Abstract
The autonomous transcription of integrated retroviruses strongly depends on genetic and epigenetic effects of the chromatin at the site of integration. These effects are mostly suppressive and proviral activity can be finally silenced by mechanisms, such as DNA methylation and histone modifications. To address the role of the integration site at the whole-genome-scale, we performed clonal analysis of provirus silencing with an avian leucosis/sarcoma virus-based reporter vector and correlated the transcriptional silencing with the epigenomic landscape of respective integrations. We demonstrate efficient provirus silencing in human HCT116 cell line, which is strongly but not absolutely dependent on the de novo DNA methyltransferase activity, particularly of Dnmt3b. Proviruses integrated close to the transcription start sites of active genes into the regions enriched in H3K4 trimethylation display long-term stability of expression and are resistant to the transcriptional silencing after over-expression of Dnmt3a or Dnmt3b. In contrast, proviruses in the intergenic regions tend to spontaneous transcriptional silencing even in Dnmt3a−/− Dnmt3b−/− cells. The silencing of proviruses within genes is accompanied with DNA methylation of long terminal repeats, whereas silencing in intergenic regions is DNA methylation-independent. These findings indicate that the epigenomic features of integration sites are crucial for their permissivity to the proviral expression.
INTRODUCTION
In the course of retrovirus infection, the integration of proviral DNA and its subsequent transcription into viral mRNAs are important steps, when the host cell regulatory mechanisms interfere with virus propagation. The host-cell control of provirus transcription can eliminate the deleterious effects of retroviruses but, on the other hand, it has to be taken into account in retrovirus-mediated gene transfer, transgenesis, and gene therapy where stable and long-term provirus expression is required. The cellular DNA sequences adjacent to the integrated retrovirus can influence the proviral transcriptional activity. In general, transcriptionally active regions are permissive for virus-gene expression while integration into heterochromatin dis-favors virus transcriptional activity (1,2).
Multiple studies analyzed retrovirus integration patterns at the genome-wide scale and revealed virus-specific differences in integration preferences. Human immunodeficiency virus type 1 (HIV-1) preferentially targets transcriptionally active genes and, correspondingly, gene-rich and GC-rich chromosomal regions (3,4). Along the transcription units (TU), there is no preference either for introns, exons, CpG dinucleotides (CpGs) islands, or transcription start sites (TSSs) (5). This integration pattern is directed by the tethering of LEDGF/p75 with HIV-1 integrase and open chromatin components (6–8). In striking contrast, integrations of gamma-retroviruses and spuma-retroviruses are over-represented around TSSs and CpG islands (5,9,10), which might be the cause of documented genotoxicity and leukemia induction by a murine leukemia virus (MLV)-derived vector in a gene therapy trial (11). Cellular factor(s) channeling MLV to integrate close to TSSs are not known, although several transcription factors and chromatin-associated proteins interacting with MLV integrase are good candidates (12). Avian sarcoma and leukosis viruses (ASLV) display weak preferences for TUs but not for TSSs (5,13,14), and mouse mammary tumor virus integrates randomly across the host genome (15).
Only few studies describe non-random sets of integration sites with either transcriptionally active or silenced proviruses. For example, Lewinski et al. (16) separated cells infected with an HIV-based reporter vector into populations with stable provirus expression and with proviruses whose expression depended on the stimulation by TNFα. Both populations showed similar over-representation of integration sites in genes, but proviruses with TNFα–dependent activity were more frequently found in centromeric alphoid repeats, in long-intergenic regions, and in very highly expressed genes (16). Similarly, the transcriptional interference was observed in an in vitro model of HIV-1 latency where most latent proviruses integrated in introns of highly transcribed genes with a modest preference for the same orientation as the host gene (17). Second, proviruses in tumors induced by Rous sarcoma virus (RSV)-derived vectors (18) represent transcriptionally active copies and accumulated in TUs, CpG islands, and around TSSs. Most strikingly, almost all genic integrations were found in the genes expressed in multiple tissues, whereas tissue-specifically expressed genes were avoided. Both studies pointed to some chromosomal features promoting or repressing the integrated proviruses but exact analysis of individually characterized proviruses is lacking.
Transcriptional provirus silencing was described in many experimental settings and multiple suppressive mechanisms evolved probably as a protection from the deleterious outcomes of retrovirus infection and mobilization of endogenous retroviruses. For example, the zinc finger protein ZFP809 of the Kruppel-associated box (KRAB) family together with the transcriptional co-repressor KRAB-associated protein 1 (KAP-1/Trim28) bind in a sequence-specific manner the repressor-binding site present in the primer-binding site of MLV (19–21). This binding explains the potent silencing of MLV in murine embryonic stem cells (22,23) and the release of silencing of the murine stem cell virus, which evolved different primer-binding specificity (24,25).
The executive mechanisms of transcriptional silencing include proviral de novo DNA methylation and marking the provirus-associated nucleosomes by repressive histone modifications. DNA methylation of long terminal repeats (LTRs) was demonstrated to accompany the silenced MLV (26–29), Rous sarcoma virus (30), HIV-1 (31–33), HTLV-1 (34,35), and various families of human endogenous retroviruses (36–39). Furthermore, mutation of CpGs within the retroviral LTRs reduces provirus silencing (40), and insertion of a CpG island core sequence into or upstream to the 5′LTR is an efficient anti-silencing strategy (41,42). On the other hand, provirus silencing occurs even in cells deficient in de novo DNA methyltransferases Dnmt3a/b (29,43), and DNA methylation is dispensable for the silencing in embryonic stem cells (44). These facts point to the repressive histone marks as an alternative mechanism of provirus silencing. Particularly, di- or tri-methylation of the H3K9 by lysine methyltransferases G9a and Eset has been correlated with transcriptional repression of newly integrated and endogenous retroviruses (45–48). Recent siRNA-based knock-down screen identified a handful of epigenetic factors participating in a non-redundant silencing network in HeLa cells (49).
Taken together, the interplay of major suppressive factors in establishment and maintaining the silent provirus remains to be clarified. We suggest here that clonal analysis of the silencing of individual proviruses in context with their chromatin environment and chromosomal positions are urgently needed for this purpose. To better understand the role of de novo DNA methyltransferases in the silencing process, we compared the expression of individual proviruses in cells with intact or deleted DNA methyltransferase genes. In this study, we found that only a defined subset of provirus integrations is fully resistant to transcription silencing and prone to the long-term expression of transduced genes.
MATERIALS AND METHODS
Construction of the retrovirus vector
We constructed the pAG3 replication-defective reporter retrovirus vector by replacement of the gag, pol, and env genes in the replication-competent vector RCASBP(A) (50) with the GFP-coding sequence. pRACSBP(A) was amplified with primers RV3-ClaI(2) and RV3-R2 (Supplementary Table S1), which span from 3′UTR across the plasmid backbone to position +634 in the gag, and the product was self-ligated. The gag initiation ATG codon and the inner gag ATG codon 120 were destroyed by introduction of point mutations using the Transformer site-directed mutagenesis kit (Clontech). Mutagenesis was performed according to the manufacturer’s protocol with mutagenic primers mutATGgag and RV3-mTAG, and selection primers select PstI/SacII and select ScaI/BglII (Supplementary Table S1) for selection with PstI or ScaI restriction enzymes, respectively. The resulting construct pRV3 represents the vector backbone comprising ASLV LTRs and necessary packaging sequences. The linker from adaptor plasmid pCla12 (50) was cloned into the unique ClaI restriction site of the pRV3 vector. The EGFP coding sequence was then cloned from the plasmid pEGFP (Clontech) via XbaI restriction sites in the Cla12 linker and the resulting retroviral vector pAG3 was used for the virus propagation.
Cell culture and virus propagation
The packaging AviPack cell line (18) was maintained in D-MEM/F12 Eagle’s modified medium (Sigma) supplemented with 5% of newborn calf serum, 5% of fetal calf serum, 1% of chicken serum (all Gibco BRL), and penicillin/streptomycin (100 mg/ml each, Sigma) in a 3% CO2 atmosphere at 37°C. HCT116 human colorectal carcinoma cell line and its subclones with knock-outs of DNA methyltransferases HCT116 Dnmt1−/−, HCT116 Dnmt1−/− Dnmt3b−/−, HCT116 Dnmt3b−/−, and HCT116 Dnmt3a−/− Dnmt3b−/− (51–53) were obtained from Bert Vogelstein, Johns Hopkins University School of Medicine, Baltimore, Maryland, and maintained in the same conditions except for supplementation with chicken serum. The AviPack packaging system was utilized for the virus propagation and pseudo-typing with vesicular stomatitis virus protein G (VSV-G) as described in (18). Briefly, 107 AviPack cells plated on a 150 mm Petri dish were cultured and co-transfected with 50 µg of pAG3 and 10 µg of pVSV-G (Clontech) plasmids by calcium phosphate precipitation after 24 h. The fresh cultivation medium was supplemented with 100 mM glucose 24 h post-transfection (p.t.) and collected twice 48 h and 72 h p.t. Obtained viral stocks were clarified by centrifugation at 200 × g for 10 min at 4°C, supernatants were collected and centrifuged at 23 000 rpm for 150 min at 4°C in rotor SW28, Beckman Optima100 (Beckman). The pellet was resuspended in a culture medium containing 10% newborn calf serum, frozen, and stored in −80°C. Titration of the infectious virus stock was performed by its serial dilution and subsequent infection of DF-1 cells. Two days post-infection (p.i.), the number of GFP-positive cells or cell clusters was counted. The titrated stock was used for infection of HCT116 cells.
Infection and subcloning of HCT116 cells
We plated 106 cells of the wild-type (wt) HCT116 cell line and its DNA methyltransferase-deficient derivatives per 100 mm Petri dishes and infected them with the AG3 replication-deficient retroviral vector at multiplicity of infection (MOI) 0.02 24 h after plating. Virus AG3 was passed through 0.2 µm SFCA filter (Corning) and 600 µl of the suspension was applied per dish and allowed to adsorb for 40 min at room temperature. After adsorption, 12 ml of fresh medium was added and cells were cultured at 37°C and 3% CO2. Three to six days p.i., the percentage of GFP-positive cells was analyzed by flow cytometry, and GFP-positive cells were sorted in single-cell sort mode with FACSVantage SE (Becton–Dickinson) into 96-well tissue culture plates to obtain single-cell clones. Expanded clones were sub-cultured and percentages of GFP-positive cells were assessed in one-week intervals with the LSR II cytometer (Becton–Dickinson).
Over-expression of Dnmt3a and Dnmt3b
Vectors for ectopic over-expression of de novo DNA methyltransferases were created from pVitro expression construct (Invivogene) by replacement of the GFP coding sequence with tdTomato fluorescence marker in the first expression cassette to allow tracking of efficiently transfected cells. This vector was denoted pVitroT and was used as a mock-transfection control. The second expression cassette of the pVitroT was used for insertions of the particular de novo DNA methyltransferase coding sequence. The Dnmt3a and Dnmt3b molecular clones were obtained from IMAGE Consortium Library. The DNA methyltransferase-coding regions were amplified by Pfu DNA polymerase (Promega) with primers hDNMT3A-BglII-L and hDNMT3A-NheI-R (Dnmt3a) or hDNMT3B-BglII-L and hDNMT3B-NheI-TAG (Dnmt3b) (Supplementary Table S1). The PCR products were cloned into the pGEM-T Easy vector (Promega) and subsequently inserted into the pVitroT vector as BglII–NheI fragments. The resulting pVitro3AT and pVitro3BT plasmids were sequenced to exclude occurrence of point mutations. The transfection was performed with Fugene HD-transfection reagent (Roche) according to the manufacturer’s protocol. The cells were then cultivated for 7 days in order to manifest potential changes in provirus expression and DNA methylation pattern, and expression of GFP in the transfected tdTomato-positive cells was analyzed with the LSR II cytometer (Becton–Dickinson).
To quantify the level of Dnmt3a and Dnmt3b expression, the transfected wt HCT116 cell culture was harvested on day 4 p.t., and tdTomato-positive cells were sorted by FACSVantage SE (Becton–Dickinson). Total RNA was isolated from the collected cells using RNAzol RT (Molecular Research Center, Inc.). One microgram of the isolated RNA was treated for 20 min with Dnase I (New England Biolabs). Dnase I-treated RNA samples were subjected to reverse transcription using M-MLV reverse transcriptase (Promega) and oligo dT primers in 50 µl reaction volume. One microliter of the resulting cDNA was used for the triplicate qPCR using the MESA GREEN qPCR MasterMix Plus for SYBR Assay Kit (Eurogentec) in a total volume of 20 µl with 400 nM concentration of primers. We used the following primers designed against human DNA methyltransferases: hDNMT3a-FW and hDNMT3A-RV designed for exons 20–22 of human Dnmt3a, hDNMT3B-FW and hDNMT3b-RV designed for exons 19–20 of human Dnmt3b (Supplementary Table S1). The size of the PCR product was 200 bp in both cases. The RNA polymerase IIa (POLR2a) amplified with primers POLR2a-FW and POLR2a-RV (Supplementary Table S1) was used as a reference housekeeping gene. The calibration curves were set according to the amplification of cDNA of the following genes: Dnmt3a, Dnmt3b, and housekeeping gene POLR2a. These products were cloned into the pGEM-T Easy vector (Promega) and 10-fold diluted in the range 10–107 molecules per one RT reaction. PCR reactions were run for 40 cycles in the Chromo4 system for RT-PCR thermocycler (Bio-Rad) with an annealing temperature of 60°C. The reaction products were resolved on 2% agarose gel. The results were normalized to 105 molecules of POLR2a.
Reactivation of silenced proviruses by Dnmt and histone deacetylase (HDAC) inhibitors
Each clone was split into four wells and separately treated with reactivation agents. The culture medium was supplemented with 4 µM 5-azacytidine (5-azaC, Sigma) and 2 mM sodium butyrate (Sigma), alone or in combination. The inhibitor concentrations used for the reactivation were titrated and consequently, set as a compromise between reactivation efficiency and minimum toxicity. The clones were treated for 2 days and subsequently collected and analyzed by flow cytometry. Prolonged treatment did not lead to stronger reactivation but more distinctive cell toxicity.
Analysis of DNA methylation by bisulfite sequencing
The genomic DNA isolated by phenol–chloroform extraction from the infected cells was treated with sodium bisulfite using the EpiTect bisulfite kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. The nested PCR of the upper strand was performed with bisRV-LTR-LO, bisRV-LTR2-L, bisRV-LTR2-Router, and bisRV-LTR2-Rinner primers complementary to the U3 region of the ASLV LTR and the leader region encompassing all but one CpG within the LTR (Supplementary Table S1). PCR reactions were carried out with 200 ng of bisulfated DNA by 35 cycles of 95°C for 30 s, 58°C for 2 min, and 72°C for 90 s. The PCR products were cloned into the pGEM-T Easy vector (Promega) and sequenced by the universal pUC/M13 forward primer.
Cloning and sequencing of provirus integration sites
The provirus-cell DNA junction sequences were amplified using the splinkerette-PCR method (54). The genomic DNA was isolated by phenol–chloroform extraction from individual clones and cleaved with either of subsequent restriction enzymes Sau3AI, DpnII, or MseI. The restriction fragments were ligated overnight at 15°C with a 10-fold molar excess of adaptors formed by the annealing of HMspAa and HMspBb–Sau3AI or HMspBb–MseI oligonucleotides (Supplementary Table S1) complementary to the particular cleavage site of the enzyme used for the DNA digestion. The ligation products were subsequently cleaved with Bsu36I to destroy undesirable products of adaptor ligation to the 3′LTRs. The resulting mixture of fragments was then purified by phenol–chloroform extraction and used as a template for nested-PCR reaction with primers specific for the retrovirus LTR and the splinkerette adaptor (Supplementary Table S1). Primary PCR was performed with primers Splink1 and spPCR-AG3-R as follows: 94°C for 3 min, 2 cycles of 94°C 15 s, 68°C 30 s, 72°C 2 min and 31 cycles of 94°C 15 s, 62°C 30 s, 72°C 2 min, and final polymerization 72°C for 5 min. The secondary PCR used primers Splink2 and spinPCR-AG3-R with program setting: 94°C 3 min, 30 cycles 94°C 15 s, 60°C 30 s, 72°C 2 min, and final 72°C 5 min. The specific PCR products were sequenced and the resulting sequences adjacent to the 5′LTR were aligned to the Human Genome assembly version hg19.
Genomic analysis of provirus integration sites
All junction sequences containing the end of 5′LTR and the unique cellular DNA sequence at least 30 bp in length were used for more detailed analysis. The sequences of the integration sites were mapped onto the annotated human-genome assembly hg19 of February 2009 (GRCh37/hg19) using BLAT. The genes/transcription units hit by the provirus integration were identified according to the UCSC Genes track using the University of California at Santa Cruz browser available at http://genome.ucsc.edu/cgi-bin/hgGateway. The UCSC Genes track shows gene predictions based on the data from RefSeq, Genbank, CCDS, and UniProt. The distance of the integration site from the transcription start site was measured according to the SwitchGear Genomics Transcription Start Sites database. Identification of the CpG islands was done based on the GRCh37/hg19 assembly available at the UCSC Genome Browser. The H3K4me3 histone modification data of the HCT116 cell line obtained by ChIP-seq were produced by the ENCODE project at University of Washington and are accessible through the ENCODE June 2010 Freeze (http://genome.ucsc.edu/ENCODE/). The same source provided data on gene transcription level of targeted TUs (Affymetrix Exon Array, ENCODE/University of Washington).
RESULTS
De novo DNA methylation is required for efficient ASLV provirus silencing
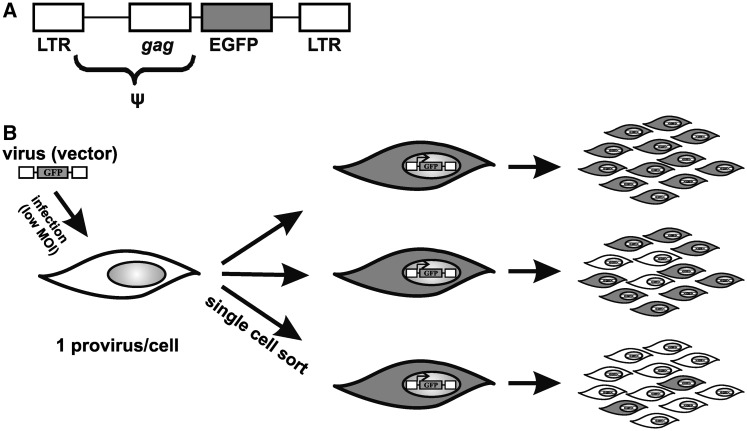
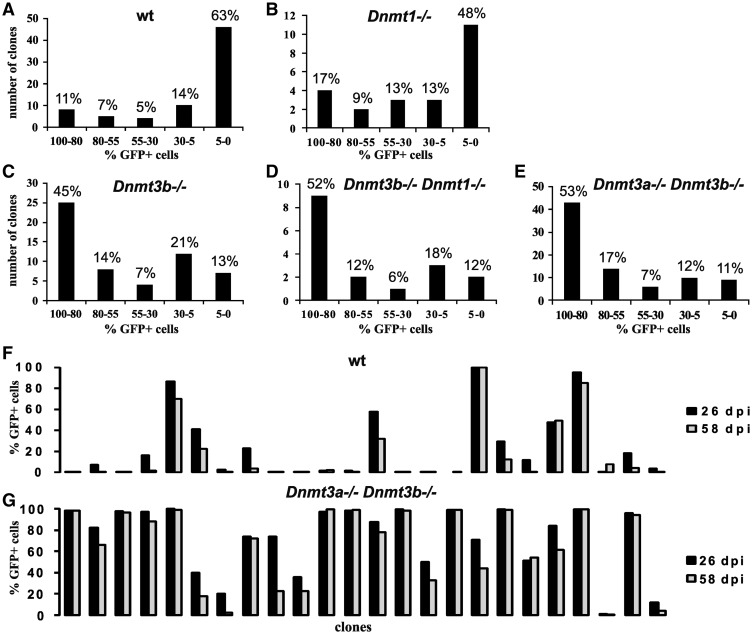
To examine the role of DNA methyltransferases in transcriptional repression of ASLV-derived vectors newly integrated in the human genome, we infected wt HCT116 tumor cells and, in parallel, their derivatives with single or combined knock-outs of Dnmt1, Dnmt3a, and Dnmt3b with an ASLV-based vector, AG3. The AG3 vector transduces green-fluorescent protein (GFP) driven by ASLV LTR (Figure 1A). AG3 is replication-deficient, which, together with very low MOI, ensures that each infected cell contains only one provirus integrated in a distinct site of the host genome. Three to six days p.i., the GFP-positive cell clones were single-cell sorted by flow cytometry, and single cell clones representing individual sites of provirus integration were established and expanded. In this way, we omit the proviruses that have already been silenced immediately after integration. We isolated 73, 23, 56, 17, and 82 clones of GFP-positive wt HCT116, HCT116 Dnmt1−/−, HCT116 Dnmt3b−/−, HCT116 Dnmt1−/− Dnmt3b−/−, and HCT116 Dnmt3a−/− Dnmt3b−/−, respectively, and followed the stability of the provirus expression for up to 4 months (Figure 1B). We observed a striking difference in provirus silencing between the wt and Dnmt1−/− HCT116 cells on the one hand and de novo methyltransferase-deficient HCT116 cells on the other hand (Figure 2). In wt HCT116 cells, we found 46 out of 73 clones strongly silenced with 0–5% of GFP-positive cells and only eight clones displaying no or very weak silencing with 80–100% of GFP-positive cells 60 days p.i. (Figure 2A). Similarly, the majority of clones were strongly silenced in HCT116 Dnmt1−/− cells (Figure 2B). In contrast, among the clones of de novo DNA methyltransferase-deficient cells, about half of the clones exhibited weak or zero silencing and only rare clones displayed strong silencing with 0–5% of GFP-positive cells 60 days p.i. (Figure 2C–E). The dynamics of silencing is shown by percentages of GFP-positive cells in a representative subset of clones derived from wt HCT116 and HCT116 Dnmt3a−/− Dnmt3b−/− cells at the end of fourth and eighth week p.i. (Figure 2F and G). The vast majority of wt HCT116 clones were largely silenced already in the fourth week p.i. and only rare clones retained the GFP expression unaffected. In HCT116 Dnmt3a−/− Dnmt3b−/− cells, there were numerous clones with a stably high percentage of GFP-positive cells and no detectable progress to silencing. Clones subjected to a certain degree of silencing represented approximately one half of the clones. We conclude that de novo DNA methyltransferase activity is important for efficient provirus silencing and the absence of Dnmt3b alone and especially in combination with Dnmt3a increases the probability of long-term and unsilenced provirus expression. The absence of maintenance methyltransferase Dnmt1 did not significantly alleviate provirus silencing. In any case, exceptional clones keep stable provirus expression even in the presence of de novo DNA methyltransferases and, vice versa, multiple clones tend to the silencing even in their absence. This behavior might be caused by genomic and epigenomic features of the respective sites of proviral integration.
Figure 1.
Experimental schema. (A) Schematic representation of the AG3 retrovirus vector. (B) Schema of HCT116 cell infection, establishment of single-cell clones and clonal analysis of provirus silencing. White and grey cells represent the GFP-negative and GFP-positive cells, respectively, and ψ, encapsidation signal.
Figure 2.
Clonal analysis of AG3 provirus silencing. Individual clones of infected GFP-positive wt HCT116 (A), HCT116 Dnmt1−/− (B), HCT116 Dnmt3b−/− (C), HCT116 Dnmt1−/− Dnmt3b−/− (D), and HCT116 Dnmt3a−/− Dnmt3b−/− (E) cells were examined for the provirus silencing 60 days p.i. and subdivided into five categories according to the percentage of GFP-positive cells. The categories are defined arbitrarily and the interval 0–5% means extremely strong silencing, whereas 80–100% means zero or very weak silencing. Data are presented as percentages of clones falling into the defined categories. Stability in time of the provirus expression in individual clones of wt HCT116 (F) and HCT116 Dnmt3a−/− Dnmt3b−/− (G) is shown as the percentage of GFP-positive cells 26 (black columns) and 58 (gray columns) days p.i. Representative sets of clones are shown.
Rescue of the silencing by expression of de novo DNA methyltransferases
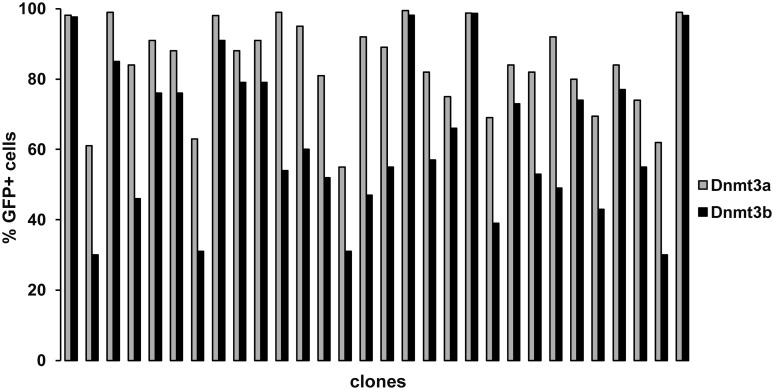
To confirm that the absence of de novo DNA methyltransferases is specifically responsible for the inefficient provirus silencing, we conducted a rescue experiment with ectopic over-expression of cloned human Dnmt3a or Dnmt3b. Clones of the HCT116 Dnmt3a−/− Dnmt3b−/−cells with stable and non-silenced provirus expression were separately transfected with vectors pVitro3AT and pVitro3BT expressing the human DNA methyltransferases Dnmt3a and Dnmt3b, respectively, and the control vector pVitroT. The levels of Dnmt3a and Dnmt3b ectopic expression were comparable as assayed by quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR) in an independent transfection experiment (data not shown). Seven days p.t. of the de novo DNA methyltransferase constructs, the majority of clones were silenced at least to some extent and the percentage of GFP-positive cells dropped substantially in multiple clones (Figure 3). The vast majority of clones exhibited more extensive loss of GFP-positive cells after the Dnmt3b transfection or, rarely, the effect of Dnmt3b and Dnmt3a was equal (Figure 3). Although the differences in rescue efficiency between Dnmt3b and Dnmt3a are small in several clones, the uniformity of the trend suggests that Dnmt3b is a more potent silencer of proviruses. Only a small fraction of clones were resistant to the de novo DNA methyltransferase over-expression with not affected percentage of GFP-positive cells (Figure 3). The frequency of these Dnmt3a/b-resistant clones corresponded approximately to the frequency of stable clones in the wt HCT116 cells. We therefore tested the Dnmt3a/b resistance in the stable clones of wt HCT116 cells in an analogous rescue experiment. All seven tested stable clones of wt HCT116 cells turned out to be resistant to Dnmt3a/b over-expression (data not shown), whereas clones with weak and slow silencing were sensitive and displayed substantial loss of GFP expression upon Dnmt3a/b transfection (Supplementary Figure S1). This suggests again that this stability of provirus expression even in the presence of de novo DNA methyltransferases is a result of integration into specific target sites, and the probability of such integration is the same in wt and de novo DNA methyltransferase-deficient HCT116 cells.
Figure 3.
Rescue of provirus silencing by ectopic over-expression of Dnmt3a and Dnmt3b DNA methyltransferases. Representative HCT116 Dnmt3a−/− Dnmt3b−/− cell clones with stable and non-silenced provirus expression were transfected with vectors expressing either Dnmt3a (gray bars) or Dnmt3b (black bars) and the percentage of GFP-positive cells was measured by flow cytometry 7 days p.t. The percentage of GFP-positive cells in mock-transfected clones remained at 97–100% of all cells (not shown). A representative set of clones is shown.
CpG methylation-independent silencing in de novo DNA methyltransferase-deficient cells
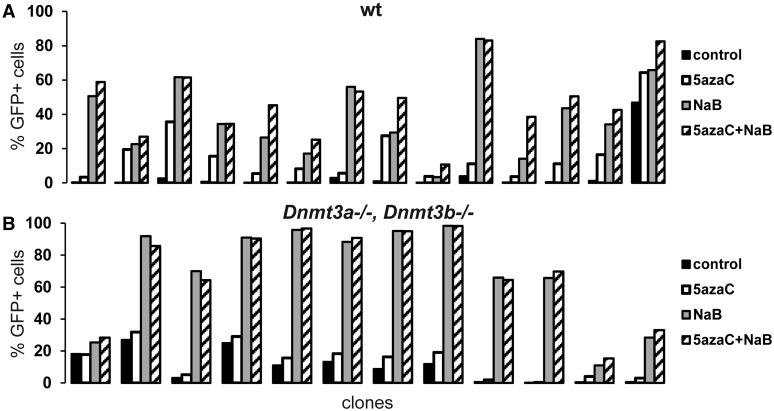
As the cells lacking Dnmt3a or Dnmt3b exhibited limited but still detectable provirus silencing, we examined the mechanism of this transcriptional suppression. First, we used 5-azaC and sodium butyrate, inhibitors of DNA methyltransferases and HDAC, respectively, to reactivate the GFP expression in clones of wt HCT116 and HCT116 Dnmt3a−/− Dnmt3b−/− cells with silenced proviruses. The silenced proviruses in the wt HCT116 cells were partially reactivated by the treatment with either 5-azaC or sodium butyrate (Figure 4A). The effect of sodium butyrate was more profound, and in combination of both drugs, the additive effect was observed in multiple clones. We observed quite a different situation in the silenced HCT116 Dnmt3a−/− Dnmt3b−/− clones (Figure 4B). The effect of 5-azaC was marginal, without any addition to that of sodium butyrate. In comparison, sodium butyrate alone reactivated provirus expression efficiently.
Figure 4.
Reactivation of silenced proviruses in clones treated with inhibitors of DNA methyltransferases and HDAC. (A) wt HCT116 cell clones, (B) HCT116 Dnmt3a−/− Dnmt3b−/− cell clones. Representative selection of clones were treated with 5-azaC (white columns), sodium butyrate (gray columns), or a combination of both (dashed columns) and the percentage of GFP-positive cells was measured by flow cytometry after 2 days of the treatment. Black columns represent the percentages of GFP-positive cells in mock-treated cells.
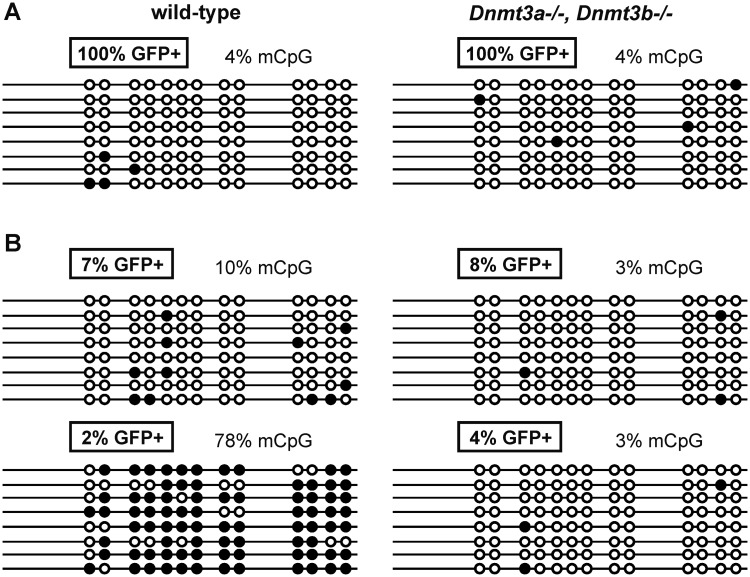
Secondly, we examined the DNA methylation status of the promoter region of either active or silenced proviruses in the wt HCT116, HCT116 Dnmt3b−/−, and HCT116 Dnmt3a−/− Dnmt3b−/− cells. The region analyzed by the bisulphite sequencing spans from the 5′end of the 5′LTR to the position +200 in the leader sequence. Three representative examples of 5′LTR CpG methylation are shown in Figure 5. All 17 active proviruses tested showed unmethylated or only sporadically methylated 5′LTRs in wt as well as DNA methyltransferase-deficient cells (Figure 5A). In the majority (9 of 14) of silenced clones of wt HCT116 cells, the proviral 5′LTRs were heavily methylated (Figure 5B). This abundance of heavily methylated 5′LTRs slightly dropped among the silenced proviruses in the HCT116 Dnmt3b−/− cells (not shown). In contrast, we did not find any significantly methylated provirus in the silenced clones of HCT116 Dnmt3a−/− Dnmt3b−/− cells (Figure 5B) and all proviruses integrated in these cells were unmethylated regardless of their transcriptional state.
Figure 5.
CpG methylation status of the 5′LTRs in wt HCT116 and HCT116 Dnmt3a−/− Dnmt3b−/− clones. Representative cell clones with non-silenced (A) and silenced (B) proviruses were chosen and CpG methylation was investigated by bisulfite sequencing. Methylated CpG dinucleotides are indicated by solid circles, non-methylated CpGs are indicated by open circles. Numbers indicate the percentages of GFP-positive cells in particular cell clones (boxed numbers) and the percentages of methylated CpG dinucleotides.
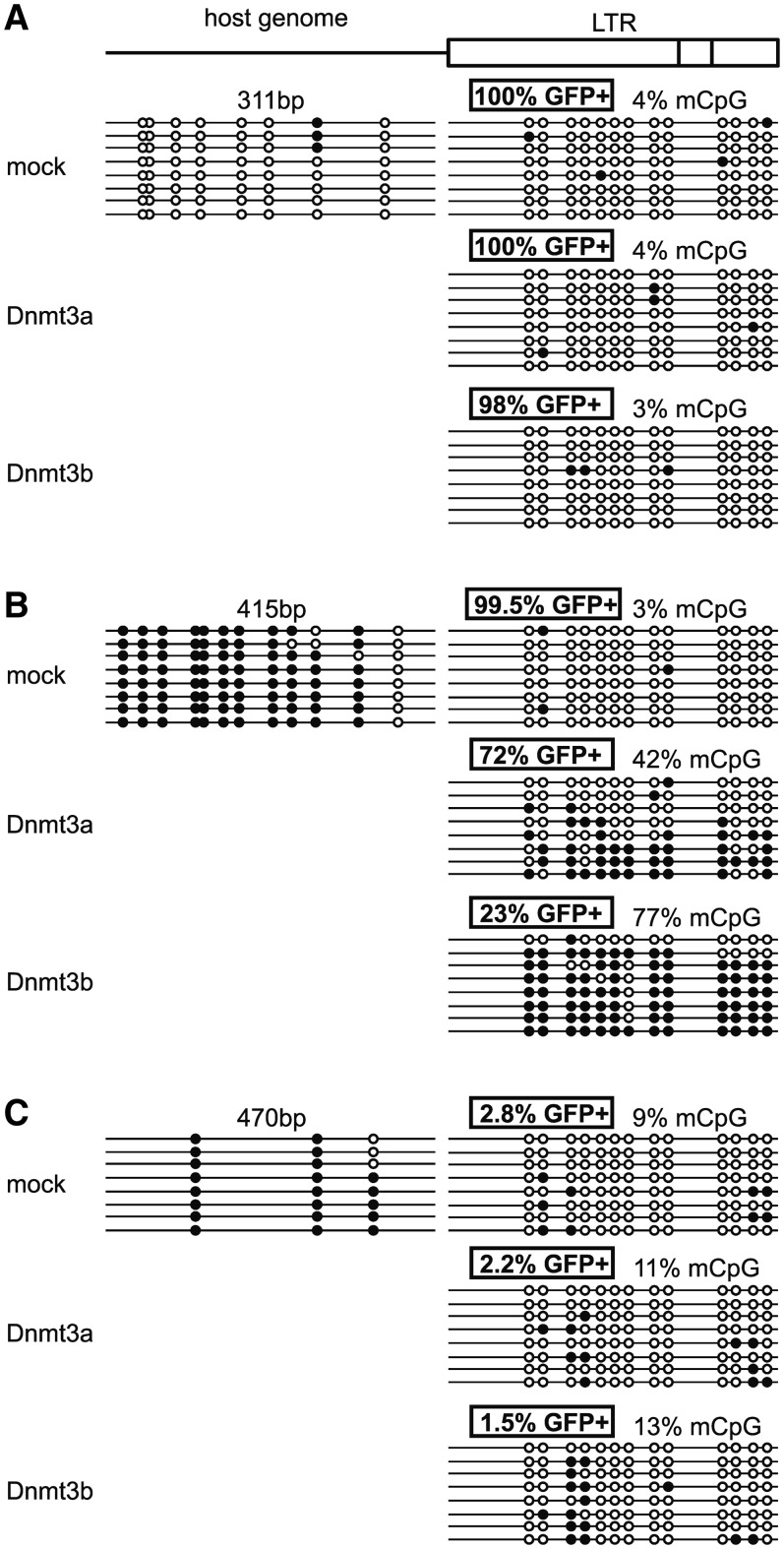
We analyzed the DNA methylation status of proviral LTRs in several clones subjected to the silencing rescue experiments. The analysis revealed that the loss of the GFP expression upon Dnmt3a/b transfection was accompanied by heavy DNA methylation of the proviral 5′LTRs. The extent of the LTR CpG methylation correlated with the loss of GFP-positive cells. The over-expression of Dnmt3a led to a lower level of DNA methylation in comparison with Dnmt3b, which corresponded to the different efficiency of de novo DNA methyltransferases in the silencing rescue experiment (for details see the chapter ‘The DNA methyltransferase-sensitive proviruses are integrated in the regions of methylated DNA’ and Figure 8).
Figure 8.
DNA methylation of the provirus and adjacent host cell sequences. CpG methylation status of the 5′LTRs and 300–600 bp of the genomic DNA upstream to the 5′LTR (not in scale) in representative clones of HCT116 Dnmt3a−/− Dnmt3b−/− cells with stable (A), stable but de novo Dnmt-sensitive (conditionally stable) (B), and silenced (C) provirus expression. CpG methylation was investigated by bisulfite sequencing after transfection of the empty vector (mock) or vectors expressing either Dnmt3a or Dnmt3b. CpG methylation status of the genomic DNA upstream to the 5′LTR is indicated only in mock-transfected cells. Methylated CpG dinucleotides are indicated by solid circles, non-methylated CpGs are indicated by open circles. Numbers indicate the percentages of GFP-positive cells in particular cell clones and the percentages of methylated CpG dinucleotides. The length of the integration site analyzed is indicated.
Epigenomic features of integration sites permissive for the stable proviral expression
The low-frequency occurrence of clones with stable and Dnmt3a/b-resistant provirus expression, as well as our previous analysis of retrovirus integration sites in virus-induced sarcomas (18), suggested the importance of the integration site for either silencing or maintenance of provirus expression. We, therefore, characterized the integration sites of the proviruses from clones with stable versus silenced proviral expression by the splinkerette-PCR technique and BLATing against the human genomic assembly GRCh37, version hg19. For the integration site analysis, we selected clones with stable GFP expression (95–100% GFP-positive-cells), mostly from the sets of clones described in Figure 2A and E but also from an additional set of clones derived from pre-selected GFP-positive cells. In total, we obtained 113 unambiguously mapped provirus integration sites from wt HCT116 and HCT116 Dnmt3a−/− Dnmt3b−/− cells (Table 1). The whole data set of integration sites together with characteristics of respective cell clones is given in Supplementary Table S2. Targeting the annotated human genes, either untranslated regions, intronic, or exonic parts, according to the USCS Genes track was regarded as integration into TU.
Table 1.
Overview of clones with characterized sites of provirus integration subdivided according to the cell line, stable provirus expression versus silencing, and localization in or outside TUs
| Cell line | Provirus expression | No. of genic insertions | No. of intergenic insertions |
|---|---|---|---|
| wt HCT116 | Stable | 7 | 0 |
| wt HCT116 | Silenced | 10 | 6 |
| HCT116 Dnmt3a−/− Dnmt3b−/− | Stable | 61a | 1a |
| HCT116 Dnmt3a−/− Dnmt3b−/− | Silenced | 8 | 20 |
aIn addition to the 33 clones with 95–100% of GFP-positive cells from the experiment described in Figure 2E, further 29 independent clones obtained from pre-selected GFP-positive HCT116 Dnmt3a−/− Dnmt3b−/− cells were included into the integration site analysis.
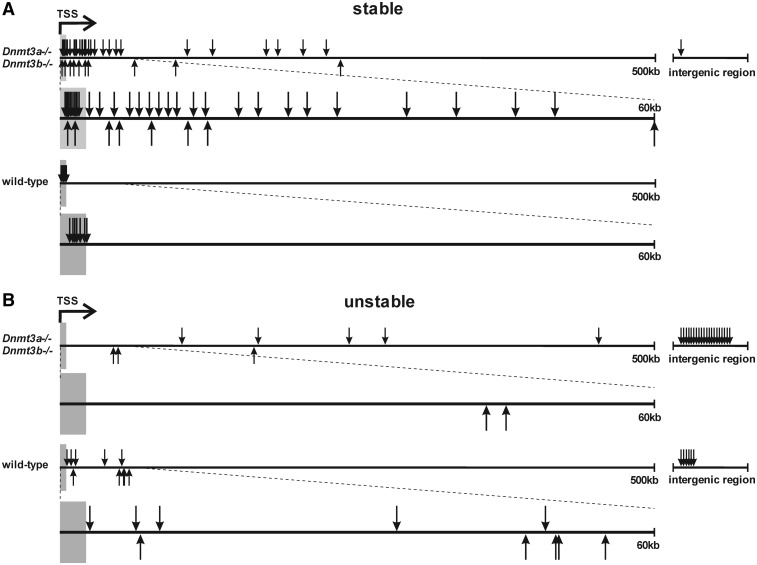
The proviruses with stable expression were found to be integrated almost exclusively into TUs in both wt HCT116 and HCT16 Dnmt3a−/− Dnmt3b−/− cells (Table 1). However, the distribution of these integration sites along the whole TUs differed between the wt HCT116 and HCT116 Dnmt3a−/− Dnmt3b−/− cells (Figure 6). The seven stable proviruses in the wt HCT116 cells were uniformly found close, not more than 2.7 kb downstream, to the TSSs. Even more striking was the absolute overlap of these integration sites with the regions enriched in lysine 4 trimethylation of histone 3 (H3K4me3) as identified in the ENCODE project database. This chromatin modification is a hallmark of regions proximal to TSSs of transcriptionally active genes. In contrast, the stably expressed proviruses in the HCT116 Dnmt3a−/− Dnmt3b−/− cells were distributed throughout the whole TUs without significant accumulation in the vicinity of the TSSs. Interestingly, integrations matching with the H3K4me3-rich regions of their respective genes corresponded to the proviruses resistant to silencing rescue by Dnmt3a/b over-expression. An example of a TU targeted by provirus integration is given in Supplementary Figure S2.
Figure 6.
Distribution of provirus integration sites along the targeted transcription units. Positions of provirus integration sites in cell clones with stable (non-silenced) provirus expression (A) and cell clones with unstable (silenced) provirus expression (B) in wt HCT116 and HCT116 Dnmt3a−/− Dnmt3b−/− cells are shown as vertical arrows in the absolute distance from the TSS up to 500 kbp. Proviruses proximal to the TSSs are shown in the enlarged 60 kbp regions below. The numbers of intergenic integrations are shown out of scale. Downward arrows, proviruses integrated in the same orientation as transcription of the targeted gene; upward arrows, proviruses integrated in antisense orientation. Grey areas, the maximum range of the H3K4me3-rich region.
The distribution of integration sites in clones with unstable (silenced) provirus expression also differed between wt HCT116 and HCT116 Dnmt3a−/− Dnmt3b−/− cells. In the wt HCT116 clones, integration sites were found either within or outside of TUs. When in TUs, the integrations were found throughout the TUs except for the proximal gene regions enriched in H3K4me3. In HCT116 Dnmt3a−/− Dnmt3b−/− cells, the majority of silenced proviruses were detected outside of TUs and the intra-genic insertions were rare, confined to distal regions of extremely large TUs, 43–450 kb from the TSS. Of note, the provirus silencing was less efficient here. Moreover, the unstable proviruses closer than 100 kb to the TSS were in the antisense orientation to the transcription of the respective gene (Figure 6).
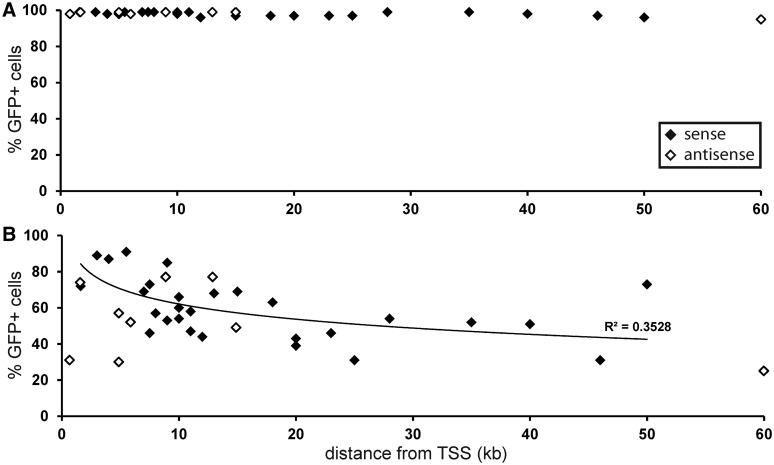
The protective effect of TSS and the associated H3K4me3-rich region for the maintenance of long-term provirus expression was further highlighted in silencing rescue experiment (Figure 7). Proviruses integrated into genes in the HCT116 Dnmt3a−/− Dnmt3b−/− cells are stably expressed with no or only a negligible level of silencing even as far as 60 kb from the respective TSS (Figure 7A). Upon the Dnmt3b over-expression, however, the sensitivity of provirus expression to the Dnmt3b increased with the distance from the TSS, most strikingly within the first 20 kb (Figure 7B). Another factor affecting the silencing can be the provirus orientation in relation to the transcription of the targeted gene. This is apparent from the distribution of proviruses integrated within genes. Among the silenced proviruses, both in wt HCT116 and in HCT116 Dnmt3a−/− Dnmt3b−/− cells, there were approximately the same numbers of sense and anti-sense integrations. The stable proviruses, however, tended to be in sense, particularly in wt HCT116 cells (Figure 6). The proviruses integrated in anti-sense orientation were more susceptible to silencing even in shorter distance from the TSS (Figure 7B).
Figure 7.
Relation between the distance of provirus integration from TSS and the sensitivity to Dnmt3b over-expression. The level of silencing upon the Dnmt3b over-expression is shown in clones of HCT116 Dnmt3a−/− Dnmt3b−/− cells. Cell clones with proviruses integrated in TUs outside of regions enriched in H3K4 trimethylation, whose expression is silenced upon Dnmt3b ectopic expression (conditionally stable). The percentages of GFP-positive cells in cell clones 7 days after mock (A) and Dnmt3b (B) over-expression (y axis) and the distances between the TSSs of targeted genes and the proximal proviral LTR (x axis). Filled diamonds represent proviruses integrated in the same orientation as the transcription of the targeted gene; Open diamonds represent proviruses integrated in antisense orientation. The trendline was calculated from the distribution of proviruses integrated in sense orientation.
Altogether, provirus expression and silencing are interconnected with transcription of the targeted genes and the H3K4me3-enriched regions are of particular importance for the protection from DNA methyltransferase-dependent silencing.
The DNA methyltransferase-sensitive proviruses are integrated in the regions of methylated DNA
The aforementioned data raised the question about the interplay between the DNA methylation at the site of provirus integration and de novo DNA methylation of proviral regulatory sequences. There is a possibility that provirus integration into hypermethylated regions can determine the de novo DNA methylation and transcriptional silencing of the provirus. In order to answer this question, we analyzed the DNA methylation status within 300–600 bp of the genomic DNA adjacent to the proviral 5′LTR in a number of representative clones of HCT116 Dnmt3a−/− Dnmt3b−/− cells. We found that stably expressed and de novo DNA methyltransferase-resistant proviruses are integrated in unmethylated genomic DNA (Figure 8A, Supplementary Figure S3). In contrast, the silenced proviruses and active proviruses sensitive to the Dnmt3a/b over-expression (conditionally stable) are integrated in hypermethylated DNA regions (Figure 8B and C). The over-expression of Dnmt3a/b in conditionally stable clones resulted in the expansion of surrounding methylation patterns into the proviral LTR promoter (Figure 8B, Supplementary Figure S4), which was accompanied by the loss of provirus expression. However, the conditionally stable and silenced proviruses integrated in intergenic regions were not methylated with the same efficiency. The conditionally stable and rare silenced intra-genic proviruses were very efficiently methylated upon Dnmt3a/b ectopic expression. Dnmt3b appeared to serve as a more efficient methylation effector than Dnmt3a, which corresponded to the differences in the silencing efficiency. The silenced proviruses in intergenic regions were methylated with very low efficiency (Figure 8C, Supplementary Figure S5).
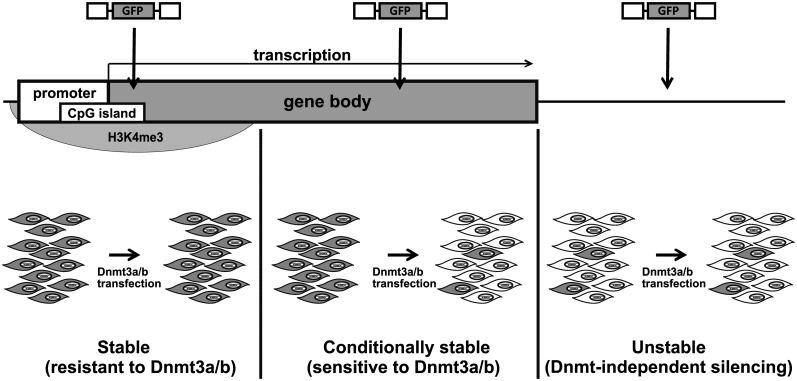
Based on the data, we suggest a model where retroviruses integrated to the close vicinity of transcriptionally active cellular promoters have the potential for absolutely stable expression. Outside of such regions, the proviruses are subjected to Dnmt3a/b-dependent (proviruses in transcribed genomic regions) or Dnmt3a/b-independent (predominantly proviruses integrated outside of genes) silencing (Figure 9).
Figure 9.
Expression of proviruses integrated in different genomic localizations, an integrative model. Proviruses integrated close to the TSS within the H3K4 trimethylation region are stably expressed and insensitive to over-expression of de novo Dnmt (left). Proviruses integrated within the gene bodies outside of the H3K4me3 regions are stably expressed in HCT116 Dnmt3a−/− Dnmt3b−/− cells but silenced after ectopic expression of either Dnmt3a or Dnmt3b (conditionally stable expression; middle). Intergenic insertions result in rapid silencing of proviral expression, which is independent of de novo Dnmts (right). Provirus expression is indicated by a picture with gray and white cells.
DISCUSSION
CpG methylation of provirus DNA and repressive histone methylation of associated nucleosomes are well-established as epigenetic mechanisms inhibiting retroviral expression at the level of transcription and leading to variegation and provirus silencing. Neither of these branches can satisfactorily explain all aspects of provirus silencing, although there are experimental settings where histone methyltransferases mediate silencing independently of DNA methyltransferases and vice versa. We demonstrate that provirus silencing occurs in the context of flanking cellular DNA, and both activating and suppressive influences of the flanking chromatin features must be considered. We present the first analysis of provirus silencing in single-cell clones with characterized chromosomal positions of proviruses. Furthermore, integration into genomes of cells deficient or proficient in de novo DNA methyltransferases provided information about the involvement of DNA methylation in retrovirus silencing at certain genomic positions. We found that retrovirus integration into TUs close to the TSSs and within the regions enriched in H3K4me3 permitted long-term unsilenced provirus expression and protected the provirus regulatory sequences from CpG methylation even under Dnmt3a/b over-expression. Proviruses integrated into the transcribed parts of genes outside of H3K4me3 regions were silenced by DNA hypermethylation of LTRs, whereas proviruses inserted in intergenic regions were efficiently silenced without accumulation of methylated CpGs.
Our analysis confirmed the significance of de novo DNA methylation for the retrovirus silencing because the absence of Dnmt1 did not lead to any significant silencing defect and the silencing was comparable in HCT116 Dnmt3b−/− and HCT116 Dnmt1−/− Dnmt3b−/− cell lines. De novo DNA methylation, however, is not inevitably necessary for provirus silencing. Proviruses integrated in intergenic regions or extremely far from TSSs in long TUs remain silenced even in HCT116 Dnmt3a−/− Dnmt3b−/− cells, and intergenic proviral insertions are not CpG methylated by ectopically expressed Dnmt3b or Dnmt3a. The comparison of provirus silencing in HCT116 Dnmt3b−/−, HCT116 Dnmt3a−/− Dnmt3b−/−, and HCT116 Dnmt1−/− Dnmt3b−/− cell lines also excluded the influence of overall genome methylation and the probability of proviral integration into densely methylated host cell DNA. These cell lines contain 97, 80, and <5%, respectively, of total genomic methylation of wt HCT116 (51) but reached similar efficiencies of the provirus silencing. Because the single knock-out of Dnmt3a was not available, we can only speculate about its silencing phenotype. Dnmt3a was reported as a potent provirus silencer in mouse embryonic stem cells (48). In HCT116 Dnmt3a−/− Dnmt3b−/− cells, however, the absence of Dnmt3a meant only a slight additional decrease in silencing efficiency in comparison with the knock-out of Dnmt3b alone and Dnmt3a scored weaker than Dnmt3b in silencing rescue experiments (Figure 3). This difference can be explained by the low Dnmt3a expression in the wt HCT116 cell line (51), lower DNA methyltransferase activity of Dnmt3a in comparison with Dnmt3b (55), and the dependence of Dnmt3a on the guidance and stimulation by Dnmt3L (56), which is not expressed in HCT116 cells.
The main finding of our study is that proviruses integrated close to the TSSs within the H3K4me3-enriched regions remain stably expressed and cannot be silenced even in cells with artificially increased expression of Dnmt3a or Dnmt3b. H3K4 trimethylation marks the 5′ parts of transcriptionally active or at least poised genes and usually forms broader surroundings of CpG islands and polymerase II-enriched regions (57,58). Mechanistically, at least Dnmt3a was shown to prefer non-methylated H3K4 under the guidance by Dnmt3L (59) and being expelled from H3K4me3 (60). In wt HCT116 cells, stably expressed proviruses were integrated exclusively in H3K4me3-enriched regions, whereas the silenced proviruses were distributed in quite opposite way, in the rest of gene bodies and in intergenic regions (Figure 6). We suggest that integrations in gene bodies normally result in provirus silencing because of increased levels of H3K36me3, which recruits de novo DNA methyltransferases (61). However, this control is leaky in de novo methyltransferase-deficient cells and in HCT116 Dnmt3a−/− Dnmt3b−/− cells, stable provirus expression is also permitted more distantly in the gene bodies outside the H3K4me3-rich regions. Accordingly, silenced proviruses in these cells were integrated almost exclusively outside of TUs and rarely scattered in distant parts of extremely large TUs 43–440 kb from the TSSs. We conclude that intergenic regions are for the most part non-permissive to the stable ASLV provirus expression and this non-permissiveness is independent of DNA methyltransferases.
The exceptional character of integrations into the H3K4me3 regions was even underlined by ectopic over-expression of Dnmt3a/3 b because these proviruses in wt HCT116 cells kept their stability in these artificial conditions. The same treatment of stable clones isolated from HCT116 Dnmt3a−/− Dnmt3b−/− cells and HCT116 Dnmt3b−/− cells led in most cases to the rescue of silencing with the exception of few resistant clones (Figure 3). The frequency of these clones was comparable with stable clones in the wt HCT116 cells and they all harbored proviruses integrated into the H3K4me3 regions. These results clearly show that the H3K4me3 environment permits autonomous expression of newly introduced DNA sequences and protects them from epigenetic silencing. The silencing in other genomic positions causes that only insertions into the H3K4me3 regions are observed when selection for the stable proviral expression is applied in cells with normal de novo DNA methyltransferase composition.
Stable expression of proviruses integrated close to the TSSs associated with CpG islands is not surprising. CpG islands were shown to protect adjacent promoters from DNA methylation (62) and this capacity has already been employed in design of a silencing-resistant and DNA methylation refractory retroviral vector (41,42). However, the protective effects do not extend far towards the bodies of active genes, which are enriched in H3K36me3 and DNA methylation (63,64). The efficiency of the silencing rescue after Dnmt3a/b over-expression increased with the distance from the TSS (Figure 7B). The functional dependence best fits the geometric distribution with variance probably produced by variable promoter strength, variable chromatin structure at exon–intron junctions, etc. The general decline of pro-transcriptional histone modification along the gene bodies was shown, e.g. for the lateral H3K79me2 (65). Proviruses transcribed in antisense orientation to the host gene tended to be more sensitive to de novo DNA methyltransferases and were not included in calculation of the trend line. This situation resembles the regulation of many imprinted loci, where the increase of DNA and H3K27 methylation and the decrease of H3K4 methylation are guided by non-coding antisense transcripts of imprinting centers (66).
Our analysis of DNA methylation found all proviruses unmethylated in HCT116 Dnmt3a−/− Dnmt3b−/− cells regardless of their expression status. This is convincing evidence that provirus silencing can be established and maintained even without DNA methylation. The stably expressed proviruses in H3K4me3-enriched regions appeared to be enclosed by unmethylated CpGs and this hypomethylated state did not change even after over-expression of Dnmt3a/3 b, evidencing the resistance of H3K4me3-enriched regions (59). In striking contrast, proviruses integrated in gene bodies keep unmethylated LTRs surrounded by highly methylated DNA sequences in HCT116 Dnmt3a−/− Dnmt3b−/− cells. CpG methylation of DNA within gene bodies must be maintained by Dnmt1 as it survives even in the double knock-out of Dnmt3a/3 b. After Dnmt3a/3 b ectopic expression, LTRs of proviruses integrated in gene bodies adopt dense CpG methylation, which positively correlates with the level of provirus silencing. The highly efficient methylation of provirus DNA in actively transcribed genes implicates a model where histone methyltransferase HYPB/Setd2 interacts with the processive RNA polymerase II and co-transcriptionally methylates H3K36 in the gene body together with proviral LTR promoters (67–69). The H3K36 trimethylation subsequently serves as a signal for de novo DNA methylation and thus provirus transcriptional silencing (61). In HCT116 Dnmt3a−/− Dnmt3b−/− cells, the DNA methylation cannot be adjusted to the local epigenetic environment. According to this hypothesis, the localization in the bodies of actively transcribed genes exposes the integrated provirus to repressive epigenetic environment and pre-determines subsequent DNA methyltransferase-dependent suppression. The intergenic provirus insertions are silenced in all cell lines independent of DNA methylation, and the silencing is highly probably driven solely by the repressive histone marks. The flanking DNA sequences are almost fully methylated, but the density of CpGs is low in intergenic regions. Actually, we found two unsilenced but Dnmt3a/3 b-sensitive proviruses outside of TUs but close to an active gene terminus. Both proviruses were found to be methylated upon Dnmt3a/3 b expression. Their proximity of 0.5 and 1.5 kb to the gene terminus enables the read-through from adjacent genes, as confirmed by the ENCODE Exon Array data of HCT116 cells, and the passing transcription complex could start the H3K36me3-dependent DNA methylation. The available ChIP-seq data detect the RNA polymerase II and H3K36me3 modification in such regions. Proviruses integrated closely upstream to active promoters were found to be transcriptionally silent but were not efficiently methylated after Dnmt3a/b over-expression.
In conclusion, we propose a model of the provirus transcriptional crosstalk with surrounding chromatin at the site of integration, where the long-term provirus expression or gradual provirus silencing are to a great part pre-determined by local epigenomic features (Figure 9). Proviruses integrated within the H3K4me3-enriched regions connected with promoters of active, mostly housekeeping genes keep their transcription activity and cannot be efficiently silenced by DNA methylation. Proviruses integrated in the bodies of transcribed genes are silenced, but their silencing depends on the de novo DNA methylation capacity of the host cell. Proviruses integrated in intergenic regions are strongly silenced in a DNA methylation-independent way. Provirus silencing is a general phenomenon; nevertheless, two extraordinary aspects of our study should be considered in the future. First, the speed and extent of silencing are species-specific and the validity of our model based on ASLV-derived vector should be further tested with various retroviral groups in different cell types. ASLVs are susceptible to efficient silencing and CpG methylation in mammalian cells (30,70–72), which together with an almost random integration into the host genome makes them an ideal model for the study of retrovirus silencing at various chromosomal loci. For HIV-1-derived lentiviral vectors, the provirus silencing was described as well (73,74) despite the complex transcriptional regulation and the presence of Sp1 sites in HIV-1 LTR. The phenomenon of HIV-1 persistence in transcriptionally latent state further underlines the importance of epigenetic silencing in the course of retrovirus infection (75,76). In our preliminary experiments, MLV-derived vectors in HCT116 cells are less susceptible to the provirus silencing (data not shown), probably due to their integration preference for TSSs (5). We assume that the epigenomic pre-determination of provirus silencing will be weaker for MLV and HIV-1 in mammalian cells and also for ASLV in permissive avian cells. Another aspect of our study, to be considered, is the early silencing occurring in the process of or immediately after provirus integration when the DNA lesion triggers an extensive chromatin response at the site of integration. We sorted the GFP-positive cells several days p.i. assuming that many proviruses had already been silenced at that time. The proportion of ab initio silenced proviruses cannot be determined in our experimental setup, but it was previously estimated to be ca. 80% for HIV-1-based vectors in human T cells (77). Our findings provide a valuable contribution to the retrovirus-mediated gene therapy concerning the efficiency, long-term effects, and safety issues of retrovirus integration. It becomes clear that efficient retroviral vectors for gene transfer require specific protective modifications averting the mostly repressive influence of the surrounding chromatin.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–2 and Supplementary Figures 1–5.
FUNDING
The Czech Science Foundation (grants Nos. 301/09/P667 and P502/11/2207) and Academy of Sciences of the Czech Republic (grant No. AV0Z50520514). Funding for open access charge: Czech Science Foundation (grant P502/11/2207).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Bert Vogelstein and Kenneth W. Kinzler for providing Dnmt-deficient HCT116 cell lines, Dana Kucerova and Dalibor Miklik for their technical support and Jan Svoboda for helpful comments.
REFERENCES
- 1.Jordan A, Defechereux P, Verdin E. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J. 2001;20:1726–1738. doi: 10.1093/emboj/20.7.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jordan A, Bisgrove D, Verdin E. HIV reproducibly establishes a latent infection after acute infection of T cells. EMBO J. 2003;22:1868–1877. doi: 10.1093/emboj/cdg188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elleder D, Pavlicek A, Paces J, Hejnar J. Preferential integration of human immunodeficiency virus type 1 into genes, cytogenetic R bands and GC-rich regions: insight from the human genome sequence. FEBS Lett. 2001;517:285–286. doi: 10.1016/s0014-5793(02)02612-1. [DOI] [PubMed] [Google Scholar]
- 4.Schroder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 5.Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC, Ecker JR, Bushman FD. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2004;2:e234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ciuffi A, Llano M, Poeschla E, Hoffmann C, Leipzig J, Shinn P, Ecker JR, Bushman F. A role of LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 2005;11:1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- 7.Shun MC, Raghavendra NK, Vandegraaff N, Daigle JE, Hughes S, Kellam P, Cherepanov P, Engelman A. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 2007;21:1767–1778. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meehan AM, Saenz DT, Morrison JH, Garcia-Rivera JA, Peretz M, Llano M, Poeschla EM. LEDGF/p75 proteins with alternative chromatin tethers are functional HIV-1 cofactors. PLoS Pathog. 2009;5:e1000522. doi: 10.1371/journal.ppat.1000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu X, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for \MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 10.Trobridge GD, Miller DG, Jacobs MA, Allen JM, Kiem HP, Kaul R, Russell DW. Foamy virus vector integration sites in normal human cells Proc. Natl Acad. Sci. USA. 2006;103:1498–1503. doi: 10.1073/pnas.0510046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hacein-Bey-Abina S, Garrigue A, Wang GP, Soulier J, Lim A, Morillon E, Clappier E, Caccavelli L, Delabesse E, Beldjord K, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Studamire B, Goff SP. Host proteins interacting with the Moloney murine leukemia virus integrase: multiple transcriptional regulators and chromatin binding factors. Retrovirology. 2008;5:e48. doi: 10.1186/1742-4690-5-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Narezkina A, Taganov KD, Litwin S, Stoyanova R, Hayashi J, Seeger C, Skalka AM, Katz RA. Genome-wide analyses of avian sarcoma virus integration sites. J. Virol. 2004;78:11656–11663. doi: 10.1128/JVI.78.21.11656-11663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barr SD, Leipzig J, Shinn P, Ecker JR, Bushman FD. Integration targeting by avian sarcoma-leukosis virus and human immunodeficiency virus in the chicken genome. J. Virol. 2005;79:12035–12044. doi: 10.1128/JVI.79.18.12035-12044.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faschinger A, Rouault F, Sollner J, Lukas A, Salmons B, Günzburg WH, Indik S. Mouse mammary tumor virus integration site selection in human and mouse genomes. J. Virol. 2008;82:1360–1367. doi: 10.1128/JVI.02098-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewinski MK, Bisgrove D, Shinn P, Chen H, Hoffmann C, Hannenhalli S, Verdin E, Berry CC, Ecker JR, Bushman FD. Genome-wide analysis of chromosomal features repressing human immunodeficiency virus transcription. J. Virol. 2005;79:6610–6619. doi: 10.1128/JVI.79.11.6610-6619.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shan L, Yang H-C, Rabi A, Bravo HC, Shroff NS, Irizarry RA, Zhang H, Margolick JB, Siliciano JD, Siliciano RF. Influence of host cell transcriptional level and orientation on HIV-1 latency in a primary-cell model. J. Virol. 2011;85:5384–5393. doi: 10.1128/JVI.02536-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plachy J, Kotab J, Divina P, Reinisova M, Senigl F, Hejnar J. Proviruses selected for high and stable expression of transduced genes accumulate in broadly transcribed genome areas. J. Virol. 2010;84:4204–4211. doi: 10.1128/JVI.02511-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolf D, Goff SP. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell. 2007;131:46–57. doi: 10.1016/j.cell.2007.07.026. [DOI] [PubMed] [Google Scholar]
- 20.Wolf D, Goff SP. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature. 2009;458:1201–1204. doi: 10.1038/nature07844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Lieschinget H, Verp S, Marquis J, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463:237–240. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- 22.Niwa O, Yokota Y, Ishida H, Sugahara T. Independent mechanisms involved in suppression of the Moloney leukemia virus genome during differentiation of murine teratocarcinoma cells. Cell. 1983;32:1105–1113. doi: 10.1016/0092-8674(83)90294-5. [DOI] [PubMed] [Google Scholar]
- 23.Petersen R, Kempler G, Barklis E. A stem cell-specific silencer in the primer-binding site of a retrovirus. Mol. Cell. Biol. 1991;11:1214–1221. doi: 10.1128/mcb.11.3.1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grez M, Akgün E, Hilberg F, Ostertag W. Embryonic stem cell virus, a rekombinant murine retrovirus with expression in embryonic stem cells. Proc. Natl Acad. Sci. USA. 1990;87:9202–9206. doi: 10.1073/pnas.87.23.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Challita PM, Skelton D, el-Khoueiry A, Yu XJ, Weinberg K, Kohn DB. Multiple modifications in cis elements of the long terminal repeat of retroviral vectors lead to increased expression and decreased DNA methylation in embryonic carcinoma cells. J. Virol. 1995;69:748–755. doi: 10.1128/jvi.69.2.748-755.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stewart CL, Stuhlmann H, Jähner D, Jaenisch R. De novo methylation, expression, and infectivity of retroviral genomes introduced into embryonal carcinoma cells. Proc. Natl Acad. Sci. USA. 1982;79:4098–4102. doi: 10.1073/pnas.79.13.4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lorincz MC, Schübeler D, Goeke SC, Walters M, Groudine M, Martin DI. Dynamic analysis of proviral induction and de novo methylation: implications for a histone deacetalase-independent, methylation density-dependent mechanism of transcriptional repression. Mol. Cell. Biol. 2000;20:842–850. doi: 10.1128/mcb.20.3.842-850.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lorincz MC, Schübeler D, Groudine M. Methylation-mediated proviral silencing is associated with MeCP2 recruitment and localized histone H3 deacetylation. Mol. Cell. Biol. 2001;21:7913–7922. doi: 10.1128/MCB.21.23.7913-7922.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lorincz MC, Schübeler D, Hutchinson SR, Dickerson DR, Groudine M. DNA methylation density influences the stability of an epigenetic imprint and Dnmt3a/b-independent de novo methylation. Mol. Cell. Biol. 2002;22:7572–7580. doi: 10.1128/MCB.22.21.7572-7580.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hejnar J, Plachy J, Geryk J, Machon O, Trejbalova K, Guntaka RV, Svoboda J. Inhibition of the Rous sarcoma virus long terminal repeat-driven transcription by in vitro methylation: different sensitivity in permissive chicken cells versus mammalian cells. Virology. 1999;255:171–181. doi: 10.1006/viro.1998.9597. [DOI] [PubMed] [Google Scholar]
- 31.Bednarik DP, Cook JA, Pitha PM. Inactivation of the HIV LTR by DNA CpG methylation: evidence for a role in latency. EMBO J. 1990;9:1157–1164. doi: 10.1002/j.1460-2075.1990.tb08222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blazkova J, Trejbalova K, Gondois-Rey F, Halfon P, Philibert P, Guiguen A, Verdin E, Olive D, Van Lint C, Hejnar J. CpG methylation controls reactivation of HIV from latency. PLoS Pathog. 2009;5:e1000554. doi: 10.1371/journal.ppat.1000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kauder SE, Bosque A, Lindqvist A, Planelles V, Verdin E. Epigenetic regulation of HIV-1 latency by cytosine methylation. PLoS Pathog. 1999;5:1000495. doi: 10.1371/journal.ppat.1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koiwa T, Hamano-Usami A, Ishida T, Okayama A, Yamaguchi K, Kamihira S, Watanabe T. 5′-long terminal repeat-selective CpG methylation of latent human T-cell leukemia virus type 1 provirus in vitro and in vivo. J. Virol. 2002;76:9389–9397. doi: 10.1128/JVI.76.18.9389-9397.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taniguchi Y, Nosaka K, Yasunaga J, Maeda M, Mueller N, Okayama A, Matsuoka M. Silencing of human T-cell leukemia virus type 1 gene transcription by epigenetic mechanisms. Retrovirology. 2005;2:e64. doi: 10.1186/1742-4690-2-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lavie L, Kitova M, Maldener E, Meese E, Mayer J. CpG methylation directly regulates transcriptional activity of the human endogenous retrovirus family HERV-K (HML-2) J. Virol. 2005;79:876–883. doi: 10.1128/JVI.79.2.876-883.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matouskova M, Blazkova J, Pajer P, Pavlicek A, Hejnar J. CpG methylation suppresses transcriptional activity of human syncytin-1 in non-placental tissues. Exp. Cell. Res. 2006;312:1011–1020. doi: 10.1016/j.yexcr.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 38.Gimenez J, Montgiraud C, Oriol G, Pichon JP, Ruel K, Tsatsaris V, Gerbaud P, Frendo JL, Evain-Brion D, Mallet F. Comparative methylation of ERVWE1/syncytin-1 and other human endogenous retrovirus LTRs in placenta tissues. DNA Res. 2009;16:195–211. doi: 10.1093/dnares/dsp011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trejbalova K, Blazkova J, Matouskova M, Kucerova D, Pecnova L, Vernerova Z, Heracek J, Hirsch I, Hejnar J. Epigenetic regulation of transcription and splicing of syncytins, fusogenic glycoproteins of retroviral origin. Nucleic Acids Res. 2011;39:8728–8739. doi: 10.1093/nar/gkr562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swindle CS, Kim HG, Klug CA. Mutation of CpGs in the murine stem cell virus retroviral vector long terminal repeat represses silencing in embryonic stem cells. J. Biol. Chem. 2004;279:34–41. doi: 10.1074/jbc.M309128200. [DOI] [PubMed] [Google Scholar]
- 41.Hejnar J, Hajkova P, Plachy J, Elleder D, Stepanets V, Svoboda J. CpG island protects Rous sarcoma virus-derived vectors integrated into nonpermissive cells from DNA methylation and transcriptional suppression. Proc. Natl Acad. Sci. USA. 2001;98:565–569. doi: 10.1073/pnas.98.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Senigl F, Plachy J, Hejnar J. The core element of a CpG island protects avian sarcoma and leukosis virus-derived vectors from transcriptional silencing. J. Virol. 2008;72:7818–7827. doi: 10.1128/JVI.00419-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pannell D, Osborne CS, Yao S, Sukonnik T, Pasceri P, Karaiskakis A, Okano M, Li E, Lipshitz HD, Ellis J. Retrovirus vector silencing is de novo methylase independent and marked by a repressive histone code. EMBO J. 2000;19:5884–5894. doi: 10.1093/emboj/19.21.5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yao S, Sukonnik T, Kean T, Bharadwaj RR, Pasceri P, Ellis J. Retrovirus silencing, variegation, extinction, and memory are controlled by a dynamic interplay of multiple epigenetic modifications. Mol. Therapy. 2004;10:27–36. doi: 10.1016/j.ymthe.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 45.Mikkelsen TS, Kum M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dong KB, Maksakova IA, Mohn F, Leung D, Appanah R, Lee S, Yang HW, Lam LL, Mager DL, Schübeler D, et al. DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. EMBO J. 2008;27:2691–2701. doi: 10.1038/emboj.2008.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz CM, Shinkai Y. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- 48.Leung DC, Dong KB, Maksakova IA, Goyal P, Appanah R, Lee S, Tachibana M, Shinkai Y, Lehnertz B, Mager DL, et al. Lysine methyltransferase G9a is required for de novo DNA methylation and the establishment, but not the maintenance, of proviral silencing. Proc. Natl Acad. Sci. USA. 2011;108:5718–5723. doi: 10.1073/pnas.1014660108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poleshko A, Einarson MB, Shalginskich N, Zhang R, Adams P, Skalka AM, Katz RA. Identification of a functional network of human epigenetic silencing factors. J. Biol. Chem. 2010;285:422–433. doi: 10.1074/jbc.M109.064667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Federspiel MJ, Hughes SH. Retroviral gene delivery. Methods Cell. Biol. 1997;52:179–214. [PubMed] [Google Scholar]
- 51.Rhee I, Jair KW, Yen RW, Lengauer C, Herman JG, Kinzler KW, Vogelstein B, Baylin SB, Schuebel KE. CpG methylation is maintained in human cancer cells lacking DNMT1. Nature. 2000;404:1003–1007. doi: 10.1038/35010000. [DOI] [PubMed] [Google Scholar]
- 52.Rhee I, Bachman KE, Park BH, Jair KW, Yen RW, Schuebel KE, Cui H, Feinberg AP, Lengauer C, Kinzler KW, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 53.Jair KW, Bachman KE, Suzuki H, Ting AH, Rhee I, Yen RW, Baylin SB, Schuebel KE. De novo CpG island methylation in human cancer cells. Cancer Res. 2006;66:682–692. doi: 10.1158/0008-5472.CAN-05-1980. [DOI] [PubMed] [Google Scholar]
- 54.Uren AG, Mikkers H, Kool J, van der Weyden L, Lund AH, Wilson CH, Rance R, Jonkers J, van Lohuizen M, Berns A, et al. A high-throughput splinkerette-PCR method for the isolation and sequencing of retroviral insertion sites. Nat. Protoc. 2009;4:789–798. doi: 10.1038/nprot.2009.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen L, Gao G, Zhang Y, Zhang H, Ye Z, Huang S, Huang J, Kang J. A single amino acid substitution confers enhanced methylation activity of mammalian Dnmt3b on chromatin DNA. Nucleic Acids Res. 2010;38:6054–6064. doi: 10.1093/nar/gkq456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Holz-Schietinger C, Reich NO. The inherent processivity of the human DNA methyltransferase 3 a (DNMT3A) is enhanced by DNMT3L. J. Biol. Chem. 2010;285:29091–29100. doi: 10.1074/jbc.M110.142513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Okitsu CY, Hsieh JC, Hsieh CL. Transcriptional activity affects the H3K4me3 level and distribution in the coding region. Mol. Cell. Biol. 2010;30:2933–2936. doi: 10.1128/MCB.01478-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes Nat. Rev. Genet. 2011;12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 59.Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu H, Coskun V, Tao J, Xie W, Ge W, Yoshikawa K, Li E, Zhang Y, Sun YE. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neuronic genes. Science. 2010;329:444–448. doi: 10.1126/science.1190485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dhayalan H, Rajavelu A, Rathert P, Tamas R, Jurkowska RZ, Ragozin S, Jeltsch A. The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J. Biol. Chem. 2010;285:26114–26120. doi: 10.1074/jbc.M109.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Macleod D, Charlton J, Mullins J, Bird AP. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev. 1994;8:2282–2292. doi: 10.1101/gad.8.19.2282. [DOI] [PubMed] [Google Scholar]
- 63.Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schübeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 64.Hodges E, Smith AD, Kendall J, Xuan Z, Ravi K, Rooks M, Zhang MQ, Ye K, Bhattacharjee A, Brizuela L, et al. High definition profiling of mammalian DNA methylation by array capture and single molecule bisulfite sequencing. Genome Res. 2009;19:1593–1605. doi: 10.1101/gr.095190.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jacinto FV, Ballestar E, Esteller M. Impaired recruitment of the histone methyltransferase DOT1L contributes to the incomplete reactivation of tumor suppressor genes upon DNA demethylation. Oncogene. 2009;28:4212–4224. doi: 10.1038/onc.2009.267. [DOI] [PubMed] [Google Scholar]
- 66.Yang PK, Kuroda MI. Noncoding RNAs and intracellular positioning in monoallelic gene expression. Cell. 2007;128:777–786. doi: 10.1016/j.cell.2007.01.032. [DOI] [PubMed] [Google Scholar]
- 67.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 68.Yoh SM, Lucas JS, Jones KA. The Iws1:Spt6:CTD complex controls cotranscriptional mRNA biosynthesis and HYPB/Setd2-mediated histone H3K36 methylation. Genes Dev. 2008;22:3422–3434. doi: 10.1101/gad.1720008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lickwar CR, Rao B, Shabalin AA, Nobel AB, Strahl BD, Lieb JD. The Set2/Rpd3S pathway suppresses cryptic transcription without regard to gene length or transcription frequency. PloS One. 2009;4:e4886. doi: 10.1371/journal.pone.0004886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Searle S, Gillespie DAF, Chiswell DJ, Wyke JA. Analysis of the variations in proviral cytosine methylation that accompany transformation and morphological reversion in a line of Rous sarcoma virus-infected Rat-1 cells. Nucleic Acids Res. 1984;12:5193–5210. doi: 10.1093/nar/12.13.5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hejnar J, Svoboda J, Geryk J, Fincham VJ, Hak R. High rate of morphological reversion in tumor cell line H-19 associated with permanent transcriptional suppression of the LTR, v-src, LTR provirus. Cell Growth Differ. 1994;5:277–285. [PubMed] [Google Scholar]
- 72.Poleshko A, Palagin I, Zhang R, Boimel P, Castagna C, Adams PD, Skalka AM, Katz RA. Identification of cellular proteins that maintain retroviral epigenetic silencing: evidence for an antiviral response. J. Virol. 2008;82:2313–2323. doi: 10.1128/JVI.01882-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.He J, Yang Q, Chang L-J. Dynamic DNA methylation and histone modifications contribute to lentiviral transgene silencing in murine embryonic carcinoma cells. J. Virol. 2005;79:13497–13508. doi: 10.1128/JVI.79.21.13497-13508.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ellis J. Silencing and variegation of gammaretrovirus and lentivirus vectors. Hum. Gene Therapy. 2005;16:1241–1246. doi: 10.1089/hum.2005.16.1241. [DOI] [PubMed] [Google Scholar]
- 75.Trono D, Van Lint C, Rouzioux C, Verdin E, Barré-Sinoussi F, Chun TW, Chomont N. HIV persistence and the prospect of long-term drug-free remissions for HIV-infected individuals. Science. 2010;329:174–180. doi: 10.1126/science.1191047. [DOI] [PubMed] [Google Scholar]
- 76.Keedy KS, Margolis DM. Therapy for persistent HIV. Trends Pharmacol. Sci. 2010;31:206–211. doi: 10.1016/j.tips.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mok HP, Javed S, Lever A. Stable gene expression occurs from a minority of integrated HIV-1-based vectors: transcriptional silencing is present in the majority. Gene Therapy. 2007;14:741–751. doi: 10.1038/sj.gt.3302923. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.