Abstract
The ARF tumour suppressor stabilizes p53 by negatively regulating the E3 ubiquitin ligase MDM2 to promote cell cycle arrest and cell death. However, ARF is also able to arrest cell proliferation by inhibiting ribosome biogenesis. In greater part this is achieved by targeting the transcription termination factor I (TTF-I) for nucleolar export, leading to an inhibition of both ribosomal RNA synthesis and processing. We now show that in the absence of ARF, TTF-I is ubiquitinylated by MDM2. MDM2 interacts directly with TTF-I and regulates its cellular abundance by targeting it for degradation by the proteasome. Enhanced TTF-I levels inhibit ribosome biogenesis by suppressing ribosomal RNA synthesis and processing, strongly suggesting that exact TTF-I levels are critical for efficient ribosome biogenesis. We further show that concomitant with its ability to displace TTF-I from the nucleolus, ARF inhibits MDM2 ubiquitinylation of TTF-I by competitively binding to a site overlapping the MDM2 interaction site. Thus, both the sub-nuclear localization and the abundance of TTF-I are key regulators of ribosome biogenesis.
INTRODUCTION
Ribosome biogenesis, the synthesis and assembly of ribosomes, is an essential task for any proliferative cell and, as might be expected, it is highly responsive to environmental changes and to various forms of stress (1). It is therefore not surprising to find that many tumour suppressors and oncogenes, including Rb, ARF, p53 and MDM2, work in collusion or opposition to maintain a level of ribosome biogenesis appropriate to the cellular state, whether it be proliferative, cell cycle arrested, differentiated or apoptotic. Although the rate of ribosome biogenesis was demonstrated to determine passage through the cell cycle checkpoint ‘Start’ and the commitment of a cell to proliferation (2), there is still a dearth of information on how exactly this control is achieved.
The catalytic core of the ribosome is formed by the 28S and 18S ribosomal RNAs (rRNAs), which along with the small 5.8S rRNA are processed from a single 45S (47S) precursor transcript. There is good evidence that a key regulator of ribosome biogenesis is the rate at which these rRNAs are synthesized (reviewed in refs 1,3,4). In human and mouse, the 200 or so gene copies encoding the 45S pre-rRNA are organized in five tandem arrays on the short arms of acrocentric chromosomes. These genes are transcribed by RNA polymerase I (RPI), and the RPI-specific basal factors SL1/TIF-IB and UBF (1,5–10). The 45S rRNA is cotranscriptionally assembled with pre-ribosomal proteins before undergoing processing via major 32S and 20S intermediates. Prior to cleavage the pre-rRNA is also extensively modified by methylation and pseudouridinylation, a process that is directed by several hundred small nucleolar RNAs (snoRNAs).
The ARF tumour suppressor has been implicated in regulating the production of the rRNAs (11,12). While searching for novel ARF interactor proteins we recently identified transcription termination factor I (TTF-I), a nucleolar factor able to terminate RPI transcription of the rRNA genes (13). ARF was found to control the sub-nuclear localization of TTF-I, and it was shown that in fact TTF-I shuttles rapidly between nucleoplasm and nucleolus with the aid of the chaperone NPM/B23 and a nucleolar localization sequence within its N-terminal regulatory domain. ARF inhibits the nucleolar import of TTF-I by binding to and inhibiting this nucleolar localization sequence, causing the accumulation of TTF-I in the nucleoplasm. Conditional depletion of TTF-I further demonstrated for the first time that it is in fact an essential factor for transcription of the rRNA genes but, more surprisingly, also for processing of the precursor rRNA.
TTF-I was originally identified by its ability to terminate RPI transcription of the pre-rRNA in vitro (14–16). Mouse TTF-I contains a C-terminal DNA-binding domain with homology to the DNA-binding domain or the Myb oncogene and binds to multiple sites both upstream and downstream of the rRNA genes. Although TTF-I was clearly shown to terminate RPI transcription in vitro and to bind to sites downstream of the rRNA genes (14–16), it is still uncertain whether this is its major role. TTF-I binding to a conserved upstream promoter proximal site on the rRNA genes has been shown to be important in regulating rRNA gene activity. Binding to this site has been shown to phase nucleosomes on the RPI promoter and thus activate transcription (17). TTF-I was shown to recruit the ATP-dependent nucleosome remodelling complex NoRC (18) and in this way can alter nucleosome positioning. However, recruitment of this same chromatin remodelling complex along with the deacetylase HDAC1 and a DNA methyltransferase has also been shown to repress the rRNA genes (19,20). Thus, the in vivo functions of TTF-I still remain somewhat obscure. We now show that the abundance of TTF-I is a key factor in determining pre-rRNA synthesis and processing. TTF-I levels are regulated by the E3-ubiquitin ligase MDM2 via direct ubiquitinylation, a function that is directly competed by ARF. Our data identify TTF-I as a target of the ARF-MDM2 tumour suppressor–oncogene balance and a key regulator of the endogenous levels of ribosome biogenesis.
MATERIALS AND METHODS
Plasmid constructs
Full-length p19ARF (mARF) a.a 1–169 was cloned into pcDNA3 (Invitrogen). N-terminal FLAG or YFP tagged mouse TTF-I (TTF-I) were expressed using pFLAGCMV2 (Invitrogen), pRevTRE (Clontech) or pEYFP-C1 (Clontech). The expression vectors for GFP-UBF, HA-Ubiquitin (21), pCOC-X2-MDM2WT and C642A (22,23), Myc-MDM2 (pDWM659) (24) were provided respectively by T. Mistelli, J.-Y. Masson, S. Laín and C. Blattner.
Antibodies
Rabbit anti-TTF-I#3 (13), and other antibodies were: anti-FLAG (F7425, Sigma-Aldrich), anti-fibrillarin (MMS-581S, Covance), anti-HA (ab9134, Abcam), anti-HA (ab9110, Abcam), anti-MDM2 (3G5, 4B11 and 2A10) (25), anti-ubiquitin (FK2 #ST1200, Calbiochem), anti-actin (#A2228 Clone AC-74, Sigma), IgG mouse (#10400C, Invitrogen), anti-YFP (632460, Clontech), anti-Myc (71D10, Cell Signaling), anti-C23/nucleolin (MS-3; sc8031, Santa Cruz) and anti-His (631212, Clontech).
Cell lines
NIH3T3 and HEK293T cells were obtained from ATCC. NIH3T3 cells were transfected with linearized pRevTREFLAG-TTF-I and pTet-OffΔNeo (Clontech). Clones were selected with hygromycin (100 µg/ml) in the presence of doxycycline (Dox; 50 µg/ml). Clones B6 and F9 were identified as displaying a significant level of FLAG-TTF-I expression in the absence of Dox. Cell lines were maintained in high glucose Dulbecco's modified eagle's Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 100 u.ml−1 each of penicillin and streptomycin (PS). Tet-Off clones B6 and F9 were maintained in the same medium with the addition of 50 µg/ml Dox. Where indicated, MT-ARF (26) cells were treated with 80 µM ZnSO4, clones B6 and F9 were grown in the absence of doxycylin and HEK293T and NIH3T3 cells were treated respectively with 10 or 20 µM MG132 for 12 h to inhibit the 26S proteasome or with 50 mM cycloheximide (Chx) for indicated times to inhibit protein synthesis.
In vivo RNA labelling, extraction and analysis
These were performed as previously described (27,28). Briefly, NIH3T3 (ATCC) cells and cell clones were grown in the presence or absence of Dox and labelled for 3 h by the addition of 2.5 µCi.ml−1 [3H]-uridine (Perkin Elmer), before RNA extraction in 1 ml Trizol (Invitrogen) according to the manufacturer's protocol. Two to four micrograms of this RNA was loaded onto 1% formaldehyde gels and electrophoresed overnight at 2.5 V/cm in MOPS running buffer. After EtBr staining, gels were photographed, UV-irradiated for 5 min at maximum energy (UV-cross-linker, Hoefer) and RNA transferred to Biodyne B membranes (Pall) by vacuum transfer (Biometra). To provide a visual image of the labelling, the membrane was UV-crosslinked (70 mJ.cm−2), treated with ENHance (Perkin-Elmer) and fluorographed at −80°C for 24–72 h. Finally, to quantitate RNA labelling, the [3H]-labelled RNA bands were cut from the membrane, using the fluorograph as guide, and were scintillation counted in ScintiVerse (Fisher) using external standard correction. All experiments were performed and analysed throughout in triplicate.
Methionine labelling
MT-ARF cells were plated in a 60-mm dish and induced for 0, 6, 12 and 24 h. Cells were washed twice with prewarmed PBS and incubated in 2 ml of prewarmed methionine-free DMEM supplemented with 10% FBS at 37°C and 5% CO2. After 60-min incubation the medium was changed for 2 ml of prewarmed methionine-free DMEM supplemented with 10% FBS and 1 mCi of [35S]-methionine and incubation continued for 90 min at 37°C and 5% CO2. Then cell extracts were analysed by western blotting and membrane was cut to estimate total protein synthesis by scintillation counting in ScintiVerse (Fisher).
Protein extracts
For total protein extracts, cells were washed with 3 ml cold PBS, scraped into 1 ml cold PBS, centrifuged 30 s at 14 000 r.p.m. and resuspended in sodium dodecyl sulphate (SDS) loading buffer. After fractionation on 8% or 5–15% gradient SDS–polyacrylamide gel electrophoresis (PAGE), cell extracts were analysed by standard western blotting procedures.
Transfection
1.25 × 106 HEK293T cells were seeded onto poly-L-lysine-treated (1 mg.ml−1, Sigma-Aldrich) 60-mm culture dishes 24 h prior to transfection. Transfection used CaPO4/chloroquine (29) and 8 or 12 μg total DNA per 60 mm culture dish. NIH3T3, MT-ARF, Clone B6 and Clone F9 cells were plated at ∼1.5 × 105 per 35 mm dish (∼25% confluency) for GFP constructs, or ∼2.5 × 105 per 35 mm dish (∼40% confluency) for immunofluorescence (IF) using Ex-GEN 500 (Fermentas).
Immunoprecipitation
Cells were scraped into IP buffer [25 mM Tris–HCl, pH 7.5, 1 mM EDTA, 0.1 mM EGTA, 5 mM MgCl2, 150 mM NaCl, 10% glycerol, 1% NP-40 (Igepal. Sigma-Aldrich), 0.1% SDS, 1% TritonX100 and 1 μg/ml each pepstatin, leupeptin and aprotinin (Sigma-Aldrich)], kept on ice for 15 min and then sonicated 4 × 20 s (S-450 Branson Ultrasonics or Sonic Dismembrator Model 100 Fisher Scientific) at maximum power or continuous power 1, respectively. Cell lysates were cleared, 14 000 r.p.m., 1 min and incubated with first antibody for 3 h at 4°C. Immunoprecipitates were recovered on Protein A-Sepharose (Amersham Biosciences) and washed once in IP buffer and three times in IP buffer less detergents. After fractionation on 8%, 12%, or 5–15% gradient SDS–PAGE, immunoprecipitates and total cell lysates were analysed by standard western blotting procedures.
Pull-down assays
His-TTF-I (a.a. 471–859) was expressed in Escherichia coli BL21-CodonPlus (Stratagene) and isolated on Ni-NTA Agarose (Qiagen) following the manufacturer's protocol. Control Ni-NTA Agarose beads were prepared in the same way but using an extract from untransformed E. coli BL21-CodonPlus cells. Myc-MDM2 was expressed in vitro using the TNT Coupled Reticulocyte Lysate System (Promega). Equal aliquots of Myc-MDM2 were transferred to binding buffer [25 mM Tris–HCl pH 7.5, 150 mM NaCl, 1% IGEPAL (Sigma)] and preabsorbed using Ni-NTA beads. The Control or His-TTF-I Ni-NTA beads were then added to the preabsorbed Myc-MDM2 and incubated at 4°C for 4 h. The beads were then recovered and washed five times with the binding buffer before gel analysis by western blot using anti-His and anti-Myc antibodies.
Immunofluorescence microscopy
For immunofluorescence cells were fixed with 4% paraformaldehyde/PBS for 15 min (only in the case of IF in Supplementary Figure S5, cells were then heated to 100°C for 10 min in 10 mM Na-citrate pH 6.0), permeabilized with 0.5% Triton/PBS for 5 min, incubated with primary antibodies (100 µl) in 5% goat serum/PBS for 1 h at room temperature (RT), stained with anti-rabbit/mouse IgG AlexaFluor 488/568 (Molecular Probes) and counter-stained with DAPI. After mounting in 50% glycerol/50% 0.2 M Na-glycine, 0.3 M NaCl and observed using a Leica DMI6000 B and OpenLab and Volocity software (Perkin-Elmer Improvision).
S1 mapping
The termination site of the 45S was mapped using a 3′ 32P-labelled probe extending from +13 169 to +13 696 covering the first five Sal boxes (T1–5) essentially as (30,31).
Psoralen crosslinking and Southern blotting
Psoralen crosslinking and Southern blotting were performed as described elsewhere (32,33).
RESULTS
The abundance of TTF-I is critical for ribosome biogenesis
In a previous study we showed that ARF arrested cell division by inhibiting the nucleolar localization of TTF-I and in so doing suppressed both 45S pre-rRNA transcription and its subsequent processing into the mature rRNAs (13). This same study also showed that the conditional depletion of TTF-I phenocopied ARF induction by inhibiting both pre-rRNA transcription and processing. These data showed that TTF-I is an essential factor in rRNA synthesis. The data also suggested that TTF-I abundance might be a positive regulator of rRNA synthesis and hence of ribosome biogenesis. To test this latter possibility we sought to establish cell lines in which TTF-I levels could be upregulated.
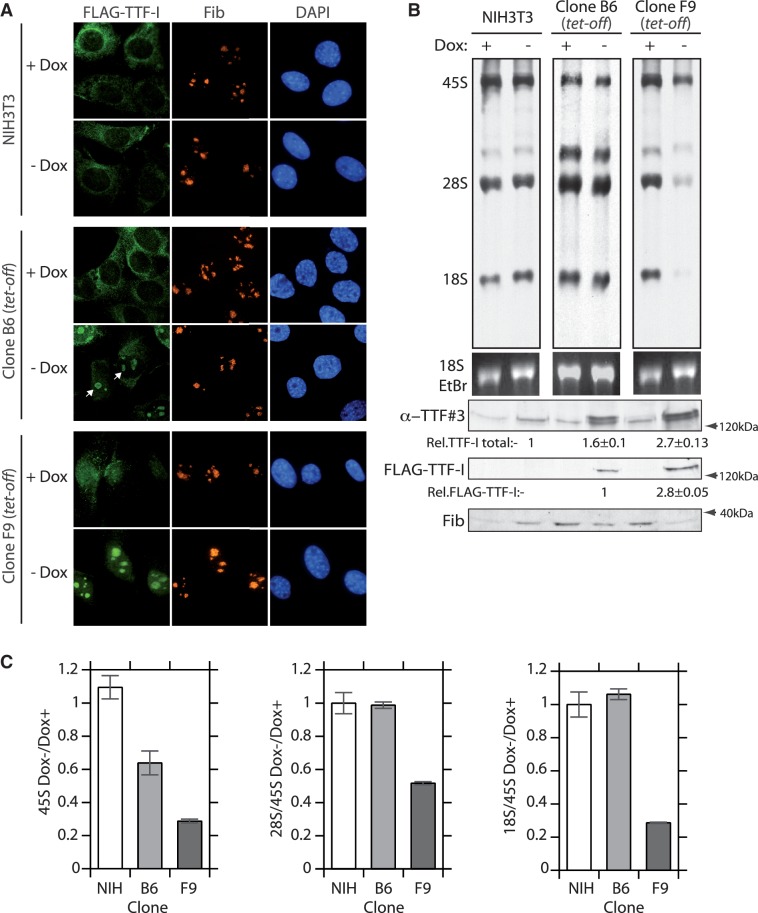
An epitope-tagged mouse TTF-I transgene was introduced into NIH3T3 cells under the negative control of Dox (tet-off) and stable cell clones screened for transgene expression after removal of doxycycline. Two cell clones, B6 and F9, displaying different levels of FLAG-TTF-I induction after Dox withdrawal were selected for further study. As expected, the exogenous FLAG-TTF-I was found to be present within the nucleolus, and while clone B6 displayed a low level of exogenous TTF-I expression, expression in clone F9 was significantly more robust (Figure 1A). In both cell clones the bulk levels of TTF-I were clearly enhanced above those in the parent NIH3T3 cells, although this enhancement was significantly more apparent in the F9 clone (Figure 1A and lower panels of 1B). However, metabolic labelling of newly synthesized rRNA revealed that the enhanced TTF-I levels did not enhance rRNA synthesis in either clone. Indeed, synthesis of the 45S pre-rRNA was suppressed in both clones in proportion to their TTF-I expression levels (Figure 1B, upper panel, Figure 1C and Supplementary Figure S1D). As mentioned above, TTF-I has been implicated not only in gene activation, but also in gene silencing. Hence it was possible that increased TTF-I levels had increased the number of silent rRNA genes. However, the psoralen crosslinking technique (32,34) revealed no change in the fraction of active rRNA genes that could explain the reduction in 45S synthesis (Supplementary Figure S1A and S1B).
Figure 1.
Enhanced expression of TTF-I is inhibitory to rRNA synthesis and processing. (A) Conditional expression of FLAG-TTF-I in tet-off regulated cell clones B6 and F9, and localization of endogenous fibrillarin (Fib) before and after induction of the UBF transgene by withdrawal of doxycyclin (Dox+, Dox–). (B) B6, F9 cell clones and the NIH3T3 cell line were maintained in the presence or absence of doxycyclin were subjected to a 3-h [3H]-uridine pulse-labelling and incorporation into rRNA determined in parallel with the immunoblot analysis of total TTF-I (anti-TTF#3), FLAG-TTF-I and fibrillarin protein levels. (C) Quantitation of three independent pulse-label analyses as in B. 45S rRNA synthesis is shown in the left-hand panel and the ratios of 28S:45S and 18S:45S rRNAs before and after doxycycline removal are shown in the middle- and right-hand panels for cell clones B6 and F9 and NIH3T3 cells.
Surprisingly, processing of the 45S rRNA to yield the mature 28S and 18S rRNAs was also suppressed by enhanced levels of TTF-I. Metabolic labelling revealed a severe and differential reduction in the synthesis of mature 28S and 18S rRNAs relative to the 45S precursor (Figure 1B and C). This was also found to be a characteristic of TTF-I depletion (13) and indeed endogenous TTF-I is found to be broadly distributed throughout the nucleolus and is certainly not limited to the rRNA genes, suggesting that it may also play a role in maturation of the rRNA (Supplementary Figure S2A and S2B).
TTF-I is ubiquitinylated and its abundance is limited by MDM2
Since both depletion and augmentation of TTF-I levels suppress 45S rRNA synthesis and processing, the levels of this protein must normally be finely tuned to cellular needs. Since protein abundance is a balance between synthesis and degradation, we determined the rate of turnover of the TTF-I protein. Clone B6 was grown in the absence of Dox until exogenous TTF-I reached steady-state levels. Dox was then added to the culture to inactivate the transgene and TTF-I levels followed over time. Exogenous FLAG-TTF-I decayed to about 25% of the steady-state level 12 h after transgene inactivation and its depletion was essentially complete by 24 h (Figure 2A). Assuming an exponential decay, this means that the TTF-I protein has a half-life of between 6 and 7 h, much shorter than the cell cycle. TTF-I degradation is therefore a likely determinant of its cellular abundance.
Figure 2.
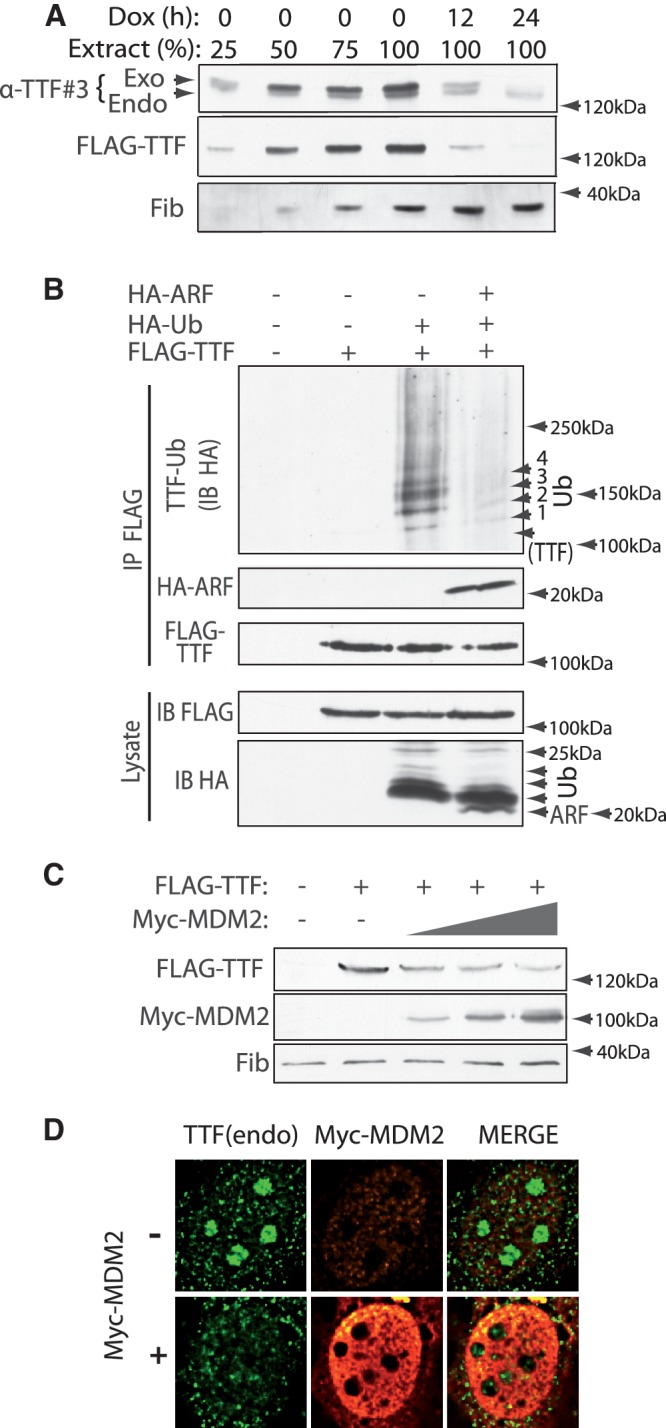
TTF-I is ubiquitinylated and its abundance is regulated by MDM2. (A) After re-addition of doxycycline to inactivate transgene expression in induced (Dox-) B6 cells, FLAG-TTF-I levels decay from the steady state with a half-time of between 6 and 7 h. Relative TTF-I levels were estimated in comparison with 25, 50, 75 and 100% loading of the steady-state expressing cells before re-addition of doxycyclin. (B) TTF-I is poly-ubiquitinylated and this ubiquitinylation is inhibited by ARF. FLAG-TTF-I was expressed along with HA-tagged Ubiquitin (Ub) and HA-mARF in HEK293T cells, complexes were immunoprecipitated (IP) with anti-FLAG antibody and immunoblotted (IB) using anti-HA and anti-FLAG antibodies. Lysate refers to the whole-cell lysates before IP. Arrowheads in the upper panel indicate discrete mono-, di-, tri- and tetra-ubiquitinylations (1–4 Ubiq) of TTF-I. (C) Expression of increasing levels of Myc-tagged MDM2 limits the accumulation of FLAG-TTF-I. Whole-cell lysates from transiently transfected HEK293T cells were immunoblotted using anti-FLAG, anti-Myc and anti-fibrillarin (Fib) antibodies. (D) Transient expression of Myc-MDM2 in NIH3T3 cells strongly decreases nucleolar level of endogenous TTF-I. Non-transfected and Myc-MDM2 transfected cells were fixed and processed for immunofluorescence with anti-TTF#3, anti-MDM2 (2A10) as described in ‘Materials and Methods’ section (see also Supplementary Figure S3).
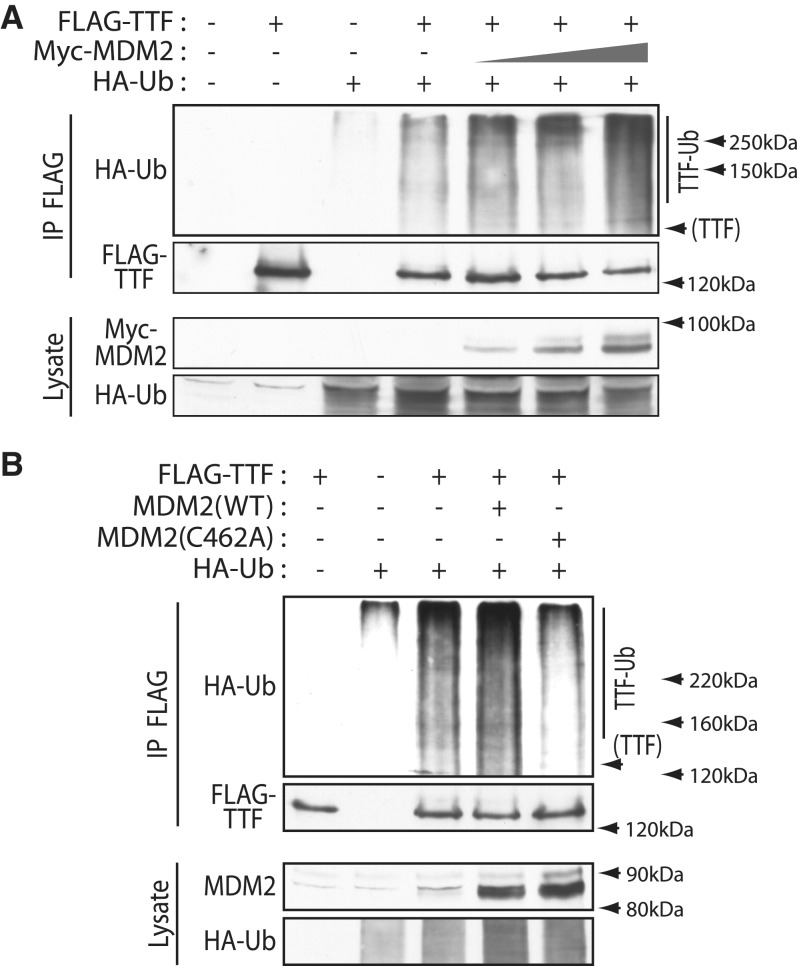
Regulated degradation is often the result of poly-ubiquitinylation, which targets proteins to the proteasome (35). Indeed, when TTF-I was co-expressed with epitope-tagged ubiquitin, it was found to be poly-ubiquitinylated, suggesting that this modification was important in determining its rate of degradation (Figure 2B). Most strikingly, co-expression of ARF almost completely repressed this ubiquitinylation. Since ARF is known to inactivate the E3 ubiquitin ligase MDM2, this suggested that TTF-I might be a direct target of this E3 ligase. Consistent with this, co-expression of increasing levels of MDM2 reduced the accumulation of FLAG-TTF-I (Figure 2C). Further, expression of MDM2 reduced the level of endogenous TTF-I within the nucleoli of these cells (Figure 2D and Supplementary Figure S3).
MDM2 induces ubiquitinylation of TTF-I
To determine if MDM2 could induce the ubiquitinylation of TTF-I, both proteins were co-expressed in vivo with epitope-tagged ubiquitin, and cells treated with the proteasome inhibitor MG132 to prevent degradation of ubiquitinylated TTF-I. Wild-type MDM2 expression induced a significant enhancement of TTF-I ubiquitinylation at the expense of unmodified TTF-I (Figure 3A). To test if the E3 ligase activity of MDM2 was required, the ability of wild-type MDM2 and the inactive C462A MDM2 mutant to induce ubiquitinylation of TTF-I was compared (Figure 3B). Co-expression of MDM2 again enhanced TTF-I ubiquitinylation, while co-expression of mutant MDM2 reduced ubiquitinylation to near background level. Thus, ubiquitinylation of TTF-I is dependent on the presence of catalytically active MDM2.
Figure 3.
MDM2 induces ubiquitinylation of TTF-I dependent on its E3 ubiquitin ligase activity. (A) Increasing levels of MDM2 induce TTF-I poly-ubiquitinylation. FLAG-TTF-I, HA-Ubiquitin (HA-Ub) and increasing levels of Myc-MDM2 were co-expressed in HEK293T cells, in the presence of the proteasome inhibitor MG132, and FLAG immunoprecipitates (IP) probed with anti-FLAG and anti-HA antibodies in comparison with the whole-cell lysates. (B) TTF-I ubiquitinylation is abrogated by the C462A point mutation of MDM2 that inactivates the ubiquitin ligase activity (MDM2(C462A)) (22,23). FLAG-TTF-I, HA-Ubiquitin and wild type (WT) or inactive MDM2 (C462A) were co-expressed as in (A) in the presence of MG132, and FLAG immunoprecipitates probed with anti-FLAG and anti-HA antibodies.
MDM2 interacts directly with TTF-I
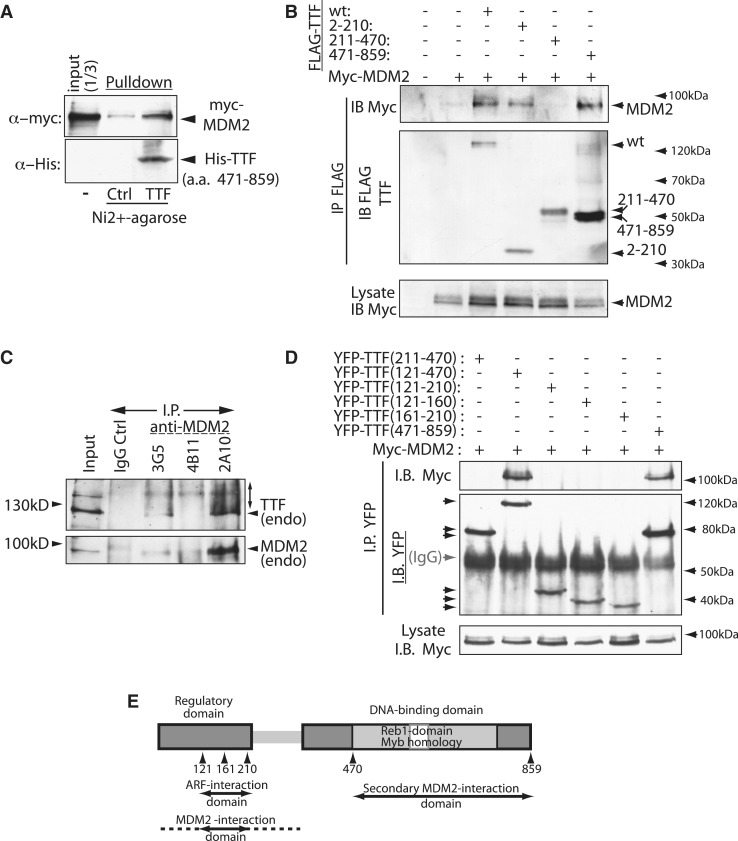
The ability of MDM2 to induce ubiquitinylation of TTF-I suggested that TTF-I was also a direct substrate of this E3-ligase. Since MDM2 usually interacts specifically with its substrates, as do other E3-ligases (36), we asked if it also interacts with TTF-I. In vitro expressed Myc-MDM2 was pulled down by the DNA-binding domain of TTF-I, showing that the two proteins are able to interact directly and specifically with each other (Figure 4A). When co-expressed in HEK293T cells, wild-type FLAG-TTF-I and MDM2 specifically co-immunoprecipitated and hence also interacted in vivo (Figure 4B). Expression of sub-fragments of TTF-I showed independent interactions of MDM2 with both the N-terminal regulatory domain (a.a. 2–210) and the C-terminal DNA-binding domain (a.a. 471–859), but not with the central domain (see again Figure 4B). Endogenous TTF-I was also found to co-immunoprecipitate with endogenous MDM2 from NIH3T3 cells treated with the proteasome inhibitor MG132 (Figure 4C). Three independent monoclonal antibodies displayed significantly different efficiencies of immunoprecipitation of MDM2, and these efficiencies correlated with the levels of TTF-I co-immunoprecipitation, underlining the specificity of the endogenous interaction. Further, the endogenous TTF-I fraction immunoprecipitated with endogenous MDM2 under the conditions of proteasome inhibition displayed a very distinct smearing towards higher molecular weights. This was not detected in the bulk endogenous (input) TTF-I and was consistent with extensive ubiquitinylation. Finer mapping of the interaction within the regulatory N-terminal domain, this time using YFP as epitope tag, confirmed the N- and C-terminal interactions and showed that a.a. 2–210 and 121–470 interact with MDM2, but that a.a. 121–210 was insufficient (Figure 4B and D). These data suggested that the key N-terminal interaction sites lay between a.a. 121 and 210, but that immediately flanking sequences also played a role in stabilizing the interaction (summarized in Figure 4E).
Figure 4.
MDM2 directly interacts with TTF-I. (A) Recombinant His-tagged TTF-I (a.a. 471–859) was expressed in E. coli BL21, immobilized on Ni2+ NTA-Agarose and used in pull-down assays with myc-tagged MDM2 expressed in vitro by coupled transcription/translation. Leftmost tracks show the input myc-MDM2 and the rightmost and central tracks respectively show the recovery of MDM2 on immobilized TTF-I (TTF) and on Ni2+-agarose pre-incubated with an extract from non-expressing BL21 E. coli (Ctrl). (B) MDM2 interacts in vivo with both N- and C-terminal domains of TTF-I. Full-length and truncated FLAG-TTF-I proteins (a.a. 2–210, 211–470 and 471–859) were co-expressed with Myc-MDM2 in HEK293T cells, and FLAG immunoprecipitates (IP) probed with anti-FLAG and anti-Myc antibodies. (C) Endogenous TTF-I also interacts with endogenous MDM2 in NIH3T3 cells. Cells were treated with 20 µM MG132 for 12 h and whole-cell protein extracts immunoprecipitated with each of three MDM2-specific antibodies (3G5, 4B11 and 2A10) or a control non-specific antibody (IgG), and immunoprecipitates were probed for MDM2 and TTF (anti-TTF#3). The higher-molecular-weight “smear” seen for immunoprecipitated endogenous TTF-I is consistent with its ubiquitinylation. (D) To map the MDM2 interaction sites on TTF-I, YFP fusion proteins were co-expressed with Myc-MDM2 in HEK293T cells, and YFP immunoprecipitates probed with anti-Myc and anti-YFP antibodies. (E) Summary of the functional sub-domains of TTF and the interactions with MDM2 and ARF. The N-terminal activation/repression and autoregulatory domain and the sequence-specific DNA-binding domain are indicated as are the Reb1 and Myb homologies.
ARF competes with the MDM2 interaction and inhibits TTF-I ubiquitinylation
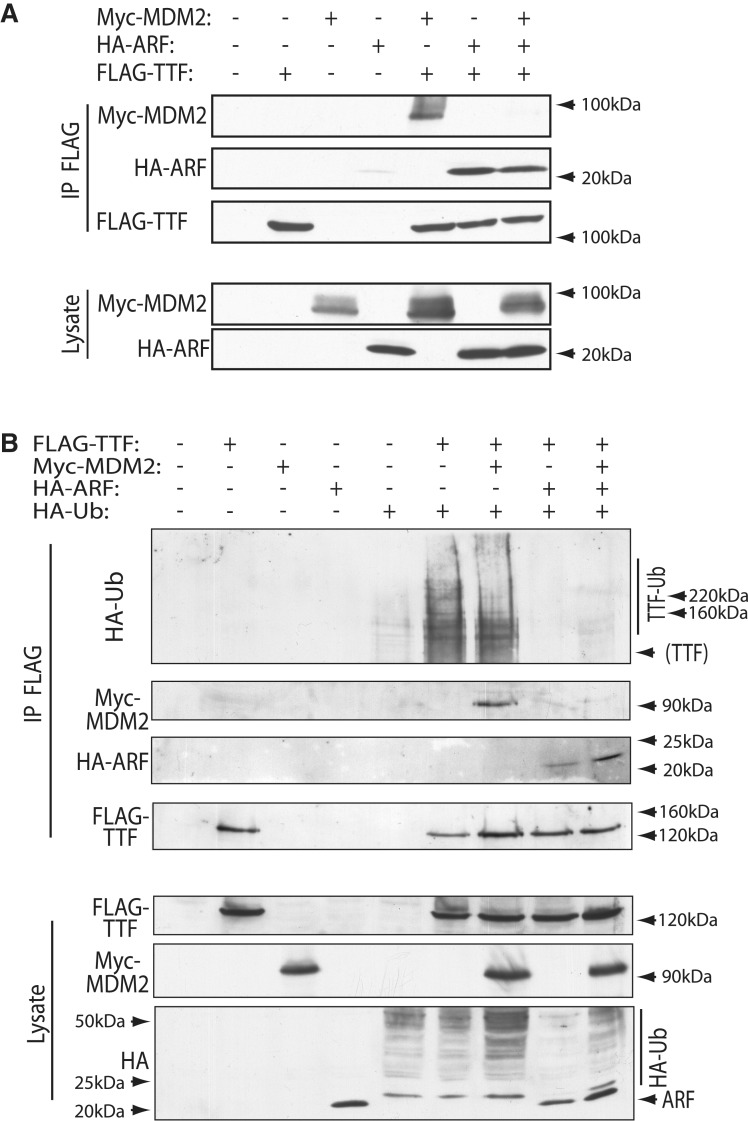
Surprisingly, the N-terminal MDM2 interaction domain of TTF-I overlapped its previously mapped ARF interaction domain (Figure 4E). This potentially explained why ARF was able to inhibit TTF-I ubiquitinylation (Figure 2B). In fact, ARF effectively competed the interaction of MDM2 with TTF-I and formed its own complex with TTF-I (Figure 5A). MDM2 or ARF expression with TTF-I produced the expected MDM2-TTF and ARF–TTF complexes. But when both ARF and MDM2 were co-expressed with TTF-I, the ARF–TTF complex was formed at the expense of the MDM2–TTF complex. This displacement of MDM2 by ARF also corresponded with the inhibition of TTF-I ubiquitinylation (Figure 5B). In the absence of proteasome inhibition, TTF-I was ubiquitinylated and interacted with MDM2 as expected. However, when ARF was introduced both ubiquitinylation of TTF-I and its interaction with MDM2 were inhibited. This again supports the notion that TTF-I is a direct target of MDM2 ubiquitinylation.
Figure 5.
MDM2 and ARF competitively regulate the ubiquitinylation of TTF-I. (A) ARF displaces MDM2 from TTF-I. FLAG-TTF-I, HA-ARF and Myc-MDM2 were co-expressed in HEK293T cells, and FLAG immunoprecipitates (IP) were probed with anti-FLAG, anti-HA and anti-Myc antibodies. (B) ARF displaces MDM2 from TTF-I and inhibits its ubiquitinylation. FLAG-TTF-I, HA-Ubiquitin, HA-ARF and Myc-MDM2 were co-expressed in HEK293T cells, and FLAG immunoprecipitates probed with anti-FLAG, anti-HA and anti-Myc antibodies. Ubiquitinylation of TTF-I is indicated in the upper panel (TTF-Ub).
DISCUSSION
In a previous study, we showed that the tumour suppressor ARF (p19ARF/p14ARF) interacts directly with the rRNA gene factor TTF-I in both mouse and human (13). The study demonstrated that TTF-I is an essential factor for transcription of the rRNA genes and for processing of their primary transcripts 47/45S pre-rRNA. ARF inhibits both rRNA gene transcription and processing by inducing the displacement of TTF-I from the nucleolus. In the absence of ARF, TTF-I shuttles between nucleolus and nucleoplasm with the aid of the chaperone NPM/B23. In the present study, we find that the abundance of TTF-I is critical for ribosome biogenesis. Conditional over-expression of TTF-I phenocopied its depletion and caused both the inhibition of 45S rRNA synthesis and processing. Thus, the exact cellular abundance of TTF-I is a critical factor in determining the rate of ribosome biogenesis. We find that this abundance is regulated by MDM2, and assays strongly support the notion that this involves a direct interaction with, and ubiquitinylation of, TTF-I (Figure 6A). Firstly, increased levels of MDM2 enhance TTF-I ubiquitinylation and reduce its cellular abundance as well as its concentration in the nucleolus. Secondly, MDM2 interacts with TTF-I both in vitro and in vivo. Hence, our data strongly suggest that MDM2 regulates ribosome biogenesis by determining the abundance of TTF-I. This is certainly not the only mechanism that implicates MDM2 in ribosome biogenesis. For example, the state of ribosome biogenesis has been shown to regulate the activity of MDM2 via interactions with free ribosomal proteins (see ref. 37 for a review). However, to our knowledge it is the first demonstration that MDM2 may itself determine the rate of ribosome biogenesis.
Figure 6.
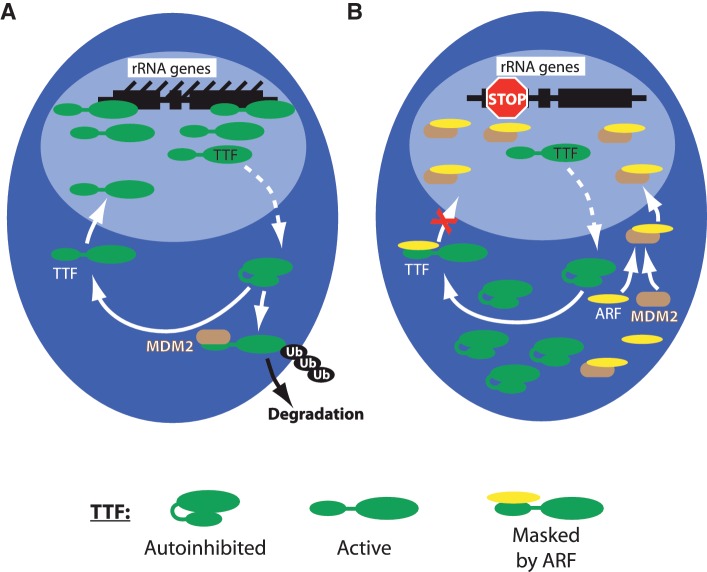
MDM2 in the control of TTF-I. (A) TTF-I normally shuttles between nucleoplasm and nucleolus. [This occurs with the aid of the chaperone NPM/B23 and a nucleolar localization sequence within the N-terminal regulatory domain of TTF-I (13).] While cycling through the nucleoplasm, TTF-I may interact with MDM2 which then ubiquitinylates it and targets it for degradation. (B) Induction of ARF by oncogenic stress or by differentiation will inhibit MDM2 function, in part by sequestering it in the nucleolus. ARF also prevents TTF-I from re-entering the nucleolus and MDM2-dependent ubiquitinylation by competitive binding to a common site on TTF-I. The darker blue region represents the nucleoplasm and the lighter blue region the nucleolus.
ARF is known to inhibit MDM2 via a direct interaction and, at least in part, by sequestering MDM2 in the nucleolus (38). This latter mode of inhibition has been the subject of some controversy (39–41); however, it may be explained by the reduced accessibility of MDM2 to antibody detection once it is displaced to the nucleolus (see Supplementary Figure S5 and Figure 5B of ref. 13). As we now show, ARF is also able to compete with MDM2 for binding to TTF-I. Hence, ARF most likely suppresses the ubiquitinylation of TTF-I both by displacing MDM2 from it and by inhibiting MDM2 function. The ability of ARF to prevent re-entry of TTF-I into the nucleolus (13) could provide yet another mechanism to inhibit its interaction with MDM2 (Figure 6B). Thus, it might be expected that ARF would enhance the cellular abundance of TTF-I by preventing its ubiquitinylation and consequent degradation. Despite this, bulk TTF-I levels remain constant during ARF induction (Supplementary Figure S4A). We do detect a mild degree of stabilization of existing TTF-I protein in the presence of ARF (Supplementary Figure S4B). However, this stabilization is balanced by the suppression of new protein synthesis exerted by ARF (Supplementary Figure S4C). Hence, TTF-I levels in cells expressing ARF are probably maintained constant by the opposing effects of enhanced TTF-I stability and reduced synthesis.
Suppression of TTF-I ubiquitinylation may then be an unavoidable by-product of ARF's functions in inhibiting MDM2 and hence stabilizing p53 (42–45), and of inhibiting ribosome biogenesis by displacing TTF-I from the nucleolus (13). However, the ability to maintain a constant TTF-I level by preventing its degradation during ARF induction could also play an important biological role, since it would allow a rapid re-establishment of normal ribosome biogenesis when ARF expression is no longer required, such as for example after the successful repair of cell damage.
Previous data have suggested that TTF-I regulates rRNA gene activation/silencing and mediates RPI transcription termination (46–48). In our study conditional over-expression of TTF-I had no significant effect on these functions. The number of active rRNA genes remained constant over many days (Supplementary Figure S1A and S1B), despite a very significant reduction in rRNA synthesis (Figure 1B and C). TTF is known to bind to the promoter proximal and 45S rRNA termination sites and we have confirmed these observations both in vitro (Lessard,F., unpublished data) and in vivo (13). However, we also did not detect any significant change in the distribution and level of 45S rRNA 3′ termini after conditional over-expression of TTF-I (Supplementary Figure S1C). Thus, despite TTF-I being an essential factor for rRNA gene transcription and its abundance being a critical regulator of both 45S pre-rRNA transcription and processing, its mechanism of action is still unclear and will require further study.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5.
FUNDING
Operating grants from the Cancer Research Society (CRS/SRC); the Canadian Institutes of Health Research (CIHR) (grant #MOP-12205); and, most recently, from the Canadian Cancer Society Research Institute (CCSRI) (grant # 2010-700477). The Research Centre of the CHUQ, in which the Laval University Cancer Research Centre is housed, is supported by a grant from the FRSQ (Québec). Funding for open access charges: Operating grant from CIHR.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We wish to thank Dr C. J. Sherr for providing p19ARF constructs and the MT-ARF cell line, Drs L. I. Rothblum and I. Grummt for providing TTF-I cDNAs, Dr C. Blattner for providing the Myc-MDM2 constucts, S. Laín for providing MDM2(WT) and MDM2(C462A), T. Mistelli for GFP-RPA43, R Hannan for GFP-UBF, J.-Y. Masson for HA-Ubiquitin and A.J. Levine for providing anti-MDM2(2A10, 3G5 and 4B11). We also thank G. Ferbeyre for his advice and help in the use of MDM2 antibodies.
REFERENCES
- 1.Moss T, Langlois F, Gagnon-Kugler T, Stefanovsky V. A housekeeper with power of attorney: the rRNA genes in ribosome biogenesis. Cell. Mol. Life Sci. 2007;64:29–49. doi: 10.1007/s00018-006-6278-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jorgensen P, Rupes I, Sharom JR, Schneper L, Broach JR, Tyers M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004;18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chedin S, Laferte A, Hoang T, Lafontaine DL, Riva M, Carles C. Is ribosome synthesis controlled by pol I transcription? Cell Cycle. 2007;6:11–15. doi: 10.4161/cc.6.1.3649. [DOI] [PubMed] [Google Scholar]
- 4.Drygin D, Rice WG, Grummt I. The RNA polymerase I transcription machinery: an emerging target for the treatment of cancer. Annu. Rev. Pharmacol. Toxicol. 2010;50:131–156. doi: 10.1146/annurev.pharmtox.010909.105844. [DOI] [PubMed] [Google Scholar]
- 5.Moss T. At the crossroads of growth control; making ribosomal RNA. Curr. Opin. Genet. Dev. 2004;14:210–217. doi: 10.1016/j.gde.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Moss T, Stefanovsky VY. At the center of eukaryotic life. Cell. 2002;109:545–548. doi: 10.1016/s0092-8674(02)00761-4. [DOI] [PubMed] [Google Scholar]
- 7.Moss T, Stefanovsky V, Langlois F, Gagnon-Kugler T. A new paradigm for the regulation of the mammalian ribosomal RNA genes. Biochem. Soc. Trans. 2006;34:1079–1081. doi: 10.1042/BST0341079. [DOI] [PubMed] [Google Scholar]
- 8.Birch JL, Zomerdijk JC. Structure and function of ribosomal RNA gene chromatin. Biochem. Soc. Trans. 2008;36:619–624. doi: 10.1042/BST0360619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grummt I. Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus. Genes Dev. 2003;17:1691–1702. doi: 10.1101/gad.1098503R. [DOI] [PubMed] [Google Scholar]
- 10.Sanij E, Hannan RD. The role of UBF in regulating the structure and dynamics of transcriptionally active rDNA chromatin. Epigenetics. 2009;4:374–382. doi: 10.4161/epi.4.6.9449. [DOI] [PubMed] [Google Scholar]
- 11.Sugimoto M, Kuo ML, Roussel MF, Sherr CJ. Nucleolar Arf tumor suppressor inhibits ribosomal RNA processing. Mol. Cell. 2003;11:415–424. doi: 10.1016/s1097-2765(03)00057-1. [DOI] [PubMed] [Google Scholar]
- 12.Itahana K, Bhat KP, Jin A, Itahana Y, Hawke D, Kobayashi R, Zhang Y. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol. Cell. 2003;12:1151–1164. doi: 10.1016/s1097-2765(03)00431-3. [DOI] [PubMed] [Google Scholar]
- 13.Lessard F, Morin F, Ivanchuk S, Langlois F, Stefanovsky V, Rutka J, Moss T. The ARF tumor suppressor controls ribosome biogenesis by regulating the RNA polymerase I transcription factor TTF-I. Mol. Cell. 2010;38:539–550. doi: 10.1016/j.molcel.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 14.Sander EE, Mason SW, Munz C, Grummt I. The amino-terminal domain of the transcription termination factor TTF-I causes protein oligomerization and inhibition of DNA binding. Nucleic Acids Res. 1996;24:3677–3684. doi: 10.1093/nar/24.19.3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evers R, Smid A, Rudloff U, Lottspeich F, Grummt I. Different domains of the murine RNA polymerase I-specific termination factor mTTF-I serve distinct functions in transcription termination. EMBO J. 1995;14:1248–1256. doi: 10.1002/j.1460-2075.1995.tb07108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuhn A, Bartsch I, Grummt I. Specific interaction of the murine transcription termination factor TTF I with class-I RNA polymerases. Nature. 1990;344:559–562. doi: 10.1038/344559a0. [DOI] [PubMed] [Google Scholar]
- 17.Langst G, Blank TA, Becker PB, Grummt I. RNA polymerase I transcription on nucleosomal templates: the transcription termination factor TTF-I induces chromatin remodeling and relieves transcriptional repression. EMBO J. 1997;16:760–768. doi: 10.1093/emboj/16.4.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Strohner R, Nemeth A, Jansa P, Hofmann-Rohrer U, Santoro R, Langst G, Grummt I. NoRC – a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J. 2001;20:4892–4900. doi: 10.1093/emboj/20.17.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Santoro R, Li J, Grummt I. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat. Genet. 2002;32:393–396. doi: 10.1038/ng1010. [DOI] [PubMed] [Google Scholar]
- 20.Zhou Y, Santoro R, Grummt I. The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. EMBO J. 2002;21:4632–4640. doi: 10.1093/emboj/cdf460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Treier M, Staszewski LM, Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell. 1994;78:787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- 22.Geyer RK, Yu ZK, Maki CG. The MDM2 RING-finger domain is required to promote p53 nuclear export. Nat. Cell Biol. 2000;2:569–573. doi: 10.1038/35023507. [DOI] [PubMed] [Google Scholar]
- 23.Xirodimas DP, Stephen CW, Lane DP. Cocompartmentalization of p53 and Mdm2 is a major determinant for Mdm2-mediated degradation of p53. Exp. Cell Rese. 2001;270:66–77. doi: 10.1006/excr.2001.5314. [DOI] [PubMed] [Google Scholar]
- 24.Hay TJ, Meek DW. Multiple sites of in vivo phosphorylation in the MDM2 oncoprotein cluster within two important functional domains. FEBS Lett. 2000;478:183–186. doi: 10.1016/s0014-5793(00)01850-0. [DOI] [PubMed] [Google Scholar]
- 25.Chen J, Marechal V, Levine AJ. Mapping of the p53 and mdm-2 interaction domains. Mol. Cell. Biol. 1993;13:4107–4114. doi: 10.1128/mcb.13.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuo ML, Duncavage EJ, Mathew R, den Besten W, Pei D, Naeve D, Yamamoto T, Cheng C, Sherr CJ, Roussel MF. Arf induces p53-dependent and -independent antiproliferative genes. Cancer Res. 2003;63:1046–1053. [PubMed] [Google Scholar]
- 27.Stefanovsky VY, Langlois F, Gagnon-Kugler T, Rothblum LI, Moss T. Growth factor signaling regulates elongation of RNA polymerase I transcription in mammals via UBF phosphorylation and rchromatin remodeling. Mol. Cell. 2006;21:629–639. doi: 10.1016/j.molcel.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 28.Stefanovsky VY, Pelletier G, Hannan R, Gagnon-Kugler T, Rothblum LI, Moss T. An immediate response of ribosomal transcription to growth factor stimulation in mammals is mediated by ERK phosphorylation of UBF. Mol. Cell. 2001;8:1063–1073. doi: 10.1016/s1097-2765(01)00384-7. [DOI] [PubMed] [Google Scholar]
- 29.Luthman H, Magnusson G. High efficiency polyoma DNA transfection of chloroquine treated cells. Nucleic Acids Res. 1983;11:1295–1308. doi: 10.1093/nar/11.5.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berk AJ, Sharp PA. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease digested hybrids. Cell. 1977;12:721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- 31.Read C, Larose AM, Leblanc B, Bannister AJ, Firek S, Smith DR, Moss T. High resolution studies of the Xenopus laevis ribosomal gene promoter in vivo and in vitro. J. Biol. Chem. 1992;267:10961–10967. [PubMed] [Google Scholar]
- 32.Conconi A, Widmer RM, Koller T, Sogo JM. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell. 1989;57:753–761. doi: 10.1016/0092-8674(89)90790-3. [DOI] [PubMed] [Google Scholar]
- 33.Stefanovsky VY, Moss T. Regulation of rRNA synthesis in human and mouse cells is not determined by changes in active gene count. Cell Cycle. 2006;5:735–739. doi: 10.4161/cc.5.7.2633. [DOI] [PubMed] [Google Scholar]
- 34.Lucchini R, Sogo JM. In: Transcription of Ribosomal Genes by Eukaryotic RNA Polymerase I. Paule MR, editor. Austin, TX: Landes Bioscience; 1998. pp. 255–276. [Google Scholar]
- 35.Hershko A, Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 36.Fang S, Weissman AM. A field guide to ubiquitylation. Cell. Mol. Life Sci. 2004;61:1546–1561. doi: 10.1007/s00018-004-4129-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deisenroth C, Zhang Y. Ribosome biogenesis surveillance: probing the ribosomal protein-Mdm2-p53 pathway. Oncogene. 2010;29:4253–4260. doi: 10.1038/onc.2010.189. [DOI] [PubMed] [Google Scholar]
- 38.Sherr CJ. Divorcing ARF and p53: an unsettled case. Nat. Rev. Cancer. 2006;6:663–673. doi: 10.1038/nrc1954. [DOI] [PubMed] [Google Scholar]
- 39.Lohrum MA, Ashcroft M, Kubbutat MH, Vousden KH. Identification of a cryptic nucleolar-localization signal in MDM2. Nat. Cell Biol. 2000;2:179–181. doi: 10.1038/35004057. [DOI] [PubMed] [Google Scholar]
- 40.Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat. Cell Biol. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- 41.Llanos S, Clark PA, Rowe J, Peters G. Stabilization of p53 by p14ARF without relocation of MDM2 to the nucleolus. Nat. Cell Biol. 2001;3:445–452. doi: 10.1038/35074506. [DOI] [PubMed] [Google Scholar]
- 42.Pomerantz J, Schreiber-Agus N, Liegeois NJ, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee HW, et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 43.Kamijo T, Weber JD, Zambetti G, Zindy F, Roussel MF, Sherr CJ. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc. Natl Acad. Sci. USA. 1998;95:8292–8297. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stott FJ, Bates S, James MC, McConnell BB, Starborg M, Brookes S, Palmero I, Ryan K, Hara E, Vousden KH, et al. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17:5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 46.Grummt I, Pikaard CS. Epigenetic silencing of RNA polymerase I transcription. Nat. Rev. Mol. Cell. Biol. 2003;4:641–649. doi: 10.1038/nrm1171. [DOI] [PubMed] [Google Scholar]
- 47.Kuhn A, Grummt I. 3′-end formation of mouse pre-rRNA involves both transcription termination and a specific processing reaction. Genes Dev. 1989;3:224–231. doi: 10.1101/gad.3.2.224. [DOI] [PubMed] [Google Scholar]
- 48.Li J, Langst G, Grummt I. NoRC-dependent nucleosome positioning silences rRNA genes. EMBO J. 2006;25:5735–5741. doi: 10.1038/sj.emboj.7601454. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.