angle | Description |
|---|---|---|---|---|---|
| Na15 | G3/G4 | 8 × O | 4 × 3.20 (G3) | +22° | Approx. Square Planar |
| 4 × 2.39 (G4) | |||||
| Na14 | G2/G3 | 8 × O | 4 × 2.80 (G2) | +35° | Twisted Square Anti-prism |
| 4 × 2.63 (G3) | |||||
| Na13 | G1/G2 | 8 × O | 4 × 2.77 (G1) | −93° | Square Prism |
| 4 × 2.88 (G2) | |||||
| Na20 | 4 × H2O | 4 × 2.84 (50%) | Square Planar | ||
| 4 × 2.36 (50%) | |||||
| Mg22 | 5 × H2O | 4 × 2.15 | Square Pyramid | ||
| 1 × 2.38 | |||||
| Na16 | G5/G6 | 8 × O | 4 × 2.77 (G5) | −89° | Square Prism |
| 4 × 2.86 (G6) | |||||
| Na17 | G6/G7 | 8 × O | 4 × 2.80 (G6) | +34° | Twisted Square Anti-prism |
| 4 × 2.63 (G7) | |||||
| Na18 | G7/G8 | 8 × O | 4 × 3.20 (G7) | +23° | Approx. Square Planar |
| 4 × 2.38 (G8) | |||||
| Na19 | 4 × H2O | 4 × 2.44 (75%) | Square Planar | ||
| 4 × 2.44 (25%) | |||||
| Mg21 (25%) | 4 × H2O | 4 × 1.98 (25%) | Square Planar |
The metal-coordinating water molecules in the table above were all located and individually refined as part of the total set of 112 waters.
CONCLUSIONS
This study adds to the small number of ligand/quadruplex structures, which have, thus far, been determined using X-ray diffraction techniques. The structure of d(GGGG)/daunomycin is most logically compared with that of d(TGGGGT)/daunomycin (25). In both cases, the G-4 quadruplexes sandwich layers of daunomycin molecules, but the ligand arrangements and DNA geometries are vastly different. In the present d(GGGG)/daunomycin structure, the crystals exhibit much higher symmetry, the DNA adopts an unusual syn, anti, anti, anti conformation and unique interlayer guanine rotations, the four grooves in each quadruplex are all equivalent, there are no daunosamine groove insertions, and the daunomycin layer structures are entirely different.
The perfect 4-fold symmetry of the two independent quadruplexes in the present structure also contrasts with the low-symmetry quadruplexes in all other unligated d(TGGGGT) crystal structures (18–24). NMR studies show that unligated parallel tetrameric quadruplexes usually display higher symmetries in solution (27), and it is evident that the full role played by packing forces when forming a particular polymorph is not yet understood.
The presence of syn glycosyl conformations in one layer of each of the two independent quadruplexes refutes the commonly expressed assertion (4,28,34,35) that these conformations are invariably anti, anti, anti, anti in parallel intermolecular DNA G-4 quadruplexes ‘which do not display significant structural polymorphism’ (35). Our contrary finding, albeit in the solid state, will inform future research especially as rules are presently under development for predicting topographical and conformational features of quadruplex structures as an aid in the design of more effective drug ligands, and for better understanding quadruplex interactions with proteins and other DNA and RNA polymorphic constructs (34). We note that all-syn tetrads have recently been found in quadruplexes derived from chemically modified guanines (36). The syn/anti energy differences are probably small, and we consider it likely that the conformational change in the present structure is an artefact of crystal packing forces resulting from the elaborate packing arrangements of the guanines in contact with the daunomycin layers.
DISCUSSION
In retrospect, it is clear that our inability to solve the crystal structure employing molecular replacement techniques can be directly attributed to the fact that we could not generate an adequate starting model, despite our many attempts. The massive structural change that accompanies the syn glycosyl torsion of Guanine 1 negates phasing by the other guanines. Also, we note that the daunomycin molecules contribute a very significant proportion of the total X-ray scattering matter, and the d(GGGG)/daunomycin crystal has two entirely differently arranged layers of four daunomycins, both of which are totally different again from the layers of three daunomycins in the putative model d(TGGGGT)/daunomycin structure. Hence, phasing by daunomycin contributions was not possible. We are pleased to acknowledge that the structure was successfully solved, albeit with difficulty, by SHELXD (33).
ACCESSION NUMBERS
The model and X-ray data have been deposited in the Research Collaboratory for Structural Bioinformatics PDB with identity code 3TVB.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–4.
FUNDING
Funding for open access charge: The University of Auckland.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENT
This work was supported by grant UOA912 from the Marsden Fund of the Royal Society of New Zealand.
REFERENCES
- 1.Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR. A highly conserved repetitive DNA sequence (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl. Acad. Sci., USA. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bibby MC. Introduction to telomeres and telomerase. Methods Mol. Biol. 2002;191:1–12. doi: 10.1385/1-59259-189-2:01. [DOI] [PubMed] [Google Scholar]
- 3.Gardano L, Harrington L. Cellular senescence and tumor suppression. 2010. Telomere biology and biochemistry. Part 1, 3–43. [Google Scholar]
- 4.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morin GB. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesises TTAGGG repeats. Cell. 1989;59:521–529. doi: 10.1016/0092-8674(89)90035-4. [DOI] [PubMed] [Google Scholar]
- 6.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 7.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 8.Neidle S. Human telomeric G-quadruplex: The current status of telomeric G-quadruplexes as therapeutic targets in human cancer. FEBS J. 2009;277:1118–1125. doi: 10.1111/j.1742-4658.2009.07463.x. [DOI] [PubMed] [Google Scholar]
- 9.Gunaratnam M, Neidle S. An evaluation cascade for G-quadruplex telomere targeting agents in human cancer cells. Methods Mol. Biol. 2010;613:303–313. doi: 10.1007/978-1-60327-418-0_19. [DOI] [PubMed] [Google Scholar]
- 10.Campbell NH, Parkinson GN, Reszka AP, Neidle S. Structural basis of DNA quadruplex recognition by an acridine drug. J. Am. Chem. Soc. 2008;130:6722–6724. doi: 10.1021/ja8016973. [DOI] [PubMed] [Google Scholar]
- 11.Collie G, Reszka AP, Haider SM, Gabelica V, Parkinson GN, Neidle S. Selectivity in small molecule binding to human RNA and DNA quadruplexes. Chem. Commun. 2009:7482–7484. doi: 10.1039/b901889a. [DOI] [PubMed] [Google Scholar]
- 12.Neidle S. The structures of quadruplex nucleic acids and their drug complexes. Curr. Opin. Struct. Biol. 2009;19:239–250. doi: 10.1016/j.sbi.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 13.Cang X, Šponer J, Cheatham TE., III Insight into G-DNA structural polymorphism and folding from sequence and loop connectivity through free energy analysis. J. Am. Chem. Soc. 2011;133:14270–14279. doi: 10.1021/ja107805r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaushik M, Kaushik S, Bansal A, Saxena S, Kukreti S. Structural diversity and specific recognition of four stranded G-quadruplex DNA. Curr. Mol. Med. 2011;11:744–769. doi: 10.2174/156652411798062421. [DOI] [PubMed] [Google Scholar]
- 15.Collie GW, Haider SM, Neidle S, Parkinson GN. A crystallographic and modelling study of a human telomeric RNA (TERRA) quadruplex. Nucleic Acids Res. 2010;38:5569–5580. doi: 10.1093/nar/gkq259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collie GW, Sparapani S, Parkinson GN, Neidle S. Structural basis of telomeric RNA quadruplex-acridine ligand recognition. J. Am. Chem. Soc. 2011;133:2721–2728. doi: 10.1021/ja109767y. [DOI] [PubMed] [Google Scholar]
- 17.Garner TP, Williams HEL, Gluszyk KI, Roe S, Oldham NJ, Stevens MFG, Moses JE, Searle MS. Selectivity of small molecule ligands for parallel and anti-parallel DNA G-quadruplex structures. Org. Biomol. Chem. 2009;7:4194–4200. doi: 10.1039/b910505k. [DOI] [PubMed] [Google Scholar]
- 18.Phillips K, Dauter Z, Murchie AIH, Lilley DMJ, Luisi B. The crystal structure of a parallel-stranded guanine tetraplex at 0.95 Å resolution. J. Mol. Biol. 1997;273:171–182. doi: 10.1006/jmbi.1997.1292. [DOI] [PubMed] [Google Scholar]
- 19.Gill ML, Strobel SA, Loria JP. Crystallization and characterization of the thallium form of the Oxytricha nova G-quadruplex. Nucleic Acids Res. 2006;34:4506–4514. doi: 10.1093/nar/gkl616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cáceres C, Wright G, Gouyette C, Parkinson G, Subirana JA. A thymine tetrad in d(TGGGGT) quadruplexes stabilized with Tl+/Na+ Ions. Nucleic Acids Res. 2004;32:1097–1102. doi: 10.1093/nar/gkh269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Creze C, Rinaldi B, Haser R, Bouvet P, Gouet P. Structure of a d(TGGGGT) quadruplex crystallised in the presence of Li+ ions. Acta Crystallogr. 2007;D63:682–688. doi: 10.1107/S0907444907013315. [DOI] [PubMed] [Google Scholar]
- 22.Haider S, Parkinson GN, Neidle S. Crystal structure of the potassium form of an Oxytricha nova G-quadruplex. J. Mol. Biol. 2002;320:189–200. doi: 10.1016/S0022-2836(02)00428-X. [DOI] [PubMed] [Google Scholar]
- 23.Luu KN, Phan AT, Kuryavyi V, Lacroix L, Patel DJ. Structure of the human telomere in K+ solution: an intramolecular (3 + 1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee MPH, Parkinson GN, Hazel P, Neidle S. Observation of the coexistence of sodium and calcium ions in a DNA quadruplex ion channel. J. Am. Chem. Soc. 2007;129:10106–10107. doi: 10.1021/ja0740869. [DOI] [PubMed] [Google Scholar]
- 25.Clark GR, Pytel PD, Squire CJ, Neidle S. Structure of the first parallel DNA quadruplex-drug complex. J. Am. Chem. Soc. 2003;125:4066–4067. doi: 10.1021/ja0297988. [DOI] [PubMed] [Google Scholar]
- 26.Haider S, Parkinson GN, Neidle S. Structure of a G-quadruplex-ligand complex. J. Mol. Biol. 2003;326:117–125. doi: 10.1016/s0022-2836(02)01354-2. [DOI] [PubMed] [Google Scholar]
- 27.Martino L, Virno A, Pagano B, Virgilio A, Di Micco S, Galeone A, Giancola C, Bifulco G, Mayol L, Randazzo A. Structural and thermodynamic studies of the interaction of Distamycin A with the parallel quadruplex structure [d(TGGGGT)]4. J. Am. Chem. Soc. 2007;129:16048–16056. doi: 10.1021/ja075710k. [DOI] [PubMed] [Google Scholar]
- 28.Parkinson GN, Cuenca F, Neidle S. Topology conservation and loop flexibility in quadruplex-drug recognition: crystal structures of inter- and intramolecular telomeric DNA quadruplex-drug complexes. J. Mol. Biol. 2008;381:1145–1156. doi: 10.1016/j.jmb.2008.06.022. [DOI] [PubMed] [Google Scholar]
- 29.Campbell NH, Patel M, Tofa AB, Ghosh R, Parkinson GN, Neidle S. Selectivity in ligand recognition of G-quadruplex loops. Biochemistry. 2009;48:1675–1680. doi: 10.1021/bi802233v. [DOI] [PubMed] [Google Scholar]
- 30.Hampel SM, Sidibe A, Genaratnam M, Riou J-F, Neidle S. Tetrasubstituted naphthalene diimide ligands with selectivity for telomeric G-quadruplexes and cancer cells. Bioorg. Med. Chem. Lett. 2010;20:6459–6463. doi: 10.1016/j.bmcl.2010.09.066. [DOI] [PubMed] [Google Scholar]
- 31.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 32.Navaza J. AMoRe: an automated package for molecular replacement. Acta Crystallogr. 1994;A50:157–163. [Google Scholar]
- 33.Sheldrick GM. A short history of SHELX. Acta Crystallogr. 2008;A64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 34.Tong X, Lan W, Zhang X, Wu H, Liu M, Cao C. Solution structure of all parallel G-quadruplex formed by the oncogene RET promoter sequence. Nucleic Acids Res. 2011;39:6753–6763. doi: 10.1093/nar/gkr233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cang X, Šponer J, Cheatham TE., III Explaining the varied glycosidic conformational, G-tract length and sequence preferences for anti-parallel G-quadruplexes. Nucleic. Acids Res. 2011;39:4499–4512. doi: 10.1093/nar/gkr031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tran PLT, Virgilo A, Esposito V, Citarella G, Mergny J-L, Galeone A. Effects of 8-methylguanine on structure, stability and kinetics of formation of tetramolecular quadruplexes. Biochimie. 2011;93:399–408. doi: 10.1016/j.biochi.2010.10.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.